Abstract
Mdm2 and Mdmx are oncoproteins that have essential yet nonredundant roles in development and function as part of a multicomponent ubiquitinating complex that targets p53 for proteasomal degradation. However, in response to DNA damage, Mdm2 and Mdmx are phosphorylated and protect p53 through various mechanisms. It has been predicted that Mdm2-Mdmx complex formation modulates Mdm2 ligase activity, yet the mechanism that promotes formation of Mdm2-Mdmx complexes is unknown. Here, we show that optimal Mdm2-Mdmx complex formation requires c-Abl phosphorylation of Mdm2 both in vitro and in vivo. In addition, Abl phosphorylation of Mdm2 is required for efficient ubiquitination of Mdmx in vitro, and eliminating c-Abl signaling, using c-Abl−/− knock-out murine embryonic fibroblasts, led to a decrease in Mdmx ubiquitination. Further, p53 levels are not induced as efficiently in c-Abl−/− murine embryonic fibroblasts following DNA damage. Overall, these results define a direct link between genotoxic stress-activated c-Abl kinase signaling and Mdm2-Mdmx complex formation. Our results add an important regulatory mechanism for the activation of p53 in response to DNA damage.
Keywords: DNA Damage, E3 Ubiquitin Ligase, Protein Degradation, Protein-Protein Interactions, Signal Transduction, Ubiquitination, Mdm2, Mdmx, c-Abl
Introduction
The tumor suppressor p53 plays a pivotal role in regulating cellular processes governing cell cycle arrest, apoptosis, and senescence. Central to regulation of p53 is the activity of two negative regulators, Mdm2 and Mdmx. The prevailing model for regulation of p53 suggests that Mdm2 is involved in regulating p53 protein stability whereas Mdmx regulates p53 transcriptional activity (1). Mdm2 and Mdmx elicit their effects largely based on phosphorylation status to modulate their protein interactions.
Kinase activity in response to DNA damage leads to phosphorylation of Mdm2 and Mdmx ultimately stabilizing p53 and leading to its transcriptional activation (2, 3). Phosphorylation of Mdm2 by ATM blocks nuclear export of p53 (4) and prevents polyubiquitination of p53 by inhibiting Mdm2 RING domain homodimerization (5). In addition, ATM and Chk2 phosphorylation of Mdmx rapidly destabilizes Mdmx (6–9). c-Abl kinase is activated by ATM in response to DNA damage (10) and phosphorylates Mdm2 and Mdmx. c-Abl phosphorylation of Mdm2 stabilizes p53 and promotes apoptosis (11, 12), whereas c-Abl phosphorylation of Mdmx activates p53 by blocking Mdmx-p53 complex formation (13).
Mdm2 and Mdmx associate via their RING domains (14), and phosphorylation near the RING domains is predicted to alter oligomerization and is associated with stabilization of p53. This model is supported by recent findings that Wip1, which dephosphorylates Mdm2 and Mdmx, inhibits p53 activity (15, 16). The oligomerization of Mdm2 and Mdmx has garnered attention recently as growing evidence suggests that this serves as a regulatory mechanism. Several studies have shown that Mdmx stabilizes Mdm2 in cells and prevents Mdm2-mediated degradation of p53 (17–19), whereas overexpressed Mdmx promotes p53-dependent apoptosis (20). However, the presence of Mdm2 and Mdmx together in vitro and in vivo has the potential to degrade p53 (21, 22). These seemingly conflicting results appear to depend on the stoichiometric balance of Mdm2 and Mdmx. Drawing from structural modeling and in vitro ubiquitin ligase assays, it has been suggested that E2 recruitment to Mdm2 homo-oligomers would autoubiquitinate Mdm2, whereas Mdm2-Mdmx hetero-oligomers would preferentially ubiquitinate substrates (23). Although molecular modeling provides useful insights into the process of E2 recruitment, experimental evidence to support this model has yet to be substantiated. However, these studies do point to the importance of understanding the oligomerization of Mdm2 and Mdmx.
The biochemical mechanism and functional importance for Mdm2 homo-oligomerization have been demonstrated (5), but the process that underlies the formation of Mdm2-Mdmx complexes remains unclear. Here, we investigate Mdm2-Mdmx complex formation as a consequence of DNA damage signaling. We show that Mdm2-Mdmx complex formation is augmented by c-Abl phosphorylation of Mdm2. Inhibiting c-Abl activity, through a dominant negative Abl mutant or the selective inhibitor imatinib, leads to a decrease in Mdm2-Mdmx complex formation. Study of c-Abl signaling using c-Abl−/− knock-out primary murine embryonic fibroblasts (MEFs)2 (24) recapitulated these findings and exhibited a decrease in Mdmx ubiquitination in response to DNA damage. In addition, p53 was not as robustly induced in c-Abl−/− MEFs after genotoxic stress. These data form a link between DNA damage signaling and Mdm2-Mdmx complex formation that is important for regulating Mdm2 activity.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
Mammalian cells were cultured at 37 °C in a humidified incubator with 5% CO2 in DMEM high glucose plus 10% FBS. c-Abl−/− and control primary MEFs (24) were cultured in the same media plus nonessential amino acids. Transfections were performed using Lipofectamine Plus reagent (Invitrogen). Equal amounts of DNA of human origin (HA-Mdm2 (2 μg), Myc-Mdmx (4 μg), Abl and kinase dead (KD)-Abl (2 μg), plus control plasmid) were transfected into cells for 4 h in Iscove's modified Dulbecco's medium without serum. Medium was replaced with normal growth medium and incubated 18–20 h prior to analysis. For the luciferase assays, H1299 cells were transfected with 50 ng of p53, 1 μg of HA-Mdm2, 500 ng of β-galactosidase, and 100 ng of PG13-luc plasmid using the calcium phosphate method. Reporter activity was normalized to β-galactosidase. Doxorubicin (Sigma) was used at 5 μm, and imatinib (LC Laboratories) was used at 10 μm or as indicated in DMEM/1% FBS.
