Abstract
Maturation of dsDNA bacteriophages involves assembling the virus prohead from a limited set of structural components followed by rearrangements required for the stability that is necessary for infecting a host under challenging environmental conditions. Here, we determine the mature capsid structure of T7 at 1 nm resolution by cryo-electron microscopy and compare it with the prohead to reveal the molecular basis of T7 shell maturation. The mature capsid presents an expanded and thinner shell, with a drastic rearrangement of the major protein monomers that increases in their interacting surfaces, in turn resulting in a new bonding lattice. The rearrangements include tilting, in-plane rotation, and radial expansion of the subunits, as well as a relative bending of the A- and P-domains of each subunit. The unique features of this shell transformation, which does not employ the accessory proteins, inserted domains, or molecular interactions observed in other phages, suggest a simple capsid assembling strategy that may have appeared early in the evolution of these viruses.
Keywords: Bacteriophage, DNA Viruses, Protein Structure, Virus Assembly, Virus Structure, Cryo-electron Microscopy, Image Processing, Molecular Modeling
Introduction
Viruses adopt different evolutionary ways to survive due to their presence in many different hosts, including prokaryotes, eukaryotes, and archaea (1). This variability has obscured for years the observation of commonalities in the evolutionary origin, basic design, and life cycle strategies of these viruses. Besides the well studied principles in capsid architecture (2), it is becoming clear that there are basic components of viral structures, such as the capsid and connector proteins in the case of bacteriophages, which share a surprising degree of similarity, as well as common general mechanisms of the maturation leading to the infective virion (3–5). These similarities are helpful to elucidate the evolutionary relationships previously obscured by the low sequence identity between equivalent proteins of different viruses, in turn informing virus classification (6–8).
Double-stranded DNA (dsDNA) bacteriophages (9) have been instrumental to study basic principles in assembly and maturation of viral particles (10, 11). dsDNA bacteriophages first assemble into an icosahedral prohead composed of a defined number of monomers of the main shell protein, scaffolding proteins, and a dodecameric connector. These three types of proteins must interact properly to yield a fully functional prohead with appropriate shape and size. Recent work suggests that the structure of these three basic components has been widely conserved across different bacteriophages. In particular, the fold of the shell protein was first described for the bacteriophage HK97 (12, 13) and later found in other viruses such as P22 (14), T4 (4), φ29 (15), HSV-1 (7), ϵ15 (16), T7 (17), and lambda (18). The HK97-like fold includes two domains: the P-domain, consisting of a long α-helix and an elongated three-stranded β-sheet, and the A-domain, composed of two α-helices and an additional β-sheet. Different bacteriophages incorporate additional domains in the main shell protein fold, resulting in different assembly mechanisms.
A major event in the maturation of dsDNA viruses is the packaging of the nucleic acid into the prohead. For this purpose, a complex machinery, consisting of the ATPases and pilot proteins, is assembled at the vertex of the prohead where the connector is located (3). Concomitant with the DNA packaging, the prohead undergoes a number of structural transformations (5, 11, 19) that can include expansion of the shell, in situ monomer rearrangements, release of the scaffolding proteins or incorporation of new components, such as accessory shell proteins or the virus tail, and chemical modifications such as cleavage or covalent cross-linking. The final outcome is a mature, infective virion.
Virus assembly and maturation offer a unique opportunity to dissect two correlated aspects: first, how a limited set of components is able to assemble in a complex structure; and second, the molecular basis of the process leading to the acquisition of stability necessary to search for the proper host. The existence of three transformation mechanisms to acquire shell stability has been recently reviewed (20). The most common mechanism is the incorporation of accessory proteins that establish an additional set of contacts to reinforce those built by the main shell protein in the prohead. The contact sites for the accessory proteins are usually exposed after the rearrangement of the shell protein monomers during maturation, thus avoiding premature interactions leading to abortive assemblies. This mechanism applies to HSV-1 (21) and adenovirus (22), as well as to the bacteriophages, lambda (23), T4 (24), and ϵ15 (25). A second mechanism involves the generation of new interactions between neighboring subunits by using additional domains inserted within the primitive protein sequence. This strategy is employed by phages φ29 (15) and P22, in which the presence of an inserted domain with a telokin Ig-like fold has been related to a direct assembly of the capsid from monomeric subunits instead of intermediate capsomeric subassemblies (20). The third mechanism, which appears to be unique to HK97, involves the formation of covalent intersubunit cross-linking, leading to extensive interlocking of the shell in a giant superstructure (26).
