Abstract
Bryostatin-1 (Bryo-1), a natural macrocyclic lactone, is clinically used as an anti-cancer agent. In this study, we demonstrate for the first time that Bryo-1 acts as a Toll-like receptor 4 (TLR4) ligand. Interestingly, activation of bone marrow-derived dendritic cells (in vitro with Bryo-1) led to a TLR4-dependent biphasic activation of nuclear factor-κB (NF-κB) and the unique induction of cytokines (IL-5, IL-6, and IL-10) and chemokines, including RANTES (regulated on activation normal T cell expressed and secreted) and macrophage inflammatory protein 1α (MIP1-α). In addition, EMSA demonstrated that Bryo-1-mediated induction of RANTES was regulated by NF-κB and the interferon regulatory factors (IRF)-1, IRF-3, and IRF-7 to the RANTES independently of myeloid differentiation primary response gene-88 (MyD88). Bryo-1 was able to induce the transcriptional activation of IRF-3 through the TLR4/MD2-dependent pathway. In vivo administration of Bryo-1 triggered a TLR-4-dependent T helper cell 2 (Th2) cytokine response and expanded a subset of myeloid dendritic cells that expressed a CD11chighCD8α− CD11b+CD4+ phenotype. This study demonstrates that Bryo-1 can act as a TLR4 ligand and activate innate immunity. Moreover, the ability of Bryo-1 to trigger RANTES and MIP1-α suggests that Bryo-1 could potentially be used to prevent HIV-1 infection. Finally, induction of a Th2 response by Bryo-1 may help treat inflammatory diseases mediated by Th1 cells. Together, our studies have a major impact on the clinical use of Bryo-1 as an anti-cancer and immunopotentiating agent.
Keywords: Dendritic Cell, Immunology, Inflammation, Innate Immunity, Interleukin
Introduction
Activation of the innate immune response is essential to control the initial stage of pathogen invasion and for the establishment of adaptive immunity (1). Dendritic cells (DCs)3 play a critical role in the balance between immunity and immunological tolerance (2) due to their uniqueness as cells highly specialized in the uptake, transport, processing, and presentation of antigens (3). The discrimination process leading to cell immunity or tolerance is highly regulated by pattern recognition receptors, such as the Toll-like receptor (TLR) family, which recognize pathogen-associated molecular patterns (1, 4). Immature DCs express many members of the TLR family (5). Recognition of pathogen-associated molecular pattern by DCs results in the activation of TLR family members, which in turn activate the NF-κB and MAPK signaling pathways and induce DC maturation (6, 7). Upon maturation, DCs acquire the ability to produce a wide range of cytokines/chemokines and promote the differentiation of various T helper cell phenotypes such as Th1, Th2, Th17, and Tregs, thereby controlling the activation of adaptive immune responses (6, 8).
The TLR family is the essential recognition and signaling component of the host defense (9, 10). DCs express TLR4, which mediates the recognition of Gram-negative bacterial LPS and several other pathogen-associated molecular patterns (11). Toll signaling to NF-κB occurs from the conserved Toll-IL-1 resistance (TIR) domain of TLR, which triggers the recruitment of the TIR domain-containing adaptor protein MyD88 (9). The recruitment of MyD88 to the activated TLR results in NF-κB activation via the MyD88 adaptor-like protein (Mal/TIRAP) (12) and the IκB kinase complex (13). Although most of the TLRs seem to be exclusively dependent on the expression of MyD88 for all their functions, TLR3 and TLR4 are unique in that they are capable of activating both MyD88-dependent and MyD88-independent responses (9, 14), through the TIR domain-containing adaptor-inducing IFN-β (TRIF) (15). It has been shown that stimulation of DC maturation and the induction of type 1 interferon (IFN-β) is mediated by the MyD88-independent signaling pathway (14, 15). However, whereas all TLRs activate NF-κB, not all TLRs induce IFN-β (8). Therefore, TIR domain-containing adaptors such as MyD88, TRIF, and Mal/TIRAP play crucial roles in TLR-signaling pathways, because they provide specificity to the response generated by signaling through each TLR (16).
Bryostatin-1 (Bryo-1) is a macrocyclic lactone isolated from the marine bryozoan, Bugula neritina (17). The potent anti-proliferative effects and anti-neoplastic properties of Bryo-1 against various tumor cells have led to its use as a chemotherapeutic agent. Recently, Bryo-1 has received much attention because of its immunomodulatory properties, both in vitro and in vivo. Bryo-1 has been shown to be a potent activator of human macrophages (18) and also to induce the production of immunomodulatory cytokines-IL-1, IL-6, IL-8, and TNF-α (18). It is also known that Bryo-1 induces the proliferation and activation of B and T cells (19, 20). Studies from our laboratory have shown that Bryo-1 enhances the maturation and antigen-presenting abilities of DCs in vitro (21). We have demonstrated that Bryo-1 alone or in combination with calcium ionophore could activate cord blood monocyte-derived DCs to express higher levels of MHC class II antigens, as well as the co-stimulatory molecules CD1a, CD80, CD83, and CD86. Furthermore, Bryo-1 and calcium ionophore-activated DCs were capable of inducing the proliferation of cord blood-derived alloreactive T cells and the production of IFN-γ (21). However, the molecular mechanism(s) by which Bryo-1 exerts its biological properties on DCs is not clearly understood. In this study, we investigated the involvement of TLR4 in Bryo-1-mediated effects in vitro and in vivo.
In this study, we have identified some unique properties of Bryo-1, which demonstrate that it can act as a TLR-4 ligand, thereby activating the innate immunity. Our findings are important and have major translational impact because Bryo-1 is currently being tested in humans as an anti-cancer agent. Furthermore, the ability of Bryo-1 to induce CC-chemokines, RANTES and MIP1-α, may have a major application as an inhibitor of HIV infection. Moreover, the inability of Bryo-1 to activate inflammatory cytokines such as IL-12 in DCs and its property to promote a Th2 response may find its use to treat inflammatory diseases by facilitating Th1 to Th2 switch.
EXPERIMENTAL PROCEDURES
Mice
Adult (6–8 weeks of age) female C57BL/6 (H-2b) (referred to as wild type (WT) and C57/10ScNJ (H-2b) (referred to as TLR4−/− or TLR4 KO) mice were purchased from NCI, National Institutes of Health, and The Jackson Laboratory, respectively. MyD88−/− mice were kindly provided by Dr. Wei Chao, Department of Anesthesia and Critical Care, Massachusetts General Hospital, Harvard Medical School, Boston. Mice were housed in polyethylene cages and given standard animal feed and water ad libitum. Mice were housed in rooms maintaining a temperature of 23 ± 1 °C and on a 12-h light/dark cycle.
Stable Cell Lines
HEK293 cell lines stably expressing pcDNA3, TLR3, TLR4, or both TLR4 and MD2 were gifts from D. Golenbock (University of Massachusetts Medical School, Worcester). All cell lines were maintained in DMEM-supplemented media as described previously (11).
Chemicals and Reagents
Bryostatin-1 (Biomol), LPS (Sigma), and polymyxin B sulfate (Invitrogen) were purchased and used in various in vivo and in vitro assays. The Gal4-IRF-3 and Gal4-luciferase reporter gene were a gift from T. Fujita (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). NF-κB luciferase construct ELAM was from D. Golenbock. IFNβ-RE-luciferase reporter gene was a gift from S. Kwok (Albert Einstein Medical Center, Philadelphia, PA). LPS derived from Escherichia coli strain 011:B4 and bryostatin-1 were purchased from Sigma and Biomol, respectively. Poly(IC) was obtained from Amersham Biosciences. ALL, MG132 (Calbiochem), and TAT-NBD (IKKγ NEMO binding domain) peptides were obtained from Alexis Biochemicals.
Generation of Murine Bone Marrow-derived DCs
Murine DCs were obtained from bone marrow cells by culturing with murine recombinant granulocyte macrophage colony-stimulating factor (GM-CSF; 5 ng/ml; Pharmingen) for 6 days, as described previously (22).
