Abstract
RUNX1 regulates formation of the definitive hematopoietic stem cell and its subsequent lineage maturation, and mutations of RUNX1 contribute to leukemic transformation. Phosphorylation of Ser-48, Ser-303, and Ser-424 by cyclin-dependent kinases (cdks) increases RUNX1 trans-activation activity without perturbing p300 interaction. We now find that endogenous RUNX1 interacts with endogenous HDAC1 or HDAC3. Mutation of the three RUNX1 serines to aspartic acid reduces co-immunoprecipitation with HDAC1 or HDAC3 when expressed in 293T cells; mutation of these three serines to alanine increases HDAC interaction, and mutation of each serine individually to aspartic acid also reduces these interactions. GST-RUNX1 isolated from bacterial extracts bound in vitro translated HDAC1 or HDAC3, and these interactions were weakened by mutation of Ser-48, Ser-303, and Ser-424 to aspartic acid. The ability of RUNX1 phosphorylation and not only serine to aspartic acid conversion to reduce HDAC1 binding was demonstrated using wild-type GST-RUNX1 phosphorylated in vitro using cdk1/cyclinB and by exposure of 293T cells transduced with RUNX1 and HDAC1 to roscovitine, a cdk inhibitor. Finally, RUNX1 or RUNX1(tripleD), in which Ser-48, Ser-303, and Ser-424 are mutated to aspartic acid, stimulated proliferation of transduced, lineage-negative murine marrow progenitors more potently than did RUNX1(tripleA), in which these serines are mutated to alanine, suggesting that stimulation of RUNX1 trans-activation by cdk-mediated reduction in HDAC interaction increases marrow progenitor cell proliferation.
Keywords: CDK (Cyclin-dependent Kinase), Hematopoiesis, Stem Cell, Transcription Factors, Transcription Repressor
Introduction
RUNX1 and its heterodimeric partner CBFβ direct emergence of adult hematopoietic stem cells (HSC)2 from hemogenic endothelium during embryogenesis and participate in subsequent lymphoid, myeloid, or megakaryocyte lineage maturation in adult marrow (1–3). RUNX1 is commonly mutated or inhibited in acute myeloid or lymphoid leukemias, and RUNX1 is overexpressed as well in a subset of acute lymphoblastic leukemias (4, 5).
RUNX1 also directly regulates G1 to S cell cycle progression. The myeloid oncoproteins CBFβ-SMMHC or RUNX1-ETO dominantly inhibit RUNX1 activities and slow G1 progression in hematopoietic cell lines or in murine or human marrow progenitors (6–9). cdk4, cyclin D2, or c-Myc overcome inhibition of proliferation by these CBF oncoproteins (8, 10, 11); exogenous RUNX1 stimulates G1 progression in 32Dcl3 or Ba/F3 cells (9, 10, 12), and stimulation of G1 via deletion of the p16INK4a/p19ARF locus or expression of the viral E7 protein (which inactivates retinoblastoma protein) cooperates with CBFβ-SMMHC or TEL-RUNX1 to induce acute leukemia (13, 14). Induction of cdk4 or cyclin D3 transcription may underlie stimulation of G1 progression by RUNX1 (10, 15).
Regulation of cell proliferation by RUNX proteins represents an evolutionarily conserved activity. In the sea urchin Strongylocentrotus purpuratus, depletion of the RUNX ortholog SpRunt-1 reduces blastocyst cell proliferation and inhibits expression of cyclinD, whose promoter binds SpRunt-1 in a chromatin immunoprecipitation (ChIP) assay (16), and in Caenorhabditis elegans, RNT-1 stimulates seam cell proliferation, with rnt-1 mutants having reduced numbers of seam cells and animals expressing exogenous RNT-1 having an expansion of seam cells (17, 18). Moreover, mutation of bro-1, encoding the CBFβ homolog BRO-1, similarly reduces seam cell proliferation; overexpression of BRO-1 expands seam cells, and simultaneous overexpression of BRO-1 and RNT-1 induces massive seam cell expansion (18).
Not only does RUNX1 regulate cell cycle progression, but RUNX1 levels increase as hematopoietic cells progress from G1 to S and from S to G2/M (15), which is accounted for by the finding that phosphorylation of RUNX1 on Ser-303 by cdks leads its ubiquitin-mediated degradation during G2/M (19). We developed additional evidence that cdks phosphorylate Ser-303 and found that Ser-48 and Ser-424 are also substrates of cdk1/cyclin B and cdk6/cyclin D3. Moreover, we demonstrated that phosphorylation of Ser-48, Ser-303, and Ser-424 strengthens the ability of RUNX1 to activate transcription and to stimulate proliferation of the Ba/F3 hematopoietic cell line (20). As interaction with the p300 co-activator was not affected by mutation of Ser-48, Ser-303, and Ser-424 to the phosphomimetic aspartic acid (20), we now consider the possibility that RUNX1 phosphorylation instead reduces HDAC interaction.
Exogenous RUNX1 was previously found to bind exogenous HDAC1 or HDAC3 in COS7 cells (21). We demonstrate that endogenous RUNX1 interacts with endogenous HDAC1 or HDAC3 and that GST-RUNX1 purified from bacterial extracts directly binds HDAC1 or HDAC3 generated by in vitro translation. We also provide data indicating that cdk phosphorylation of RUNX1 markedly reduces interaction with HDAC1 or HDAC3. Moreover, we find that a RUNX1 variant with Ser-48, Ser-303, and Ser-424 mutated to alanine, RUNX1(tripleA), stimulates proliferation of lineage-negative murine marrow progenitors less effectively than does RUNX1 or RUNX1(tripleD), in which these three serines are mutated to aspartic acid.