The maturation process in bacteriophage T7 is of particular interest because the virus does not have accessory proteins, inserted domains, or cross-linking between subunits. T7 infects Escherichia coli and belongs to the Podoviridae family. It has a 40-kb dsDNA, an icosahedral capsid with a triangulation number of T = 7, and a non-contractile tail (27). The shell is made of 415 copies of protein gp10A. The prohead also contains a scaffolding protein (gp9) and a dodecameric connector (gp8) that attaches a core complex formed by proteins gp14, gp15, and gp16 to one of the 12 5-fold vertices (28). The virus tail is added to this vertex in the mature virion. The packaging machinery is also located at this vertex, forming a portal by incorporating terminases that select, cleave, and push the DNA inside the viral head through the connector. More specifically, the T7 prohead interacts with terminase gp19 to package the complex built by the DNA and terminase gp18 after cutting the unit length DNA from the replicating concatemers (29). The maturation of T7 involves the disappearance of the scaffolding protein, major rearrangements of the core, the incorporation of the tail, and a dramatic transformation of the shell, which undergoes a radial expansion. The structure of the T7 prohead has been solved by three-dimensional cryo-electron microscopy (cryo-EM)2 at 2 nm resolution (27, 28), and a quasi-atomic model for the procapsid shell has also been obtained (17), revealing that the main protein gp10A has an HK97-like fold.
Here, we perform the cryo-EM reconstruction of the mature capsid together with a quasi-atomic model of its shell to elucidate the molecular details of T7 maturation. Comparing the shell of the prohead and the mature head reveals that T7 follows a maturation scheme that involves a change in the shell diameter from 51 to 60 nm and a decrease in its thickness from 4.5 to 2.3 nm due to a major rearrangement of the subunit domains. These transformations are based on the tilting, rotation, and radial translation of gp10 monomers, resulting in a higher intermonomeric interacting surface. Our results suggest that T7 might represent one of the simplest primordial capsid maturation mechanisms.
EXPERIMENTAL PROCEDURES
Sample Preparation
Mature T7 virions were purified from E. coli lysates of BL21 cells infected with wild type phage using sucrose gradient centrifugation as described elsewhere (28, 30). The composition of the purified viral particles was validated by SDS-PAGE analysis in the presence of β-mercaptoethanol. The viral DNA was released by incubation with 1 m NaClO4 at 37 °C during 16 h. The empty heads were further purified by 10–40% sucrose gradient centrifugation. The homogeneity and overall quality of the samples was tested by electron microscopy of negatively stained aliquots. The samples were finally dialyzed against 50 mm Tris-HCl, pH 7.8, 10 mm MgCl2, and 0.1 m NaCl before cryo-electron microscopy.
Electron Microscopy and Image Processing
The samples were frozen in liquid ethane and transferred to an FEI TecnaiG2 FEG200 cryo-electron microscope. Image acquisition and processing were performed as described previously (30). More specifically, the micrographs were recorded on Kodak SO-163 film under low dose conditions at a nominal magnification of 50,000×, digitized in a Zeiss scanner (Photoscan TD, Z/I Imaging Corp.) using a 7-μm step, and sampled to 0.14 nm/pixel. A total of 5100 particles were used to compute mean intensities and variances and to remove linear background gradients. The three-dimensional reconstruction was performed using the software packages Bsoft (31), Spider (32), and Xmipp (33, 34). Images were automatically sorted into defocus groups covering a range between −0.5 and −3 μm.
Icosahedral symmetry was imposed throughout the refinement process. The final resolution for the reconstruction of the mature capsid was 1.08 nm using the 0.5 Fourier shell correlation criterion and 0.9 nm for 0.3 Fourier shell correlation (supplemental Fig. S1). Automated B-factor determination and compensation were performed with the EM-BFACTOR program, with a value for Brestore of −5.29 nm2 (35).
Segmentation and Structural Analysis
Segmentation of the individual subunits from the reconstructed density was performed manually using the University of California San Francisco (UCSF) Chimera software (36). The segmentation was improved iteratively (17).
The electrostatic potential was calculated with the Coulombic Surface Coloring option in Chimera. Protein-protein interaction areas were determined using the PISA package (37). They were defined as one-half of the difference between the total accessible area in isolated monomers and that of the monomers assembled in the complex (38). Residues were considered interacting if any of the atoms were within 0.4 nm of each other, employing the CCP4i package (39). The interactions were cross-checked using Chimera with similar results.
