Abstract
Thyroglobulin (Tg) is secreted by thyroid epithelial cells. It is essential for thyroid hormonogenesis and iodine storage. Although studied for many years, only indirect and partial surveys of its post-translational modifications were reported. Here, we present a direct proteomic approach, used to study the degree of iodination of mouse Tg without any preliminary purification. A comprehensive coverage of Tg was obtained using a combination of different proteases, MS/MS fragmentation procedures with inclusion lists and a hybrid mass high-resolution LTQ-Orbitrap XL mass spectrometer. Although only 16 iodinated sites are currently known for human Tg, we uncovered 37 iodinated tyrosine residues, most of them being mono- or diiodinated. We report the specific isotopic pattern of thyroxine modification, not recognized as a normal peptide pattern. Four hormonogenic sites were detected. Two donor sites were identified through the detection of a pyruvic acid residue in place of the initial tyrosine. Evidence for polypeptide cleavages sites due to the action of cathepsins and dipeptidyl proteases in the thyroid were also detected. This work shows that semi-quantitation of Tg iodination states is feasible for human biopsies and should be of significant medical interest for further characterization of human thyroid pathologies.
Keywords: Mass Spectrometry (MS), Post-translational Modification, Proteomics, Thyroid, Tyrosine, Cathepsin, Hormogenic Sites, Iodination, Isotopic Pattern, Thyroglobulin
Introduction
Thyroglobulin (Tg)3 is one of the most abundant proteins produced by the thyroid gland and comprises two identical subunits of ∼330 kDa. This prohormonal glycoprotein is secreted by thyroid epithelial cells. It is stored in the lumen of thyroid follicles in a highly condensed and covalently cross-linked form with numerous disulfide bridges. There, Tg is used as a scaffold for thyroid hormonogenesis and as an iodine reservoir. Many post-translational modifications are known to occur on this protein such as glycosylation, sulfation, and iodination (1). A signal peptide is processed during its export to the lumen. The most specific Tg post-translational modification is certainly its iodination and 3,5,3′-triiodothyronine (T3) and thyroxine (T4) thyroid hormone synthesis. Indeed, through the combined action of NADPH oxidase and thyroperoxidase on the outer surface of the thyrocyte apical membrane, iodide ions that are also transported to the follicular lumen are covalently linked to some Tg tyrosyl residues to form mono- or diiodotyrosines (MIT or DIT, respectively). Coupling of two of these DIT residues then leads to the formation of T4 at an acceptor site, whereas coupling one MIT and one DIT moiety essentially forms T3 (2). In this reaction, the release of an iodotyrosyl moiety at the donor site leaves an “empty” side chain and the fate of this unusual residue is controversial. Some authors claim that a dehydroalanine is left instead of the tyrosine, whereas others observed pyruvic acid associated with cleavage of the polypeptide chain (3–7). Finally, the protein is proteolytically processed in the lumen, followed by endocytosis in lysosomes, to release the hormones. The principal proteases involved include the cathepsins (B, D, K, L, and S, with more or less specific roles ranging from solubilizing the highly cross-linked forms of Tg to releasing the hormone moiety), aminopeptidase N and dipeptidylpeptidases (8–12).
Studies of the post-translational modification of Tg began in the 1950s using radioactive iodide. Researchers attributed the radioactivity to different Tg fractions according to their sedimentation coefficient or molecular weight on SDS-PAGE gel. Later, Tg fragments obtained by proteolysis with trypsin or other proteases were separated by reversed-phase HPLC. The amount of radioactivity and the chemical form of the iodinated residues (MIT, DIT, T3, or T4) were then analyzed (13–15). Most of the studies were performed on purified Tg or Tg fragments from bovine, rat, guinea pig, rabbit, or human tissues, after in vitro chemical iodination or not. Significant progress was made in the 1980s when the first polypeptide sequence was reported for Bos taurus (16). The tyrosine residues involved were then identified more accurately using Edman amino acid sequencing and, later, mass spectrometry (MS) to identify the peptides. These experiments led to the establishment of a list of a number of iodinated tyrosine residues. Nonetheless, except for peptide identification, little has been done concerning MS-based characterization of tyrosine iodination. An initial attempt was made using purified (1237–1610) bovine Tg tryptic fragment (full sequence numbering). After endoproteinase Asp-N digestion, the resulting peptides were resolved by reversed-phase liquid chromatography and analyzed by electrospray and fast atom bombardment MS (6). The post-translational modifications of all seven tyrosine residues on this fragment were characterized at an unprecedented level of resolution, highlighting Tyr1310 as a new T3 and T4 acceptor site, and unveiling a dehydroalanine at position 1394 as a donor site residue. After proteolysis and reversed-phase separation as well, Dunn and co-workers (3) used MS to identify another donor site (Tyr149) on bovine Tg with the presence of pyruvic acid. Finally, in a more technical study in 2005, Salek and Lehmann (17) tackled the problem of iodotyrosine identification. They showed that peptides containing a mono- or diiodotyrosine residue generated specific markers during collisional-induced dissociation (CID) fragmentation.
To study post-translational modifications of Tg in the thyroid and their heterogeneity, we proposed to analyze them using the most direct strategy possible, avoiding all purification steps and their inherent bias, and without the need for radioactivity. We dissected thyroid glands from mice and directly analyzed Tg using a shotgun-based MS/MS approach. We revealed as many tyrosine positions as possible by using a combination of different proteases and different MS/MS fragmentation procedures, involving analysis with inclusion lists, and using a high-resolution LTQ-Orbitrap XL hybrid mass spectrometer. We identified 37 modified tyrosine residues in mouse tissue, whereas only 16 sites are known for human Tg. Of these, we clearly identified 4 hormonogenic sites and two donor sites by identifying a pyruvic residue instead of the initial tyrosine. The analysis of cathepsin and dipeptidyl cleavage sites provided information of what happens in the cells. Our panoramic results bring new challenging data relative to Tg post-translational modifications.
EXPERIMENTAL PROCEDURES
Obtaining Tg from Mouse Thyroid Lobes
The study protocol was approved by the Committee for Animal Studies and was in accordance with the guidelines of the National Institute of Health principles of animal laboratory care (NIH publication 86-23, revised 1995). Three mice (mixed C57BL6/129Sv background) were sacrificed. Per animal, a thyroid lobe was removed and quickly cut into 6–8 small pieces. NuPage lithium dodecyl sulfate sample buffer (Invitrogen) supplemented with 15% β-mercaptoethanol was immediately added resulting in direct protein solubilization. The samples were incubated for 5 min at 99 °C. After 30 s of centrifugation, the soluble material was used for SDS-PAGE.