DC Analysis in Vivo
Twenty four hours after Bryo-1 (75 μg/kg body weight, i.p.) injection, WT and TLR4−/−mice were sacrificed and spleens removed. The RBCs were lysed, and the cell numbers were adjusted to 1 × 106 cells/ml in RPMI 1640 medium supplemented with 10% FCS. The cells were labeled for various DC activation markers and analyzed for the different DC populations (myeloid, lymphoid, and plasmacytoid).
Cell Surface Antigen Detection with Monoclonal Antibodies Using Flow Cytometry
Phenotypic analysis of DCs was carried out by double or triple staining with phycoerythrin (PE)-conjugated, allophycocyanin-conjugated, or fluorescein isothiocyanate (FITC)-conjugated mAbs following incubation with Fc-block (anti-CD16/CD32 mAb; Pharmingen) to avoid nonspecific binding. The following mAbs were used: FITC-anti-CD40, PE-anti-CD80, PE-anti-CD86, allophycocyanin-anti-CD11c, FITC-anti-CD11b, FITC-anti-B220, FITC-anti-CD4, and PE-anti-CD8α (Pharmingen). Cells were analyzed by flow cytometry (EPICS FC500; Coulter Electronics, Miami, FL).
Bio-Plex Immunoassay
Various cytokines and chemokines were assayed in the serum and supernatants of BMDCs from WT (TLR4+/+) and TLR4−/− mice, treated with vehicle, LPS, or Bryo-1. DCs from WT and TLR4−/− mice were treated with Byro-1 (10 ng/ml) for 24 h in vitro, after which the cells were spun down and the supernatants collected. Supernatants from LPS (100 ng/ml) and vehicle-treated BMDCs were used as positive and negative controls. For serum samples, blood was drawn at 24 h after Bryo-1 (75 μg/kg body weight), and serum was separated. Vehicle treated and untreated mice were used as controls. The serum was assayed using the multiplex bead-based assays (Bio-Plex assay, Bio-Rad), designed to quantitate multiple cytokine analysis, as described previously (23). We looked at a panel of 18 cytokines and chemokines, including IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12(p40), IL-12(p70), IL-17, G-CSF, GM-CSF, TNF-α, IFN-γ, KC, MIP1-α, and RANTES.
ELISA
The supernatants from the cultures were harvested and analyzed using ELISA kit for the production of RANTES (PeproTech Inc., NJ) and IFN-β (PBL Biomedical Laboratories, NJ).
Electrophoretic Mobility Shift Assay
BMDCs from WT and TLR4 KO mice were obtained as described above and subsequently treated with either LPS (100 ng/ml) or Bryo-1 (10 ng/ml). At the indicated times, nuclear extracts were prepared as described previously (24), and electrophoretic mobility shift analysis was performed probing for the DNA binding activity of NF-κB. Briefly, 5 μg of nuclear extract was incubated with 1 μg of poly(dI-dC)-poly(dI-dC) (Amersham Biosciences) for 10 min at room temperature. To this mixture, 2 × 104 cpm of a 32P-labeled oligonucleotide (NF-κB, 5′-ATGTGAGGGGACTTTCCCAGGC-3′, Santa Cruz Biotechnology; RANTES-ISRE-IRF-1-RE, 5′-ACACAAAATGGAAAACTGAAATCACCCTTGG-3′; and IFNβ-PRD III-I, 5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′, Integrated DNA Technologies) was added (binding site is underlined) in a buffer consisting of 4 mm Tris-HCl, pH 7.9, 12 mm Hepes-KOH, 60 mm KCl, 12% glycerol, 0.5 mm EDTA, and 1 mm dithiothreitol and incubated for an additional 20 min. Complexes were resolved on a nondenaturing 5% polyacylamide gel and subsequently exposed on Hyperfilm (Amersham Biosciences). For supershifts, antibodies raised against specific subunits of NF-κB, p65, p50 (nuclear localization signal), c-Rel, and RelB, or the IRF family members, IRF-1, IRF-3 (H-246), and IRF7 (FL-425) (Santa Cruz Biotechnology), were preincubated for 10 min at room temperature before the addition of 1 mm phenylmethylsulfonyl fluoride and 1 μg of poly(dI-dC)-poly(dI-dC).
Luciferase Reporter Gene Assays
HEK293 cells (1 × 106) were seeded into 6-well plates and transiently transfected 24 h later using Lipofectamine transfection reagent (Invitrogen), following the recommendations of the manufacturer. For NF-κB and IFNβ reporter assays, cells were transfected with 0.5 μg of either ELAM or IFN-β, respectively, and 0.1 μg of CMV-β-Gal reporter vectors. For IRF-3 reporter assay, HEK293 cells (4 × 104) were seeded into 96-well plates and transfected with 40 ng/well of the Gal4-luciferase reporter gene. At 36 h following transfection, cells were treated with Bryo-1 (50 ng/ml) and LPS (100 ng/ml) for 6 h or left untreated. Cell lysates were prepared, and reporter gene activities were measured using the Dual-Luciferase reporter system (Applied Biosystems). Data were normalized for transfection efficiency and expressed as the mean relative stimulation ± S.D.
Detection of RANTES in DCs from Myd88−/− Mice
To detect expression of RANTES in DCs from Myd88−/− mice, we performed intracellular staining using Cytofix/CytopermTM fixation/permeabilization kit following the recommendations of the manufacturer (BD Biosciences).
Western Blot Analysis of IRF3
Western blotting was performed using antibodies against total IRF3 and phosphorylated IRF3 (P-IRF3 Ser-396; Santa Cruz Biotechnology) at a dilution of 1:1000 and β-actin at dilution of 1:5000. HRP-conjugated secondary Ab was used at 1:4000 dilution (Cell Signaling). In brief, lysates from vehicle- or Bryo-1-treated BMDCs were prepared, and protein concentration was measured using standard Bradford assay (Bio-Rad). The proteins fractionated in SDS-PAGE were transferred onto PVDF membranes using a dry blot apparatus (Bio-Rad). The membrane was first incubated in blocking buffer for 1 h at room temperature, followed by incubation in primary antibody at 4 °C overnight. The membrane was incubated for 1 h in HRP-conjugated secondary antibody (Cell Signaling Technology, Danvers, MA) in blocking buffer after washing three times (10–15 min) with washing buffer (PBS + 0.2% Tween 20). The membranes were incubated in developing solution (ECL Western blotting detection reagents, GE Healthcare), and signal was detected using ChemiDoc System (Bio-Rad). Densitometric analyses of the Western blots were performed using ChemiDoc software (Bio-Rad).
Statistical Analysis
The statistical comparisons between different study groups were carried out using Student's t test and GraphPad software and differences of p < 0.05 were considered to be significant. Each experiment was repeated at least three times.