EXPERIMENTAL PROCEDURES
Cell Culture and Transduction
293T cells were cultured in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum (HI-FBS). Jurkat cells were cultured in RPMI 1640 medium with 10% HI-FBS with 100 mm glutamine and 50 μm β-mercaptoethanol. M1 cells were cultured with RPMI 1640 medium with 10% heat-inactivated horse serum. Ba/F3 cells were cultured in RPMI 1640 medium with 10% HI-FBS and 1 ng/ml murine IL-3 (PeproTech). For co-immunoprecipitation studies, 293T cells on 100-mm dishes were transiently transfected with CMV expression plasmids (4 μg/DNA) using 20 μl of Lipofectamine 2000 (Invitrogen). Roscovitine (Calbiochem) was utilized at 80 μm; U0126 or PP2 were used at 10 μm and LY294002 at 50 μm (Cell Signaling). For retroviral vector packaging, 293T cells were transiently transfected with 15 μg of pBABEpuro vectors and 4 μg of pkat2ecopac using 35 μl of Lipofectamine 2000 (22), and supernatants were collected 2 and 3 days later. Murine marrow cells isolated from the femurs of C57BL/6 mice treated 6 days earlier with 5-fluorouracil (150 mg/kg) were subjected to red cell lysis with NH4Cl and placed in Iscove's modified Dulbecco medium at 5 × 105 cells/ml with 10% HI-FBS, 10 ng/ml murine IL-3, 10 ng/ml murine IL-6, and 10 ng/ml murine stem cell factor (PeproTech). After 24 h, 1 ml of viral supernatant was added per ml of cells with 4 μg/ml Polybrene. Puromycin (2 μg/ml) was added 72 h later, and after an additional 24 h, viable cells were isolated using a Lympholyte-M polysucrose density gradient (Cedarlane Labs), subjected to lineage depletion using immunomagnetic beads and a mixture of lineage antibodies (Stem Cell Technologies), and placed in Iscove's modified Dulbecco medium with 10% HI-FBS, IL-3, IL-6, and stem cell factor with or without 200 nm 4HT. Viable cell counts were enumerated using trypan blue dye and a hemocytometer.
Co-immunoprecipitation and Western Blotting
Jurkat cells were washed with phosphate-buffered saline (PBS), resuspended in 10 mm KCl, 1.5 mm MgCl2, 10 mm Hepes, pH 7.9, with 1 mm PMSF, 1 mm DTT, and a mixture of protease inhibitors (Sigma) and homogenized to break the cell membrane. After centrifugation at 2,000 × g, the nuclear pellet was resuspended in low salt buffer (20 mm KCl, 1.5 mm MgCl2, 0.2 mm EDTA, 20% glycerol, 20 mm Hepes, pH 7.9) and homogenized. An equal volume of high salt buffer (1.2 m KCl, 1.5 mm MgCl2, 0.2 mm EDTA, 20% glycerol, 20 mm Hepes, pH 7.9) was then added dropwise and rotated at 4 °C for 30 min. After centrifugation at 14,000 × g, the supernatant was collected and dialyzed against 100 mm KCl, 0.2 mm EDTA, 20% glycerol, 20 mm Hepes, pH 7.9. Two days after transient transfection, 293T cells were collected and washed with PBS. Cell lysates were then prepared by incubation at 4 °C for 30 min with 400 μl of 20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, 0.2% Nonidet P-40, 0.2% Triton X-100, 0.2% deoxycholate, 10% (w/v) glycerol, 1 mm NaF, 1 mm sodium orthovanadate, 1 mm PMSF, and protease inhibitors. After clarification at 14,000 × g for 15 min, aliquots of cell extracts were saved as “input,” and supernatants were precleared used 50 μl of 50% protein A-Sepharose. 200 μg of 293T or 400 μg of Jurkat or M1 total protein was then added to 1 ml of IP buffer (180 mm KCl, 0.05% Nonidet P-40, 1 mm DTT, 1 mm PMSF) and incubated with 8 μg of rabbit IgG, rabbit anti-Myc A-14 antiserum (Santa Cruz Biotechnology) for 3 h or with rabbit anti-RUNX1 antiserum (Active Motif), mouse anti-HDAC1 clone 2E10, or HDAC3 clone 3G6 (Millipore) overnight at 4 °C, followed by addition of 50 μl of protein A-Sepharose. The beads were then washed three times with IP buffer, and the samples were eluted in Laemmli sample buffer at 95 °C and subjected to Western blotting using mouse 9E10 anti-Myc (Santa Cruz Biotechnology), mouse M2 anti-FLAG (Sigma), rabbit anti-HDAC1 ab7028 (Abcam), mouse anti-HDAC3 clone 3G6, or rabbit anti-RUNX1 antibodies as described previously (6). Western analyses were also conducted using rabbit anti-ERα (MC-20) antiserum or mouse anti-β-actin AC-15 antibody (Sigma), mouse anti-RUNX1 antibody (kindly provided by N. Speck), or phospho-RUNX1(Ser-424) antiserum (20). Band intensities were quantified using ImageJ software (National Institutes of Health).
Bacterial Protein Isolation, in Vitro Translation, Kinase Reaction, and Binding Assay
GST-RUNX1 fusion proteins were expressed in Escherichia coli, extracted, and bound to GST-Bind resin (Novagen) as described previously (20). pcDNA3-FLAG-HDAC1 or pcDNA3-FLAG-HDAC3 was subjected to in vitro transcription and translation in the presence of [35S]methionine per the manufacturer's instructions (Promega). Equivalent aliquots were then incubated with resin-bound GST-RUNX1 proteins in the presence of 20 mm Hepes, pH 7.9, 180 mm KCl, 0.5 mm EDTA, 0.1% Nonidet P-40, 1 mg/ml bovine serum albumin, 1 mm DTT, 1 mm PMSF for 2 h at 4 °C; washed three times with the same solution; subjected to SDS-PAGE followed by incubation of the gel with Enhance (PerkinElmer Life Sciences); dried, and autoradiographed. In some experiments, resin-bound GST-RUNX1 samples were subjected to in vitro kinase assay with or without cdk1/cyclin B, as described but with 3-fold increased cdk1/cyclin B to favor complete reaction (20), prior to incubation with 35S-labeled HDAC1 or HDAC3. Phosphorylation of RUNX1(Ser-424) was assessed by Western blotting using anti-phospho-Ser-424 antiserum (20), and total GST-RUNX1 protein input to the HDAC-binding reactions was assessed either by SDS-PAGE followed by Coomassie Blue dye staining or by Western blot analysis with mouse anti-RUNX1 antibody (kindly provided by N. Speck). The Student's t test was used for statistical comparisons.