One-dimensional SDS-PAGE and In-gel Proteolysis
Proteins were resolved on NuPAGE Novex 4–12% BisTris gels (Invitrogen) run with MES buffer (Invitrogen). The gels were stained with Coomassie Blue Safe stain (Invitrogen). Each lane was cut into 2.5-mm slices from top to the bottom. Each sample was deposited in triplicate for subsequent proteolysis with trypsin, chymotrypsin, or GluC. For proteolysis, the protein bands were treated as previously described (18). For trypsin digestion, dry gel pieces were rehydrated with 12 ng/ml of trypsin (Roche Applied Science) in 25 mm NH4HCO3 (pH 8.5) containing 1% CaCl2 and incubated overnight at 37 °C. Sequencing grade chymotrypsin (Roche Applied Science) was used as previously described (19). GluC (Roche Applied Science) was reconstituted in distilled water. After proteolysis, the peptides were extracted, first with 100% HCOOH, then with 56% CH3CN, 1% HCOOH, and finally with 100% CH3CN. The resulting pools were dried completely in a vacuum and stored at −20 °C until needed for MS analysis.
Nano-LC-MS/MS Analysis
LC-MS/MS experiments were performed on a LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher) coupled to an UltiMate 3000 LC system (Dionex-LC Packings). The peptide mixtures (0.1–2 pmol) were loaded and desalted online in an LC Packings Acclaim Pepmap 100 C18 reversed-phase precolumn (5 μm bead size, 100-Å pore size, 5 mm × 300 μm). They were resolved on an LC Packings nanoscale Acclaim Pepmap 100 C18 (3 μm bead size, 100-Å pore size, 15 cm × 75 μm) at a flow rate of 0.3 μl/min. The peptides were separated using a 90-min gradient (5 to 60% solvent B) with aqueous solvent A (0.1% HCOOH) and solvent B (0.1% HCOOH, 80% CH3CN). The column was then washed for 10 min with 100% B and re-equilibrated for 20 min with 5% B. Full-scan mass spectra were measured from m/z 300 to 1800. The LTQ-Orbitrap XL mass spectrometer was run in data-dependent mode using TOP5 strategy with a Fourier transform mass spectrometer resolution set at 30,000 as previously reported (18, 20). In brief, a scan cycle was started with a full scan of high mass accuracy in the Orbitrap, followed by MS/MS scans in the linear ion trap on the 5 most abundant precursor ions with dynamic exclusion of previously selected ions. This dynamic exclusion consisted in two acquisitions of MS/MS spectra of the most abundant ion over 30 s and then excluding this ion for the fragmentations over the next 60 s. The activation type normally used was CID with a standard normalized collision energy set at 30. Alternatively the Higher Energy Collision Dissociation (HCD) mode was used. In this case, a TOP3 strategy was used with Fourier transform mass spectrometer resolution set at 30,000. MS/MS scans in the linear ion trap were then performed at 7,500 resolution and a normalized energy set at 35. In some experiments a specific parent mass list set-up was used to detect all the expected iodine post-translational modifications including dehydroalanine or pyruvic acid at donor sites. This parent mass list comprised 1335 values.
Polypeptide Database Mining
Peak lists were generated using Matrix Science MASCOT DAEMON software (version 2.2.2) from the ThermoFisher Xcalibur FT (version 2.0.7, extract_msn.exe) data import filter. Data import filter options were set at 400 (minimum mass), 5000 (maximum mass), 0 (grouping tolerance), 0 (intermediate scans), and 1000 (threshold). Using the MASCOT (2.2.06) search engine (Matrix Science), we searched all MS/MS spectra against a homemade mouse database built using the VARSPLIC algorithm (21). The VARSPLIC tool generates all natural variants or sequence conflicts described in the SwissProt data base (version 57.7). This database comprises 925,162 polypeptide sequences, totaling 1,244,593,373 amino acids and was used for producing supplemental Tables S1–S6. A restricted database was used to analyze post-translational modifications as explained under “Results.” It contains 1294 peptide sequences, totaling 630,319 amino acids. The list of proteins in this subdatabase is shown in supplemental Table S7. In all cases, peptide searches were performed with the following parameters: a mass tolerance of 7 ppm on the parent ion and 0.5 Da for the MS/MS, static modifications of carbamidomethylated Cys (+57.0215), dynamic modification of oxidized Met (+15.9949), tyrosine post-translational modification: +125.8966 for monoiodotyrosine; +251.7933 for diiodotyrosine; +469.7162 for triiodothyronine; +595.6128 for thyroxine; and −94.0419 for dehydroalanine. The maximum number of missed cleavages was set at 3 for the semi-chymotrypsin search and 2 for all other searches. All peptide matches with a Peptide Score above the peptide identity threshold set at p < 0.01 for the selected data base, and rank 1, were filtered using IRMa 1.22.4 software (22). The proteins were validated (supplemental Tables S2, S4, and S6) when at least two peptides were identified above the identity threshold. The false-positive rate for protein identification was estimated using the appropriate decoy data base below 0.7% in the worse case with these parameters.
MS Data Deposition
MS data were deposited in the PRIDE PRoteomics IDEntifications data base (23) under accession numbers 12019 to 12032 (inclusive). They are freely available online.
RESULTS
Tg Is the Most Abundant Protein in Thyroid Extract
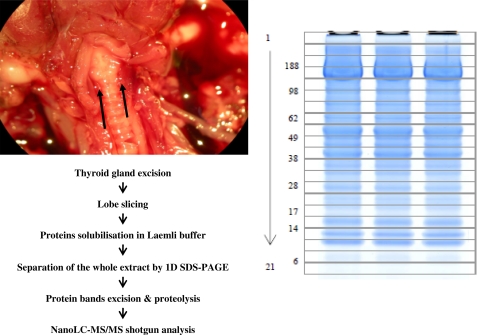
Fig. 1 shows the analytical flow chart we used to obtain the most comprehensive characterization of Tg iodinated sites based on MS. We removed a thyroid lobe from three mice. Each lobe was quickly cut into 6–8 pieces and dissolved directly in lithium dodecyl sulfate buffer. Then, total proteins were then resolved by one-dimensional SDS-PAGE and fractionated into 21 different slices on the basis of their molecular weight. In these samples, we first focused our attention on the four upper bands corresponding to proteins with molecular masses above 250 kDa, where the intact Tg polypeptide was expected. Following trypsin proteolysis of these bands from the three thyroid samples, the proteins were identified using nano-LC-MS/MS shotgun by means of an LTQ-Orbitrap XL mass spectrometer. The 12 runs produced a dataset comprising 96,569 recorded MS/MS spectra. Besides the mouse O08710 Tg sequence, several variants are currently reported in the SwissProt data base. One natural variant was described with an L2283P mutation (24) causing congenital goiter in mice. The Q2NKY1 (isoform CRA_a) with the T1327A and I1721T double modification is reported in the NCBI data base (25). A third variant was described with a similar sequence to Q2NKY1 but with 13 additional amino acids at its N-terminal (26). Moreover, Tg encompasses a 20-amino acid peptide signal. We built a specific mouse protein database taking all the currently described variants as well as the 15 sequence conflicts indicated in the SwissProt data base into account for Tg. Using this in-house data base, we used the MASCOT search engine to assign a set of 1382 unique peptides in these fractions (supplemental Table S1), corresponding to 107 different proteins (supplemental Table S2). The presence of Tg in these fractions was demonstrated with 172 confident unique peptides, covering 59% of the protein sequence. The peptide(1305–1328), which is proteotypic of the Q2NKY1 sequence was detected in abundance in all three animals, whereas no proteotypic peptides for the other variants were found (supplemental Table S1). On this basis, we assumed that the three mice with mixed C57BL6/129Sv genetic background have the Q2NKY1 Tg. Through spectral counting, we estimated that Tg was the most abundant protein. Of the 21,044 assigned MS/MS spectra, three-quarters (15,935) are Tg peptide signatures. This proportion was observed for the three animals analyzed (83, 65, and 79%). As Tg is quite an unusually huge protein, we further analyzed its abundance after normalizing the spectral count using the molecular weight of each polypeptide (supplemental Table S2). In this case, Tg is also far from the other identified polypeptides in terms of quantity (the second one is Myosin Q5SX39 identified with 148 unique peptides and 1265 MS/MS). Such abundance allowed us to further investigate Tg without the need for purification.
