Abstract
Gaucher disease is caused by mutations in the enzyme acid β-glucosidase (GCase), the most common of which is the substitution of serine for asparagine at residue 370 (N370S). To characterize the nature of this mutation, we expressed human N370S GCase in insect cells and compared the x-ray structure and biochemical properties of the purified protein with that of the recombinant human GCase (imiglucerase, Cerezyme®). The x-ray structure of N370S mutant acid β-glucosidase at acidic and neutral pH values indicates that the overall folding of the N370S mutant is identical to that of recombinant GCase. Subtle differences were observed in the conformation of a flexible loop at the active site and in the hydrogen bonding ability of aromatic residues on this loop with residue 370 and the catalytic residues Glu-235 and Glu-340. Circular dichroism spectroscopy showed a pH-dependent change in the environment of tryptophan residues in imiglucerase that is absent in N370S GCase. The mutant protein was catalytically deficient with reduced Vmax and increased Km values for the substrate p-nitrophenyl-β-d-glucopyranoside and reduced sensitivity to competitive inhibitors. N370S GCase was more stable to thermal denaturation and had an increased lysosomal half-life compared with imiglucerase following uptake into macrophages. The competitive inhibitor N-(n-nonyl)deoxynojirimycin increased lysosomal levels of both N370S and imiglucerase 2–3-fold by reducing lysosomal degradation. Overall, these data indicate that the N370S mutation results in a normally folded but less flexible protein with reduced catalytic activity compared with imiglucerase.
Keywords: Circular Dichroism (CD), Enzyme Mutation, Lysosomal Storage Disease, Protein Folding, Protein Stability, X-ray Crystallography, Gaucher Disease, N370S Mutation, Acid beta-Glucosidase, Glucocerebrosidase
Introduction
Gaucher disease is a rare autosomal recessive genetic disorder caused by mutations in the GBA gene leading to a deficiency of the lysosomal enzyme acid β-glucosidase, EC 3.2.1.45 (GCase).2 Deficiency of the enzyme leads to accumulation of the glycolipid glucosylceramide in the macrophages of the reticuloendothelial system (1, 2). Although over 200 mutations have been identified in the GBA gene leading to Gaucher disease, a few common mutations predominate, the most prevalent being a missense mutation resulting in the substitution of a serine for asparagine at amino acid residue 370 (N370S) (3). Patients homozygous for the N370S mutation have an attenuated form of Gaucher disease with residual enzyme activities 10–15% of normal in in vitro assays (4, 5). How in vitro activity relates to in vivo activity is not clear as GCase can be activated by the protein saposin C (6, 7) as well as a variety of phospholipids (8, 9) and detergents (10). The extent of activation by these compounds differs significantly depending on the assay conditions chosen (2). The variety of assay conditions used as well as the use of either partially purified enzyme preparations or cell extracts has led to conflicting reports to the effect of the N370S mutation on the biochemical and structural properties of GCase. Studies have demonstrated both normal (11) and altered activation by phospholipids and/or saposin (12, 13), normal (11, 12, 14) and reduced (15) stability, as well as normal (13, 16) and altered post-translational processing (17, 18). Studies on recombinantly expressed forms of N370S GCase, however, indicate that this mutation results in a stable but catalytically deficient enzyme. Ohashi et al. (12) found that cells transfected with the N370S mutant cDNA had comparable RNA and protein levels to cells transfected with wild type cDNA, and glycosylation and processing were also comparable with the wild type enzyme. In that study, the specific activity was 5% of normal, and the enzyme had a 15-fold higher Km value for the substrate 4-methylumbelliferyl-β-d-glucoside and a 7.8-fold higher IC50 for the inhibitor conduritol B epoxide. Stability toward heat denaturation was also normal compared with both wild type GCase expressed in NIH 3T3 cells and purified placental enzyme (12). In a similar study, Grace et al. (4, 5) expressed the N370S GCase mutant in Sf9 insect cells and found the N370S mutant protein to have a specific activity 10–20% of normal, a higher IC50 value for inhibitors, normal processing, and thermal stability. These studies concluded that the N370S mutant is stable but catalytically defective. Additional studies on recombinant N370S GCase have demonstrated reduced enzymatic activity, normal stability, and normal binding to LIMP-2, the protein responsible for transporting GCase to the lysosome (5, 11, 12, 19). These results support earlier investigations on patient fibroblasts and purified enzyme from type 1 Gaucher patients, which demonstrated normal processing and stability but decreased activity. However, it should be noted that these studies were conducted prior to the sequencing of the GBA gene, and it is not known whether the patients were homozygous for the N370S mutation (6).
Recently, it has been proposed that the N370S mutation gives rise to an unstable protein that, although catalytically competent, is misfolded at neutral pH, accumulates, and/or is degraded in the endoplasmic reticulum before it can be transported to the lysosome (20). This is based on the observation that binding of competitive inhibitors can stabilize both normal and N370S GCase against thermal denaturation and in cell culture lead to an increase in lysosomal levels of the enzymes. This has resulted in the development of several competitive inhibitors as “chemical chaperones” with the goal of promoting folding of the mutant protein in the ER and facilitating transport to the lysosome where, under lysosomal pH conditions, the inhibitor dissociates and the enzyme regains activity (17, 18, 21–23).
Although the crystal structure of recombinant human GCase has been solved by several investigators under a variety of conditions, these three-dimensional structures do not provide insight into the biochemical or structural defect caused by the N370S mutation (11, 24–30). The N370S mutation site is located in the catalytic domain of the enzyme, but it is remote from the active site lying at the interface of domains II and III and is not directly involved in the catalytic activity. It is also not clear given this location why the N370S mutation would result in an unstable protein (25).
To resolve the conflicting reports regarding the nature of the N370S GCase mutation and its effect on activity and stability, we have expressed the N370S mutant in insect cells and solved the x-ray structure of the mutant at acidic and neutral pH values. To confirm the observations made from the x-ray structure, we also compared the intracellular stability and biophysical properties of the mutant and wild type enzyme (imiglucerase).