RESULTS
Endogenous RUNX1 Interacts with Endogenous HDAC1 or HDAC3
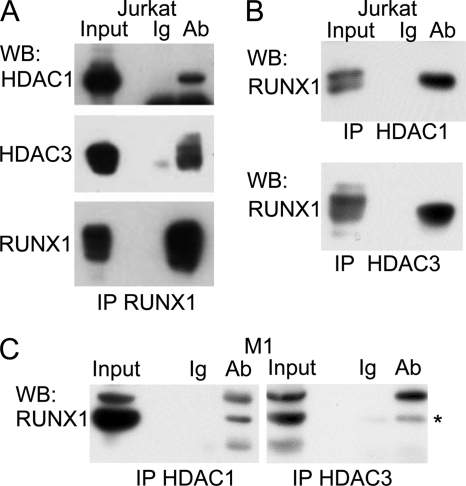
Nuclear extracts from Jurkat T cells were subject to immunoprecipitation with RUNX1 antiserum or Ig control, followed by Western blotting for endogenous HDAC1 or HDAC3 (Fig. 1A). Specific interaction between endogenous RUNX1 and HDAC1 or HDAC3 is evident. Similar extracts were subject to reciprocal immunoprecipitation with HDAC1 or HDAC3 antibodies or Ig control, followed by Western blotting for endogenous RUNX1 (Fig. 1B). Specific interaction between RUNX1 and HDAC1 or HDAC3 was again noted. Co-immunoprecipitation also demonstrates interaction of RUNX1 with HDAC1 or HDAC3 RUNX1 in M1 myeloid cells (Fig. 1C).
FIGURE 1.
Endogenous RUNX1 binds endogenous HDAC1 or HDAC3. A, nuclear extracts corresponding to 5 × 107 Jurkat cells were subjected to immunoprecipitation (IP) with rabbit anti-RUNX1 antiserum or rabbit Ig followed by Western blotting (WB) for HDAC1, HDAC3, or RUNX1. B, similar nuclear extracts were subjected to immunoprecipitation with mouse anti-HDAC1 (H1), mouse anti-HDAC3 (H3), or mouse Ig followed by Western blotting with rabbit anti-RUNX1. C, nuclear extracts corresponding to 5 × 107 M1 cells were subjected to immunoprecipitation with rabbit anti-RUNX1 antiserum or rabbit Ig followed by Western blotting for HDAC1 or HDAC3. * indicates position of RUNX1. Input in A–C correspond to 10% of the extracts used for immunoprecipitation.
Mutation of RUNX1 Serines 48, 303, or 424 to Aspartic Acid Reduces HDAC1 or HDAC3 Interaction
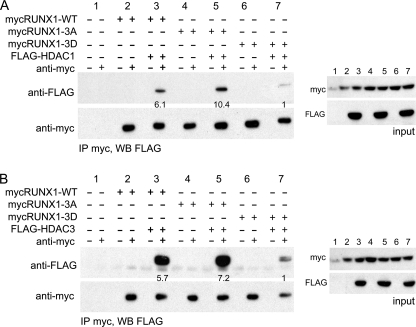
We next sought to test the idea that cdk phosphorylation of RUNX1 leads to reduced HDAC interaction, focusing on HDAC1 and HDAC3, for which RUNX1 has the highest affinity (21), and on the 480 residue RUNX1c isoform, which dominates during adult hematopoiesis (23). Myc-tagged RUNX1, RUNX1(tripleA), or RUNX1(tripleD) were co-expressed in 293T cells along with FLAG-tagged HDAC1 or HDAC3, followed by immunoprecipitation with Myc antibody or Ig control and then Western blotting with FLAG or Myc antibodies. Data representative of three repetitions are shown (Fig. 2). Mutation of Ser-48, Ser-303, and Ser-424 to aspartic acid markedly reduced interaction with either HDAC1 (mean 4-fold, p = 0.03) or HDAC3 (mean 3-fold, p < 0.01). Notably, mutation of Ser-48, Ser-303, and Ser-424 to alanine significantly increased interaction with HDAC1 (mean 2-fold, p = 0.05), and there was a trend toward increased HDAC3 interaction (mean 1.3-fold, p = 0.08). Myc-RUNX1(tripleD) expression was not reduced compared with wild-type RUNX1, as seen from analysis of either the input or co-immunoprecipitation samples with Myc antibody.
FIGURE 2.
Mutation of RUNX1 serines 48, 303, and 424 to aspartic acid to mimic cdk phosphorylation reduces HDAC1 and HDAC3 interaction in cell extracts. A, 293T cells were co-transfected with Myc-tagged wild-type RUNX1 (mycRUNX-WT), RUNX1(tripleA) (mycRUNX-3A), or RUNX1(tripleD) (mycRUNX1–3D) in the absence or presence of FLAG-tagged HDAC1 (FLAG-HDAC1), each expressed from the CMV promoter. Two days later, cell extracts were subjected to immunoprecipitation (IP) with rabbit anti-Myc antiserum or normal rabbit Ig followed by Western blotting (WB) using mouse anti-FLAG or anti-Myc antibodies (left panels). Equal amounts of cell extracts were also analyzed by Western blotting prior to immunoprecipitation to assess input protein expression (right panels). Co-immunoprecipitated FLAG-HDAC1 band intensities were quantified, as shown below each lane of upper left panel. B, similar analysis was conducted substituting FLAG-tagged HDAC3 (FLAG-HDAC3) for FLAG-HDAC1.