FIGURE 1.
Schematic illustration of analytical flow. Proteomic analysis consists in direct extraction of thyroid lobes and high-throughput nano-LC ESI-MS/MS. In the dissected mouse shown on the left, the thyroid lobes are indicated with black arrows. The SDS-PAGE gel stained with Coomassie Blue Safe stain is shown on the right. The molecular masses in the first well corresponds to 188, 98, 62, 38, 28, 17, 14, and 6 kDa. The numbering of the gel slices (1–21) is also indicated.
Following iodination in the thyroid lumen, the Tg was processed by cathepsins and exopeptidases. The various resulting fragments were resolved from intact Tg onto our SDS-PAGE. We analyzed the 17 other SDS-PAGE bands comprising proteins with molecular masses lower than 250 kDa for each of the three samples. Combining the results for all 21 slices, a total of 9935 unique peptides were confidently identified (supplemental Table S3). They correspond to the 1364 proteins listed in supplemental Table S4. In this dataset, we identified 174 different peptides corresponding to Tg fragments (supplemental Table S3). Once again, we did not detect any tryptic peptides proteotyping the other sequences apart from Q2NKY1 (supplemental Table S3).
It should be noted that all the bands corresponding to molecular masses below 250 kDa were found to contain Tg peptides. Regarding spectral counts, Tg fragments account for 29% of total proteins, whereas hemoglobin (HBB1), the second in order of importance, represents only about 8%. So Tg is really the most abundant protein in thyroid extract.
Comprehensive Coverage of Tg
To get a comprehensive coverage of intact mouse Tg, we used a multiplexed proteolytic digestion strategy. Samples from the three animals were re-applied to SDS-PAGE in duplicate to process the four upper bands containing intact and heavy fragments of Tg with chymotrypsin, and endo-GluC proteases. The 588 unique peptides in this MS/MS dataset are listed in supplemental Table S5. The proteins identified with these peptides are listed in supplemental Table S6. 172 different Tg peptides were previously found with trypsin, 250 were found with chymotrypsin, and 28 with endo-GluC. Chymotrypsin was found to be quite helpful in unveiling additional tyrosine residues because of its cleavage specificity (i.e. after aromatic and hydrophobic residues). On collecting all the results for the three proteolytic enzymes, we obtained 83% coverage of the Tg sequence. No tryptic peptides matching the Tg peptide signal described earlier (27) could be detected. This was expected, because the exported processed form accumulated in the thyroid follicle is present in significantly excessive quantities compared with the immature form. However, we confirmed that the mature sequence starts at N21, as we detected the 21–32 semi-chymotryptic, 21–39 semi-tryptic, and 21–40 semi-GluC peptides.
To search for Tg-specific post-translational modifications, as well as lumen or intracellular proteolytic events, we restricted the protein data base to the proteins detected in the extract (28). Supplemental Table S7 shows the compilation of proteins that were inserted into this subdatabase.
Identification of Iodination Sites by Pseudo-CID Fragmentation
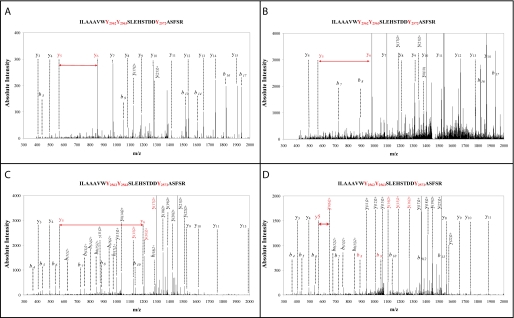
Using the raw data sets obtained for all bands using all types of enzymes, we searched the protein subdatabase for specific tyrosine modifications in the Tg polypeptide: monoiodination, diiodination, triiodothyronine, and thyroxine modification. The 1356 unique Tg peptides identified in this search are listed in supplemental Table S8. Identification was highly redundant as these peptides account for 52285 MS/MS spectra. Of the 1356 peptides, 881 were unmodified and 184 comprised at least one iodination. Tyrosine iodination was found to be a very stable post-translational modification. Pseudo-CID fragmentation of these iodinated peptides obtained in the LTQ linear trap followed the usual fragmentation pathway of protonated peptides leading to informative y or b ion series. Remarkably, we observed the four expected iodinations on Tyr2572 for the large peptide(2555–2577), whose sequence is ILAAAVWYYSLEHSTDDYAFSR. Fig. 2 shows four MS/MS spectra corresponding to these modifications at this specific position: monoiodotyrosine, diiodotyrosine, triiodotyronine, and thyroxine. These spectra were observed, as expected, at four different retention times (43, 59, 67, and 69 min, respectively) because of the increase in hydrophobicity due to the successive addition of iodine atoms. As shown in these spectra, the y and b series are complete enough for unambiguously assigning the modifications to Tyr2572 and not to one of the two other tyrosine residues at its N terminus. The y5 and y6 ions (red labels) in the first three spectra are clearly visible. Sometimes in the type D spectrum, the y6 ion is not present but the presence of the doubly charged y0(6)2+ is noted. Furthermore, the b8, b9, or doubly charged y(14)2+, y(15)2+, and y(16)2+ ions confirmed that Tyr2562 and Tyr2563 are not modified in these peptides.
FIGURE 2.
Unambiguous assignment of four different iodination patterns on Tyr2572. Representative ESI CID spectra obtained for Tg peptide(2555–2577) (ILAAAVWYYSLEHSTDDYAFSR) are presented. Monoiodotyrosine (panel A), diiodotyrosine (panel B), triiodothyronine (panel C), and thyroxine (panel D) modifications on Tyr2572 were observed. In these spectra, information from the y and b series was complete enough to assign the modifications to Tyr2572 unambiguously. Note the presence of y5 and y6 ions (red labels) in all spectra.