EXPERIMENTAL PROCEDURES
Biochemical Characterization of N370S GCase Mutant
N370S GCase was produced in a baculovirus expression system, purified, and characterized as described previously (21). To measure the Km and Vmax values, ∼2 μg ml−1 of enzyme was incubated with 0.2, 0.4, 0.6, 1, 2, 4, 6, and 8 mm p-nitrophenyl-β-d-glucopyranoside (Sigma) in 0.1 m potassium phosphate, 0.1% BSA, 0.125% sodium taurocholate, 0.162% Triton X-100, 0.02% sodium azide, pH 5.9, at 37 °C for 15 min. The reaction was stopped with 0.5 m glycine, pH 10.5, and the absorbance of the p-nitrophenol product was measured at 400 nm (ϵ400 = 18.3 mm−1 cm−1) using a SpectraMax® plate reader with SOFTmax® PRO software version 3.1.2 (Molecular Devices, Sunnyvale, CA). Data were fit using the Michaelis-Menten equation with JMP software (SAS Institute, Cary, NC).
Intracellular Half-life of GCase
In vitro measurement of GCase intracellular half-life was performed in NR8383 rat lung alveolar macrophages (ATCC, Manassas, VA) grown in Kaighn's F12/K media (Invitrogen) with 15% heat-inactivated FBS, 2 mm glutamine, 100 units ml−1 penicillin, 0.1 mg ml−1 streptomycin. For GCase stabilization experiments, cells were harvested from T150 flasks by gently scraping loosely attached cells, and 5 × 105 cells ml−1 were transferred to tubes where uptake of 25 μg ml−1 GCase was carried out over a 2-h period in F12/K media with 4 mg ml−1 BSA and 20 mm HEPES, pH 6.8. The cells were then washed with 1 mg ml−1 mannan (Sigma) and PBS, pH 7.2, and again plated out in growth media in triplicate with N-(n-nonyl) deoxynojirimycin (NN-DNJ) (Toronto Research Chemicals Inc. Ontario, Canada) at the indicated concentration. Cells were harvested at various time points, washed with PBS, and lysed by freeze/thawing in 50 mm potassium phosphate, pH 6.5, with 0.25% Triton X-100 (KP buffer) and protease inhibitors (Roche Diagnostics). The lysates were assayed for total protein with the micro BCA protein assay kit (Pierce, Thermo Scientific, Rockford, IL). GCase activity was measured using the synthetic substrate 4-methylumbelliferyl-β-d-glucoside (Sigma). The lysates were diluted in KP buffer, mixed 1:1 with 15 mm 4-methylumbelliferyl-β-d-glucoside in citrate phosphate buffer, pH 5.4, with 0.25% sodium taurocholate, 1% BSA, and 0.25% Triton X-100 in a 96-well black plate and incubated for 1 h at 37 °C. The reaction was quenched with 1 m glycine-NaOH, pH 12.5, and fluorescence was measured with a SpectraMax® Gemini XPS fluorometer at excitation wavelength 365 nm, emission wavelength 445 nm, and cut-off wavelength 420 nm. Activity was calculated using a GCase standard curve (pg/μl) and was normalized to total cellular protein (ng μg−1). A portion of each sample (20 μg) was prepared for Western blotting using a PAGEprep clean-up kit (Pierce) before loading onto a 4–12% NuPAGE Novex BisTris gel using MES SDS running buffer (Invitrogen). Protein was transferred to a PVDF membrane and blotted using a biotinylated polyclonal anti-GCase antibody. ECL was performed using the SuperSignal West Pico chemiluminescent substrate (Pierce), and the amount of GCase remaining at each time point was determined by densitometry (GE Healthcare).
N370S Crystallization and Structure Determination
N370S GCase was partially deglycosylated with peptide:N-glycosidase (30 units/mg protein in 1% Nonidet P-40, 2 mm tris(2-carboxyethyl)phosphine/PBS, pH 7.0, for 48 h at 2.5 mg ml−1 protein concentration). The deglycosylated protein was then purified and exchanged into 10 mm MES, 1 mm tris(2-carboxyethyl)phosphine, 0.1 m NaCl, pH 6.5 buffer, using a Superdex 200 10/300 column (GE Healthcare). Fractions were collected and concentrated to 5–7 mg ml−1 before setting up crystal drops. Crystals were grown by sitting drop vapor diffusion using a Phoenix drop setter robot (Art Robbins Instruments, Sunnyvale, CA). More specifically, 0.5 μl of protein mixed with 0.5 μl of well solution of either 0.7 m potassium phosphate dibasic, 0.7 m sodium phosphate monobasic, 0.1 m HEPES, pH 7.4, with a final pH of 7.1 (neutral), or 1 m ammonium sulfate, 0.2 m potassium chloride, 0.05 m guanidine hydrochloride, 0.1 m sodium citrate, pH 5.4 (acidic). Crystals appeared within 3 days and were cryoprotected with 20% glycerol and flash-frozen in liquid nitrogen. Data were collected at ALS503 (Advanced Light Source at Lawrence Berkeley National Laboratory, Berkeley, CA) and processed using MOSFLM (31) and Scala (32). The structures were solved by molecular replacement using Molrep (33) with Protein Data Bank 1OGS as the search model. Iterative manual model building was carried out with Coot (34), coupled with refinement using Refmac5 (35). The coordinates and structure factors were deposited in the Protein Data Bank (code 3KE0 and 3KEH).
Circular Dichroism Spectroscopy (CD)
Samples were dialyzed into 10 mm citrate-phosphate, 150 mm NaCl at either pH 5.4 or 7.1 using a 10-kDa molecular mass cutoff Slide-A-Lyzer (Pierce). CD spectra were obtained at 0.4 mg ml−1 protein in a 1-cm quartz cuvette at 25 °C using a Jasco J-810 spectropolarimeter (Jasco Inc, Easton, MD). For each spectrum, two accumulations were made over a 250–350 nm wavelength range using a 1 nm data pitch, 10 nm/min scan speed, 8-s response, and a 2.5 nm bandwidth. A corresponding buffer spectrum was subtracted from each sample spectrum.
Differential Scanning Calorimetry (DSC)
DSC analysis was performed using a CAP-VP-DSC microcalorimeter (GE Healthcare). Samples were prepared to a final concentration of ∼16 μm after a dialysis as described above. Thermal denaturation curves were generated for both molecules at pH 5.4 and pH 7.1 in 10 mm citrate-phosphate, 150 mm NaCl buffer. Samples were heated from 10 to 100 °C at a scan rate of 90 °C h−1, cooled at 90 °C h−1, and reheated to 100 °C to evaluate unfolding reversibility. Data analysis was performed in Origin 7.0 (OriginLab Corp, Northampton, MA) equipped with the DSC analysis add-on (GE Healthcare). Each protein excess heat capacity curve was corrected by reference subtraction of a matching buffer scan followed by concentration normalization of the data. Base-line correction was performed by manual selection of pre- and post-transition base lines, followed by linear interpolation of the base line in the transition region. The transition midpoint (apparent Tm) was determined by calculating the apex of the thermal denaturation curve from the first derivative of the normalized data.