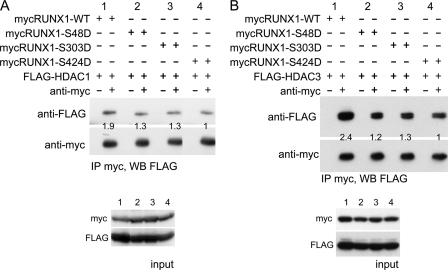
To identify the specific serine residue or residues responsible for reduced affinity of RUNX1(tripleD) for HDAC1 or HDAC3, similar analysis was conducted comparing their affinity for RUNX1, RUNX1-S48D, RUNX1-S303D, or RUNX1-S424D. Data representative of four repetitions with HDAC1 and three with HDAC3 are shown (Fig. 3). Mutation of each serine reduced interaction with HDAC1 or HDAC3, with mutation of Ser-424 having the greatest effect. On average, S48D reduced interaction 1.3-fold with HDAC1 (p = 0.07) and 2.3-fold with HDAC3 (p = 0.02); S303D reduced interaction 1.5-fold with HDAC1 (p = 0.05) and 1.6-fold with HDAC3 (p = 0.05); and S424D reduced interaction 2.2-fold with HDAC1 (p = 0.01) and 2.1-fold with HDAC3 (p = 0.02).
FIGURE 3.
Mutation of RUNX1 serine 48, 303, or 424 to aspartic acid reduces HDAC1 and HDAC3 interaction in cell extracts. A, 293T cells were co-transfected with Myc-tagged RUNX1, mycRUNX1-S48D, mycRUNX1-S303D, or mycRUNX1-S424D in the presence of FLAG-tagged HDAC1. Two days later, cell extracts were subjected to immunoprecipitation (IP) with rabbit anti-Myc antiserum or normal rabbit Ig followed by Western blotting (WB) using mouse anti-FLAG or anti-Myc antibodies (top panels). Equal amounts of cell extracts were also analyzed by Western blotting prior to immunoprecipitation to assess input protein expression (bottom panels). Co-immunoprecipitated FLAG-HDAC1 band intensities were quantified, as shown below each lane of the upper panel. Images of lanes from the same gels were positioned adjacent to each other. B, similar analysis was conducted substituting FLAG-tagged HDAC3 for FLAG-HDAC1.
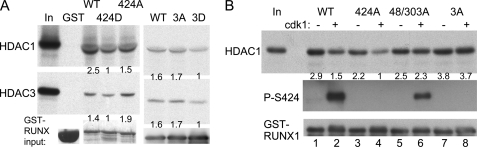
As interactions between proteins co-expressed in mammalian cells might be mediated by additional proteins, we sought to confirm the importance of RUNX1(Ser-424) for HDAC interactions using more purified proteins. HDAC1 or HDAC3 was generated by coupled in vitro transcription and translation in the presence of [35S]methionine, and GST, GST-RUNX1, GST-RUNX1(S424D), or GST-RUNX1(S424A), as well as GST-RUNX1(tripleA) or GST-RUNX1(tripleD), were expressed in E. coli. Affinity of the HDACs for the GST proteins isolated via interaction with glutathione-agarose beads was then assessed. Data representative of three repetitions is shown (Fig. 4A). GST-RUNX1(S424D) had reduced affinity for HDAC1 or HDAC3 compared with GST-RUNX1 or GST-RUNX1(S424A), and GST alone did not interact with either HDAC. On average, the S424D alteration reduced affinity for HDAC1 2.1-fold compared with wild-type RUNX1 (p = 0.05) and reduced HDAC3 affinity 1.4-fold (p = 0.04), whereas the S424A mutation had minimal effect. Similarly, the tripleD mutation of Ser-48, Ser-303, and Ser-424, on average, reduced affinity for 1.9-fold for HDAC1 (p = 0.02) and 1.5-fold for HDAC3 (p = 0.04).
FIGURE 4.
Mutation of RUNX1 serine 424 to aspartic acid or phosphorylation by cdk1 reduces HDAC interaction using partially purified proteins. A, GST, GST-RUNX1 (WT), GST-RUNX1-S424D (424D), GST-RUNX1-S424A (424A), GST-RUNX1(tripleA) (3A), or GST-RUNX1(tripleD) (3D) proteins bound to glutathione-agarose beads were incubated with 35S-labeled HDAC1 or HDAC3 generated by in vitro transcription and translation, followed by washing, SDS-PAGE, and autoradiography. Equal volumes of input GST protein samples were also subjected to SDS-PAGE followed by Coomassie Blue dye staining (left panels) or RUNX1 Western blotting (right panels) to assess total protein expression. HDAC1 or HDAC3 band intensities relative to input GST proteins were quantified, as shown below each lane. B, GST-RUNX1, GST-RUNX1-S424A, GST-RUNX1-S48A/303A, or GST-RUNX1(tripleA) (3A) bound to agarose beads were incubated with or without cdk1/cyclin B in the presence of kinase buffer and ATP. The beads were then incubated with 35S-labeled HDAC1 followed by washing, SDS-PAGE, and autoradiography. Phosphorylation of Ser-424 was confirmed by Western blotting with anti-phospho-Ser-424 RUNX1 antiserum (middle panel), and GST-RUNX1 input was assessed by Western blot analysis of equal volumes of input samples using anti-RUNX1 antibody (bottom panel). HDAC1 band intensities relative to GST-RUNX1 were quantified, as shown below each lane of the top panel.