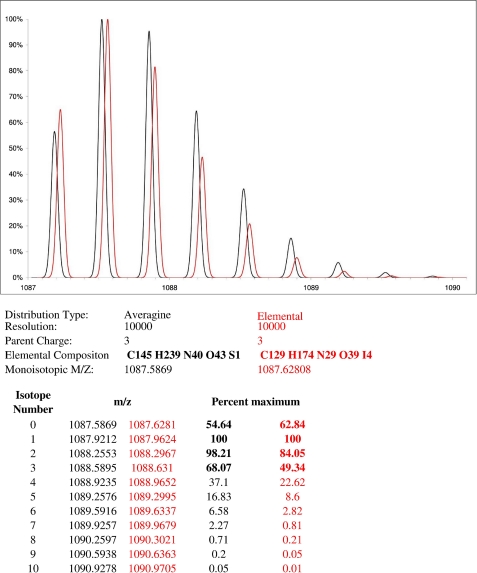
Regarding the thyroxine modification, we pointed out that the specificities of its isotopic pattern should be carefully taken into consideration when analyzing the spectra. With a high resolution mass spectrometer such as the LTQ-Orbitrap XL, monoisotopic mass, i.e. nomination of the correct peak of an isotopically resolved group of peptides peak as a monoisotopic peak (29), is determined using an algorithm based on the study reported by Senko et al. (30). These authors proposed the concept of a virtual amino acid, Averagine, constructed using the statistical occurrences of amino acids in the PIR protein data base (31). Such an Averagine contains the normal isotope variants of carbon (C), hydrogen (H), nitrogen (N), oxygen (O), and sulfur (S). This Averagine can then be used to obtain an isotopic pattern for any peptide, with precise masses and typical relative heights. The iodine atom (53, its atomic mass is 126.904) is inadequately present in the PIR data base and was simply neglected in this algorithm. In our study, the four iodine atoms in the thyroxine modification particularly influence the relative heights of the peaks in the isotopic pattern. Fig. 3 shows the experimental isotopic pattern recorded for peptide(2555–2577) containing a thyroxine modification at Tyr2572 exhibiting a theoretical monoisotopic molecular mass of 1087.6808, as well as the relative heights of the first isotopes of an “Averagine peptide” (black line) with a closely related molecular mass of 1087.5869. This comparison shows a clear difference between both profiles. We therefore concluded that the effect of four iodine atoms should not be ignored or this type of iodinated pattern would not be recognized as a normal peptide pattern. Being aware of this problem, we found a total of 3 different thyroxine modifications as described below.
FIGURE 3.
Specific ion pattern for peptide containing the thyroxine modification. This figure was constructed using Protein Prospector software with default resolution set to 10,000. This parameter has no effect on the ratios of the two isotopic patterns shown here. The relative height is shown for the first isotopes of an “Averagine peptide” (black line) with a molecular mass of 1087.5869 and the iodinated peptide (red line) (ILAAVWYYSLEHSTDDYAFSR) containing a thyroxine modification at Tyr2572 with a molecular mass of 1087.6808. The elemental composition and characteristics of these two chemical entities are given below, as well as the calculated values for isotopic distribution. The isotopic distribution of iodinated peptide is different from that of noniodinated peptide with a similar mass. The effect of 4 iodine atoms should not be ignored, otherwise this type of iodinated pattern would not be recognized as a peptide pattern.
Table 1 shows a summary of all the iodinated tyrosines detected using our methodology. The corresponding peptide characteristics (supplemental Table S8) and mapping of unmodified and modified peptides onto the mouse Tg sequence (supplemental Fig. S1) are given as supplementary data. A total of 36 and 24 tyrosines were found to be monoiodinated (MIT) and diiodinated (DIT), respectively. Thirteen tyrosines were only monoiodinated. Of the diiodinated residues, 23 were also found to be monoiodinated. Thus, one position, namely Tyr810, was only seen as a diiodinated tyrosine. T3 acceptor sites are sites where 3′,3,5-triiodothyronine is formed by coupling one DIT and one MIT. The 3′,3,5-triiodotyronine modification was detected on three tyrosine residues: 993, 1310, and 2572. T4 acceptor sites are sites where thyroxine is formed by coupling two DITs. This type of modification was detected on three tyrosines: 25, 1310, and 2572. It is worth noting that Tyr1310 and Tyr2572 positions were also found to be triiodinated. We found that all these T3 and T4 acceptor positions were also found as modified mono and diiodo forms. Interestingly, Tyr1310 was found to be modified (monoiodo, diiodotyrosine, 3′,3,5-triiodothyronine, and thyroxine) on the peptide that is proteotypic of Q2NKY1 (supplemental Fig. S2), confirming once more that this isoform is encoded in the genome of the mice used here. This could be another illustration of the proteogenomics concept (32). Taking these iodinations into account, tyrosine sites had 91% Tg coverage when trypsin, chymotrypsin, and endo-GluC proteolysis results were merged. Our data show a reproducible iodination pattern as we found 1484, 1047, and 1596 iodinated peptides for each of the 3 mice. As currently recommended in shotgun analysis (33), the standardized values obtained by dividing these numbers by the total number of peptides observed in each mouse are quite consistent: 7.8, 7.5, and 8.3% of the total peptides in each case. 80% of mono- and diiodotyrosine modifications were observed in all three mice.
TABLE 1.
Iodinated tyrosines in mouse and human Tg
| Residuesa | Mouseb | Humanb |
|---|---|---|
| Our results | Literature | |
| Y25 | M, D, Thyr. | M, Tri.2,Thyr.1,2 (24) |
| Y150 | M, D | M, D2 (149) |
| Y235 | M | Not detected |
| Y237 | M | Not conserved |
| Y259 | M, D | M2 (258) |
| Y377 | M | Not detected |
| Y383 | M, D | Not detected |
| Y430 | M, D | Not conserved |
| Y632 | M, D | Not conserved |
| Y692 | M, D | Not conserved |
| Y704 | M, D | M, D,Tri.2,Thyr.1,2 (704) |
| Y785 | M, D | M2 (785) |
| Y810 | D | Not detected |
| Y840 | M | Not detected |
| Y867 | M | M, D2 (866) |
| Y884 | M | D2 (883) |
| Y993 | M, D, Tri. | M, D2 (991) |
| Y1006 | M, D | Not detected |
| Y1115 | M | Not detected |
| Y1166 | M, D | Not detected |
| Y1310 | M, D, Tri., Thyr. | M, Tri.2,Thyr.1,2 (1309) |
| Y1384 | M, D | Not conserved |
| Y1424 | M, D | Not conserved |
| Y1446 | M, D | Not detected |
| Y1466 | Not conserved | M, D2 (1466) |
| Y1480 | M | Not detected |
| Y1681 | M | Not conserved |
| Y1913 | M | Not detected |
| Y2180 | M, D | Not conserved |
| Y2182 | M | M2 (2183) |
| Y2477 | M, D | Not detected |
| Y2486 | M, D | Not conserved |
| Y2539 | M, D | Iodinated?2 |
| Y2572 | M, D, Tri., Thyr. | M, D, Tri.2,Thyr.1,2 (2572) |
| Y2586 | M, D | M2,Thyr.1 (2586) |
| Y2616 | Not detected | M2 (2616) |
| Y2629 | M | Not conserved |
| Y2657 | M | Not detected |
| Y2669 | M, D | Not detected |
| Y2696 | Not detected | D2 (2696) |
| Y2764 | Not detectedc | M, D2,Tri.1,2,Thyr.2 (2765) |
a The residue positions correspond to the mouse Tg sequence. Only tyrosine residues iodinated in at least one species are shown.
b Information on iodinated positions for human Tg was collected from (1) the SwissProt database (2) (49). Not detected means tyrosine was present at this position but not iodinated. Not conserved means tyrosine was not present at this position, M stands for monoiodination, D for diiodination, Tri for triiodothyronine modification, and Thyr for thyroxine.
c This position was only detected with 3 unmodified peptides and no definitive conclusion should be drawn.