Site-directed mutagenesis was performed by PCR, and constructs were sequenced in their entirety. shRNA to c-Abl (5′-GACCAACTTGTTCAGCGCC-3′) and scramble control sequences (25) were cloned into the pLVTHM vector (Addgene) and used to make virus for subsequent infection. A population of positive cells was used for experiments.
Protein Analysis and Immunoprecipitation
Whole cell extracts (WCEs) were prepared in Nonidet P-40 lysis buffer (25 mm Tris, pH 8.0, 150 mm NaCl, 0.5 mm EDTA, 0.5 mm EGTA, 1% Nonidet P-40, 1 mm sodium orthovanadate, 1 mm DTT) and supplemented with protease inhibitor mixture set III (Calbiochem) at 1:100 and incubated on ice for 30 min. Debris was collected by centrifugation, and equal amounts of supernatant protein were resuspended in SDS loading buffer and boiled for 5 min. Protein was fractionated by SDS-PAGE and transferred to PVDF membrane (Amersham Biosciences). Antibodies used for Western blotting were: Mdm2 (IF2 and 2A10) and c-Abl (24-21) (Calbiochem); p53 (DO-1), ubiquitin (P4D1), tubulin (TU-02), GAPDH (8C2), and β-actin (C4) (Santa Cruz Biotechnology); Mdmx (Bethyl Laboratories); phosphotyrosine (4G10) (Upstate); phospho-c-Abl Tyr245 (Cell Signaling); Myc (9E10) and HA (12CA5) (Roche Applied Science). Immunoprecipitation was performed using 500 ng of either Myc-tagged or HA-tagged antibody from WCE overnight at 4 °C in 700 μl of PBS. Protein G-Sepharose (Pierce) was added to the mixture and incubated an additional 1 h at 4 °C, precipitates were washed three times in PBS, and samples were resuspended in SDS loading buffer and boiled for 5 min. Immunoprecipitation of endogenous Mdm2 from human cells used either IF2 and 2A10 antibodies or SMP14 antibody, and immunoprecipitation from MEFs used polyclonal anti-Mdm2 (C-18; Santa Cruz). Immunoprecipitation of endogenous Mdmx used polyclonal anti-Mdmx (A300–287A; Bethyl). Endogenous ubiquitinated proteins from MEFs were purified using Tandem Ubiquitin Binding Entities (LifeSensors) following the manufacturer's instructions.
Recombinant Proteins and in Vitro Kinase Reactions
Recombinant full-length human Mdm2 and Mdmx proteins were purified from Escherichia coli BL-21 following induction with IPTG. In vitro Abl kinase reactions were performed at 37 °C for 30 min in kinase buffer (25 mm Tris, pH 7.4, 10 mm MgCl2, 1 mm MnCl2, 0.5 mm DTT, 10 μm ATP) and 0.5 μg of Abl or heat-inactivated Abl (100 °C for 5 min) (Invitrogen). After in vitro kinase reactions, protein was purified to remove Abl from downstream applications. ELISAs were performed using standard techniques. 250 μg of target protein was bound to the ELISA plate. Mdm2 binding was detected using SMP14 antibody (Santa Cruz Biotechnology) and TMB One (Promega).
RESULTS
DNA Damage Augments Mdm2-Mdmx Complex Formation
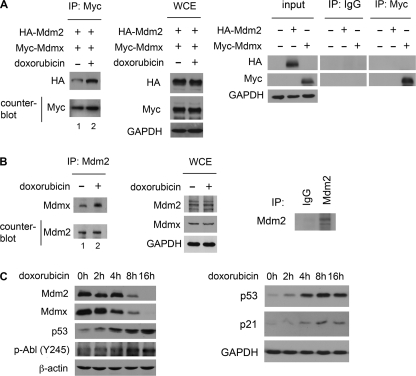
Mdm2 and Mdmx are capable of forming hetero-oligomeric complexes. Studies have suggested somewhat conflicting results for the functional role of Mdm2-Mdmx complexes in regulation of p53. However, the requirement for the formation of Mdm2-Mdmx complexes in cell-based assays has not been addressed. c-Abl has been shown to phosphorylate Mdm2 and lead to stabilization of p53 (11, 12). We hypothesized that phosphorylation of Mdm2 by c-Abl would affect Mdm2-Mdmx complex formation as a mechanism for functional inactivation of Mdm2. To test this hypothesis, we examined the conditions that favor the formation of Mdm2-Mdmx complexes. We employed a co-immunoprecipitation approach following treatment with 5 μm doxorubicin, a topoisomerase II inhibitor that results in single and double strand DNA breaks and is known to activate c-Abl. It has been shown that Mdm2 levels are independent of p53 regulation under these conditions (26) and other forms of severe genotoxic stress (27). This level of DNA damage allowed us to investigate Mdm2 phosphorylation without p53-mediated transcription of Mdm2. 293T cells were transfected with expression plasmids for HA-Mdm2 and Myc-Mdmx. Immunoprecipitation of Myc-Mdmx showed a significant increase in the level of co-purification with HA-Mdm2 after DNA damage (Fig. 1A, lane 2). Counterblotting for Myc showed an equivalent immunoprecipitation of Myc-Mdmx. The WCEs showed no difference in expression levels after 2 h of doxorubicin. Control immunoprecipitations using normal mouse IgG demonstrate specificity in our assays. To examine endogenous Mdm2-Mdmx complex formation, Mdm2 was immunoprecipitated from control and doxorubicin-treated MCF-7 cells. Consistent with the overexpression experiments, a significant increase in endogenous Mdm2-Mdmx co-purification was observed in response to DNA damage (Fig. 1B, lane 2). Counterblotting for Mdm2 showed equivalent immunoprecipitation. Again, the WCEs showed no difference in protein expression after 2 h of doxorubicin, and control immunoprecipitations using normal mouse IgG show specificity in these experiments.