cdk Phosphorylation of RUNX1 Reduces HDAC1 Affinity
To confirm that cdk phosphorylation and not only serine to aspartic acid mutation reduces HDAC affinity, GST-RUNX1 was incubated in the presence or absence of cdk1/cyclin B in a reaction mixture containing ATP. Sufficient cdk1/cyclin B was included to favor complete reaction. Phosphorylation of Ser-424 was confirmed by Western blotting, and interaction with 35S-labeled HDAC1 was assessed. Incubation of purified GST-RUNX1 with cdk1/cyclin B led to Ser-424 phosphorylation and to reduced interaction with HDAC1 (mean 2-fold, p = 0.05), and data representative of three independent assessments are shown (Fig. 4B, lanes 1 and 2). To map serines relevant to cdk alteration of HDAC1 affinity in vitro, similar analysis was then conducted using GST-RUNX1(S424A), again reduced HDAC1 interaction was seen upon cdk1 addition (Fig. 4B, lanes 3 and 4), with mean reduction 1.7-fold (p = 0.05), suggesting that phosphorylation of Ser-48 and Ser-303 reduces HDAC1 binding. However, no significant reduction in HDAC1 affinity was evident when GST-RUNX1(S48A/S303A) was used as substrate (Fig. 4B, lanes 5 and 6), even though Ser-424 was still phosphorylated. Potential explanations for this finding will be discussed. Finally, no reduction in HDAC1 binding was seen when GST-RUNX1(tripleA) was used as substrate (Fig. 4B, lanes 7 and 8). Thus, modification of two cdk consensus sites within RUNX1 (Ser-48 and Ser-303), but not other serines including nonconsensus serines, reduces HDAC1 affinity in this in vitro kinase assay.
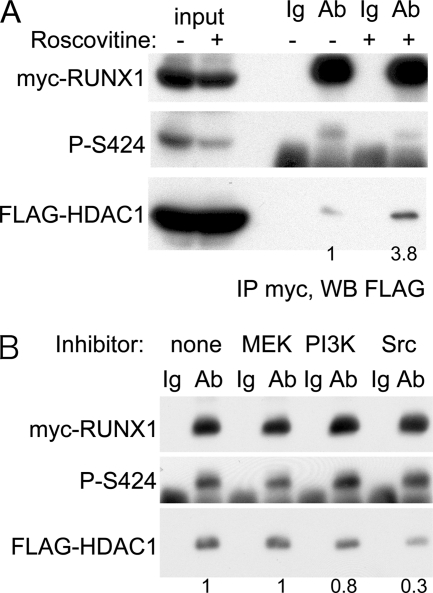
To assess the relevance of RUNX1 cdk phosphorylation to HDAC1 interaction in the 293T mammalian cell line, co-immunoprecipitation of exogenous myc-RUNX1 and FLAG-HDAC1 was assessed in the absence or presence of roscovitine, a cdk inhibitor (Fig. 5A). Addition of roscovitine for 3 h reduced phosphorylation of RUNX1 Ser-424, and likely also Ser-48 and Ser-303 based on our prior use of this agent (20), and increased HDAC1 interaction with RUNX1 almost 4-fold. In contrast, addition of MEK, PI3K, or Src kinase inhibitors did not increase HDAC1 interaction with RUNX1, with the Src inhibitor reducing interaction (Fig. 5B).
FIGURE 5.
Inhibition or RUNX1 cdk phosphorylation in mammalian cells increases HDAC1 interaction. A, 293T cells transiently transfected with myc-RUNX1 and FLAG-HDAC1 42 h previously were cultured in the absence or presence of a cdk inhibitor (roscovitine) for 3 h. Cell extracts were then prepared and subjected to immunoprecipitation (IP) with rabbit anti-Myc antiserum or normal rabbit Ig followed by Western blotting (WB) with mouse anti-Myc antiserum (top panel), rabbit anti-phospho-Ser-424 RUNX1 antiserum (middle panel), or mouse anti-FLAG antibody (bottom panel). Immunoprecipitated FLAG-HDAC1 was quantified as indicated below the bottom panel. B, similar analysis was conducted with cells cultured in the absence or presence of a MEK inhibitor (U0126), a PI3K inhibitor (LY294002), or a Src inhibitor (PP2) for 3 h prior to harvest. FLAG-HDAC1 band intensities were quantified, as shown below each lane.
RUNX1 cdk Phosphorylation Increases Marrow Progenitor Proliferation
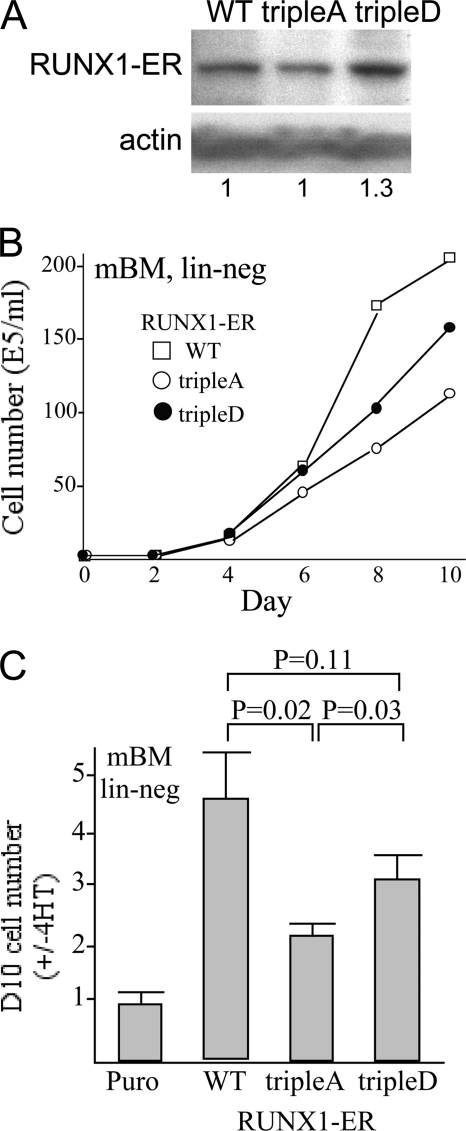
We previously found that RUNX1 or RUNX1(tripleD) linked to the estradiol receptor (ER) ligand-binding domain stimulates Ba/F3 proliferation more effectively than RUNX1(tripleA)-ER (20). We now demonstrate related findings using normal marrow progenitors. Marrow mononuclear cells were transduced with RUNX1-ER, RUNX1(tripleA)-ER, RUNX1(tripleD)-ER, or the empty pBABEpuro vector, lineage-depleted after puromycin selection, and then cultured with either 4-hydroxytamoxifen (4HT) or the ethanol vehicle. Lineage depletion after transduction and drug selection provides a starting population consisting mainly of myeloid progenitors expressing the ER fusion protein in an inactive state and avoids biasing the cell population during transduction. At the time of 4HT addition, similar expression of the three transgenes, relative to β-actin, was evident (Fig. 6A). Representative growth data after 4HT addition are shown (Fig. 6B), as is the ratio of viable cell numbers in the presence versus the absence of 4HT on day 10 (Fig. 6C). Both wild-type RUNX1 and RUNX1(tripleD) stimulated proliferation to a significantly greater extent than RUNX1(tripleA). Although not reaching statistical significance, there was a trend toward weaker stimulation by the tripleD variant compared with RUNX1, perhaps reflecting a high level of endogenous RUNX1 cdk phosphorylation and inability of aspartic acid to fully mimic the effect of serine phosphorylation. No significant cell death was evident in these cultures, and surface analysis of Mac-1 and Gr1 expression indicated that 4HT activation of each construct led to a similar 10–20% increase in Mac-1+Gr1− monocytic cells (not shown). Finally, we also compared RUNX1-ER with RUNX1(S424D)-ER and RUNX1(S424A)-ER, and we found that each stimulated marrow progenitor proliferation similarly, perhaps reflecting increased HDAC binding by RUNX1(tripleA) compared with RUNX1(S424A) (data not shown).