The Search for Immonium Ions Recorded Poor Results
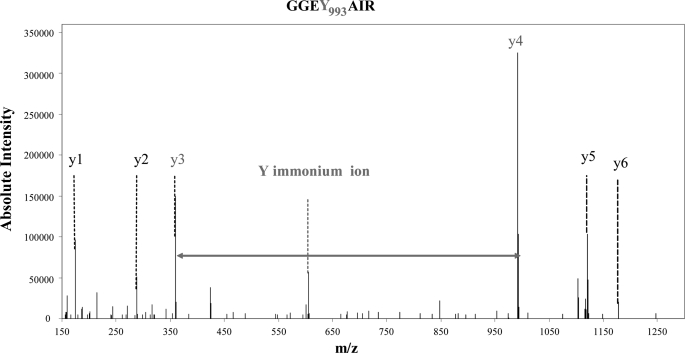
Precursor ion scanning is used to recognize modified peptides on the basis of residue-specific fragment ions. Salek and Lehman (17) noticed that peptides containing a mono- or diiodotyrosine residue generate abundant immonium ions under CID at m/z 261.97 and 387.87, respectively. These residue-specific marker ions were suggested as possibly helpful in discovering new iodination sites. These immonium ions are generated at rather “high” m/z and can sometimes be observed after CID fragmentation, depending on the size and charge of the fragmented peptide. Besides CID, we used HCD mode to obtain more information in the lower m/z part of the spectra. This HCD fragmentation takes place in a dedicated octopole collision cell in the LTQ-Orbitrap XL hybrid mass spectrometer. Fig. 4 shows the spectrum obtained after HCD fragmentation for peptide(990–996), the sequence of which is GGEYAIR, modified with a triiodothyronine. For such a small peptide we obtained an almost complete y series, particularly y3 and y4 ions with a mass difference of 632.780 Da (for a theoretical mass difference of 632.892). The triiodothyronine immonium ion, which is clearly present on the spectra (m/z at 605.792) is a residue-specific marker ion that confirms the assignment. These immonium ions have been observed in some of our MS/MS spectra for mono- or diiodotyrosines. However, in most HCD and CID spectra, these signature ions have not been found. Because mono- and diiodotyrosine, triiodothyronine, and thyroxine were identified in the set of HCD data with a complete series of y (and sometimes b) ions, we wonder if these immonium ions appear only under specific experimental conditions.
FIGURE 4.
HCD fragmentation spectrum showing unambiguous attribution of peptide(990–996) with a triiodothyronine modification on Tyr993. HCD fragmentation provides information from m/z = 100 Thomson. For such a small peptide, peptide(990–996), we obtained an almost complete y series. The y3 and y4 ions with a mass difference of 632.780 Da (for a theoretical mass difference of 632.892) indicated the presence of the modification. The triiodothyronine immonium ion, which is clearly present on the spectra (605.792), is a residue-specific marker ion that confirms the complete y series.
Fate of the Lost Side Chain during Thyroid Hormonogenesis
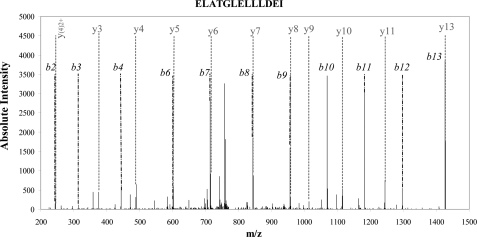
During thyroid hormone synthesis, the iodophenyl moiety of the iodotyrosine residue (donor site) is cleaved and transferred to the other tyrosine residue (acceptor site). As shown in Table 1, many potential donor sites exist, as we found numerous monoiodinated and diiodinated tyrosines. Whether dehydroalanine or pyruvic acid (with a polypeptide break) replace Tyr residues at these sites was investigated using our large experimental dataset. We did not detect any peptides with a dehydroalanine residue replacing a tyrosine, even including the specific MS/MS records we obtained by including the parent mass list for such events (data not shown). However, we did find evidence of the existence of at least two donor sites (Tyr259 and Tyr2539) in which the tyrosine is replaced by pyruvic acid and the polypeptide chain is broken. For example, Tyr259 is usually encompassed by a large Tg peptide(245–279) with the sequence ELAETGLELLLDEIY259DTIFAGLDQASTFTQSTMYR (detected unmodified and with a monoiodo or diiodo modification at Tyr259). Fig. 5 shows the spectrum assigned to a shorter peptide(259–279) corresponding to a subset of this sequence, Y*DTIFAGLDQASTFTQSTMYR, where the first tyrosine residue had been replaced with pyruvic acid (detected 55 times with a maximum confidence score of 75.64). Moreover, we also specifically detected the complementary peptide(245–258) with the sequence ELAETGLELLLDEI in our dataset (19 times with a maximum confidence score of 100.76). For Tyr2539, the Y*QALQNSLGGEDSDAR peptide with the first tyrosine residue replaced by pyruvic acid was detected 25 times with a maximum confidence score of 108.76. In that case the complementary TAF peptide(2536–2538) was too short to be detected. It is worth mentioning that the two tyrosine residues found as donor sites (Tyr259 and Tyr2539) were both detected as mono- or diiodotyrosine residues, a logical prerequisite to be a donor tyrosyl residue.
FIGURE 5.
CID fragmentation spectra of peptide(245–258) (ELATGELLLDEI). The almost complete b and y series obtained from a non-noisy spectrum (35 fragment ions using 42 most intense peaks) gave a Mascot ion score of 101. This spectrum assignment indicates the break on the peptide bond after an isoleucine, which cannot be explained enzymatically but by the transformation of a monoiodo (or diiodo) tyrosine into a pyruvic acid residue.
Evidence for Cathepsins and Other Proteases in Processed Tg
Many proteolytic events take place in the lumen and intracellular compartments during the processing of iodinated Tg. We analyzed our MS/MS data to list all proteolytic events detected. Supplemental Table S8 reports all semi-trypsin, semi-chymotrypsin, and semi-GluC peptides belonging to Tg. Of these, those arising from nonspecific MASCOT protease patterns (758) were extracted. In terms of spectral count, these represented ∼20% of all the Tg peptides detected during the whole study. The abundant semi-tryptic peptide(426–432) (AVEHYQR) was found to be diiodinated. This peptide is a signature of a cellular proteolytic event at Ile425. Note that some proteolytic events were found after two different proteolytic digestive processes: Lys520, Cys1748, Cys1894, Ser2075, and Ile2688. Other major proteolytic events were detected at Tyr237, Ile425, Gln670, Cys730, Arg1160, Ser1259, Gln1601, Ile1747, Gln2079, His2163, Tyr2639, and Lys2762. Some Tg peptide motives seem very sensitive to proteolytic attack: Gln1096-Arg1097-Asn1098-Pro1099, Ala2388-Asp2389-Val2390-Ala2391, and Gln2430-Gln2431 are worth mentioning. The resulting peptides are evidence of sites of Tg processing by cathepsins and aminopeptidase/dipeptidylpeptidase trimming.