FIGURE 1.
DNA damage augments Mdm2-Mdmx complex formation. A, 293T cells expressing HA-Mdm2 and Myc-Mdmx were treated with 5 μm doxorubicin for 2 h. Myc-Mdmx was immunoprecipitated (IP) using the Myc tag, and HA-Mdm2 co-purification was analyzed by Western blotting with HA antibody. The counterblot for Myc shows equal Myc-Mdmx immunoprecipitation. WCEs were probed as indicated. Control immunoprecipitations using normal mouse IgG and Myc antibody demonstrate specificity in our assays. B, MCF-7 cells were treated with doxorubicin as in A. Endogenous Mdm2 was immunoprecipitated, and Mdmx co-purification was analyzed by Western blotting. The counterblot for Mdm2 shows equal immunoprecipitation of Mdm2. WCEs show cellular levels of proteins after doxorubicin treatment. Immunoprecipitations using normal mouse IgG and Mdm2 antibody are included as controls. C, MCF-7 cells were treated with doxorubicin as in A for the time points indicated. Cell lysates were prepared and analyzed by Western blotting for Mdm2, Mdmx, p53, p21, and phosphorylated c-Abl (p-Abl (Y245)).
Mdm2 and Mdmx are substrates of c-Abl, and c-Abl plays an important role in the DNA damage response (28). In MCF-7 cells treated with doxorubicin, c-Abl is activated by 4 h with a maximal response at 8 h, as determined by phosphorylation at Tyr245 (29). Under these conditions, Mdm2 and Mdmx proteins are present at high levels at 2 h (Fig. 1C). Additionally, activation of c-Abl preceded destabilization of both Mdm2 and Mdmx and was concomitant with accumulation of p53 (30) (Fig. 1C). Further, p53 induced the downstream target, p21 under these conditions. These data show that DNA damage leads to a significant increase in Mdm2-Mdmx complex formation and that under these conditions c-Abl is activated.
Abl Phosphorylation of Mdm2 Increases Mdm2-Mdmx Complex Formation in Vitro
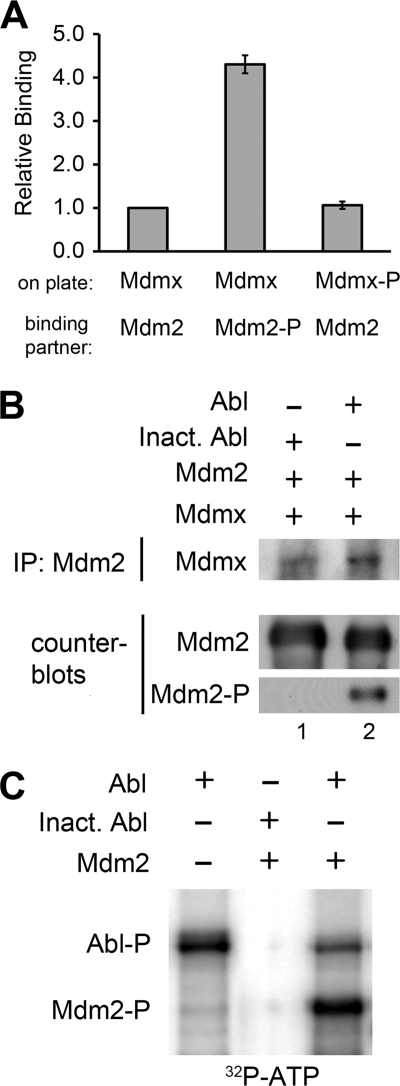
To test directly the importance of Abl phosphorylation of Mdm2 in Mdm2-Mdmx complex formation, we employed an ELISA-based approach to measure the relative binding of recombinant Mdm2 and Mdmx proteins. Abl-phosphorylated Mdm2 (Mdm2-P) exhibited a >4-fold increase in binding to Mdmx over unphosphorylated Mdm2. Whereas Abl phosphorylation of Mdm2 played a role in complex formation, phosphorylation of Mdmx (Mdmx-P) did not affect complex formation (13) (Fig. 2A). Co-immunoprecipitation of recombinant Mdm2 with Mdmx recapitulated the increase in complex formation following Abl phosphorylation of Mdm2 (Fig. 2B, lane 2). Counterblotting showed equivalent immunoprecipitation of Mdm2. To confirm the presence of Abl modification on Mdm2 we also reprobed the blot for phosphotyrosine residues. 32P incorporation using [γ-32P]ATP in our kinase reactions confirmed that Abl efficiently phosphorylated Mdm2. Autophosphorylation of Abl served as an internal control for the reaction (Fig. 2C). These data define a direct link between Abl phosphorylation of Mdm2 and an increase in Mdm2-Mdmx complex formation in vitro.