FIGURE 6.
RUNX1 cdk phosphorylation increases induction of murine marrow progenitor proliferation in vitro. A, murine marrow cells isolated from mice exposed to 5-fluorouracil were transduced with pBABEpuro vector (Puro), or the same vector expressing RUNX1-ER (WT), RUNX1(tripleA)-ER, or RUNX1(tripleD)-ER, followed by puromycin selection, isolation of viable cells, and lineage depletion. Total protein samples from lineage-depleted marrow cells transduced with wild-type RUNX1 (WT), the tripleA variant, or the tripleD variant were subjected to Western blotting use ER antiserum and β-actin antibody. Expression of RUNX1 or its variants relative to actin was quantified as indicated below the panels. B, lineage-negative (lin-neg) murine bone marrow (mBM) cells were then cultured with or without 4-HT in medium containing FBS and IL-3, IL-6, and stem cell factor. Viable cell numbers in 4HT on days 0–10 after lineage depletion are shown from a representative experiment. C, ratio of viable cell counts on day 10 (with 4HT divided by without 4HT) is shown (mean ± S.E. of three determinations).
DISCUSSION
RUNX1 has the capacity to either activate or repress transcription. Its impact on a particular target gene in a given cellular context necessarily depends on several factors, including expression levels of relevant co-activators or co-repressors, interaction with additional transcription factors that favor their combined interaction with activating or repressive complexes, and by post-translational modifications of RUNX1 that influence these interactions. Here, we demonstrate that cdk phosphorylation of RUNX1 at Ser-48, Ser-303, and Ser-424 reduces RUNX1 affinity for HDAC1 or HDAC3. These results provide a mechanistic basis for our prior finding that cdks enhance trans-activation by RUNX1 (20).
Notch signaling stimulates cyclin D3 transcription and induces cdk4 and cdk6 (24), and Wnt signals induce cyclin D1 transcription and stabilize cyclin D1 protein (25), suggesting that displacement of HDACs from RUNX1 as a result of Notch- or Wnt-mediated activation of cdks may activate RUNX1 to enable adult HSC development and expansion. On the other hand, zebrafish embryos lacking HDAC1 do not develop adult HSC (26), suggesting that specification of adult HSC by RUNX1 may occur in a relatively quiescent state when RUNX1 is dephosphorylated at Ser-424 and so is more capable of interacting with HDAC1.
Hematopoietic cytokine receptor signals also elevate cyclin D expression (27). Consequent cdk phosphorylation may allow RUNX1 to favor marrow progenitor proliferation, consistent with our finding that RUNX1(tripleD) stimulates progenitor expansion more effectively than RUNX1(tripleA). On the other hand, the majority of adult HSC exists in a quiescent state, suggesting that the marrow HSC niche favors RUNX1 interaction with HDACs and repression rather than activation of target genes that stimulate cell proliferation. We have reproduced the finding that RUNX1 markedly suppresses progenitor engraftment 4 or 16 weeks after RUNX1-transduced cells are transplanted into syngeneic recipients (28), and we find that RUNX1(tripleD) produces similar strong suppression, whereas RUNX1(tripleA) is less effective.3 Therefore, in this in vivo experimental paradigm, RUNX1 trans-activation mediated by p300 rather than trans-repression mediated by HDACs may produce the observed lack of engraftment, perhaps primarily reflecting effects of exogenous RUNX1 on differentiation rather than proliferation. On the other hand, culture of marrow cells transduced with RUNX1-ER in vitro evidently allows short term stimulation of proliferation in response to 4HT; this stimulation occurred more strongly with RUNX1 or RUNX1(tripleD) than with the tripleA variant, suggesting that RUNX1-mediated gene activation enhances cell cycle progression in normal marrow progenitors. Inability of RUNX1(tripleD) to stimulate proliferation more effectively than RUNX1 may reflect significant endogenous RUNX1 cdk phosphorylation in the cells assayed. Future experiments will further pursue the role of RUNX1 cdk phosphorylation on the regulation of hematopoietic stem/progenitor proliferative status in vivo versus differentiation.
We demonstrate for the first time that endogenous RUNX1 interacts with endogenous HDAC1 or HDAC3. Moreover, we provide evidence that this interaction is direct, as GST-RUNX1 isolated from bacteria binds HDAC1 or HDAC3 generated by in vitro translation. HDAC1 exists in several co-repressor complexes, including an HDAC1-mSin3A complex and the NuRD complex, which includes chromatin remodeling ATPases (29). mSin3A also interacts with RUNX1, and this interaction is regulated by ERK phosphorylation of Ser-276/Ser-293 and by arginine methylation of Arg-206/Arg-210 (30, 31). Thus, several biochemical pathways converge to relieve transcriptional repression mediated by RUNX1. Interaction of a transcription factor with more than one component of a co-repressor complex is not without precedent; for example, PLZF interacts with both mSin3A and HDAC1 (32).