DISCUSSION
Tg is a polypeptide matrix necessary for the synthesis of thyroid hormones. It is a very large glycoprotein consisting of two identical chains of 330 kDa. The numerous studies carried out so far have only led to a partial and indirect survey of its degree of iodination. Table 1 shows the positions of the iodinated tyrosines in both mouse and human Tg. A more general view of the status of Tg modification has been attempted through review work and many results have been deduced similarity with other established sequences, but were not always demonstrated (6, 34–36). Palumbo et al. (37) noted that of the 140 tyrosines in the Tg dimer in rats, only 25–30 are normally iodinated and a much smaller number undergo coupling to form thyroid hormones (T3 and T4). Dunn et al. (38, 39) mentioned the presence of four essential hormonogenic sites in rabbit Tg. Those sites, designated A, B, C, and D, were found either experimentally or by homology when the cDNA sequences were established (16, 40, 41). These sites correspond to tyrosyl numbers 5, 2553, 2746, and 1290, respectively (this numbering applies to the mature Tg rabbit molecule, omits the peptide signal, and corresponds to positions 25, 2572, 2764, and 1310 in the mouse sequence with its presequence). Sites A and B are T4 and T3 sites; D is exclusively T4, and C mainly T3. As summarized in Table 1, our proteomic approach provides the broadest panoramic survey ever achieved using native Tg. The sequence coverage for Tg is very high (89% when iodination modifications are included). In the present study, we detected mouse equivalent hormonogenic sites A, B, and D. In our case, sites B and D carry T4 and T3, whereas site A carries only T4. By analogy with other Tg mammals, site C (Tyr2764, mouse numbering) is located at the C terminus. In our proteomic strategy, the SYSK peptide containing Tyr2764 generated by trypsin was too small to be detected by the mass spectrometer unless a limited proteolysis strategy was used. As far as triiodothyronine is concerned, we demonstrated its presence at positions Tyr2572 (site B), Tyr1310 (site D), and also at Tyr993 (mouse numbering) as mentioned in Table 1. This latter position has not previously been described as a hormonogenic site. It should be noted that we did not detect triiodothyronine at position 25 (site A).
We have already mentioned that the sequence coverage obtained for Tg was very high. We wondered why some regions were not covered and noted that most of the undetected peptides carry a potential glycosylation site as predicted by bioinformatic tools based on homology with other species. The number of tyrosyl residues detected (71 of 76), particularly the number of mono- or diiodinated tyrosines, 36 and 24, respectively, is higher than had ever been found before (37). If we consider that only a few positions (4 in our study) are involved in hormonogenesis we might think that the coupling process is highly regioselective and probably driven by constraints because of the native three-dimensional structure of Tg. The abundant iodination found in mouse Tg is probably a storage process for iodine, which can be mobilized when iodine intake becomes limited. More iodination sites were found in mouse Tg compared with those previously identified in the thyroids of larger mammals. This difference may be related to the more active thyroid metabolism in smaller animals or to the greater sensitivity of our methodology.
Dunn et al. (39) proposed a classification of iodinated tyrosyl groups into three consensus families: ((D/E)Y), ((S/T)YS)), and (EXY) motives. (i) The ((D/E)Y) di-amino acid consensus was associated with T4 at sites A, B, and D. This is also the case in mouse Tg as supported by our results. It should be noted that the novel T3 site characterized here at residue 993 belongs to this EY consensus. Moreover, this consensus sequence is found iodinated but with apparently no hormone formation at four other locations Tyr383 (DY), Tyr1115 (EY), Tyr2486 (DY), and Tyr2586 (DY). (ii) The ((S/T)YS) tri-amino acid consensus was associated with triiodothyronine synthesis at site C. In the present study, we noticed that of the 36 mono- or 24 diiodotyrosines none could be linked to this consensus group because the sequence ((S/T)YS) is only present at position 2764 (mouse numbering). (iii) The consensus sequence (EXY) appeared to favor iodination. Of 8 possibilities of this consensus in mouse Tg, 6 are iodinated sites as shown here. We can increase this figure to 9 of 12 if we include an extended ((E/D)XY) consensus sequence. This ratio is significantly higher than the mean ratio of 36 iodinated positions of 71 detectable tyrosine residues. Our results confirmed that the presence of an acidic residue located two amino acids upstream of a tyrosine promotes iodination.
Identifying donor tyrosyl residues has been a difficult challenge for years. At donor sites, the coupling process should leave dehydroalanine, which may be converted to alanine, pyruvic acid, or acetic acid during subsequent isolation steps (5, 42). Marriq et al. (36, 43) first reported a cleavage in the peptide bond with the appearance of pyruvate instead of tyrosine. In 1997, Gentile et al. (6) noted that the identification of donor tyrosyl residues was indirect in all the cases reported so far. In their study, they claimed to provide the first direct identification of tyrosine 1375 in mature bovine Tg (not conserved in humans or mice) as a donor residue. This result was obtained by taking into account the fact that tyrosine was converted into dehydroalanine. They used electrospray MS with 250–500 ppm precision. In 1998, Dunn et al. (3) using also electrospray MS demonstrated a cleavage in the polypeptidic chain in bovine Tg, and identified a peptide in which Tyr130 (Tyr150 in mouse numbering) was replaced by pyruvate. In our large dataset, we were unable to detect any peptide displaying a dehydroalanine modification, whereas working to 5 ppm precision, even when the parent mass list was included. We identified two donor tyrosine residues at N-terminal and C-terminal fragments of the Tg molecule with peptide bond cleavage, and demonstrated the presence of pyruvic acid replacing the tyrosine residue. Our thyroid-wide proteomic analysis suggests that the residual side chain is converted to pyruvic acid in vivo or in vitro after iodotyrosyl transfer. We do not have any evidence that this cleavage occurs in the thyroid in vivo but we wonder whether the acidic conditions used during subsequent experimental manipulations were severe enough to break the peptide bond. In any case, detection of this cleavage event provides clear evidence for donor site identification.
Although MS is widely used in the analysis of post-translational protein modifications (44–46), it has rarely been used to identify iodinated tyrosyl residues in Tg and only on fragmented Tg (3, 6, 17). Here, we report the first detailed analysis of the entire Tg molecule from a direct, fresh extract of the mouse thyroid gland. Using this strategy, we obtained unambiguous characterization of 71 tyrosyl residues of this huge protein, confirmed the presence of three thyroxine sites and three triiodothyronine sites, and detected one new T3 site (Tyr993). From the same dataset, we were able to characterize two hormonogenic donor tyrosines at each end of the molecule. Based on spectral count, the ratio of the number of detected spectra for modified to unmodified peptides can be calculated for each modified tyrosine site. Because each peptide has its own ionization characteristics, this ratio is not very informative per se but it could become very enlightening when comparing Tg iodination levels under different conditions such as pathophysiological states. The use of isotope synthetic peptides carrying the different iodinated forms using a stable isotope dilution strategy (47) could also be used to record the absolute quantity of each major modification.