RESULTS
Treatment of BMDCs with Bryo-1 in Vitro Leads to TLR4-dependent Expression of Chemokines, Cytokines, and Up-regulation of Co-stimulatory Molecules
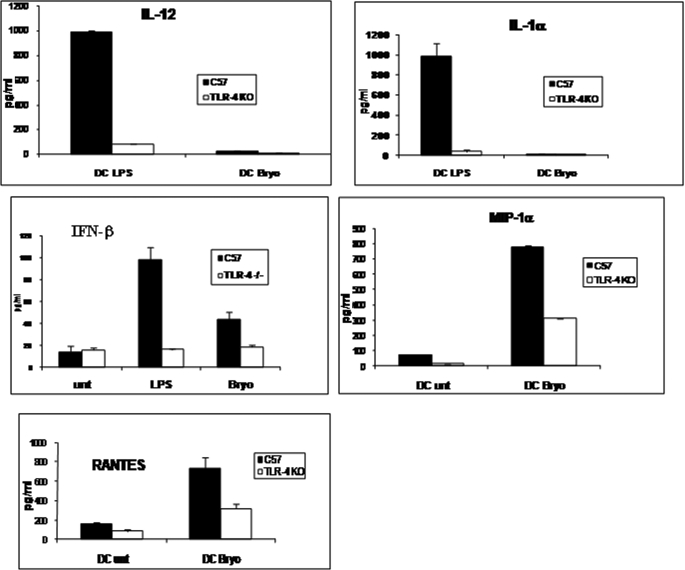
Earlier studies from our laboratory have shown that Bryo-1 is capable of inducing maturation of DCs (21). To determine the cytokine/chemokine profile induced by Bryo-1, immature BMDC from WT and TLR4−/− mice were treated with Bryo-1 or vehicle. Next, supernatants were evaluated for the presence of cytokines and chemokines by ELISA, as described under “Experimental Procedures.” Specifically, we studied cytokines and chemokines that are induced following activation of DCs through TLRs, including IL-1α, IFN-β, IFN-γ, IL-12, TNF-α, IL-6, MIP1-α, KC, and RANTES. We observed that activation of BMDCs with LPS from WT mice led to significant induction of IL-12 and IL-1α as well as low levels of IFN-β (Fig. 1), and furthermore, these cytokines were dramatically reduced in LPS-activated BMDCs from TLR4 KO mice. Interestingly, Bryo-1 activated BMDCs produced little or no IL-12 and IL-1α and low levels of IFN-β. Moreover, of all the cytokines and chemokines screened, Bryo-1 activated BMDCs from wild-type mice produced only MIP1-α and RANTES (also known as CCL5) (Fig. 1). It was noted that Bryo-1-induced production of IFN-β, MIP1-α, and RANTES in DCs was significantly higher in WT BMDCs when compared with BMDCs from TLR4−/− mice, suggesting that the induction of IFN-β, MIP1-α, and RANTES by Bryo-1 was also regulated, at least in part, through TLR4 activation.
FIGURE 1.
Bryo-1 acts as a TLR-4 ligand and triggers cytokine and chemokine production in murine BMDCs, which is significantly different from that induced by LPS. Supernatants from BMDCs from C57BL/6 wild-type or TLR4−/− mice treated with vehicle, LPS, or Bryo-1 for 24 h were examined for cytokines and chemokines as described under “Experimental Procedures.”
Bryo-1 Induces NF-κB Activation through TLR-4
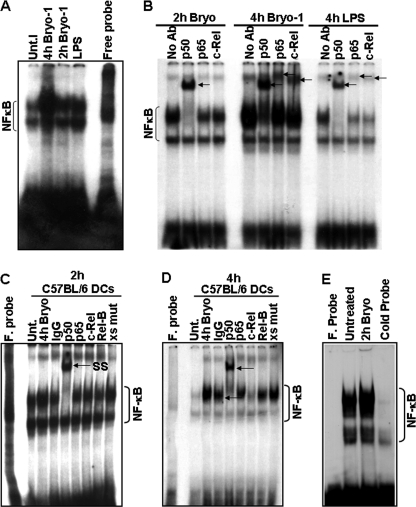
Because all TLR signaling pathways culminate in the activation of NF-κB transcription factor, which in turn regulates the induction of cytokines, we investigated whether Bryo-1-induced activation of DCs involved modulation of NF-κB binding activity. To explore this possibility, nuclear extracts from Bryo-1 (10 ng/ml) or LPS (100 ng/ml)-stimulated BMDCs were prepared, and the NF-κB binding activity was examined by EMSA analysis, using a specific 32P-labeled NF-κB oligonucleotide. As shown in Fig. 2A, Bryo-1 induced NF-κB activation at 2 and 4 h following treatment. We used LPS-treated BMDCs as a positive control. To further evaluate the composition of the NF-κB complexes induced by Bryo-1, supershift analysis using antibodies against the NF-κB members p50, p65, c-Rel, and RelB was performed. As shown in Fig. 2B, the majority of the NF-κB complexes was composed of a p50 homodimer after a 2-h treatment with Bryo-1 and of p50/c-Rel and p50/p65 heterodimers following a longer (4 h) incubation time with Bryo-1. To determine the binding specificity of these complexes, competition analysis using a mutated NF-κB oligonucleotide was performed. As shown in Fig. 2, C and D, binding of the nuclear complexes to the DNA probe was specific because Bryo-1-mediated stimulation of DNA-protein complexes was not blocked by excess of unlabeled mutated NF-κB oligonucleotide (9th lane). Competition assays using 100-fold molar excess of unlabeled DNA probe as specific competitor further demonstrated the specificity of the band corresponding to NF-κB (Fig. 2E, cold probe lane).
FIGURE 2.
Effect of Bryo-1 on the activation of NF-κB in BMDCs. A, BMDCs from C57BL/6 mice were treated with Bryo-1 (10 ng/ml) or LPS (100 ng/ml). At the indicated time points, NF-κB DNA binding activity in nuclear extracts was determined by EMSA. Unt, untreated. B, specific formation of complexes, supershifted by anti-p50, anti-p65, and anti-c-Rel antibodies, are indicated by arrows. C and D, mutated oligonucleotide did not compete out the NF-κB complexes. F probe, free probe. E, competition assays demonstrated competing out of labeled NF-κB complexes in the presence of unlabeled wild-type NF-κB oligonucleotides.
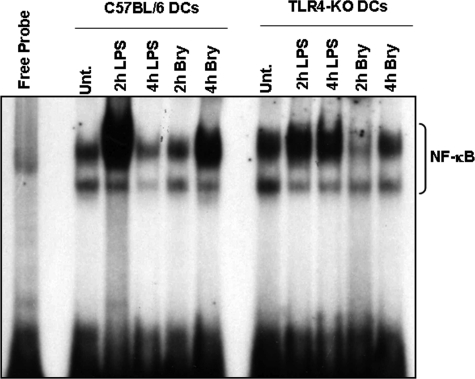
To determine whether TLR4 was implicated in Bryo-1-mediated activation of NF-κB, we examined the ability of BMDC nuclear extracts from C57BL/6 and TLR4−/− mice to bind to the NF-κB probe in response to Bryo-1. Interestingly, we found that the NF-κB binding activity of nuclear extracts from TLR4−/− BMDCs was significantly reduced relative to that of wild-type cells suggesting that TLR4 was in fact involved in Bryo-1 signaling in BMDCs (Fig. 3).
FIGURE 3.
Bryo-1-induced activation of NF-κB is TLR4-mediated. BMDCs from C57BL/6 or TLR4−/− mice were treated with Bryo-1 (10 ng/ml) or LPS (100 ng/ml). At the indicated time points, NF-κB DNA binding activity in nuclear extracts was determined by EMSA. Activated complexes were examined by autoradiography. Unt., untreated.
Bryo-1-mediated Induction of RANTES Is Regulated by Transcription Factors NF-κB and IRFs
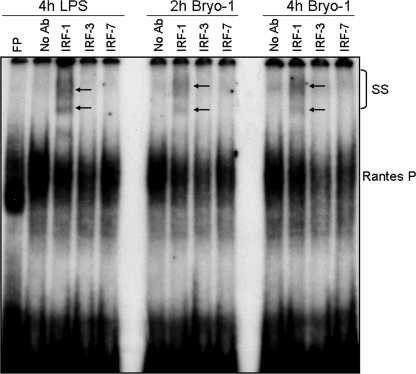
We next addressed the molecular mechanism(s) by which Bryo-1 triggers the production of RANTES by DCs. To determine whether the increased RANTES production induced by Bryo-1 is mediated by IRFs, the binding of nuclear extracts from Bryo-1 or vehicle-treated BMDCs to a 32P-labeled oligonucleotide containing the IRF-binding motif in the RANTES gene promoter was examined by EMSA analysis, as described previously (24). As in LPS-stimulated BMDCs, binding of nuclear extracts to the IRF motif of the RANTES promoter was observed in response to Bryo-1 treatment of BMDCs (Fig. 4A). Furthermore, supershift analysis of the IRF complexes, using antibodies against the IRF-1, IRF-3, and IRF-7 proteins, revealed IRF-1 as the protein bound to the RANTES promoter in both Bryo-1- and LPS-stimulated BMDCs (Fig. 4A). Interestingly, IRF-3 (Fig. 4A, 8th and 12th lanes) and IRF-7 (9th and 13th lanes) also caused a gel shift when compared with the binding activity induced by Bryo-1 in the absence of antibody (Fig. 4A, 6th and 10th lanes) at both time points (2 and 4 h) suggesting that IRF-1 and possibly IRF-3 and IRF-7 may be the modulators of Bryo-1-mediated RANTES gene expression.