FIGURE 2.
Abl phosphorylation of Mdm2 increases Mdm2-Mdmx complex formation in vitro. A, ELISA-based protein binding assay using recombinant Mdm2 and Mdmx. Mdmx was bound to an assay plate followed by Mdm2. Proteins were subjected to in vitro Abl kinase reactions as indicated (-P). B, co-purification of recombinant Mdm2 and Mdmx following in vitro Abl kinase reactions with either active Abl or heat-inactivated Abl (100 °C 10 min). Mdm2 was immunoprecipitated (IP) and Western blot-probed for Mdmx. Counterblots show equal immunoprecipitation of Mdm2 and in vitro Abl activity (Mdm2-P) via phosphotyrosine antibody. C, autoradiograph of 32P incorporation in the in vitro Abl kinase reaction demonstrating phosphorylation of Mdm2 (Mdm2-P). Autophosphorylation of Abl served as an internal control for the reaction.
c-Abl Phosphorylation of Mdm2 Increases Mdm2-Mdmx Complex Formation in Vivo
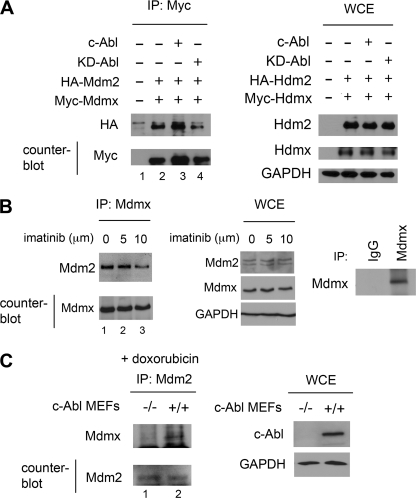
The increase in Mdm2-Mdmx complex formation and c-Abl activation occurs early in response to DNA damage in MCF-7 cells (Fig. 1C). To investigate the cellular role of c-Abl in Mdm2-Mdmx complex formation, 293T cells were transfected with HA-Mdm2, Myc-Mdmx, and either wild-type Abl or KD-Abl (K290R dominant negative) (31). Immunoprecipitation of Myc-Mdmx showed a significant decrease in the level of HA-Mdm2 co-purification when KD-Abl was expressed (Fig. 3A, compare lanes 2 and 4). Counterblotting showed equivalent immunoprecipitation of Myc-Mdmx in these samples. To extend this result and test the role of c-Abl in endogenous Mdm2-Mdmx complex formation, we used the selective inhibitor, imatinib (32). MCF-7 cells were treated with increasing concentrations of imatinib for 2 h. At high dose, imatinib led to a modest decrease in the level of endogenous Mdm2-Mdmx complex formation (Fig. 3B, lane 3). The levels of Mdm2 and Mdmx in WCEs used for immunoprecipitation remained unchanged. Control immunoprecipitations using normal rabbit IgG demonstrate specificity in this assay.
FIGURE 3.
c-Abl phosphorylation of Mdm2 increases Mdm2-Mdmx complex formation in vivo. A, 293T cells expressing HA-Mdm2, Myc-Mdmx, and either Abl or dominant negative KD-Abl (K290R) are shown. Myc-Mdmx was immunoprecipitated (IP) using the Myc tag and analyzed by Western blotting for co-purification of HA-Mdm2. The counterblot for Myc shows equal Myc-Mdmx immunoprecipitation. WCEs were probed as indicated. B, MCF-7 cells were treated with imatinib for 2 h as indicated, and endogenous Mdmx was immunoprecipitated and analyzed by Western blotting for co-purification of Mdm2. The counterblot for Mdmx shows equal immunoprecipitation. WCEs were analyzed by Western blotting to show cellular levels of indicated proteins after imatinib treatment. Immunoprecipitations using normal rabbit IgG and Mdmx antibody are included as controls. C, c-Abl−/− and wild-type MEFs were treated with 5 μm doxorubicin for 2 h. Endogenous Mdm2 was immunoprecipitated and Western blot-probed for Mdmx co-purification. The counterblot shows the level of Mdm2 immunoprecipitation from MEFs. WCEs were probed as indicated.
To address the importance of c-Abl signaling in vivo further, c-Abl−/− knock-out primary MEFs (24) were used to probe Mdm2-Mdmx complex formation. In response to DNA damage, co-immunoprecipitation of endogenous Mdm2 with Mdmx was reduced significantly in MEFs lacking c-Abl compared with control MEFs (Fig. 3C, lane 1). Counterblotting for Mdm2 showed an equivalent level of Mdm2 immunoprecipitation in the MEFs. These data show, in a cellular context, a direct link between c-Abl kinase activity and an increase in Mdm2-Mdmx complex formation.