The release of thyroid hormones from the Tg prohormone requires the presence of different cathepsins, B, D, and L (9). Friedrichs et al. (11) noted that this rather complex process involves sequential proteolytic events with some interaction with different cathepsins, B, K, and L (48). Covalently cross-linked Tg is stored in the thyroid follicular lumen where limited proteolysis occurs. Tg processing continues under intracellular conditions after Tg fragment internalization (12). Our organ-oriented data were obtained from a complex mixture of partially or fully degraded Tg. Novel proteomic strategies, such as those developed to label N-terminal extremities for degradosome or proteogenomic studies (19), could be developed to follow the in vivo degradation process in thyroid follicules more accurately.
In conclusion, in this study, we established the iodination status of Tg in mice. Our direct organ to mass spectrometer shotgun approach revealed a more comprehensive set of iodination sites in Tg. Our methodology may be applied to analyze whether potential alteration in the process of Tg iodination could be detected. This study could be carried out on thyroid extracts from various mouse models, such as Tg processing or iodine uptake using genetically modified mutants. Drugs that modify the Tg iodination status could also be investigated with this type of monitoring. The approach does not require large samples. It should be straightforward in application to human thyroid punctures, which could be important for clinical studies on iodination dysfunction.
Supplementary Material
Acknowledgments
We thank C. Bruley (CEA-Grenoble, iRTSV) for kindly providing the IRMa 1.21.0 parser, G. Imbert and O. Pible (CEA-Marcoule, iBEB) for help with data processing, and E. Quéméneur (CEA-Marcoule, iBEB) for continuous support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S2 and Tables S1–S8.
- Tg
- thyroglobulin
- MIT
- monoiodotyrosine
- DIT
- diiodotyrosine
- T3
- triiodothyronine
- T4
- thyroxine
- endo-GluC
- endoproteinase GluC
- HCD
- higher energy collision dissociation
- CID
- collision-induced dissociation
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Deshpande V., Venkatesh S. G. (1999) Biochim. Biophys. Acta 1430, 157–178 [DOI] [PubMed] [Google Scholar]
- 2. Taurog A., Dorris M. L., Doerge D. R. (1996) Arch. Biochem. Biophys. 330, 24–32 [DOI] [PubMed] [Google Scholar]
- 3. Dunn A. D., Corsi C. M., Myers H. E., Dunn J. T. (1998) J. Biol. Chem. 273, 25223–25229 [DOI] [PubMed] [Google Scholar]
- 4. Gavaret J. M., Cahnmann H. J., Nunez J. (1981) J. Biol. Chem. 256, 9167–9173 [PubMed] [Google Scholar]
- 5. Gavaret J. M., Nunez J., Cahnmann H. J. (1980) J. Biol. Chem. 255, 5281–5285 [PubMed] [Google Scholar]
- 6. Gentile F., Ferranti P., Mamone G., Malorni A., Salvatore G. (1997) J. Biol. Chem. 272, 639–646 [DOI] [PubMed] [Google Scholar]
- 7. Ohmiya Y., Hayashi H., Kondo T., Kondo Y. (1990) J. Biol. Chem. 265, 9066–9071 [PubMed] [Google Scholar]
- 8. Brix K., Linke M., Tepel C., Herzog V. (2001) Biol. Chem. 382, 717–725 [DOI] [PubMed] [Google Scholar]
- 9. Dunn A. D., Crutchfield H. E., Dunn J. T. (1991) J. Biol. Chem. 266, 20198–20204 [PubMed] [Google Scholar]
- 10. Dunn A. D., Crutchfield H. E., Dunn J. T. (1991) Endocrinology 128, 3073–3080 [DOI] [PubMed] [Google Scholar]
- 11. Friedrichs B., Tepel C., Reinheckel T., Deussing J., von Figura K., Herzog V., Peters C., Saftig P., Brix K. (2003) J. Clin. Invest. 111, 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jordans S., Jenko-Kokalj S., Kühl N. M., Tedelind S., Sendt W., Brömme D., Turk D., Brix K. (2009) BMC Biochem. 10, 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gavaret J. M., Dème D., Nunez J., Salvatore G. (1977) J. Biol. Chem. 252, 3281–3285 [PubMed] [Google Scholar]
- 14. Lejeune P. J., Marriq C., Rolland M., Lissitzky S. (1983) FEBS Lett. 156, 77–82 [DOI] [PubMed] [Google Scholar]
- 15. Turner C. D., Chernoff S. B., Taurog A., Rawitch A. B. (1983) Arch. Biochem. Biophys. 222, 245–258 [DOI] [PubMed] [Google Scholar]
- 16. Mercken L., Simons M. J., Swillens S., Massaer M., Vassart G. (1985) Nature 316, 647–651 [DOI] [PubMed] [Google Scholar]
- 17. Salek M., Lehmann W. D. (2005) Proteomics 5, 351–353 [DOI] [PubMed] [Google Scholar]
- 18. de Groot A., Dulermo R., Ortet P., Blanchard L., Guérin P., Fernandez B., Vacherie B., Dossat C., Jolivet E., Siguier P., Chandler M., Barakat M., Dedieu A., Barbe V., Heulin T., Sommer S., Achouak W., Armengaud J. (2009) PLoS Genet. 5, e1000434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baudet M., Ortet P., Gaillard J. C., Fernandez B., Guerin P., Enjalbal C., Subra G., de Groot A., Barakat M., Dedieu A., Armengaud J. (2010) Mol. Cell Proteomics 9, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zivanovic Y., Armengaud J., Lagorce A., Leplat C., Guérin P., Dutertre M., Anthouard V., Forterre P., Wincker P., Confalonieri F. (2009) Genome Biol. 10, R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kersey P., Hermjakob H., Apweiler R. (2000) Bioinformatics 16, 1048–1049 [DOI] [PubMed] [Google Scholar]