RESULTS
Biochemical Characterization of the N370S Mutant
The biochemical characterization of recombinant human GCase (imiglucerase, Cerezyme®) and N370S mutant GCase expressed in baculovirus system are shown in Table 1. Imiglucerase, which is currently used in enzyme replacement therapy for Gaucher disease, contains a histidine at position 495. This differs from the sequence of the native human GCase sequence, which has an Arg at this position. The Arg to His substitution at 495 does not alter the protein structure or enzymatic activity (30). As reported previously (12), the N370S mutation results in reduced Vmax and ∼3.5-fold higher Km values using the artificial substrate p-nitrophenyl-β-d-glucopyranoside compared with imiglucerase. The N370S mutant also has a 10-fold higher IC50 for the competitive inhibitors NN-DNJ and isofagomine. To evaluate any potential effect on intracellular transport of the mutant enzyme, we previously measured binding to the luminal domain of LIMP-2, which is the protein responsible for the normal intracellular trafficking of GCase to the lysosome (19). The N370S mutant and imiglucerase have similar affinities for LIMP-2, as measured by surface plasmon resonance.
TABLE 1.
Biochemical properties of N370S compared with imiglucerase
| Sample | Km | Vmax | Kd for LIMP-2 (19) | IC50 for IFG | IC50 for NN-DNJ |
|---|---|---|---|---|---|
| mm | μmol min−1mg−1 | nm | μm | μm | |
| Imiglucerase | 0.9 | 49.6 | 120 | 0.05 | 2.2 |
| N370S | 3.3 | 17.7 | 130 | 0.5 | 20 |
Crystal Structures of N370S GCase
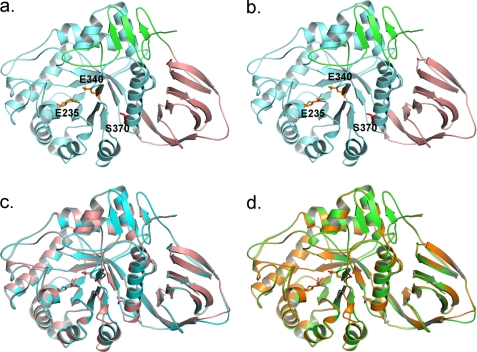
N370S GCase crystals appeared readily in a focused screen based on the published conditions for imiglucerase at acidic (25) and neutral pH (26). The N370S GCase crystallized in the space group C2221 at both pH values with two molecules in each asymmetric unit, and it has nearly identical cell dimensions compared with several previously published GCase structures (Table 2). The structures of N370S GCase at the two pH values are very similar to each other and to the apo-GCase structures deposited in the Protein Data Bank (1OGS, 3GX1, 3GXD, 2F61, 2NT1, 2J25, and 2WKL). N370S GCase has the same three-domain structure as other recombinant GCase as follows: domain I containing residues 1–27 and 383–414; domain II, an Ig fold containing residues 30–75 and 431–497; and domain III, the catalytic domain of residues 76–381 and 416–430, which form a (α/β)8 TIM barrel (Fig. 1, a and b). There are two amino acid changes in the N370S GCase sequence compared with imiglucerase. One is the mutation N370S and the second one is H495R, which is the same sequence as the native GCase. Both changes can be seen clearly in the electron density map (supplemental Fig. 1). The root mean square deviation over Cα is 0.5 Å or less, indicating the two sequence differences do not change the overall structure of the N370S mutant.
TABLE 2.
Data collection and refinement statistics
| N370S, pH 5.4 | N370S, pH 7.1 | |
|---|---|---|
| Data collection | ||
| Space group | C2221 | C2221 |
| Cell dimensions | ||
| a, b, c | 109.5, 285.1, 92.2 Å | 107.8, 285.9, 92.0 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° |
| Resolutiona | 50-2.7 Å (2.78-2.70 Å) | 50-2.8 Å (2.88-2.80 Å) |
| Rsymb | 14.7% (51.5%) | 15.8% (35.5%) |
| I/s〈I〉 | 7.3 (2.5) | 8.4 (2.3) |
| Completeness | 99.5 (94.1) | 94.7 (91.1) |
| Redundancy | 6.1 (2.7) | 5.2 (2.4) |
| Refinement | ||
| Resolution | 50-2.7 Å | 50-2.8 Å |
| No. of reflections | 37,753 | 31,773 |
| Rcrystc/Rfreed | 17.2/22.9% | 21.4/27.3% |
| Root mean square deviation | ||
| Bond length | 0.019 Å | 0.015 Å |
| Bond angles | 1.7° | 1.6° |
| No. of refined atoms | ||
| Protein | 7902 | 7902 |
| Water | 252 | 85 |
| Sugar | 42 | 42 |
| Ramachandran outliers | 0.2% | 0.8% |
a Numbers in parentheses refer to the highest resolution shell.
b Rsym = Σij|(Ii(j) − 〈I(j)〉|/Σij Ii(j), where Ii(j) is the intensity of the ith observation of reflection j. Ii(j) is the weighted mean of all measurements of j.
c Rcryst = Σj|Fo(j)| − |Fc(j)|/Σj|Fo(j)|, where Fo and Fc are the observed and calculated structure factors.
d Rfree was calculated as for Rcryst, but on 5% of data excluded before refinement.
FIGURE 1.
Structure of N370S GCase at neutral and acidic pH values. a, N370S GCase at pH 7.1. b, N370S GCase at pH 5.4. Green, domain I; pink, domain II; light blue, domain III. Catalytic residues are shown as orange sticks. Ser-370 is shown as red sticks. c, overlay of pH 7.1 N370S GCase structure (cyan) with imiglucerase at neutral pH (2NT1, pink). d, overlay of pH 5.4 N370S GCase structure (green) with imiglucerase at acidic pH (3GXI, orange).