Multisite Phosphorylation of Mdm2 Is Required for Maximal Mdm2-Mdmx Complex Formation
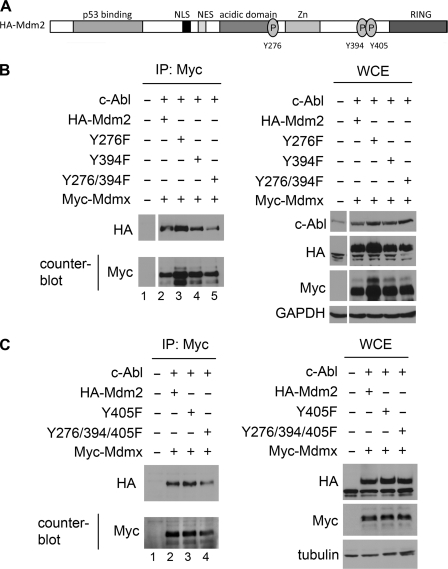
Mdm2 has three known c-Abl phosphorylation sites (Tyr276, Tyr394, and Tyr405) (11, 12). Considering that two of these lie proximal to the RING domain, we tested the importance of these three sites in Mdm2-Mdmx complex formation. A series of phenylalanine mutants of HA-Mdm2 were generated to each site individually (Y276F, Y394F, and Y405F) or in combination (Y276F/Y394F and Y276F/Y394F/Y405F) to target c-Abl phosphorylation sites (Fig. 4A). If c-Abl phosphorylation is important for Mdm2-Mdmx complex formation then we would expect to see a decrease in Mdm2-Mdmx co-purification with these mutants. Expression of these mutants with Myc-Mdmx and c-Abl in 293T cells revealed a cooperative effect of Tyr276 and Tyr394 in formation of Mdm2-Mdmx complexes. Immunoprecipitation of Myc-Mdmx exhibited a significant decrease in the level of HA-Mdm2 co-purification with the double mutant Y276F/Y394F compared with wild-type HA-Mdm2 (Fig. 4B, lane 5). The counterblot shows equivalent immunoprecipitation for HA-Mdm2 and the Y276F/Y394F mutant. The control WCE loading was 1/15 the quantity of protein used for immunoprecipitation. HA-Mdm2 Y405F did not affect co-purification with Myc-Mdmx (Fig. 4C, lane 3), whereas the triple mutant HA-Mdm2 Y276F/Y394F/Y405F reduced co-purification to a level similar to that observed for HA-Mdm2 Y276F/Y394F (Fig. 4C, lane 4). These data show that multisite phosphorylation of Mdm2 by c-Abl (Y276F/Y394F) is important for optimal Mdm2-Mdmx complex formation. Importantly, these data do not rule out contributing factors provided by additional signaling events that could augment Mdm2-Mdmx complex formation.
FIGURE 4.
Multisite phosphorylation of Mdm2 is required for maximal Mdm2-Mdmx complex formation. A, schematic of the c-Abl phosphorylation sites (Tyr276, Tyr394, and Tyr405) in Mdm2. Tyrosine to phenylalanine mutants to each site individually (Y276F, Y394F, and Y405F) or in combination (Y276F/Y394F and Y276F/Y394F/Y405F) were generated in HA-Mdm2. B, 293T cells expressing Myc-Mdmx, Abl, and wild-type HA-Mdm2 or HA-Mdm2 point mutants (Y276F, Y394F, and Y276/394F). Myc-Mdmx was immunoprecipitated (IP) using the Myc tag, and HA-Mdm2 co-purification was analyzed by Western blotting with HA antibody. The counterblot shows the level of Myc-Mdmx immunoprecipitated. WCEs were probed as indicated. C, 293T cells expressing Myc-Mdmx, Abl and HA-Mdm2, or HA-Mdm2 point mutants (Y405F and Y276F/Y394F/Y405F). Myc-Mdmx was immunoprecipitated using the Myc tag, and HA-Mdm2 co-purification was analyzed by Western blotting with HA. The counterblot shows the level of Myc-Mdmx immunoprecipitated. WCEs were probed as indicated.
c-Abl Promotes Mdmx Ubiquitination
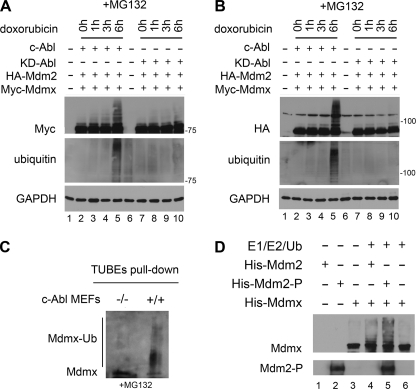
Mdm2 oligomerization in response to DNA damage has been shown to alter the Mdm2 E3 ligase function (5), suggesting that phosphorylation of Mdm2 can modulate complex formation to form a unique ligase complex. Examination of the role for c-Abl in ubiquitination of Mdm2 and Mdmx following DNA damage was done by expression of either Abl or KD-Abl with HA-Mdm2 and Myc-Mdmx in 293T cells. Transfected cells were treated with 5 μm doxorubicin to induce DNA damage and 25 μm MG132 to block proteasome activity. Our results revealed an abundance of ubiquitin-modified Mdmx in cells expressing Abl but not in cells expressing KD-Abl (Fig. 5A, compare lanes 5 and 10). The blot was reprobed for ubiquitin, suggesting that the laddering was ubiquitination of Mdmx. We also found that Mdm2 was efficiently ubiquitinated in cells expressing c-Abl but not in cells expressing KD-Abl (Fig. 5B, compare lanes 5 and 10). The lack of ubiquitination in cells expressing KD-Abl illustrates the importance of c-Abl in response to genotoxic stress. To investigate ubiquitination of endogenous protein, we used c-Abl−/− MEFs and Tandem Ubiquitin Binding Entities, which efficiently bind ubiquitinated proteins while also protecting against deubiquitination. Our results showed a significant decrease in ubiquitination of endogenous Mdmx in the primary c-Abl−/− MEFs compared with control MEFs in the presence of 25 μm MG132 (Fig. 5C). This result is representative of two independent experiments.