Generation of the gp10A Structure in the Head and Prohead from the Cryo-EM Maps
To obtain the structure of the gp10A monomer in the mature capsid of T7, we started with a sequence alignment against the protein gp5 in the mature capsid of HK97 (Protein Data Bank (PDB) ID 1OHG). Segment Leu100–Glu345 in gp10A was aligned to segment Leu133–Pro373 in gp5 with PSI-BLAST (40). Given the low sequence similarity between the two segments, we improved the alignment in Chimera by considering secondary structure computed with PSIPRED version 2.6 (41), using the alignment score weight of 20%. The resulting alignment conserved secondary structure and had the sequence identity of 14%. A comparative model was obtained with MODELLER (42) using standard parameters. The model was fit rigidly into the cryo-EM electron density by maximizing the cross-correlation using Chimera. Residues Leu126–Pro147 in the E-loop of gp10A were removed because the corresponding density in the cryo-EM map could not be determined unambiguously. Next, flexible fitting was performed. The model α-helices were first moved manually into the corresponding areas of the cryo-EM map using Chimera. The fit was refined by the Flex-EM program (43), keeping the helices and sheets as rigid bodies, and optimized with the conjugated gradients (CG) option. The resulting model was further improved in a second Flex-EM run, refining all atoms independently from each other using a molecular dynamics optimization. Only 2% of residues in the resulting model were outliers in the Ramachandran plot. Seven copies of this model were fit sequentially as rigid bodies into the cryo-EM density of the whole asymmetric unit. Subsequently, the previously deleted E-loop was added to each of the seven model copies followed by a simultaneous relaxation of the loop atoms in the cryo-EM map, using five steps of molecular dynamics with Flex-EM (see Fig. 2C). Finally, a complete model of the mature shell was then obtained by applying the icosahedral symmetry to the model of the asymmetric unit followed by a refinement of atomic positions with the URO program (44) that allowed us to retain the icosahedral symmetry during the refinement. The correlation coefficient between the complete model and the cryo-EM map is 0.70, whereas the local cross-correlation coefficient is 0.81.
FIGURE 2.
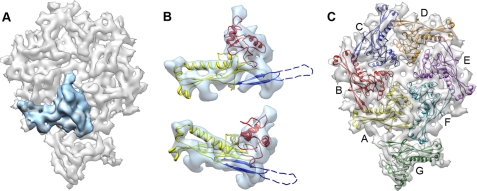
Segmentation of the asymmetric unit and model of the shell protein gp10A. A, one gp10A monomer subunit is shown in blue within the asymmetric unit in gray. B, gp10A model fitted in the averaged density of the hexameric monomers. Top, outer side view; bottom, perspective view. The ribbon model is colored for the different motifs: A-domain, red; P-domain, yellow; and E-loop, blue. Part of the E-loop (dashed line) is omitted as no hints for the corresponding densities are defined in the average density. C, a pseudo-atomic model of the asymmetric unit of the mature head. The unit contains an entire hexamer (subunits A–F) and one pentamer subunit (subunit G). The color code of the monomers in this figure is maintained throughout the entire report.
The new structure for gp10A in the prohead was generated from gp5 in the prohead II of HK97 (PDB ID 3E8K). The same approach used for the mature head was employed, and the new model was basically identical to that published before (17), but it presented some minor reassignments of residues for the secondary structure elements. In this new structure, the E-loops were modeled, first manually adjusted, and refined later with molecular dynamics. Visual inspection of the fitting showed that E-loops are in the electron density previously assigned to them (17), although we consider their fitting less reliable than in the mature capsid.
Graphical Data
All graphical representations were rendered using the UCSF Chimera software.
RESULTS AND DISCUSSION
Structure of the T7 Capsid Shell from Cryo-electron Microscopy
An earlier study of T7 at 2 nm resolution using cryo-EM and three-dimensional reconstruction without imposing symmetry constraints revealed the overall changes undergone by the shell and the core-connector complex during the transformation of the prohead to the mature virus (28). An improvement in the resolution to 1 nm by imposing icosahedral symmetry resulted in a quasi-atomic model of the prohead showing that the shell is formed by gp10A subunits arranged in an HK97-like conformation (17). In the current study, we have solved the structure of the mature capsid by a combination of single particle cryo-EM three-dimensional reconstruction using icosahedral symmetry and protein modeling. Then, we compared this structure with the T7 procapsid to learn about the molecular basis of this maturation process.
Preliminary reconstructions from purified mature T7 virions showed that the presence of DNA inside the capsid did not allow us to define the precise boundaries between the inner shell face and the outermost DNA layer. Thus, to obtain an accurate segmentation of the shell electron density and subsequently perform molecular modeling, we first eliminated the DNA from the virions by treating the preparation with a solution of NaClO4 until total DNA ejection. After DNase treatment and further purification with sucrose gradient centrifugation, we obtained mature empty capsids composed of the main shell protein gp10A with only minor contents of gp10B and traces of the tail and core components (supplemental Fig. S2).
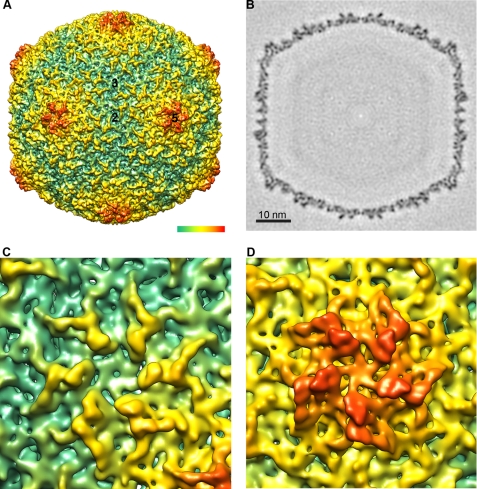
The electron density map (Fig. 1) of the empty capsid obtained by cryo-EM and three-dimensional reconstruction (see “Experimental Procedures”) had a resolution of ∼1 nm and was fully consistent with those obtained previously at lower resolution (Fig. 1A). The shell had an overall diameter of 60 nm with an average thickness of 2.5 nm, with the outer face being more corrugated than the inner face (Fig. 1B). The outer surface of the shell was smoother than in the prohead, and the gp10A protein formed almost symmetrical hexamers (Fig. 1C), with noticeable pentamers at the capsid vertices (Fig. 1D).