FIGURE 4.
Bryo-1 stimulation of BMDCs induces nuclear binding of IRF-1 to the promoter of RANTES gene. Nuclear extracts were prepared 2 and 4 h after Bryo-1 or LPS (4 h) treatment, and the presence of RANTES-IRF-1 binding activity was determined by EMSA. Complex formations (supershifted (SS) band) following incubation with anti-IRF-1, anti-IRF-3, and anti-IRF-7 were visualized by autoradiography and are indicated by arrows. FP, free probe.
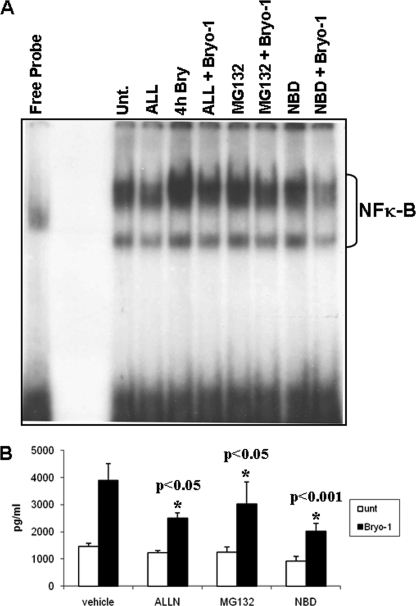
NF-κB Inhibitors Decrease Bryo-1-induced NF-κB Binding Activity
The promoter of the RANTES gene carries IRF and NF-κB cis-acting elements both of which may be critical for RANTES gene expression (25). To further examine the specificity of Bryo-1-mediated increase in NF-κB binding activity, a series of proteosome inhibitors were used. Briefly, DCs were preincubated for 2 h with the NF-κB inhibitors ALL (10 μm), MG132 (1 μm), TAT-NBD peptide (200 μm), or left untreated followed by Bryo-1 treatment for 4 h. DCs were harvested for nuclear extract preparation, and the supernatant was collected for cytokine analysis by ELISA, as described under “Experimental Procedures.” The nuclear extracts were examined for their ability to bind to a 32P-labeled oligonucleotide containing the NF-κB consensus sequence. In the presence of nuclear extracts from vehicle-treated BMDCs, constitutive NF-κB complex formation was observed (Fig. 5, 2nd lane). Bryo-1 treatment, however, further increased protein complex binding to the NF-κB site (Fig. 5, 4th lane). Preincubation of BMDCs with various NF-κB inhibitors resulted in a decreased DNA binding activity of nuclear extracts relative to that of Bryo-1 alone. Furthermore, the NF-κB inhibitors ALL and NBD peptide were more effective at reducing protein-DNA complex formation than MG132 under our experimental conditions.
FIGURE 5.
Effect of NF-κB inhibitors on Bryo-1-induced NF-κB binding activity. BMDCs were incubated for 2 h with the NF-κB inhibitors, ALL (10 μm), MG132 (1 μm), and NBD peptide (200 μm), prior to Bryo-1 stimulation for 4 h and the NF-κB binding activity in the nuclear extracts was examined by EMSA and detected by autoradiography (A). The supernatants were assayed for the presence of RANTES by a sandwich ELISA (B). Vertical bars in B represent mean ± S.E. of three independent experiments, and asterisks represent statistically significant (p < 0.05) suppression of RANTES expression in the presence of NF-κB inhibitors. Unt, untreated.
In addition, the supernatants were also collected and analyzed for the presence of RANTES by ELISA (Fig. 5B). It was noted that Bryo-1-mediated induction of the NF-κB binding activity was considerably decreased in the presence of the proteosome inhibitors, ALL, TAT-NBD peptide, and to a lesser extent MG132. In addition, preincubation of BMDCs with the NF-κB inhibitors resulted in a decrease in the amount of RANTES chemokine released into the medium (Fig. 5B), suggesting that NF-κB was involved in the production of RANTES. Altogether, these data indicated that NF-κB and IRF-1 may be involved in the induction of RANTES in BMDCs in response to Bryo-1 stimulation.
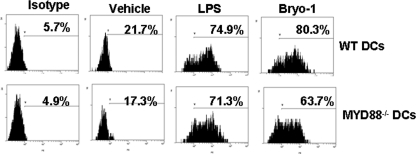
Role of MyD88 in RANTES Production following Activation of BMDCs by Bryo-1
TLR4 activates both the MyD88- and the TRIF-dependent pathways. To investigate further the role of MyD88, immature BMDCs from WT and MyD88-deficient mice were cultured with Bryo-1 and analyzed for intracellular RANTES induction. The data indicated that BMDCs from MyD88 KO mice produced significant levels of RANTES comparable with the WT mice thereby suggesting that Bryo-1-induced RANTES production was independent of MyD88 (Fig. 6). These data were also consistent with the above observation that RANTES induction by Bryo-1 involved IRFs whose activation is independent of MyD88.
FIGURE 6.
Production of RANTES by DCs from MyD88−/− mice. BMDCs from C57BL/6 wild-type or MyD88−/− mice treated with vehicle, LPS, or Bryo-1 for 24 h in vitro and stained with Abs against RANTES and the cells were examined by flow cytometry.
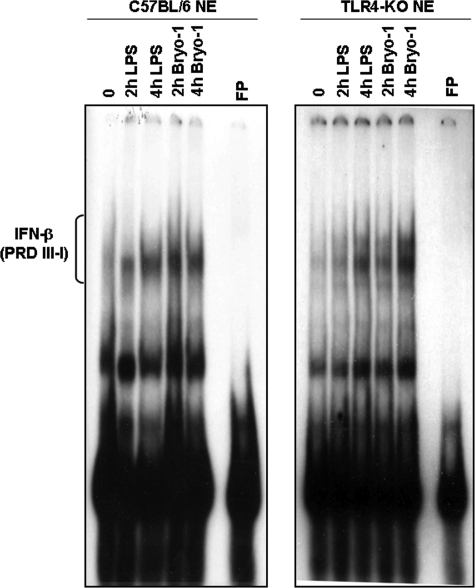
Role of TLR-4 in Bryo-1-mediated Induction of IFN-β in DCs
There is increasing evidence that supports a key role for DCs in the production of type I interferons and the regulation of innate and adaptive immune responses (26, 27). We noted that activation of DCs with Bryo-1 failed to induce a majority of the cytokines characteristic of DCs except low levels of IFN-β. To further corroborate the role played by TLR-4 in the induction of IFN-β by Bryo-1, EMSA analysis of nuclear extracts from Bryo-1 (10 ng/ml) or LPS (100 ng/ml) stimulated WT or TLR4−/− BMDCs using a specific 32P-labeled DNA oligonucleotide containing the positive regulatory domains (PRDI-III) of the IFN-β gene promoter was performed. The data demonstrated that although Bryo-1 induced the binding of nuclear complexes to the IFN-β promoter DNA probe in the WT cells, Bryo-1-mediated stimulation of DNA-protein complex formation was significantly reduced and delayed in TLR4−/− BMDC (Fig. 7). These data suggested that TLR-4 was involved in the induction of IFN-β by Bryo-1, consistent with the observation that TLR4-deficient BMDC produced decreased levels of IFN-β upon Bryo-1 activation when compared with wild-type BMDC (Fig. 1).
FIGURE 7.
Induction of DNA binding activity and IFN-β production by Bryo-1 stimulation of BMDCs. Nuclear extracts were isolated from wild-type C57BL/6 or TLR4−/− mice treated with either Bryo-1 (10 ng/ml) or LPS (100 ng/ml) for the indicated times and subjected to EMSA using a 32P-labeled IFN-stimulated response element consensus sequence of the IFN-β gene promoter as probe. Activated complexes were detected by autoradiography. FP, free probe.