FIGURE 5.
c-Abl kinase activity promotes ubiquitination of Mdmx. A, 293T cells expressing HA-Mdm2, Myc-Mdmx, and either Abl or dominant negative KD-Abl (K290R). Cells were treated with 5 μm doxorubicin and 25 μm MG132 for the time points indicated. WCEs were analyzed by Western blotting for Myc-Mdmx, and blots were reprobed for ubiquitin. B, 293T cells treated as described in A. Western blots were probed for HA-Mdm2 and blots reprobed for ubiquitin. C, c-Abl−/− and wild-type MEFs treated with 25 μm MG132 for 4 h. GST-Tandem Ubiquitin Binding Entities (GST-TUBEs) were used to pull out ubiquitinated (Ub) proteins. The Western blot was probed for Mdmx. D, in vitro ubiquitination assay of recombinant Mdm2 or Abl phosphorylated Mdm2 (Mdm2-P) with recombinant Mdmx. The Western blot was probed for Mdmx, and the blot was reprobed for phosphotyrosine.
To investigate directly the impact of Abl phosphorylation in the ability of Mdm2 to ubiquitinate Mdmx, an in vitro ubiquitination assay was performed. Consistent with our expression system and the c-Abl−/− MEFs, our results showed that Abl-phosphorylated Mdm2 (Mdm2-P) significantly increased the abundance of ubiquitinated Mdmx compared with nonphosphorylated Mdm2 (Fig. 5D, compare lanes 4 and 5). Counterblotting confirmed the presence of phosphotyrosine modifications to Mdm2. These data show that c-Abl is important for maximizing ubiquitination of Mdmx following DNA damage.
Inactivation of c-Abl Protects Mdm2 and Mdmx and Decreases p53 Accumulation
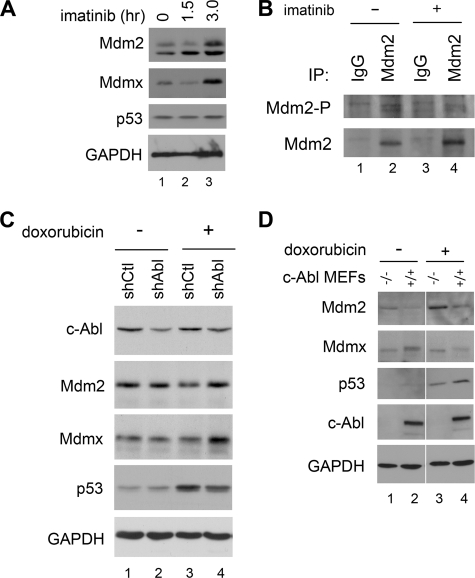
Consistent with an increase in ubiquitination of Mdm2 and Mdmx through the activity of c-Abl, imatinib-treated MCF-7 cells showed a protection of endogenous Mdm2 and Mdmx protein levels (Fig. 6A, lane 3). This suggests a role for c-Abl phosphorylation in basal maintenance of Mdm2 and Mdmx protein in unstressed cells. Under these conditions, p53 levels were unchanged, indicating that imatinib treatment alone is not sufficient to induce p53 (Fig. 6A). This observation is likely the result of the pool of Mdm2 that does not regulate p53 in the absence of DNA damage (33). To show that imatinib treatment is capable of blocking tyrosine phosphorylation of Mdm2, MCF-7 lysates were immunoprecipitated with Mdm2 antibody and probed for phosphotyrosine (Fig. 6B, lanes 2 and 4). In addition, after DNA damage, cells expressing shRNA for gene silencing of c-Abl (25) exhibited a significant increase in Mdmx protein despite only modest c-Abl protein knockdown. Mdm2 protein level increased to a lesser degree under these conditions (Fig. 6C, compare lanes 3 and 4). Further, p53 accumulates in control cells following DNA damage, whereas in cells with c-Abl protein knockdown the accumulation of p53 is significantly attenuated (Fig. 6C, lanes 3 and 4).
FIGURE 6.
Inactivation of c-Abl protects Mdm2 and Mdmx and decreases p53 accumulation. A, MCF-7 cells were treated with 10 μm imatinib for the time points indicated. WCEs were analyzed by Western blotting for Mdm2, Mdmx, and p53. B, MCF-7 cells were treated with 10 μm imatinib for 3 h, and Mdm2 was immunoprecipitated (IP) using SMP14 to detect tyrosine phosphorylation of Mdm2 using a phosphotyrosine antibody. The counterblot for total Mdm2 shows equal immunoprecipitation of Mdm2. C, MCF-7 cells stably expressing shRNA control or c-Abl silencing sequences treated with 5 μm doxorubicin for 2 h. WCEs were probed for endogenous c-Abl, Mdm2, Mdmx, and p53. D, c-Abl−/− and wild-type MEFs treated with 5 μm doxorubicin for 2 h. WCEs were probed by Western blotting for Mdm2, Mdmx, p53, and c-Abl.