FIGURE 1.
Three-dimensional reconstruction of the mature capsid. A, surface rendering viewed along a 2-fold axis of icosahedral symmetry. Numbers indicate the 5-fold, 3-fold, and 2-fold icosahedral symmetry axes. The color-coded bar represents depth measurement. B, central section through the density map. Bar represents 10 nm. C, close-up view of the hexamer. D, close-up view of the pentamer. The density is contoured at 3σ above the mean.
The asymmetric unit of the shell was segmented, and each of the seven copies of the protein gp10A was extracted manually. The segmentation was improved by an iterative alignment and averaging of the independent densities of the monomers. The resulting structures (Fig. 2A) showed that the mature subunit resembles that of the prohead shell, but important changes were also evident. The previously skewed hexamers of gp10A were more symmetric and more similar to the pentamers, which did not appear to have changed their 5-fold overall morphology. The structure of the monomer in the mature shell was similar for both the pentamer and the hexamer and was flatter than in the prohead, thus building a more extended capsomer.
Modeling of the gp10A Monomer into the Mature Shell Capsid
The observed similarity between the folds of the shell proteins gp10A in T7 and gp5 in HK97 was used to produce a quasi-atomic structure of the shell in the mature head of T7, as it was previously done for the prohead (17). We used the coordinates of gp5 in the mature shell of HK97, solved by x-ray crystallography (13, 45), to build a comparative model for gp10A that was flexibly fitted into the cryo-EM density of gp10A obtained by segmentation (see “Experimental Procedures”). Fig. 2B shows two views of the fitted model, with the A-domain (residues 210–300) in red, the P-domain (residues 100–119, 154–209, and 301–345) in yellow, and the E-loop (residues 120–153) in blue. An initial asymmetric shell unit was built using this model, after which the structure of each of the seven monomers was individually refined against its corresponding experimental density (Fig. 2C). The structural alignment of the seven different monomers showed a good overall fit, with the exception of the E-loop in the pentameric monomer, which showed a significant bend (∼30°) toward the interior of the capsid, as compared with the E-loop in the hexameric monomers. The model for the whole shell was obtained by applying icosahedral symmetry followed by global refinement. The final molecular structure reproduced well the features of the cryo-EM map, with an overall correlation coefficient of 0.70 for the fitting and a local correlation coefficient of 0.81 for the area corresponding to the modeled coordinates. The resulting model showed that the shell of the mature capsid contains extensive contacts between subunits, with the A-domain mostly involved in the interactions between monomers within capsomers and the P-domain mostly involved in the interfaces between the pentamers and the hexamers.
Structural Changes in the Shell after Maturation
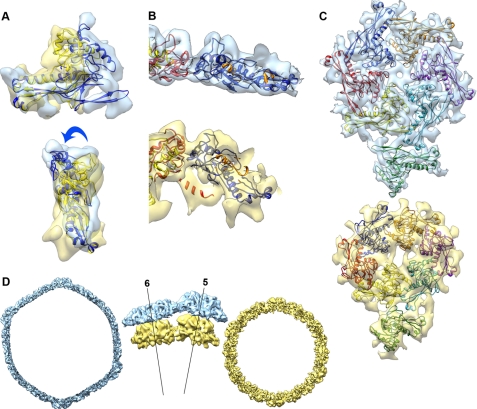
The most significant difference between the structure of gp10A in the mature head with respect to the prohead was the bending of the A-domain relative to the P-domain by 22° (Fig. 3A), which makes them more parallel to the shell surface. A similar relative movement between the A- and P-domains has previously been found in the maturation of HK97 (46) and P22 (20). Monomers were rearranged in the capsomers of the head relative to the prohead. The monomers located at different heights in the prohead hexamer were in the same plane in the mature shell. Their positions were adjusted differently depending on their previous location in the asymmetric unit, also including a change from a radial orientation to a planar shape (Fig. 3B). These changes generated a new set of contacts between monomers (Fig. 3C). The A-domain that protruded in the prohead shell became tangent to the surface (Fig. 3B), which was in turn less corrugated. There were also changes in the orientation of the monomers in the plane of the shell, as follows. The monomers of the hexamer, which were distributed following a skewed, elliptical arrangement with a pseudo-2-fold symmetry axis, underwent an average of 30° in-plane rotation that increased the intercapsomeric distance from 11 to 13.4 nm and expanded the average hexamer diameter by around 25% (supplemental Fig. S3). The pentameric subunits moved similarly in a 16° in-plane rotation, which brought the diameter of the pentamer from 9.2 to 11.4 nm (24% expansion).