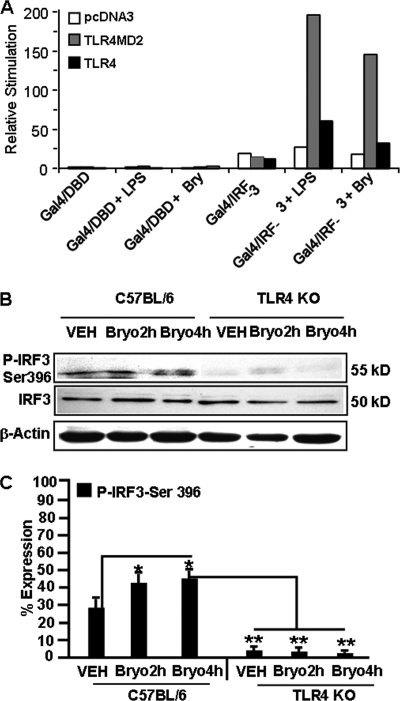
Bryo-1 Induces Transcriptional Activation of IRF-3 via TLRs
To determine whether Bryo-1 activates the transcription factor IRF-3 through TLRs, we used an in vivo assay for IRF-3 activation, in which a hybrid protein consisting of the yeast Gal 4 DNA-binding domain is fused to the IRF-3 (Gal4-IRF-3) lacking its own DNA-binding domain (28). Transcriptional activation of the Gal4 reporter gene requires activation of IRF-3 (28). When TLR3, TLR4MD2, or TLR4-expressing HEK293 cell lines were transfected with vectors encoding a luciferase reporter gene containing the Gal4 upstream activation sequence and either a vector encoding Gal4-DBD or Gal4-IRF-3 followed by 8 h of stimulation with Bryo-1, the Gal4 reporter gene was activated. Bryo-1 induced increases in luciferase activity in both TLR3 and TLR4/MD2 but not TLR4 (Fig. 8A). Taken together, these results suggested that TLRs are involved in Bryo-1-mediated activation of IRF-3. To further explore this possibility, the expression levels of total IRF-3 and phosphorylated IRF-3 (P-IRF3 Ser-396) were determined in BMDC cell lysates from C57BL/6 (TLR4+/+) and TLR4 KO (TLR4−/−) mice in the absence or presence of Bryo-1. As shown in Fig. 8, B and C, Bryo-1 stimulation led to increased phosphorylation of IRF-3 in BMDCs at both 2 and 4 h post-treatment, when compared with vehicle-treated BMDCs. There were no changes in total IRF-3 in vehicle- or Bryo-1-treated BMDCs. However, there was no phosphorylation of IRF-3 in BMDCs generated from TLR4 KO mice in the presence of Bryo-1. These data demonstrate that Bryo-1 may activate IRF-3 via TLR4 in BMDCs (Fig. 8, B and C).
FIGURE 8.
Effect of Bryo-1 on IRF-3 activation. A, pcDNA3, TLR4MD2-, and TLR4-HEK293-expressing cells were seeded in 96-well plates. After 24 h, cells were transfected with a luciferase reporter gene containing the upstream Gal4-activating sequence and Gal4/DBD (control) or Gal4/IRF-3 (50 ng) plus pCMV-β-Gal (10 ng). After 24 h, cells were stimulated with Bryo-1 (Bry) (10 ng/ml), LPS (100 ng/ml), or left untreated for 8 h, and luciferase reporter gene activity was measured. The relative stimulation value represents the ratio of firefly to β-galactosidase luciferase activities. B and C, protein from BMDCs (wild type or TLR4 KO) treated with vehicle or Bryo-1 (2 and 4 h) were fractionated in polyacrylamide gel, and expression of total IRF3 and p-IRF3-Ser-396 was analyzed by Western blotting using antibody against mouse-specific IRF3 and p-IRF3-Ser-396. Expression of p-IRF3-Ser-396 is presented as percentage of β-actin expression on the y axis, and expression of β-actin was considered to be 100%. Vertical bars represent mean ± S.E. of three independent experiments, and * represents statistically significant (p < 0.05) up-regulation of phosphorylated IRF3-Ser-396 in Bryo-1-treated groups when compared with vehicle (VEH), and ** represents statistically significant (p < 0.05) down-regulation in expression of phosphorylated IRF3-Ser-396 when Bryo-1 groups are compared between wild-type, and TLR4 KO cells are compared.
Bryo-1 Treatment in Vivo Leads to TLR4-dependent Th2 Cytokine Production
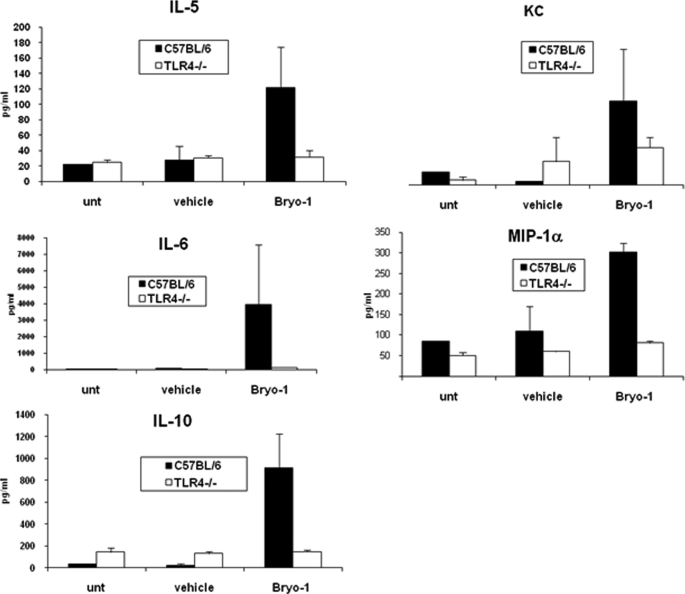
We next tested if administration of Bryo-1 would trigger cytokine production in vivo and if this would also be regulated by TLR4. To this end, serum collected from WT and TLR4−/− mice 24 h following treatment with Bryo-1 or vehicle was assayed for 18 different cytokines/chemokines, including IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12(p40), IL-12(p70), IL-17, G-CSF, GM-CSF, TNF-α, IFN-β, KC, MIP1-α, and RANTES. Surprisingly, WT mice receiving Byro-1 demonstrated induction of only certain Th2 cytokines, IL-5, IL-6, IL-10, and the chemokines, MIP1-α and KC, when compared with vehicle-treated mice (Fig. 9). In contrast, TLR4−/− mice failed to produce significant levels of these cytokines and chemokines following Bryo-1 injection (Fig. 9).
FIGURE 9.
Bryo-1 administration in vivo triggers chemokines and Th2 cytokines in WT but not TLR4−/− mice. Serum samples from WT and TLR4−/− mice either untreated or treated with vehicle or Bryo-1 (75 μg/kg body weight) were collected 24 h post-treatment. The serum was assayed for various cytokines/chemokines by bioplex assay.