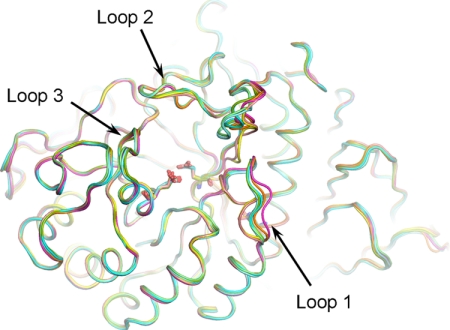
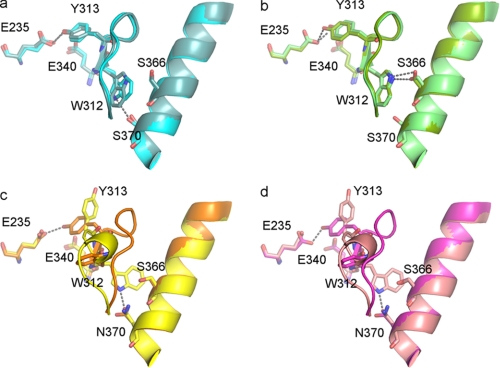
In previously published crystal structures of GCase, three loops surrounding the active site, loop 1 (amino acids 311–319), loop 2 (amino acids 342–354), and loop 3 (amino acids 394–399) presented in multiple conformations, indicating the dynamic flexibility of these three regions.3 We compared the conformations of these loops in the N370S GCase structure to those of the published GCase structures in the presence and absence of inhibitors (Table 3). Loop 3 of N370S GCase displays the identical two conformations in each asymmetric unit as those in GCase, with or without inhibitors bound, and therefore does not appear to be affected by the mutation. Loop 2 samples a range of motions in the N370S GCase structure as noted in the published structures indicating a generalized flexibility in this region. In contrast, loop 1 adopts one of two conformations, extended or α-turn. In the absence of inhibitors, loop 1 is observed in either conformation, but the extended conformation occurs more frequently (Fig. 2 and Table 3). In the presence of isofagomine, NN-DNJ, or NB-DNJ, only the α-turn conformation is observed, although in the GCase-conduritol B epoxide structure, only the extended conformation is present. In N370S GCase at either pH, we only observed the extended conformation of loop 1 (Fig. 3, a and b). In the apo-structures of GCase, if loop 1 adopts both the extended and the α-turn conformations, then Trp-312 in only the α-turn conformation forms a hydrogen bond with Asn-370, as observed in 3GXI and 2NT1 (Fig. 3, c and d). If only the extended conformation is present in the GCase apo-structure, as in the case of 1OGS and 2F61, only one of the two monomers forms a hydrogen bond between Trp-312 and Asn-370 (Table 3). In the co-crystal structures of GCase with isofagomine, NN-DNJ, or NB-DNJ, this H-bond is lost even though loop 1 has the α-turn conformation. In the structure of N370S GCase at acidic pH, Trp-312 can no longer interact with Ser-370, but rather it forms a hydrogen bond with Ser-366 on the same helix (helix 7, according to the numbering in Ref. 25). At neutral pH, Trp-312 forms a hydrogen bond with Ser-370 in one of the two conformations. The Trp-312 side chain density appears at the same position in the second conformation, but it is less well defined. Thus, the exact orientation of the side chain remains uncertain. Tyr-313 is also located on loop 1 and is observed to form a hydrogen bond with the catalytic residue Glu-235 in the extended conformation. In the α-turn conformation of GCase, Tyr-313 swings out and allows easier access to the catalytic residues. Upon binding of isofagomine, NN-DNJ, and NB-DNJ, but not conduritol B epoxide, Tyr-313 hydrogen bonds with Glu-340 rather than Glu-235. Only one orientation of Tyr-313, which hydrogen bonds with Glu-235, is observed in N370S GCase.
TABLE 3.
Comparison of crystal structures of GCase
The abbreviations used are as follows: GOL, glycerol; IFG, isofagomine; CBE, conduritol B epoxide; ND, not determined.
| PDB code | Protein source+molecule at active site | Space group | pH | Loop 1 conformation | Tyr-313 H-bond to catalytic residues | Trp-312 H-bond to helix 7a | Ref. |
|---|---|---|---|---|---|---|---|
| 1OGS | Imiglucerase | C2221 | 4.5 | Extended | Glu-235 | 1 | 25 |
| Extended | Glu-235 | ||||||
| 2NT0 | Imiglucerase | P21 | 4.5 | Extended | Glu-235 | 26 | |
| +GOL | Extended | Glu-235 | |||||
| 3GXM | Imiglucerase | P21 | 4.5 | α-Helical | 1 | 28 | |
| Extended | Glu-235 | ||||||
| 3GXD | Imiglucerase | P21 | 4.5b | α-Helical | 1 | 28 | |
| Extended | |||||||
| 3GXI | Imiglucerase | P21 | 5.5 | α-Helical | 1 | 28 | |
| Extended | Glu-235 | ||||||
| 2J25 | Imiglucerase | C2221 | 5.5 | α-Helical | 1 | 24 | |
| Extended | Glu-235 | ||||||
| 2F61 | Imiglucerase | C2221 | 6.0 | Extended | 1 | 11 | |
| Extended | Glu-235 | ||||||
| 2NT1 | Imiglucerase | P21 | 7.5c | α-Helical | 1 | 26 | |
| Extended | Glu-340 | ||||||
| 2NSX | Imiglucerase | P21 | 4.5 | α-Helical (IFG) | Glu-340 | 26 | |
| +IFG | Extended (GOL) | ||||||
| 3GXF | Imiglucerase | P21 | 7.5c | α-Helical (IFG) | Glu-340 | 28 | |
| +IFG | Extended (GOL) | ||||||
| 1Y7V | Imiglucerase | C2221 | 4.6 | Extended | Glu-235 | 27 | |
| +CBE | Extended | Glu-235 | |||||
| 2WKL | Velaglucerase alfa | C2221 | 7.0 | α-Helical | 1 | 30 | |
| Extended | Glu-235 | 1 | |||||
| 2V3F | Taliglucerase alfa | P21 | 6.5 | α-Helical | Glu-340 | 29 | |
| +BTB | α-Helical | Glu-340 | |||||
| 2V3D | Taliglucerase alfa | P21 | 6.5 | α-Helical | Glu-340 | 29 | |
| +NB-DNJ | α-Helical | Glu-340 | |||||
| 2V3E | Taliglucerase alfa | P21 | 6.5 | α-Helical | Glu-340 | 29 | |
| +NN-DNJ | α-Helical | Glu-340 | |||||
| 3KE0 | N370S GCase | C2221 | 5.4 | Extended | Glu-235 | 1 | This study |
| Extended | Glu-235 | 1 | |||||
| 3KEH | N370S GCase | C2221 | 7.1 | Extended | Glu-235 | 1 | This study |
| Extended | Glu-235 | NDd |
a Numbering of the helix is same as Ref. 25.
b This was crystallized at pH 5.5 and soaked to pH 4.5.
c A very similar condition as was used for N370S crystallization at neutral pH. The final pH is likely at pH 7.1.
d The side chain of Trp-312 in one of the confirmations has poorer density than the other one. Its density is at the same position, but the orientation is a little bit hard to define.