The RUNX1(S424D) variant has reduced affinity for HDAC1 or HDAC3, either upon co-expression in 293T cells or when the affinities of GST-RUNX1 and GST-RUNX1(S424D) for in vitro translated HDAC1 or HDAC3 are compared. RUNX1(S48D) and RUNX1(S303D) also demonstrated reduced affinity for either HDAC1 or HDAC3, when expressed in 293T cells, although the effect of these latter mutations on HDAC affinity in this context was less than when Ser-424 was changed to aspartic acid. A role for Ser-424 in interaction of RUNX1 with HDAC1 or HDAC3 is consistent with the finding that these HDACs interact with RUNX1 residues 380–432 linked to the GAL4 DNA-binding domain (33).
When GST-RUNX1 was incubated with excess cdk1/cyclin B1, interaction with HDAC1 was reduced, indicating that cdk1 phosphorylation and not only Ser to Asp mutations reduces HDAC1 affinity for RUNX1. Use of 3-fold more cdk1/cyclin B than we utilized in our prior study (20) was required to make this observation, as inclusion of lesser amounts of cdk1/cyclin B in the kinase reaction did not result in reduced HDAC1 binding (not shown), which is likely due to the lack of near stoichiometric phosphorylation. Of note, even the amount of cdk1 used may not have achieved full phosphorylation of all three cdk consensus serines. Phosphorylation of GST-RUNX1(S424A) by cdk1/cyclin B reduced HDAC1 affinity, as this variant can still be phosphorylated on Ser-48 and Ser-303, and these data provide further support for the involvement of these two serines in HDAC1 binding. In contrast, phosphorylation of GST-RUNX1(S48A/S303A) did not reduce RUNX1 interaction with HDAC1, even though this variant can still be phosphorylated on Ser-424. This finding appears inconsistent with data in Figs. 3 and 4A indicating that Ser-424 phosphorylation contributes to regulation of RUNX1-HDAC interactions. Perhaps less than stoichiometric phosphorylation of Ser-424 alone in vitro was insufficient to prevent HDAC1 binding. Also, we demonstrated that exogenous RUNX1 is at least partially phosphorylated on all three consensus serines in 293T cells (20), and our current finding that RUNX1(tripleA) increases HDAC1 binding almost 2-fold relative to RUNX1 suggests that 1–3 of these residues are phosphorylated in 293T cells such that mutation to alanine allows all rather than about half of the RUNX1 polypeptides to strongly bind HDAC1. Expression of RUNX1(S424D) in these cells would thus bias towards two or three phosphorylated serines, reducing interaction with HDAC1, whereas in vitro phosphorylation of GST-RUNX1(S48A/S303A) on only Ser-424 may not as easily prevent HDAC1 binding. In sum, our data support a role for all three consensus serines in HDAC interaction, although their relative contributions in vivo remains to be established.
Incubation of GST-RUNX1 with cdk1/cyclin B and ATP did not reduce HDAC3 affinity (data not shown), even though RUNX1-HDAC3 interaction was reduced by mutation of Ser-48, Ser-303, and Ser-424 to aspartic acid in 293T cells or in vitro. We had found that in vitro cdk1/cyclin B modifies additional serines or threonines in RUNX1 besides those that fit the more stringent (S/T)PX(R/K) cdk1 consensus (20). Perhaps these modifications, which likely occur at (S/T)P sites, increase HDAC3 affinity for RUNX1 in a manner that does not occur in vivo. On the other hand, the inability of these additional, likely artificial, modifications to prevent HDAC1 binding to GST-RUNX1(tripleA) highlights the specific role of phosphorylation of serines that fit the cdk consensus in regulating HDAC1 interaction.
We also find that the cdk inhibitor roscovitine increases interaction of exogenously expressed RUNX1 and HDAC1, consistent with the conclusion that RUNX1 cdk phosphorylation interferes with HDAC1 interaction. We could not demonstrate a similar increased co-immunoprecipitation between endogenous RUNX1 and HDAC1 or HDAC3 when the Jurkat or M1 hematopoietic cell lines were exposed to roscovitine, perhaps reflecting the increased cytotoxicity seen at the exposure time needed to reduce RUNX1 cdk phosphorylation.
In summary, our findings support the idea that in proliferating hematopoietic cells cdks modify RUNX1 on Ser-48, Ser-303, and Ser-424 to displace HDAC1 and HDAC3 to favor trans-activation of genes, including those that stimulate proliferation, whereas in quiescent cells reduced cdk activity would be expected to favor increased RUNX1-HDAC interaction to help maintain reduced proliferation. Increased or diminished RUNX1-HDAC interaction may also contribute to developmental decisions during hematopoiesis. Manipulation of RUNX1 cdk phosphorylation potentially provides a strategy to favor clinically useful adult HSC formation and expansion and to manipulate RUNX1 activities to reduce proliferation or induce differentiation of leukemia stem cells, including those harboring dominant-negative CBF oncoproteins or RUNX1 mutations.
Acknowledgments
We thank S. Hiebert for HDAC expression plasmids and N. Speck for RUNX1 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA098805, U01HL099775, and U01HL100397. This work was also supported by a grant from the Maryland Stem Cell Research Foundation (to A. D. F.).
O. Ma, H. Guo, and A. Friedman, unpublished data.
- HSC
- hematopoietic stem cell
- 4HT
- 4-hydroxytamoxifen
- cdk
- cyclin-dependent kinase
- ER
- estradiol receptor
- HDAC
- histone deacetylase
- HI-FBS
- heat-inactivated fetal bovine serum
- CBF
- core binding factor.