Phenotype of DCs Expanded in Vivo by Bryo-1
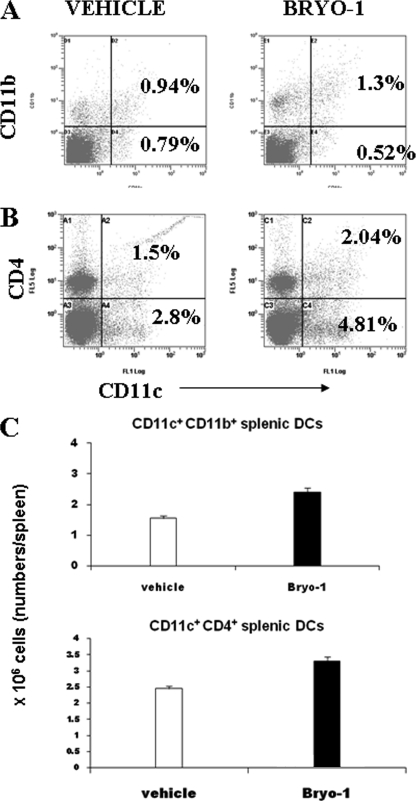
The type of cytokine(s) produced early in an immune response is key in determining if a Th1- or Th2-like immune response is generated. The subsets of DC are thought to be important in influencing the type of response (29, 30). To this end, WT mice were injected with Bryo-1, and the splenocytes were examined for the various DC lineage markers. Cells were double-stained for CD11c and CD11b, B220, CD4, and CD8α. Flow cytometric analysis demonstrated that there was a significant increase in the CD11c+ CD11b+ cell population in the Bryo-1-treated group when compared with the vehicle control (Fig. 10A). The CD11c+B220+ cell population was not enhanced significantly in the Bryo-1-treated group (data not shown). The spleen cells were also triple stained for CD11c, CD4, and CD8α. Our results demonstrated an increase in the CD11c+CD4+ (Fig. 10B) population in the Bryo-1-treated group but no increase in the CD11c+CD8α+ population (data not shown). The total cellularity of CD11b+ and CD4+ DCs in the spleen was higher in the Bryo-1-treated groups when compared with the vehicle-treated group (Fig. 10C). Taken together with the earlier lineage marker data, these findings indicated that Bryo-1 may expand a subset of the myeloid arm of DCs that are CD11c+CD8+-CD11b+CD4+.
FIGURE 10.
Effect of Bryo-1 administration in vivo on DC subpopulations in spleen. C57BL/6 mice received intraperitoneal injection of vehicle or Bryo-1, and 24 h later, the spleen cells were analyzed for the expression of CD11c, CD11b, or CD4 using flow cytometry. A and B show double staining for CD11c and CD11b, and CD11c and CD4, respectively. C depicts the total cellularity of the CD11c+CD11b+ and CD11c+CD4+ DCs in the spleens of vehicle or Bryo-1-treated C57BL/6 mice. The data represent means ± S.E. of three mice/group. The total number of cells in the Bryo-1-treated mice were significantly higher than the vehicle groups (p < 0.05).
Up-regulation of Various DC Co-stimulatory Molecules (CD40, CD80, and CD86) and Chemokine (RANTES) Are Bryo-1-specific
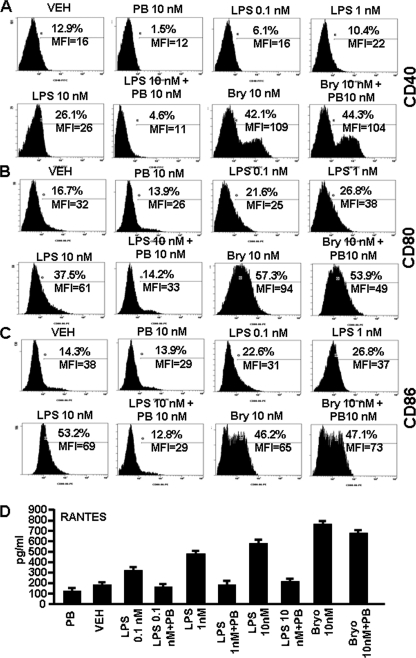
As shown by us in the current and previous study (21), LPS-induced activation of DCs showed a distinct pattern when compared with that induced by Bryo-1. To further rule out the possibility that the effect of Bryo-1 may have been mediated by low levels of endotoxin contamination, we investigated the effect of LPS and Bryo-1 on DC maturation and RANTES production in the presence of polymyxin B, which is known to block LPS-mediated effects. To this end, BMDCs generated from C57BL/6 mice were cultured in the presence of vehicle, polymyxin B sulfate (PB; 10 nm) alone, Bryo-1 (10 nm), Bryo-1 + PB, various doses of LPS (0.1, 1, and 10 nm), and LPS + PB for 24 h in a 96-well plate. Next, the cells were stained for CD40, CD80, and CD8, and the supernatants from cultures were examined for RANTES production. The data showed that Bryo-1 caused the maturation of DCs with a significant increase in the expression of CD40, CD80, and CD86 when compared with vehicle-treated groups (Fig. 11, A–C), as noted in our earlier studies (21). However, we did not observe down-regulation of these markers in the presence of PB in Bryo-1 (Bryo-1 + PB)-treated groups. LPS, when used alone, failed to induce DC maturation at 1 nm but was able to up-regulate the expression of CD40, CD80, and CD86 at 1 and 10 nm. However, at 10 nm, LPS was not as effective as 10 nm of Bryo-1 (Fig. 11, A–C). Furthermore, addition of PB to LPS cultures caused complete inhibition of up-regulation of all markers induced by LPS (Fig. 11, A–C). Also, similar results were observed when RANTES expression was analyzed in various cultures of DCs. LPS caused a dose-dependent increase in RANTES production, and PB completely inhibited RANTES induction by all doses of LPS tested. In contrast, PB failed to significantly inhibit Bryo-1-induced RANTES (Fig. 11D). Together, these data suggested that Bryo-1-induced activation of DCs was independent of LPS contamination.
FIGURE 11.
DC maturation and RANTES production are Bryo-1-specific. DCs generated from bone marrow of C57BL/6 mice were cultured in the presence of vehicle (VEH), PB, Bryo-1 (Bry), Bryo-1 + PB, LPS, or LPS + PB for 24 h in vitro. The DCs were collected and analyzed by flow cytometry after staining with mouse FITC-CD40-, PE-CD80-, or PE-CD86-specific mAbs. Supernatants of various cultures were collected and analyzed for the presence of RANTES by ELISA. A–C show the expression levels of cell surface markers CD40, CD80, and CD86, respectively. The data are expressed as the mean fluorescence intensity (MFI) and represent one of three independent experiments. D represents the production of RANTES by DCs, and data are expressed as the mean levels (pg/ml) ± S.E. of triplicate cultures. The production of RANTES in the Bryo-1-treated DCs culture was significantly higher than in the vehicle group (p < 0.05). Presence of PB in the cultures containing Bryo-1 showed no significant blocking of RANTES, whereas PB caused significant inhibition of RANTES secretion in cultures with LPS (p < 0.05).
DISCUSSION
Bryostatin 1 is a macrocyclic lactone that is known to regulate PKC activity (31). It has been shown to inhibit cell growth and angiogenesis, promote apoptosis, and induce cancer cell differentiation, and because of that, it is being screened as an anticancer agent in humans (32). Our laboratory has previously shown that Bryo-1 promotes maturation of murine and human DCs, and such DCs act as very potent allophycocyanins, better than LPS-activated DCs, to induce T cell proliferation (21). In this study, we demonstrate for the first time that Bryo-1 activates DCs through TLR4. Moreover, we report that stimulation of BMDCs by Bryo-1 resulted in the stimulation of NF-κB binding activity. The NF-κB DNA binding activity was significantly decreased in BMDCs from TLR4−/− mice, suggesting that Bryo-1 induction of NF-κB binding activity is TLR4 signal-mediated. Interestingly, unlike LPS, Bryo-1 induced a biphasic activation of NF-κB. The initial phase, which occurred 2 h after Bryo-1 treatment, was characterized by the formation of p50 homodimers, which are associated with a repressor function. By 4 h, the composition of the DNA-protein complexes switched to p50/c-Rel and p50/p65 heterodimers, which are the transcriptionally active complexes. Although the functional implications of these findings will require additional studies, it is possible that the distinct NF-κB complexes formed in response to Bryo-1 may have a role in the differential regulation of gene expression in BMDCs, as well as in the control of DC-mediated immune and inflammatory responses.