FIGURE 2.
Active site loop conformations of N370S GCase. N370S GCase structures are shown in shades of blue at pH 7.1, and shades of green at pH 5.4. Imiglucerase structures are shown in shades of pink at neutral pH (2NT1), and shades of yellow at acidic ph (3GXI). Two conformations are shown for each structure.
FIGURE 3.
Loop 1 conformations and H-bonds with active sites and helix 7. a, N370S GCase at pH 7.1 in shades of blue. b, N370S GCase at pH 5.4 in shades of green. c, imiglucerase at neutral pH (2NT1) in shades of pink. d, imiglucerase at acidic pH (3GXI) in shades of yellow. Two conformations are shown for each structure. H-bonds are shown as dotted lines.
No major structural difference is observed between the crystal structure of N370S GCase and the published structures of the three wild type recombinant GCases produced in a Chinese hamster ovary cell line (imiglucerase), a human fibrosarcoma cell line (velaglucerase alfa), or a carrot cell line (taliglucerase alfa) at either acidic or neutral pH. In addition to the conformational differences already noted in the three loops surrounding the active site, a few charged, polar, or glycine residues on the flexible loops in the Ig-like domains also showed differences in orientation between N370S and the wild type GCases (data not shown). These residues are on the surface farther away from the active site and have higher B factors, which is indicative of their higher flexibility. It is unlikely that this difference in the orientation of surface polar residues will have a gross impact on the overall structure. A more significant difference is a subtle change in the orientation of Trp-312 and its ability to hydrogen bond with the residue in position 370. How the difference in this hydrogen bonding potential affects the catalytic activity of GCase or N370S is not clear. However, the lack of different conformations for the loop 1 region indicates that the N370S mutation results in a more rigid structure, which could have an adverse effect on catalysis.
Solution Structure Comparison by Circular Dichroism
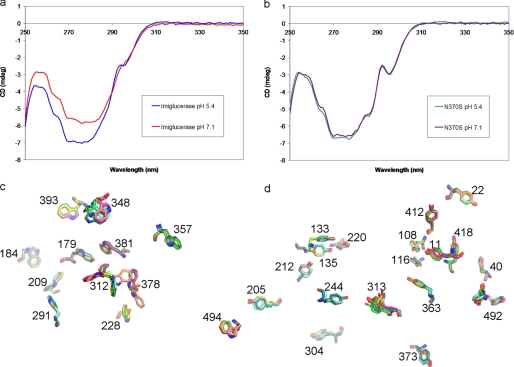
There is a high degree of similarity between the crystal structures of N370S GCase and the wild type enzyme at all pH values and in the presence and absence of a variety of inhibitors. Therefore, the possibility that the crystallization conditions were favoring a stable conformation that was not representative of the solution structure or that the alternative conformations observed in GCase structures are a result of a crystal packing artifact could not be ruled out. To further probe the secondary and tertiary structure difference between N370S GCase and imiglucerase in solution, near and far UV CD was performed at both neutral and acidic pH values. The far UV spectra for the native and N370S GCase are nearly identical at both pH values (data not shown). This is consistent with the crystallographic data that no major changes occur in the secondary structure as a result of the mutation. Although the near-UV CD spectra of imiglucerase indicates the protein is quite sensitive to pH change (Fig. 4a), the spectra for N370S GCase at the neutral and acidic pH values almost overlay completely with each other (Fig. 4b). The CD spectra for the N370S mutant at both pH values resemble that of imiglucerase at acidic pH. A distinct difference in the spectra occurs around 295 nm, a region that is associated with tryptophan side chains and is indicative of the tertiary structure surrounding these residues in the protein. When the orientation of all 12 tryptophan side chains in imiglucerase and N370S GCase crystal structures at both neural and acidic pH are compared, three tryptophan residues are found in different side chain orientations (Fig. 4c). Tryptophan 312 is observed to adopt drastically different conformations, and tryptophan 348 appears to be sampling slightly different random orientations, as occurs with loop 2 in general. Imiglucerase has two different conformations for tryptophan 378, but N370S GCase only displays one of the two imiglucerase conformations. As with the crystal structure, the difference in the CD spectra between the N370S mutant and imiglucerase correlates with a change in the orientation of tryptophan residues, in particular Trp-312 on loop 1, and possibly to a lesser extent Trp-378. Among the 19 tyrosine residues in GCase, Tyr-313 on loop 1 also adopts two alternative conformations in imiglucerase but only has one orientation in N370S (Fig. 4d and Table 3). Imiglucerase shows a clear pH-dependent change in the local environment of one or more aromatic residues. This pH-dependent change is absent with the N370S mutant. The fact that the N370S mutation does not possess alternative conformations of loop 1 in the crystal structures and does not undergo a pH-dependent change in this region again suggests that the molecule is less flexible in this area compared with imiglucerase.
FIGURE 4.
Solution structure comparison and the conformation of Tyr and Trp residues in GCase. a, CD spectra of imiglucerase at pH 7.1 and 5.4. b, CD spectra of N370S at pH 7.1 and 5.4. c, overlay of all Trp residues in imiglucerase (2NT1, neutral pH; 3GXI, acidic pH) and in N370S at pH 7.1 and 5.4. d, overlay of all Tyr residues in imiglucerase (2NT1, neutral pH; 3GXI, acidic pH) and in N370S at pH 7.1 and 5.4. Color scheme is same as in Fig. 3.