FIGURE 3.
Structural changes involved in the T7 shell maturation. A, superposition of models of gp10A for the prohead (yellow) and head (blue) obtained by alignment of the P-domain, showing the bending of the A-domain relative to the P-domain. B, detail of the head (top) and prohead (bottom) hexameric subunits shown tangential to the capsid surface. C, head (top) and prohead (bottom) asymmetric units shown from the outside of the capsid. D, cross-section view of the head (blue) and prohead (yellow) densities. The central detailed side views are shown tangential to the capsid surface, revealing the radial outward movement of the capsomers. The labeled lines represent radial axes along the center of the pentamer (5) and the hexamer (6).
The thinning of the shell during maturation involves, besides the above mentioned bending of the A-domain relative to the P-domain, a change of the radial orientation of the monomers (Fig. 3B). The tilting angle was an average of 22° within the hexamers and 10° for the monomers within the pentamers. The smaller tilting angle made the mature pentamers protrude significantly from the shell and adopt a radial orientation more similar to that of the prohead (Fig. 3D). As a whole, the hexamers moved outward by 4.6 nm, and the pentamers moved outward by 5.4 nm. The overall consequence of all the described rearrangements was an increase in the diameter of the shell of T7 from 51 to 60 nm and a decrease in its thickness from 4.5 to 2.3 nm (Fig. 3D).
Modification of the Fold in gp10A
Apart from the bending of the A-domain with respect to the P-domain, we observed other differences in the fold of gp10A. Our model for the structure of gp10A lacked the N-terminal domain (residues 1–100) due to the low sequence identity of this stretch with its HK97 counterpart. This N-terminal domain was assigned in the prohead to a defined region at the inner part of the shell using difference maps between the cryo-EM map and the model (17). The same difference map for the mature shell located in this case the domain at the outer part (Fig. 4, A and B). In the mature head, the differential area has an L-shaped form (Fig. 4C), and it can be subdivided in two almost perpendicular domains; the largest one is in the neighborhood of the N terminus position of the modeled sequence stretch (Fig. 4C), and its average volume (11 nm3 at 3σ) could accommodate 100 amino acids (assuming an average protein density of ∼1.38 g × cm3). This rearrangement of the N-terminal domain from inside the shell toward the outer part has also been found in the maturation process of other viruses, such as poliovirus and T4 (47, 48). The N-terminal domain of gp10A was tentatively implicated in the interaction with the scaffolding protein in the T7 prohead (17). A similar domain interacting with the scaffold that also changes its inner position during maturation has been described in P22 (14). It is thus reasonable to propose that the N-terminal domain of gp10A in T7 is connected to the scaffolding lattice that stabilizes the prohead. Its outward movement, together with the rearrangement of shell subunits, would promote the release of the scaffolding proteins by preventing shell-scaffold interactions.
FIGURE 4.
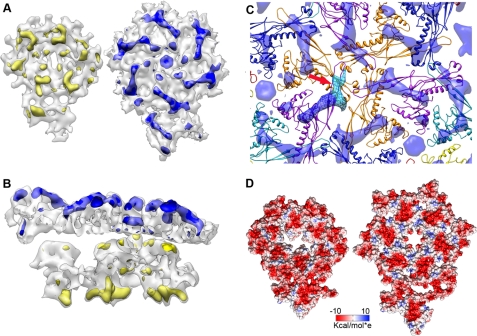
Differences between prohead and mature head capsids. A, left, difference map (yellow) between the experimental and modeled densities for the asymmetric unit of the prohead, as seen from the inner side of the shell. Right, difference map (blue) for the asymmetric unit of the mature head, as seen from the outside of the shell. The difference maps are rendered at 5σ. B, side view of the difference maps. C, difference map around a 3-fold axis involving three identical copies of monomer D (Fig. 2). The volume attributed to the N-terminal domain is colored in dark blue, and the E-loop is in light blue (red arrow). D, electrostatic potential in the surface of the asymmetric unit, as seen from the inside of the virion. Left, prohead. Right, mature head. Negative charge is in red, and positive is in blue.
Another interesting feature was the change in the E-loop observed after its threading in the density. This part of the fold was described as involved in the interaction of neighboring capsomers but was not originally included into the modeling of gp10A (17). We identified the smaller portion of the L-shaped difference volume described above with this E-loop (Fig. 4C, arrow). In this work, we modeled the E-loop starting with a preliminary fitting that did not include it until we got a final complete structure. Its new location in the hexamers suggests that it might be responsible for reinforcing the interactions of monomers around the 3-fold and quasi-3-fold symmetry axes. At this resolution, it was not possible to determine whether the modification of the fold of gp10A involves any twisting and/or bending of the α-helices or β-sheets, as recently described for HK97 (46).