It is well established that TLR4 signals through at least four adaptor molecules known as TIRAP, MyD88, TRAM, and TRIF. They associate with a diverse repertoire of signaling components, and they constitute the platforms for the initiation of different signaling pathways. It has been shown that LPS stimulation of TLR4 can activate the NF-κB signaling pathway by the following: (i) oligomerization of MyD88 (9, 26), which induces the recruitment and activation of IL-1 receptor-activated kinase (IRAK) 1 and IRAK4, or (ii) via TRAM-TRIF adaptor molecules, which induce activation of NF-κB independently of MyD88. Early stimulation of NF-κB activity by TLR4 activation results in the production of pro-inflammatory cytokines and is regulated through the MyD88-dependent pathway (6, 33). In this study, we noted that Bryo-1-activated BMDCs produced RANTES independent of MyD88 thereby suggesting that NF-κB activation in response to Bryo-1 may be MyD88-independent. Our studies show that like LPS, Bryo-1 stimulation of TLR4 induced IFN-β production (Fig. 7B) and increased binding activity to the IFN-β gene promoter (Fig. 7A). However, the binding activity was significantly reduced and delayed in BMDCs from TLR4−/−mice. Similarly, IFN-β production was markedly decreased in the supernatant of Bryo-1-treated TLR4−/− DCs. Furthermore, gene-targeting experiments have shown that LPS-mediated production of IFN-β through TLR4 occurs mostly, if not entirely, independent of MyD88 (25) but is dependent on the adaptor molecules, TRAM/TRIF (34, 35). It has recently been reported that the IRF-3 transcription factor is a key regulator of this pathway (26). Activation of IRF-3, by gene reporter assays, was observed following Bryo-1 stimulation of TLR4/MD2 HEK293-expressing cells. These results, together with a delay in the activation of NF-κB and the lack of pro-inflammatory cytokine production by Bryo-1 stimulation, begin to support the premise that Bryo-1 may be signaling through the MYD88-independent pathway of TLR4.
We also analyzed the binding activity of several IRF transcription factors to the promoter response elements of the RANTES gene. We observed that, in addition to increasing IFN-β production and activating NF-κB and IRF-3 transcription factors, Bryo-1 was able to induce DNA binding of the transcription factors IRF-1, IRF-3, and IRF-7 to the RANTES-IRF-1 site of the gene promoter. Furthermore, treatment of DCs with various NF-κB inhibitors prior to Bryo-1 stimulation not only partially decreased the DNA binding activity by EMSA analysis but resulted in a significant decrease in the production of the chemokine, RANTES (Fig. 6), suggesting that Bryo-1-mediated production of RANTES requires a functional TLR4 and may be regulated by NF-κB and IRF transcription factors. Consistent with our findings, Lee et al. (44) have shown that NF-κB and IRF-1 cooperatively activated the RANTES promoter in co-transfection studies and suggested that activation of the RANTES promoter by NF-κB and IRF-1 may occur through a multiprotein complex formation, as described previously (42). In addition, Kirchhoff et al. (36) have shown that IRF-1 could activate NF-κB, which then synergized with IRF-1 to induce IFN-β gene expression. Expression of the RANTES gene has been demonstrated to be stimulated by inflammatory cytokines (37, 38), LPS (39), T cell activation (40), and viral infection (41, 42). Recently, it has been demonstrated that IRF-3 plays a key role in the activation of RANTES transcription by viral infection (43). Interestingly, in these studies the promoter element recognized by IRF-3 mapped to the same region as the IRF-1-binding site, identified by Lee et al. (44). These findings support our observation that IRF-3 and potentially IRF-7 bound to the IRF-1 response element of the RANTES promoter. IRF-3 and IRF-7 are known to be key activators of the RANTES gene (43), which further support our hypothesis that IRF-1, IRF-3, and IRF-7 in cooperation with NF-κB may regulate Bryo-1-mediated induction of RANTES gene expression. These data indicate that different stimuli can trigger the recruitment of multiple IRF transcription factors, which then regulate binding to and promoter activity of the RANTES gene. However, what still remains to be elucidated is how different physiological stimuli induce the recruitment of selective IRF transcription factors to the same binding site of the RANTES promoter to activate gene expression.
It is known that TLR4 signaling initiates a broad range of innate and adaptive immune responses. Interestingly, our studies demonstrated that Bryo-1 activation of TLR4 in DCs in vitro led to increased production of only certain specific cytokines and chemokines. Moreover, the cytokine profiles elicited by Bryo-1 were entirely different from that induced by LPS. For example, activation of DCs with LPS led to significant induction of IL-12 and IL-1α and IFN-β. In contrast, Bryo-1-activated DCs failed to produce significant levels of IL-12 and IL-1α while producing lower levels of IFN-β when compared with LPS stimulation. Moreover, in vivo administration of Bryo-1 led to dramatic induction of IL-5 and IL-10 but not any of the Th1 cytokines tested. These findings suggested that Bryo-1 may selectively activate certain DC subsets. Previous studies have shown that CD11c highCD8α+ CD11b− CD4− cells commonly called “lymphoid” DCs secrete abundant IL-12 and stimulate a Th1 response, whereas CD11chighCD8α− CD11b+ CD4+ cells, which constitute a subset of “myeloid” DCs, do not secrete significant IL-12 but rather produce IL-10 and activate a Th2 response (45). In this study, we noted that in vivo administration of Bryo-1 induced DCs that were predominantly expressing the following markers: CD11c highCD8α− CD11b+CD4+, suggestive of the induction of a subset of myeloid DCs. These data are also consistent with our observation that such Bryo-1-activated DCs failed to produce IL-12 but induced significant levels of Th2 cytokines in vivo such as IL-5, IL-6, and IL-10. These cytokines are known to be anti-inflammatory and to increase antibody response.
Activation of murine BMDCs through various TLRs have been shown to induce a variety of chemokines such as MIP1-α, MIP1-β, MCP-1, RANTES, and KC (46). In this study, we noted that Bryo-1 induced high levels of MIP1-α and RANTES production by the DCs when compared with LPS. Moreover, the production of these chemokines was significantly decreased in TLR4-deficient mice. RANTES, also termed CCL5, is a proinflammatory CC-chemokine that plays multiple roles during inflammation. RANTES displays high affinity binding and signaling through multiple independent chemokine receptors, including CCR1, CCR3, and CCR5. Also, MIP1 belongs to CC-chemokine subfamily consisting of MIP1-α (CCL3), MIP1-β (CCL4), MIP1-δ (CCL9/10) and MIP1-γ (CCL15) that are produced by many cells, including DCs. MIP1 proteins, act via G-protein-coupled cell surface receptors CCR1, CCR3, and CCR5. Because of their ability to bind such chemokine receptors, RANTES and MIP1 are considered to be the most potent natural inhibitor of HIV-1 infection and are currently being pursued as safe and effective viral entry inhibitors (47, 48). Our findings raise an exciting possibility of the potential use of Bryo-1 as an inhibitor of HIV-1 infection.
Bryo-1 is well known for its anti-tumor activity, which has been ascribed to modulation of protein kinase C (PKC) activity. Clinical trials have suggested that Bryo-1 does not exhibit significant toxic effects such as myelosuppression, gastrointestinal toxicity, or neuropathy, except for myalgia (49, 50). In the clinical trials, Bryo-1 was also shown to induce IL-6 and TNF-α (50). This study has a major translational impact on the clinical use of Bryo-1 because it demonstrates for the first time that Bryo-1 may act as a TLR-4 ligand and may serve as a potent activator of innate immunity, which could be beneficial to trigger anti-tumor immunity. Moreover, the ability of Bryo-1 to promote a Th2 response in vivo may help treat clinical disorders that are mediated by Th1 inflammatory cells.
This work was supported, in whole or in part, by National Institutes of Health Grants P01AT003961, R01DA016545, R01ES09098, R01AI053703, R01AI058300, and R01HL05864.
- DC
- dendritic cell
- BMDC
- bone marrow dendritic cell
- PB
- polymyxin B sulfate
- TLR
- Toll-like receptor
- RANTES
- regulated on activation normal T cell expressed and secreted
- IRF
- interferon regulatory factor
- TIR
- Toll-IL-1 resistance
- PE
- phycoerythrin
- IRF
- interferon regulatory factor
- ALL
- N-acetyl-Leu-Leu-Nle-CHO
- KC
- keratinocyte chemoattractant
- NBD
- NEMO binding domain.