Differential Scanning Calorimetry
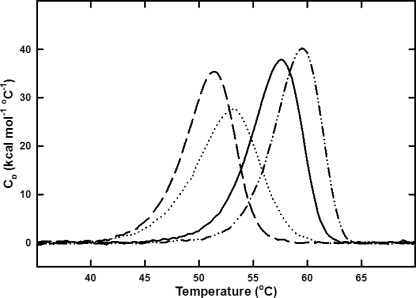
The crystallographic and CD data do not indicate a structural change in the N370S mutant that would lead to global instability. In contrast, these data suggest a more rigid structure with less flexibility, which could be expected to increase stability. To examine this, the thermodynamic stability of N370S GCase versus imiglucerase was compared by DSC at pH 5.4 and 7.1. Thermal denaturation for both N370S GCase and imiglucerase was irreversible, preventing a complete thermodynamic analysis. However, improved thermal stability at acidic pH was observed for N370S GCase and imiglucerase with an increase in the apparent melting temperature (Tm) of ∼6 °C for both proteins (Fig. 5 and Table 4). The apparent Tm of N370S GCase is nearly 2 °C higher than imiglucerase at both pH values, indicating increased thermal stability.
FIGURE 5.
Thermal denaturation curves for imiglucerase and N370S GCase as measured by DSC. Excess molar heat capacity (kcal mol−1 °C−1) plotted against temperature (°C) from 35 to 70 °C for 16 μm samples in 0.1 m citrate/phosphate buffer, imiglucerase, pH 5.4 (solid line), rGCR, pH 7.1 (dashed line), N370S, pH 5.4 (dash dot dot line) and N370S, pH 7.1 (dotted line).
TABLE 4.
Melting temperatures of imiglucerase and N370S at neutral and acid pH values
| Imiglucerase | N370S | |
|---|---|---|
| Tm ± σ °C | Tm ± σ °C | |
| pH 7.1 | 51.30 ± 0.02 | 53.05 ± 0.06 |
| pH 5.4 | 57.67 ± 0.04 | 59.54 ± 0.12 |
Intracellular Half-life
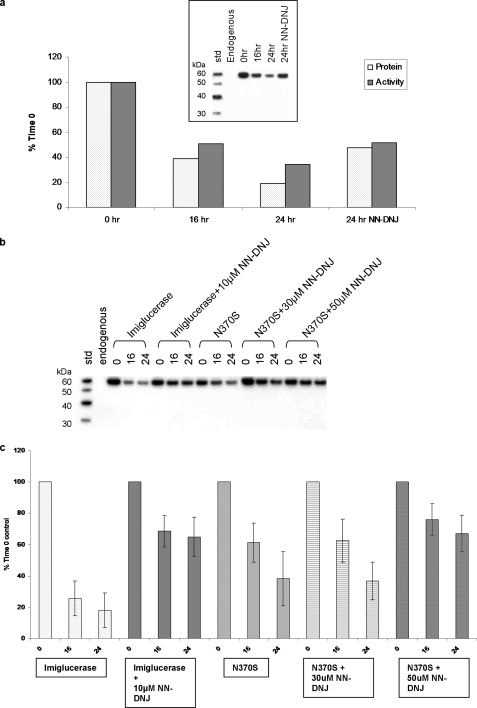
Crystallographic, biochemical, and biophysical analysis did not indicate any structural or biochemical changes that suggest the N370S mutation results in a misfolded or unstable protein nor did they explain the observation that competitive inhibitors can increase the intracellular levels of N370S and GCase in general. Although our studies demonstrate a catalytic defect that is in line with earlier observations (4, 5, 12), we were studying purified folded proteins so the possibility that competitive inhibitors could affect a population of misfolded molecules that we were not isolating could not be completely ruled out. To evaluate this possibility, we studied the effect of the competitive inhibitor NN-DNJ on the lysosomal stability of the purified and folded N370S and imiglucerase in a rat macrophage cell line. To facilitate uptake into macrophages, imiglucerase is remodeled to expose terminal mannose residues during the manufacturing process. The N370S GCase purified from insect cells naturally contains pauci-mannose glycan structures, and no further processing is required. Because imiglucerase is delivered directly to the endosomal/lysosomal compartment via the mannose receptor on the cell surface (36), it is possible to study the effect of NN-DNJ on lysosomal GCase while removing any effect on folding or ER degradation. Endogenous rat GCase is not recognized by the polyclonal antibody used in this experiment (Fig. 6, a and b), making it possible to follow the intracellular half-life of recombinant human GCase by Western blotting. Following a 2-h uptake of imiglucerase, GCase activity increases two and a half-fold above background levels (data not shown). This is then followed by a decrease in both protein and activity levels with ∼20% remaining after 24 h. Addition of 10 μm NN-DNJ results in increased lysosomal stability with ∼50–60% of protein and activity remaining after 24 h (Fig. 6a). Uptake of N370S GCase is comparable with that of imiglucerase based on Western blotting. However, due to the reduced specific activity of the N370S GCase mutant, activity levels could not be reliably determined above background levels. Interestingly, and in line with the DSC results, N370S GCase has increased intracellular stability compared with imiglucerase (40 versus 20% remaining after 24 h), and a higher concentration of NN-DNJ was required to achieve the same level of intracellular stabilization as imiglucerase (50 versus 10 μm) (Fig. 6, b and c).
FIGURE 6.
Stabilization of imiglucerase and N370S GCase by small molecule inhibitor NN-DNJ. NR8383 rat lung alveolar macrophages were incubated with imiglucerase or N370S for uptake. Following the GCase uptake, cells were treated with or without NN-DNJ and lysed at a time course. a, experiment in which the imiglucerase activities were measured with and without 10 μm NN-DNJ treatment. Percentage of enzyme activity remaining was measured for uptake of imiglucerase, subtracted by endogenous GCase activity. Inset, Western blot used for quantification of the protein level remained. The total area of the intact protein band at time 0 on the Western blot was set as 100%. b, Western blot used to quantitate the effect of NN-DNJ on the stabilization of uptaken imiglucerase and N370S GCase. c, percentage of intact protein remained was plotted against the time post uptake. The endogenous rat GCase was not detectable with the polyclonal antibody used.