To test the Mdm2 and Mdmx protein levels and p53 accumulation in a cellular system completely lacking c-Abl, we again utilized the c-Abl−/− MEFs. In response to DNA damage, the level of endogenous Mdm2 and Mdmx protein in the c-Abl−/− MEFs was more abundant than in control cells (Fig. 6D, compare lanes 3 and 4). Further, if the Mdm2-Mdmx complex is lending itself to degradation through c-Abl activity then we would expect p53 to be destabilized in the absence of c-Abl following DNA damage. Consistent with this, our results showed that p53 protein was not as robustly induced in c-Abl−/− MEFs in response to DNA damage (Fig. 6D). This is consistent with previous data showing that c-Abl protects p53 (28). In addition, expression of HA-Mdm2 Y276F/Y394F (c-Abl point mutant) showed a significant destabilization of p53 compared with expression of wild-type HA-Mdm2 (supplemental Fig. S1A). Also, p53 transcriptional activity was significantly reduced in cells treated with imatinib to block c-Abl signaling (Fig. S1B). These data show that c-Abl is important for reducing Mdm2 and Mdmx protein levels after genotoxic stress and suggest another cellular mechanism for the stabilization and activation of p53.
DISCUSSION
Previous data clearly show a role for DNA damage-induced phosphorylation of Mdm2 and Mdmx in the stabilization of p53. However, most reports have focused on modulating the direct binding of Mdm2 and Mdmx to p53 or to other effector molecules that lead to their destabilization. Recently, the oligomerization of Mdm2 and Mdmx has attracted much attention as a mechanism for modulating ligase activity. Structural modeling studies of the RING domains of Mdm2 and Mdmx have suggested that heterodimerization provides a mechanism for the recruitment of E2s to the ligase complex and directs target specificity (23). This report predicts that the Mdm2-Mdmx heterodimer would destabilize p53, but our cell-based assays show that endogenous Mdm2 and Mdmx form hetero-oligomers in response to c-Abl phosphorylation and stabilize p53. Our data also show that c-Abl phosphorylation of Mdm2 destabilizes Mdmx through an ubiquitin-dependent pathway. These results add an important regulatory mechanism to the formation and functional role of the Mdm2-Mdmx complex.
Our data suggest that multisite phosphorylation of Mdm2 by c-Abl is important for Mdm2-Mdmx complex formation (Fig. 4). Further, one of the tyrosine residues important for complex formation is proximal to the RING domain of Mdm2. The placement of this modification provides for potential RING domain structural changes required to modulate interactions. This is consistent with ATM phosphorylation of Mdm2 affecting RING domain homodimerization and activity (5). Importantly, RING domain dimerization appears to be a general requirement for active ligase complex assembly (34).
In the absence of c-Abl activity, using the c-Abl−/− MEFs, we observed a loss of Mdmx ubiquitination following genotoxic stress (Fig. 5C). Therefore, c-Abl phosphorylation is important not only for Mdm2-Mdmx complex formation but also the full transcriptional potential of p53 via destabilization of Mdmx (13). In addition, we observed a lack of Mdmx ubiquitination in response to DNA damage when KD-Abl was expressed in cells (Fig. 5A). In these experiments there is evidence of ubiquitination at 1 and 3 h, although the most abundant level of ubiquitination was not observed until 6 h. The kinetics of ubiquitination in these experiments, but not the underlying mechanism, is likely driven in part by ectopic expression of Mdm2 and Mdmx. We expected ubiquitination of Mdmx in these experiments even in the presence of KD-Abl because c-Abl is downstream of ATM in the DNA damage-signaling cascade. ATM and ATM-dependent Chk1/2 phosphorylation of Mdmx leads to ubiquitin-dependent destabilization of Mdmx (6–9). Thus, our results highlight the importance of c-Abl signaling under extreme genotoxic stress and are likely due to the type and severity of DNA damage.
Overall, our data suggest a positive effect of c-Abl phosphorylation of Mdm2 in p53 stabilization through destabilization of the Mdm2-Mdmx complex. Although the stress response is often associated with activation of Mdm2 under sublethal stress response, there are different responses for Mdm2 based on different stress conditions. In general, sublethal damage is thought to be associated with cycle arrest whereas severe stress leads to an apoptotic response (27). It has been proposed by others (5, 35) and supported by these data that modulating the oligomerization of E3 complexes provides a mechanism for control of ubiquitination. Selectively inducing Mdm2-Mdmx complex formation would provide a novel therapeutic target to protect p53 and other tumor suppressors that are negatively regulated by this complex.
Supplementary Material
Acknowledgments
We thank Dr. Hua Lu for the human HA-Mdm2 and Myc-Mdmx plasmids and Dr. Jean Y. J. Wang for the c-Abl−/− knockout and control MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA109262 (to L. D. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- MEF
- murine embryonic fibroblast
- KD
- kinase dead
- WCE
- whole cell extract
- ATM
- ataxia telangiectasia mutated.