Another consequence of the structural rearrangements involved in the T7 shell maturation is the change of the electrostatic potential at the inner face of the shell. Although the prohead shell inner face was almost totally negatively charged, this inner face in the mature head showed the presence of positively charged domains (Fig. 4D). This implies that new contacts between the inner face of the shell and the outermost layer of the packaged DNA could be established, either during the packaging process or in the final DNA packaged viral particle.
Changes in the Interactions between Monomers
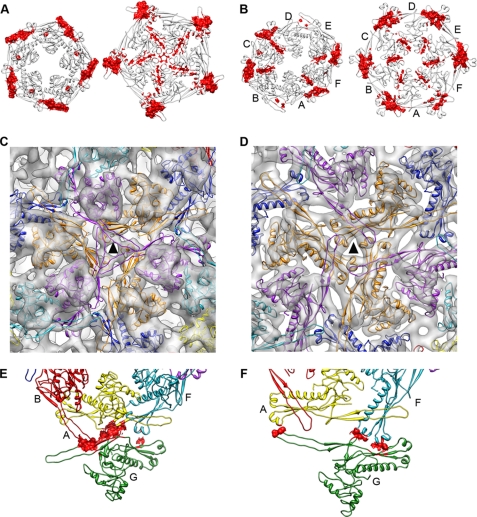
The interactions between the capsid monomers changed during the shell maturation for both intracapsomeric and intercapsomeric contacts. On the intracapsomeric side, the main contacts within the prohead pentameric monomers were established using the P-domains with mostly hydrophobic contacts (supplemental Table S1). The mature pentamers exhibited additional, mostly polar and charged contact areas also involving the A-domain (Fig. 5A). There was an overall increase of the contact area between adjacent monomers from 7.7 nm2/contact in the prohead pentamer up to 11.5 nm2 in the mature form (supplemental Table S2). In the case of the hexamer, the area of all the contacts between monomers also increased and reflected the 2-fold symmetry of the capsomer (Fig. 5B and supplemental Table S2). In the mature shell, the interactions were closer to 6-fold symmetrical, as expected. This increase of the monomer contact areas during the T7 maturation was consistent with the results obtained for HK97 (49).
FIGURE 5.
Changes in the interactions during the maturation of the T7 shell. In all cases, the figures on the left are for the prohead, and the mature head is on the right. A, contact surfaces for the pentamer (red). The view is from the outside of the capsid. B, contact surfaces for the hexamer (red). C and D, interactions around the 3-fold axis (triangle) in the prohead (C) and mature head (D). E and F, residues involved in pentameric-hexameric interactions (red) in the prohead (E) and mature head (F).
The intercapsomeric contacts in hexamers were mainly centered on the 3-fold and quasi-3-fold symmetry axes and were based on polar and charged residues. In the prohead, they involved the P-loop from one subunit (residues 172–165 and 321–328), the P-domain from the nearest neighbor, and an additional interaction of these two domains with the E-loop from an adjacent subunit (Fig. 5C). The basic bonding pattern was maintained after the maturation (Fig. 5D), although the interface area between monomers increased from 0.4 to 3.2 nm2. The intercapsomeric interactions between hexamers and pentamers were also modified (Figs. 5, E and F). In the prohead, there were three different types of contacts between monomers: A/G, F/G, and B/G, all using P-domain/P-domain interactions. In the mature head, the contacts were between monomers F/G (P-loop/P-domain) and monomers A/G (P-loop/E-loop).
Capsid Stabilization
The stabilization of the capsid in dsDNA virus follows different strategies. Certain phages incorporate accessory proteins to the shell to reinforce the contacts between capsomers and to stabilize the mature virion. T4, lambda, and ϵ15 are examples of phages that use this strategy. φ29 and P22 have additional domains inserted into the HK97-like fold to form new contacts over the basic network provided by the basal fold. In HK97, the reinforcement of the shell is attained by covalent cross-linking of neighboring subunits that secures the assembly. Underlying these differences, there is a common trend, which involves the expansion of the capsid based on a rearrangement of the capsid subunits. We have found that T7 follows a strategy to stabilize the shell that might represent a simpler, more primitive mechanism for capsid maturation. The maturation is based on a drastic rearrangement of the subunits from the prohead to the mature head that leads to a reinforcement of the bonding pattern, without intervention of any additional help. This rearrangement includes a complex set of changes at all structural levels, as follows. The fold of gp10A changes with a bending of the A-domain in respect to the P-domain, the N-terminal part of gp10A moves outwards, and the E-loop bends and further covers the monomers; the monomers perform translations, in-plane rotations, and tilting with angles that depend on their positions into the hexameric or pentameric capsomers; and the pentamers and hexamers increase their radii and symmetry. The result is an expanded, thinner shell in which the interacting areas between subunits are larger to reinforce the bonding lattice.