DISCUSSION
The crystal structure of N370S GCase is virtually indistinguishable from that of the wild type recombinant enzymes at both pH values studied. As reported for the wild type enzymes, there are subtle changes in the orientation of residues in the loop 1 region but no global changes in the overall domain structure with Cα root mean square deviation at 0.5 Å compared with the imiglucerase structures. A few exposed polar and charged residues in the Ig domain showed different conformations with high temperature factors (B factors), which indicate their inherent high dynamicity. The main effect of the N370S mutation on GCase structure is a change in the orientation of tryptophan 312 in the loop 1 region of the molecule. In the wild type enzyme, Trp-312 and loop 1 can exist in two conformations, one extended and the other α-helical. The Tyr-313 on loop 1 also adopts different orientations and can hydrogen bond with either of the catalytic residues Glu-235 and Glu-340. As with Trp-312, Tyr-313 in the N370S mutant only exists in one conformation and that resembles imiglucerase at acidic pH. Interestingly, we did not observe a pH effect on the crystal structure of N370S GCase. Similarly no effect of pH was observed on the solution structure of the N370S GCase as determined by circular dichroism spectroscopy. This differs from the effect of pH on imiglucerase in solution where a pH-dependent change in the environment of aromatic residues was observed. The CD spectra of N370S GCase at both acidic and neutral pH resembled that of imiglucerase at acidic pH.
Previous studies have suggested that residues in the loop 1 region may play a role in the access of substrate to the active site (26). The different orientations of loop 1 and Tyr-312 have been observed to change upon binding to inhibitors altering access to the catalytic site. Our results also suggest, but do not explain, an important role for this loop flexibility in the catalytic mechanism of GCase. How the different orientation of residues affects activity is not clear. Recent studies by Liou and Grabowski (37) indicate that the N370S mutation results in a catalytic defect in both the glucosylation and deglucosylation steps of substrate hydrolysis. The authors (37) proposed that the involvement of Asn-370 in the catalysis is through isolated conformational effects near the active site, rather than a global change. This is consistent with our crystal structures in that only small changes near the active site, mainly involving the conformation of loop 1 residues, were observed in N370S GCase. We also observed an increase in Km for N370S compared with imiglucerase when using an in vitro artificial substrate. Both the crystal structure and CD data point to the aromatic residues on loop 1, namely tryptophan 312 and tyrosine 313, being associated with the decreased specific activity. Tryptophan 312 can form a hydrogen bond with the α-helix where Asn-370 is located, and tyrosine 313 forms direct interactions with the catalytic residues. It is conceivable that the change in the interaction between loop 1 and Asn-370 could have a direct effect on the catalytic activity.
The crystallographic and CD data suggest that the N370S mutation results in a more rigid structure compared with imiglucerase. This is also consistent with the reduced catalytic activity and increased thermal stability at both acidic and neutral pH conditions. Our structural and biochemical data, which are consistent with earlier studies, demonstrate that the N370S mutant protein is processed normally and is present at normal levels but has reduced catalytic activity. However, they do not explain the more recent observations that competitive inhibitors increase intracellular levels of N370S GCase.
Although we, and others, have not observed any significant differences in expression between N370S GCase and wild type, the possibility that we were isolating a more stable form of the mutant and an alternative misfolded form was being degraded in the ER could not be ruled out. To evaluate this possibility, we investigated the effect of the competitive inhibitor NN-DNJ on the intracellular stability of purified N370S and imiglucerase in macrophages. It is interesting that the 2–3-fold increase in GCase levels we observed after 24 h in the presence of NN-DNJ is comparable with the increase originally reported using Gaucher patient fibroblasts (20) as well as subsequent studies in patient fibroblasts using several different competitive inhibitors (17, 21), suggesting that the mechanism(s) of action is similar in all cases. In our experiments, both proteins were already folded and targeted to lysosome via the mannose receptor, so an effect on protein folding or transport from the ER to lysosome could be ruled out in this case (36). The competitive inhibitor isofagomine has been reported to increase the activity and lysosomal level of GCase in patient fibroblasts by several mechanisms (17). The main effect is thought to be mediated through an increase in protein folding and a subsequent decrease in protein degradation, presumably in the ER. Although a direct effect on protein folding has not been demonstrated, it has been inferred from pulse-chase experiments that demonstrate an altered glycoform profile following treatment with isofagomine. Interestingly, the glycoform processing in the lysosome is also shown to be altered in the presence of isofagomine suggesting a general effect on the resistance of GCase to both proteases and glycosidases. Our data indicate that N370S GCase is correctly folded (although the rate of folding could be different) and that the main effect of competitive inhibitors is to decrease the susceptibility to proteases whether they are lysosomal or in the ER. Because a percentage of all proteins are thought to be misfolded and degraded in the ER, both an effect on ER and lysosomal degradation are likely. What is not clear is the extent to which each contributes in the case of N370S GCase. Our results showing that the effect on imiglucerase and N370S GCase is comparable in magnitude to that observed using patient cells suggest that the major (but not necessarily the only) mechanism by which competitive inhibitors increase lysosomal levels of GCase is by reducing degradation within the lysosome rather than an effect on protein folding and trafficking. A reduction in lysosomal degradation in the presence of unaltered transport to the lysosome is consistent with many of the reports demonstrating an increased ratio of lysosomal to nonlysosomal protein levels in the presence of competitive inhibitors (18, 21).
Our results support the earlier biochemical and cellular studies that demonstrated that the N370S mutation results in a catalytically deficient enzyme with normal stability that is expressed at normal or near normal levels. The N370S mutation influences the flexibility of the loop 1 region resulting in reduced catalytic activity. It is worth noting that upon inhibitor binding, wild type GCase is locked into a single conformation, displaying rigidity similar to the N370S mutant (Table 3). This increased rigidity in both the N370S mutant enzyme and the wild type enzyme in the presence of inhibitors confers an increased stability to both thermal denaturation and lysosomal degradation. The increased lysosomal stability in the presence of inhibitors is consistent with the reported effects of these compounds on both protein levels and intracellular distribution and suggests that this mechanism is responsible for the majority of the increase in protein levels observed rather than an impact on protein folding or trafficking. Although our results do not fully explain the mechanism by which the N370S mutation results in reduced catalytic activity, they do reconcile the earlier observations that the N370S mutation causes a catalytic mutant with the more recent observations on the effect of competitive inhibitors.
Supplementary Material
Acknowledgments
The N370S GCase mutant was originally cloned by David Reczek and Lori Rulli, expressed by Christine DeMaria, Jean McLarty, and Judy Jensen, and purified by Lee Sherman and Julie Pollock. The enzymatic kinetics assay was performed by Bethany Foster and Jennifer Baker-Malcolm.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The atomic coordinates and structure factors (codes 3KE0 and 3KEH) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GCase
- glucocerabrosidase
- NN-DNJ
- N-nonyl-deoxynojirimycin
- NB-DNJ
- N-butyl-deoxynojirimycin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ER
- endoplasmic reticulum
- DSC
- differential scanning calorimetry.