REFERENCES
- 1. Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., Hirai H. (2004) Nat. Med. 10, 299–304 [DOI] [PubMed] [Google Scholar]
- 2. Growney J. D., Shigematsu H., Li Z., Lee B. H., Adelsperger J., Rowan R., Curley D. P., Kutok J. L., Akashi K., Williams I. R., Speck N. A., Gilliland D. G. (2005) Blood 106, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A. (2009) Nature 457, 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedman A. D. (1999) Leukemia 13, 1932–1942 [DOI] [PubMed] [Google Scholar]
- 5. Niini T., Kanerva J., Vettenranta K., Saarinen-Pihkala U. M., Knuutila S. (2000) Haematologica 85, 362–366 [PubMed] [Google Scholar]
- 6. Cao W., Britos-Bray M., Claxton D. F., Kelley C. A., Speck N. A., Liu P. P., Friedman A. D. (1997) Oncogene 15, 1315–1327 [DOI] [PubMed] [Google Scholar]
- 7. Cao W., Adya N., Britos-Bray M., Liu P. P., Friedman A. D. (1998) J. Biol. Chem. 273, 31534–31540 [DOI] [PubMed] [Google Scholar]
- 8. Burel S. A., Harakawa N., Zhou L., Pabst T., Tenen D. G., Zhang D. E. (2001) Mol. Cell. Biol. 21, 5577–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Costa J., Chaudhuri S., Civin C. I., Friedman A. D. (2005) Leukemia 19, 921–929 [DOI] [PubMed] [Google Scholar]
- 10. Lou J., Cao W., Bernardin F., Ayyanathan K., Rauscher F. J., III, Friedman A. D. (2000) Oncogene 19, 2695–2703 [DOI] [PubMed] [Google Scholar]
- 11. Bernardin F., Yang Y., Civin C. I., Friedman A. D. (2002) Cancer Biol. Ther. 1, 492–496 [DOI] [PubMed] [Google Scholar]
- 12. Strom D. K., Nip J., Westendorf J. J., Linggi B., Lutterbach B., Downing J. R., Lenny N., Hiebert S. W. (2000) J. Biol. Chem. 275, 3438–3445 [DOI] [PubMed] [Google Scholar]
- 13. Yang Y., Wang W., Cleaves R., Zahurak M., Cheng L., Civin C. I., Friedman A. D. (2002) Cancer Res. 62, 2232–2235 [PubMed] [Google Scholar]
- 14. Bernardin F., Yang Y., Cleaves R., Zahurak M., Cheng L., Civin C. I., Friedman A. D. (2002) Cancer Res. 62, 3904–3908 [PubMed] [Google Scholar]
- 15. Bernardin-Fried F., Kummalue T., Leijen S., Collector M. I., Ravid K., Friedman A. D. (2004) J. Biol. Chem. 279, 15678–15687 [DOI] [PubMed] [Google Scholar]
- 16. Robertson A. J., Coluccio A., Knowlton P., Dickey-Sims C., Coffman J. A. (2008) PLoS ONE. 3, e3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nimmo R., Antebi A., Woollard A. (2005) Development 132, 5043–5054 [DOI] [PubMed] [Google Scholar]
- 18. Kagoshima H., Nimmo R., Saad N., Tanaka J., Miwa Y., Mitani S., Kohara Y., Woollard A. (2007) Development 134, 3905–3915 [DOI] [PubMed] [Google Scholar]
- 19. Biggs J. R., Peterson L. F., Zhang Y., Kraft A. S., Zhang D. E. (2006) Mol. Cell. Biol. 26, 7420–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L., Fried F. B., Guo H., Friedman A. D. (2008) Blood 111, 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Durst K. L., Lutterbach B., Kummalue T., Friedman A. D., Hiebert S. W. (2003) Mol. Cell. Biol. 23, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finer M. H., Dull T. J., Quin L., Farson D., Roberts M. R. (1994) Blood 83, 43–50 [PubMed] [Google Scholar]
- 23. Bee T., Liddiard K., Swiers G., Bickley S. R., Vink C. S., Jarratt A., Hughes J. R., Medvinsky A., de Bruijn M. F. (2009) Blood Cells Mol. Dis. 43, 35–42 [DOI] [PubMed] [Google Scholar]
- 24. Joshi I., Minter L. M., Telfer J., Demarest R. M., Capobianco A. J., Aster J. C., Sicinski P., Fauq A., Golde T. E., Osborne B. A. (2009) Blood 113, 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahashi-Yanaga F., Sasaguri T. (2008) Cell. Signal. 20, 581–589 [DOI] [PubMed] [Google Scholar]
- 26. Burns C. E., Galloway J. L., Smith A. C., Keefe M. D., Cashman T. J., Paik E. J., Mayhall E. A., Amsterdam A. H., Zon L. I. (2009) Blood 113, 5776–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roussel M. F., Theodoras A. M., Pagano M., Sherr C. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6837–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsuzuki S., Hong D., Gupta R., Matsuo K., Seto M., Enver T. (2007) PLoS Med. 4, e172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denslow S. A., Wade P. A. (2007) Oncogene 26, 5433–5438 [DOI] [PubMed] [Google Scholar]
- 30. Imai Y., Kurokawa M., Yamaguchi Y., Izutsu K., Nitta E., Mitani K., Satake M., Noda T., Ito Y., Hirai H. (2004) Mol. Cell. Biol. 24, 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H., Allis C. D., Tempst P., Nimer S. D. (2008) Genes Dev. 22, 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wong C. W., Privalsky M. L. (1998) J. Biol. Chem. 273, 27695–27702 [DOI] [PubMed] [Google Scholar]
- 33. Reed-Inderbitzin E., Moreno-Miralles I., Vanden-Eynden S. K., Xie J., Lutterbach B., Durst-Goodwin K. L., Luce K. S., Irvin B. J., Cleary M. L., Brandt S. J., Hiebert S. W. (2006) Oncogene 25, 5777–5786 [DOI] [PubMed] [Google Scholar]