- 22. Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., Bruley C. (2009) Bioinformatics 25, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 23. Vizcaíno J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) Nucleic Acids Res. 38, D736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim P. S., Hossain S. A., Park Y. N., Lee I., Yoo S. E., Arvan P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9909–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strausberg R. L., Feingold E. A., Grouse L. H., Derge J. G., Klausner R. D., Collins F. S., Wagner L., Shenmen C. M., Schuler G. D., Altschul S. F., Zeeberg B., Buetow K. H., Schaefer C. F., Bhat N. K., Hopkins R. F., Jordan H., Moore T., Max S. I., Wang J., Hsieh F., Diatchenko L., Marusina K., Farmer A. A., Rubin G. M., Hong L., Stapleton M., Soares M. B., Bonaldo M. F., Casavant T. L., Scheetz T. E., Brownstein M. J., Usdin T. B., Toshiyuki S., Carninci P., Prange C., Raha S. S., Loquellano N. A., Peters G. J., Abramson R. D., Mullahy S. J., Bosak S. A., McEwan P. J., McKernan K. J., Malek J. A., Gunaratne P. H., Richards S., Worley K. C., Hale S., Garcia A. M., Gay L. J., Hulyk S. W., Villalon D. K., Muzny D. M., Sodergren E. J., Lu X., Gibbs R. A., Fahey J., Helton E., Ketteman M., Madan A., Rodrigues S., Sanchez A., Whiting M., Madan A., Young A. C., Shevchenko Y., Bouffard G. G., Blakesley R. W., Touchman J. W., Green E. D., Dickson M. C., Rodriguez A. C., Grimwood J., Schmutz J., Myers R. M., Butterfield Y. S., Krzywinski M. I., Skalska U., Smailus D. E., Schnerch A., Schein J. E., Jones S. J., Marra M. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mural R. J., Adams M. D., Myers E. W., Smith H. O., Miklos G. L., Wides R., Halpern A., Li P. W., Sutton G. G., Nadeau J., Salzberg S. L., Holt R. A., Kodira C. D., Lu F., Chen L., Deng Z., Evangelista C. C., Gan W., Heiman T. J., Li J., Li Z., Merkulov G. V., Milshina N. V., Naik A. K., Qi R., Shue B. C., Wang A., Wang J., Wang X., Yan X., Ye J., Yooseph S., Zhao Q., Zheng L., Zhu S. C., Biddick K., Bolanos R., Delcher A. L., Dew I. M., Fasulo D., Flanigan M. J., Huson D. H., Kravitz S. A., Miller J. R., Mobarry C. M., Reinert K., Remington K. A., Zhang Q., Zheng X. H., Nusskern D. R., Lai Z., Lei Y., Zhong W., Yao A., Guan P., Ji R. R., Gu Z., Wang Z. Y., Zhong F., Xiao C., Chiang C. C., Yandell M., Wortman J. R., Amanatides P. G., Hladun S. L., Pratts E. C., Johnson J. E., Dodson K. L., Woodford K. J., Evans C. A., Gropman B., Rusch D. B., Venter E., Wang M., Smith T. J., Houck J. T., Tompkins D. E., Haynes C., Jacob D., Chin S. H., Allen D. R., Dahlke C. E., Sanders R., Li K., Liu X., Levitsky A. A., Majoros W. H., Chen Q., Xia A. C., Lopez J. R., Donnelly M. T., Newman M. H., Glodek A., Kraft C. L., Nodell M., Ali F., An H. J., Baldwin-Pitts D., Beeson K. Y., Cai S., Carnes M., Carver A., Caulk P. M., Center A., Chen Y. H., Cheng M. L., Coyne M. D., Crowder M., Danaher S., Davenport L. B., Desilets R., Dietz S. M., Doup L., Dullaghan P., Ferriera S., Fosler C. R., Gire H. C., Gluecksmann A., Gocayne J. D., Gray J., Hart B., Haynes J., Hoover J., Howland T., Ibegwam C., Jalali M., Johns D., Kline L., Ma D. S., MacCawley S., Magoon A., Mann F., May D., McIntosh T. C., Mehta S., Moy L., Moy M. C., Murphy B. J., Murphy S. D., Nelson K. A., Nuri Z., Parker K. A., Prudhomme A. C., Puri V. N., Qureshi H., Raley J. C., Reardon M. S., Regier M. A., Rogers Y. H., Romblad D. L., Schutz J., Scott J. L., Scott R., Sitter C. D., Smallwood M., Sprague A. C., Stewart E., Strong R. V., Suh E., Sylvester K., Thomas R., Tint N. N., Tsonis C., Wang G., Wang G., Williams M. S., Williams S. M., Windsor S. M., Wolfe K., Wu M. M., Zaveri J., Chaturvedi K., Gabrielian A. E., Ke Z., Sun J., Subramanian G., Venter J. C., Pfannkoch C. M., Barnstead M., Stephenson L. D. (2002) Science 296, 1661–1671 [DOI] [PubMed] [Google Scholar]
- 27. Malthièry Y., Marriq C., Bergé-Lefranc J. L., Franc J. L., Henry M., Lejeune P. J., Ruf J., Lissitzky S. (1989) Biochimie 71, 195–209 [DOI] [PubMed] [Google Scholar]
- 28. Tharakan R., Edwards N., Graham D. R. (2010) Proteomics 10, 1160–1171 [DOI] [PubMed] [Google Scholar]
- 29. Valkenborg D., Jansen I., Burzykowski T. (2008) J. Am. Soc. Mass. Spectrom. 19, 703–712 [DOI] [PubMed] [Google Scholar]
- 30. Senko M. W., Beu S. C., McLafferty F. W. (1994) Anal. Chem. 66, 415–418 [DOI] [PubMed] [Google Scholar]
- 31. Barker W. C., Garavelli J. S., McGarvey P. B., Marzec C. R., Orcutt B. C., Srinivasarao G. Y., Yeh L. S., Ledley R. S., Mewes H. W., Pfeiffer F., Tsugita A., Wu C. (1999) Nucleic Acids Res. 27, 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Armengaud J. (2010) Expert Rev. Proteomics 7, 65–77 [DOI] [PubMed] [Google Scholar]
- 33. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 34. Dunn J. T., Kim P. S., Dunn A. D., Heppner D. G., Jr., Moore R. C. (1983) J. Biol. Chem. 258, 9093–9099 [PubMed] [Google Scholar]
- 35. Kim P. S., Dunn J. T., Kaiser D. L. (1984) Endocrinology 114, 369–374 [DOI] [PubMed] [Google Scholar]
- 36. Marriq C., Lejeune P. J., Venot N., Vinet L. (1991) Mol. Cell. Endocrinol. 81, 155–164 [DOI] [PubMed] [Google Scholar]
- 37. Palumbo G., Gentile F., Condorelli G. L., Salvatore G. (1990) J. Biol. Chem. 265, 8887–8892 [PubMed] [Google Scholar]
- 38. Dunn J. T., Anderson P. C., Fox J. W., Fassler C. A., Dunn A. D., Hite L. A., Moore R. C. (1987) J. Biol. Chem. 262, 16948–16952 [PubMed] [Google Scholar]
- 39. Dunn J. T., Dunn A. D. (1999) Biochimie 81, 505–509 [DOI] [PubMed] [Google Scholar]
- 40. Malthiéry Y., Lissitzky S. (1987) Eur. J. Biochem. 165, 491–498 [DOI] [PubMed] [Google Scholar]
- 41. Musti A. M., Avvedimento E. V., Polistina C., Ursini V. M., Obici S., Nitsch L., Cocozza S., Di Lauro R. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dunn J. T. (1995) Eur. J. Endocrinol. 132, 603–604 [DOI] [PubMed] [Google Scholar]
- 43. Marriq C., Lejeune P. J., Venot N., Vinet L. (1989) FEBS Lett. 242, 414–418 [DOI] [PubMed] [Google Scholar]
- 44. Jensen O. N. (2004) Curr. Opin. Chem. Biol. 8, 33–41 [DOI] [PubMed] [Google Scholar]
- 45. Küster B., Mann M. (1998) Curr. Opin. Struct. Biol. 8, 393–400 [DOI] [PubMed] [Google Scholar]
- 46. Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 47. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tepel C., Brömme D., Herzog V., Brix K. (2000) J. Cell Sci. 113, 4487–4498 [DOI] [PubMed] [Google Scholar]
- 49. Lamas L., Anderson P. C., Fox J. W., Dunn J. T. (1989) J. Biol. Chem. 264, 13541–13545 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.