REFERENCES
- 1. Brady R. O., Kanfer J. N., Bradley R. M., Shapiro D. (1966) J. Clin. Invest. 45, 1112–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grabowski G. A., Petsko G. A., Kolodny E. H. (2006) in Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Valle D., Sly W. S., Vogelstein B., Childs B., Kinzler K. W. eds) McGraw-Hill Inc., New York [Google Scholar]
- 3. Beutler E., Gelbart T., Scott C. R. (2005) Blood Cells Mol. Dis. 35, 355–364 [DOI] [PubMed] [Google Scholar]
- 4. Grace M. E., Graves P. N., Smith F. I., Grabowski G. A. (1990) J. Biol. Chem. 265, 6827–6835 [PubMed] [Google Scholar]
- 5. Grace M. E., Newman K. M., Scheinker V., Berg-Fussman A., Grabowski G. A. (1994) J. Biol. Chem. 269, 2283–2291 [PubMed] [Google Scholar]
- 6. Ho M. W., O'Brien J. S. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 2810–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peters S. P., Coyle P., Coffee C. J., Glew R. H., Ho M. W., O'Brien J. S., Radin N. S., Erickson J. S. (1977) J. Biol. Chem. 252, 563–573 [PubMed] [Google Scholar]
- 8. Basu A., Glew R. H. (1985) J. Biol. Chem. 260, 13067–13073 [PubMed] [Google Scholar]
- 9. Dale G. L., Villacorte D. G., Beutler E. (1976) Biochem. Biophys. Res. Commun. 71, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 10. Blonder E., Klibansky C., de Vries A. (1976) Biochim. Biophys. Acta 431, 45–53 [DOI] [PubMed] [Google Scholar]
- 11. Liou B., Kazimierczuk A., Zhang M., Scott C. R., Hegde R. S., Grabowski G. A. (2006) J. Biol. Chem. 281, 4242–4253 [DOI] [PubMed] [Google Scholar]
- 12. Ohashi T., Hong C. M., Weiler S., Tomich J. M., Aerts J. M., Tager J. M., Barranger J. A. (1991) J. Biol. Chem. 266, 3661–3667 [PubMed] [Google Scholar]
- 13. Salvioli R., Tatti M., Scarpa S., Moavero S. M., Ciaffoni F., Felicetti F., Kaneski C. R., Brady R. O., Vaccaro A. M. (2005) Biochem. J. 390, 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michelin K., Wajner A., Bock H., Fachel A., Rosenberg R., Pires R. F., Pereira M. L., Giugliani R., Coelho J. C. (2005) Clin. Chim. Acta 362, 101–109 [DOI] [PubMed] [Google Scholar]
- 15. Ron I., Horowitz M. (2005) Hum. Mol. Genet 14, 2387–2398 [DOI] [PubMed] [Google Scholar]
- 16. Bergmann J. E., Grabowski G. A. (1989) Am. J. Hum. Genet. 44, 741–750 [PMC free article] [PubMed] [Google Scholar]
- 17. Steet R. A., Chung S., Wustman B., Powe A., Do H., Kornfeld S. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmitz M., Alfalah M., Aerts J. M., Naim H. Y., Zimmer K. P. (2005) Int. J. Biochem. Cell Biol. 37, 2310–2320 [DOI] [PubMed] [Google Scholar]
- 19. Reczek D., Schwake M., Schröder J., Hughes H., Blanz J., Jin X., Brondyk W., Van Patten S., Edmunds T., Saftig P. (2007) Cell 131, 770–783 [DOI] [PubMed] [Google Scholar]
- 20. Sawkar A. R., Cheng W. C., Beutler E., Wong C. H., Balch W. E., Kelly J. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15428–15433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sawkar A. R., Schmitz M., Zimmer K. P., Reczek D., Edmunds T., Balch W. E., Kelly J. W. (2006) ACS Chem. Biol. 1, 235–251 [DOI] [PubMed] [Google Scholar]
- 22. Shen J. S., Edwards N. J., Hong Y. B., Murray G. J. (2008) Biochem. Biophys. Res. Commun. 369, 1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steet R., Chung S., Lee W. S., Pine C. W., Do H., Kornfeld S. (2007) Biochem. Pharmacol. 73, 1376–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brumshtein B., Wormald M. R., Silman I., Futerman A. H., Sussman J. L. (2006) Acta Crystallogr. D. Biol. Crystallogr. 62, 1458–1465 [DOI] [PubMed] [Google Scholar]
- 25. Dvir H., Harel M., McCarthy A. A., Toker L., Silman I., Futerman A. H., Sussman J. L. (2003) EMBO Rep. 4, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lieberman R. L., Wustman B. A., Huertas P., Powe A. C., Jr., Pine C. W., Khanna R., Schlossmacher M. G., Ringe D., Petsko G. A. (2007) Nat. Chem. Biol. 3, 101–107 [DOI] [PubMed] [Google Scholar]
- 27. Premkumar L., Sawkar A. R., Boldin-Adamsky S., Toker L., Silman I., Kelly J. W., Futerman A. H., Sussman J. L. (2005) J. Biol. Chem. 280, 23815–23819 [DOI] [PubMed] [Google Scholar]
- 28. Lieberman R. L., D'aquino J. A., Ringe D., Petsko G. A. (2009) Biochemistry 48, 4816–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brumshtein B., Greenblatt H. M., Butters T. D., Shaaltiel Y., Aviezer D., Silman I., Futerman A. H., Sussman J. L. (2007) J. Biol. Chem. 282, 29052–29058 [DOI] [PubMed] [Google Scholar]
- 30. Brumshtein B., Salinas P., Peterson B., Chan V., Silman I., Sussman J. L., Savickas P. J., Robinson G. S., Futerman A. H. (2010) Glycobiology 20, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leslie A. G. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 32. Evans P. R. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 33. Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36. Piepenhagen P. A., Vanpatten S., Hughes H., Waire J., Murray J., Andrews L., Edmunds T., O'Callaghan M., Thurberg B. L. (2010) Microsc. Res. Tech. 73, 694–703 [DOI] [PubMed] [Google Scholar]
- 37. Liou B., Grabowski G. A. (2009) Mol. Genet. Metab. 97, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.