REFERENCES
- 1. Montoya C. J., Jie H. B., Al-Harthi L., Mulder C., Patiño P. J., Rugeles M. T., Krieg A. M., Landay A. L., Wilson S. B. (2006) J. Immunol. 177, 1028–1039 [DOI] [PubMed] [Google Scholar]
- 2. Banchereau J., Briere F., Caux C., Davoust J., Lebecque S., Liu Y. J., Pulendran B., Palucka K. (2000) Annu. Rev. Immunol. 18, 767–811 [DOI] [PubMed] [Google Scholar]
- 3. Zenke M., Hieronymus T. (2006) Trends Immunol. 27, 140–145 [DOI] [PubMed] [Google Scholar]
- 4. Medzhitov R., Janeway C., Jr. (2000) Trends Microbiol. 8, 452–456 [DOI] [PubMed] [Google Scholar]
- 5. Kaisho T., Akira S. (2001) Trends Immunol. 22, 78–83 [DOI] [PubMed] [Google Scholar]
- 6. O'Neill L. A. (2002) Trends Immunol. 23, 296–300 [DOI] [PubMed] [Google Scholar]
- 7. Yan X., Xiu F., An H., Wang X., Wang J., Cao X. (2007) Life Sci. 80, 307–313 [DOI] [PubMed] [Google Scholar]
- 8. Wang Z. Y., Yang D., Chen Q., Leifer C. A., Segal D. M., Su S. B., Caspi R. R., Howard Z. O., Oppenheim J. J. (2006) Exp. Hematol. 34, 1115–1124 [DOI] [PubMed] [Google Scholar]
- 9. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 10. Medzhitov R., Preston-Hurlburt P., Janeway C. A., Jr. (1997) Nature 388, 394–397 [DOI] [PubMed] [Google Scholar]
- 11. Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. (2005) Immunity 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 12. Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 13. Karin M., Ben-Neriah Y. (2000) Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 14. Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. (2003) Nat. Immunol. 4, 161–167 [DOI] [PubMed] [Google Scholar]
- 15. Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 16. Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 17. Ramsdell J. S., Pettit G. R., Tashjian A. H., Jr. (1986) J. Biol. Chem. 261, 17073–17080 [PubMed] [Google Scholar]
- 18. Bosco M. C., Rottschafer S., Taylor L. S., Ortaldo J. R., Longo D. L., Espinoza-Delgado I. (1997) Blood 89, 3402–3411 [PubMed] [Google Scholar]
- 19. Scheid C., Prendiville J., Jayson G., Crowther D., Fox B., Pettit G. R., Stern P. L. (1994) Cancer Immunol. Immunother. 39, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hess A. D., Vogelsang G. B., Silanskis M., Friedman K. A., Beschorner W. E., Santos G. W. (1988) Transplant. Proc. 20, Suppl. 3, 487–492 [PubMed] [Google Scholar]
- 21. Do Y., Mainali E., Nagarkatti P. S., Nagarkatti M. (2004) Cell. Immunol. 231, 8–13 [DOI] [PubMed] [Google Scholar]
- 22. Do Y., Hegde V. L., Nagarkatti P. S., Nagarkatti M. (2004) Cancer Res. 64, 6756–6765 [DOI] [PubMed] [Google Scholar]
- 23. Singh N. P., Hegde V. L., Hofseth L. J., Nagarkatti M., Nagarkatti P. (2007) Mol. Pharmacol. 72, 1508–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fiorentino L., Stehlik C., Oliveira V., Ariza M. E., Godzik A., Reed J. C. (2002) J. Biol. Chem. 277, 35333–35340 [DOI] [PubMed] [Google Scholar]
- 25. Hoshino K., Kaisho T., Iwabe T., Takeuchi O., Akira S. (2002) Int. Immunol. 14, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 26. Honda K., Yanai H., Takaoka A., Taniguchi T. (2005) Int. Immunol. 17, 1367–1378 [DOI] [PubMed] [Google Scholar]
- 27. Tailor P., Tamura T., Ozato K. (2006) Cell Res. 16, 134–140 [DOI] [PubMed] [Google Scholar]
- 28. Maniatis T., Falvo J. V., Kim T. H., Kim T. K., Lin C. H., Parekh B. S., Wathelet M. G. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 609–620 [DOI] [PubMed] [Google Scholar]
- 29. Kelso A. (1995) Immunol. Today 16, 374–379 [DOI] [PubMed] [Google Scholar]
- 30. Mosmann T. R., Sad S. (1996) Immunol. Today 17, 138–146 [DOI] [PubMed] [Google Scholar]
- 31. Sun M. K., Alkon D. L. (2006) CNS Drug Rev. 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kortmansky J., Schwartz G. K. (2003) Cancer Invest. 21, 924–936 [DOI] [PubMed] [Google Scholar]
- 33. Akira S. (2003) J. Biol. Chem. 278, 38105–38108 [DOI] [PubMed] [Google Scholar]
- 34. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 35. Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S. O., Goode J., Lin P., Mann N., Mudd S., Crozat K., Sovath S., Han J., Beutler B. (2003) Nature 424, 743–748 [DOI] [PubMed] [Google Scholar]
- 36. Kirchhoff S., Wilhelm D., Angel P., Hauser H. (1999) Eur. J. Biochem. 261, 546–554 [DOI] [PubMed] [Google Scholar]
- 37. Stellato C., Beck L. A., Gorgone G. A., Proud D., Schall T. J., Ono S. J., Lichtenstein L. M., Schleimer R. P. (1995) J. Immunol. 155, 410–418 [PubMed] [Google Scholar]
- 38. Marfaing-Koka A., Devergne O., Gorgone G., Portier A., Schall T. J., Galanaud P., Emilie D. (1995) J. Immunol. 154, 1870–1878 [PubMed] [Google Scholar]
- 39. Shin H. S., Drysdale B. E., Shin M. L., Noble P. W., Fisher S. N., Paznekas W. A. (1994) Mol. Cell. Biol. 14, 2914–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nelson P. J., Kim H. T., Manning W. C., Goralski T. J., Krensky A. M. (1993) J. Immunol. 151, 2601–2612 [PubMed] [Google Scholar]
- 41. Saito T., Deskin R. W., Casola A., Häeberle H., Olszewska B., Ernst P. B., Alam R., Ogra P. L., Garofalo R. (1997) J. Infect. Dis. 175, 497–504 [DOI] [PubMed] [Google Scholar]
- 42. Lokuta M. A., Maher J., Noe K. H., Pitha P. M., Shin M. L., Shin H. S. (1996) J. Biol. Chem. 271, 13731–13738 [DOI] [PubMed] [Google Scholar]
- 43. Lin R., Heylbroeck C., Genin P., Pitha P. M., Hiscott J. (1999) Mol. Cell. Biol. 19, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee A. H., Hong J. H., Seo Y. S. (2000) Biochem. J. 350, 131–138 [PMC free article] [PubMed] [Google Scholar]
- 45. Pulendran B., Tang H., Denning T. L. (2008) Curr. Opin. Immunol. 20, 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dearman R. J., Cumberbatch M., Maxwell G., Basketter D. A., Kimber I. (2009) Immunology 126, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cocchi F., DeVico A. L., Garzino-Demo A., Arya S. K., Gallo R. C., Lusso P. (1995) Science 270, 1811–1815 [DOI] [PubMed] [Google Scholar]
- 48. Vangelista L., Secchi M., Lusso P. (2008) Vaccine 26, 3008–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clamp A., Jayson G. C. (2002) Anticancer Drugs 13, 673–683 [DOI] [PubMed] [Google Scholar]
- 50. Philip P. A., Rea D., Thavasu P., Carmichael J., Stuart N. S., Rockett H., Talbot D. C., Ganesan T., Pettit G. R., Balkwill F., et al. (1993) J. Natl. Cancer Inst. 85, 1812–1818 [DOI] [PubMed] [Google Scholar]