Despite these changes, there are some features in the monomer basic bonding pattern that are conserved. The contact topology around the 3-fold and quasi-3-fold symmetry axes is maintained; the P-domain/P-loop interactions between neighboring subunits are mostly the same after maturation (Fig. 5, C and D); and the E-loop swapping from adjacent subunits, although expanded, is still present. HK97 has the same conservation of the E-loop interactions, and these contacts at the 3-fold axis have been suggested as fixed points acting as hinges to secure the subunit rearrangements (46). It is probable that they play the same role in T7 as well.
In summary, our results indicate that the maturation of T7 is more similar to the maturation mechanism of HK97 than to the maturation in P22-like systems (20). Nevertheless, as no chemical cross-linking is involved in T7 maturation, it could be speculated that T7 represents the most basic system of capsid maturation. HK97 would represent a more advanced strategy that still does not employ additional folds or proteins. Other phages would have incorporated their accessory proteins later in evolution to facilitate their maturation process and/or to incorporate further functionalities.
Acknowledgments
The support, critical comments, and revisions of Andrej Sali are greatly acknowledged. We are indebted to Xabier Agirrezabala for previous work in our laboratory on the T7 virus structure and in particular for the preliminary study of mature capsids containing DNA. We also thank Nuria Verdaguer for constructive comments. We are grateful to Ron Conway, Mike Homer, Intel, Hewlett-Packard, IBM, and Netapp for computer hardware gifts to A. Sali.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM083960, R01GM54762, and PN2 EY016525 (to A. Sali). This work was also supported by Grants BFU2008-02328/BMC and Consolider CSD2007-00010 from the Direccion General de Investigacion of the Ministry of Science and Innovation (to J. L. C.), S-2009/MAT-1507 from the Direccion General de Univertsidades e Investigacion from the Madrid Government (to J. L. C.), BIO2008-02361 from the Direccion General de Investigacion of the Ministry of Science and Innovation (to J. R. C.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3IZG and 2XVR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The structure reported in this paper has been submitted to the Electron Microscopy Data Bank (EMDB) under accession number EMD-1810.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- cryo-EM
- cryo-electron microscopy.
REFERENCES
- 1. Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (eds) (2005) Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, Elsevier Academic Press, San Diego [Google Scholar]
- 2. Casjens S., Chiu W., Burnett R. M., Garcea R. L. (1997) in Structural Biology of Viruses, pp. 38–79, Oxford University Press, New York [Google Scholar]
- 3. Valpuesta J. M., Carrascosa J. L. (1994) Q. Rev. Biophys. 27, 107–155 [DOI] [PubMed] [Google Scholar]
- 4. Fokine A., Leiman P. G., Shneider M. M., Ahvazi B., Boeshans K. M., Steven A. C., Black L. W., Mesyanzhinov V. V., Rossmann M. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. (2005) Curr. Opin. Struct. Biol. 15, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bamford D. H., Grimes J. M., Stuart D. I. (2005) Curr. Opin. Struct. Biol. 15, 655–663 [DOI] [PubMed] [Google Scholar]
- 7. Baker M. L., Jiang W., Rixon F. J., Chiu W. (2005) J. Virol. 79, 14967–14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benson S. D., Bamford J. K., Bamford D. H., Burnett R. M. (2004) Mol. Cell 16, 673–685 [DOI] [PubMed] [Google Scholar]
- 9. Ackermann H. W. (1998) Adv. Virus Res. 51, 135–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ackermann H. W. (2003) Res. Microbiol. 154, 245–251 [DOI] [PubMed] [Google Scholar]
- 11. King J., Chiu W. (1997) in Structural Biology of Viruses (Chiu W., Burnett R. M., Garcea R.L., eds) pp. 288–311, Oxford University Press, New York [Google Scholar]
- 12. Hendrix R. W. (2005) Adv. Virus Res. 64, 1–14 [DOI] [PubMed] [Google Scholar]
- 13. Wikoff W. R., Liljas L., Duda R. L., Tsuruta H., Hendrix R. W., Johnson J. E. (2000) Science 289, 2129–2133 [DOI] [PubMed] [Google Scholar]
- 14. Jiang W., Li Z., Zhang Z., Baker M. L., Prevelige P. E., Jr., Chiu W. (2003) Nat. Struct. Biol. 10, 131–135 [DOI] [PubMed] [Google Scholar]
- 15. Morais M. C., Choi K. H., Koti J. S., Chipman P. R., Anderson D. L., Rossmann M. G. (2005) Mol. Cell 18, 149–159 [DOI] [PubMed] [Google Scholar]