REFERENCES
- 1. Toledo F., Wahl G. M. (2007) Int. J. Biochem. Cell Biol. 39, 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meek D. W. (2009) Nat. Rev. Cancer 9, 714–723 [DOI] [PubMed] [Google Scholar]
- 3. Waning D. L., Lehman J. A., Batuello C. N., Mayo L. D. (2010) Pharmaceuticals 3, 1576–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maya R., Balass M., Kim S. T., Shkedy D., Leal J. F., Shifman O., Moas M., Buschmann T., Ronai Z., Shiloh Y., Kastan M. B., Katzir E., Oren M. (2001) Genes Dev. 15, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng Q., Chen L., Li Z., Lane W. S., Chen J. (2009) EMBO J. 28, 3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen L., Gilkes D. M., Pan Y., Lane W. S., Chen J. (2005) EMBO J. 24, 3411–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okamoto K., Kashima K., Pereg Y., Ishida M., Yamazaki S., Nota A., Teunisse A., Migliorini D., Kitabayashi I., Marine J. C., Prives C., Shiloh Y., Jochemsen A. G., Taya Y. (2005) Mol. Cell. Biol. 25, 9608–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. LeBron C., Chen L., Gilkes D. M., Chen J. (2006) EMBO J. 25, 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pereg Y., Lam S., Teunisse A., Biton S., Meulmeester E., Mittelman L., Buscemi G., Okamoto K., Taya Y., Shiloh Y., Jochemsen A. G. (2006) Mol. Cell. Biol. 26, 6819–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baskaran R., Wood L. D., Whitaker L. L., Canman C. E., Morgan S. E., Xu Y., Barlow C., Baltimore D., Wynshaw-Boris A., Kastan M. B., Wang J. Y. (1997) Nature 387, 516–519 [DOI] [PubMed] [Google Scholar]
- 11. Dias S. S., Milne D. M., Meek D. W. (2006) Oncogene 25, 6666–6671 [DOI] [PubMed] [Google Scholar]
- 12. Goldberg Z., Vogt Sionov R., Berger M., Zwang Y., Perets R., Van Etten R. A., Oren M., Taya Y., Haupt Y. (2002) EMBO J. 21, 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuckerman V., Lenos K., Popowicz G. M., Silberman I., Grossman T., Marine J. C., Holak T. A., Jochemsen A. G., Haupt Y. (2009) J. Biol. Chem. 284, 4031–4039 [DOI] [PubMed] [Google Scholar]
- 14. Poyurovsky M. V., Priest C., Kentsis A., Borden K. L., Pan Z. Q., Pavletich N., Prives C. (2007) EMBO J. 26, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu X., Ma O., Nguyen T. A., Jones S. N., Oren M., Donehower L. A. (2007) Cancer Cell 12, 342–354 [DOI] [PubMed] [Google Scholar]
- 16. Zhang X., Lin L., Guo H., Yang J., Jones S. N., Jochemsen A., Lu X. (2009) Cancer Res. 69, 7960–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stad R., Little N. A., Xirodimas D. P., Frenk R., van der Eb A. J., Lane D. P., Saville M. K., Jochemsen A. G. (2001) EMBO Rep. 2, 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharp D. A., Kratowicz S. A., Sank M. J., George D. L. (1999) J. Biol. Chem. 274, 38189–38196 [DOI] [PubMed] [Google Scholar]
- 19. Jackson M. W., Berberich S. J. (2000) Mol. Cell. Biol. 20, 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mancini F., Gentiletti F., D'Angelo M., Giglio S., Nanni S., D'Angelo C., Farsetti A., Citro G., Sacchi A., Pontecorvi A., Moretti F. (2004) J. Biol. Chem. 279, 8169–8180 [DOI] [PubMed] [Google Scholar]
- 21. Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 22. Linares L. K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 24. Tybulewicz V. L., Crawford C. E., Jackson P. K., Bronson R. T., Mulligan R. C. (1991) Cell 65, 1153–1163 [DOI] [PubMed] [Google Scholar]
- 25. Levy D., Adamovich Y., Reuven N., Shaul Y. (2008) Mol. Cell 29, 350–361 [DOI] [PubMed] [Google Scholar]
- 26. Ma Y., Yuan R., Meng Q., Goldberg I. D., Rosen E. M., Fan S. (2000) Mol. Cell. Biol. Res. Commun. 3, 122–128 [DOI] [PubMed] [Google Scholar]
- 27. Rinaldo C., Prodosmo A., Mancini F., Iacovelli S., Sacchi A., Moretti F., Soddu S. (2007) Mol. Cell 25, 739–750 [DOI] [PubMed] [Google Scholar]
- 28. Levav-Cohen Y., Goldberg Z., Zuckerman V., Grossman T., Haupt S., Haupt Y. (2005) Biochem. Biophys. Res. Commun. 331, 737–749 [DOI] [PubMed] [Google Scholar]
- 29. Shafman T., Khanna K. K., Kedar P., Spring K., Kozlov S., Yen T., Hobson K., Gatei M., Zhang N., Watters D., Egerton M., Shiloh Y., Kharbanda S., Kufe D., Lavin M. F. (1997) Nature 387, 520–523 [DOI] [PubMed] [Google Scholar]
- 30. Stommel J. M., Wahl G. M. (2004) EMBO J. 23, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pendergast A. M., Muller A. J., Havlik M. H., Clark R., McCormick F., Witte O. N. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5927–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buchdunger E., Zimmermann J., Mett H., Meyer T., Müller M., Druker B. J., Lydon N. B. (1996) Cancer Res. 56, 100–104 [PubMed] [Google Scholar]
- 33. Momand J., Zambetti G. P. (1996) Oncogene 12, 2279–2289 [PubMed] [Google Scholar]
- 34. Kentsis A., Gordon R. E., Borden K. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang X., Orlicky S., Lin Z., Willems A., Neculai D., Ceccarelli D., Mercurio F., Shilton B. H., Sicheri F., Tyers M. (2007) Cell 129, 1165–1176 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.