- 16. Jiang W., Chang J., Jakana J., Weigele P., King J., Chiu W. (2006) Nature 439, 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Agirrezabala X., Velázquez-Muriel J. A., Gómez-Puertas P., Scheres S. H., Carazo J. M., Carrascosa J. L. (2007) Structure 15, 461–472 [DOI] [PubMed] [Google Scholar]
- 18. Lander G. C., Evilevitch A., Jeembaeva M., Potter C. S., Carragher B., Johnson J. E. (2008) Structure 16, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson J. E. (2010) Curr. Opin. Struct. Biol. 20, 210–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parent K. N., Khayat R., Tu L. H., Suhanovsky M. M., Cortines J. R., Teschke C. M., Johnson J. E., Baker T. S. (2010) Structure 18, 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trus B. L., Booy F. P., Newcomb W. W., Brown J. C., Homa F. L., Thomsen D. R., Steven A. C. (1996) J. Mol. Biol. 263, 447–462 [DOI] [PubMed] [Google Scholar]
- 22. Saban S. D., Silvestry M., Nemerow G. R., Stewart P. L. (2006) J. Virol. 80, 12049–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang F., Forrer P., Dauter Z., Conway J. F., Cheng N., Cerritelli M. E., Steven A. C., Plückthun A., Wlodawer A. (2000) Nat. Struct. Biol. 7, 230–237 [DOI] [PubMed] [Google Scholar]
- 24. Fokine A., Chipman P. R., Leiman P. G., Mesyanzhinov V. V., Rao V. B., Rossmann M. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang W., Baker M. L., Jakana J., Weigele P. R., King J., Chiu W. (2008) Nature 451, 1130–1134 [DOI] [PubMed] [Google Scholar]
- 26. Conway J. F., Duda R. L., Cheng N., Hendrix R. W., Steven A. C. (1995) J. Mol. Biol. 253, 86–99 [DOI] [PubMed] [Google Scholar]
- 27. Cerritelli M. E., Conway J. F., Cheng N., Trus B. L., Steven A. C., Chiu W., Johnson J. E. (2003) in Advances in Protein Chemistry (Richards F. M., Eisenberg D. S., Kuriyan J. eds) pp. 301–323, Academic Press, Orlando, FL [Google Scholar]
- 28. Agirrezabala X., Martín-Benito J., Castón J. R., Miranda R., Valpuesta J. M., Carrascosa J. L. (2005) EMBO J. 24, 3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujisawa H., Morita M. (1997) Genes Cells 2, 537–545 [DOI] [PubMed] [Google Scholar]
- 30. Agirrezabala X., Martín-Benito J., Valle M., González J. M., Valencia A., Valpuesta J. M., Carrascosa J. L. (2005) J. Mol. Biol. 347, 895–902 [DOI] [PubMed] [Google Scholar]
- 31. Heymann J. B., Belnap D. M. (2007) J. Struct. Biol. 157, 3–18 [DOI] [PubMed] [Google Scholar]
- 32. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 33. Scheres S. H., Núñez-Ramírez R., Sorzano C. O., Carazo J. M., Marabini R. (2008) Nat. Protoc. 3, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sorzano C. O., Marabini R., Velázquez-Muriel J., Bilbao-Castro J. R., Scheres S. H., Carazo J. M., Pascual-Montano A. (2004) J. Struct. Biol. 148, 194–204 [DOI] [PubMed] [Google Scholar]
- 35. Fernández J. J., Luque D., Castón J. R., Carrascosa J. L. (2008) J. Struct. Biol. 164, 170–175 [DOI] [PubMed] [Google Scholar]
- 36. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 38. Gretes M., Lim D. C., de Castro L., Jensen S. E., Kang S. G., Lee K. J., Strynadka N. C. (2009) J. Mol. Biol. 389, 289–305 [DOI] [PubMed] [Google Scholar]
- 39. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 40. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 42. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 43. Topf M., Lasker K., Webb B., Wolfson H., Chiu W., Sali A. (2008) Structure 16, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Navaza J., Lepault J., Rey F. A., Alvarez-Rúa C., Borge J. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1820–1825 [DOI] [PubMed] [Google Scholar]
- 45. Helgstrand C., Wikoff W. R., Duda R. L., Hendrix R. W., Johnson J. E., Liljas L. (2003) J. Mol. Biol. 334, 885–899 [DOI] [PubMed] [Google Scholar]
- 46. Gertsman I., Gan L., Guttman M., Lee K., Speir J. A., Duda R. L., Hendrix R. W., Komives E. A., Johnson J. E. (2009) Nature 458, 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Belnap D. M., Filman D. J., Trus B. L., Cheng N., Booy F. P., Conway J. F., Curry S., Hiremath C. N., Tsang S. K., Steven A. C., Hogle J. M. (2000) J. Virol. 74, 1342–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steven A. C., Bauer A. C., Bisher M. E., Robey F. A., Black L. W. (1991) J. Struct. Biol. 106, 221–236 [DOI] [PubMed] [Google Scholar]
- 49. Conway J. F., Wikoff W. R., Cheng N., Duda R. L., Hendrix R. W., Johnson J. E., Steven A. C. (2001) Science 292, 744–748 [DOI] [PubMed] [Google Scholar]