Abstract
Alternative splicing often produces effectors with opposite functions in apoptosis. Splicing decisions must therefore be tightly connected to stresses, stimuli, and pathways that control cell survival and cell growth. We have shown previously that PKC signaling prevents the production of proapoptotic Bcl-xS to favor the accumulation of the larger antiapoptotic Bcl-xL splice variant in 293 cells. Here we show that the genotoxic stress induced by oxaliplatin elicits an ATM-, CHK2-, and p53-dependent splicing switch that favors the production of the proapoptotic Bcl-xS variant. This DNA damage-induced splicing shift requires the activity of protein-tyrosine phosphatases. Interestingly, the ATM/CHK2/p53/tyrosine phosphatases pathway activated by oxaliplatin regulates Bcl-x splicing through the same regulatory sequence element (SB1) that receives signals from the PKC pathway. Convergence of the PKC and DNA damage signaling routes may control the abundance of a key splicing repressor because SB1-mediated repression is lost when protein synthesis is impaired but is rescued by blocking proteasome-mediated protein degradation. The SB1 splicing regulatory module therefore receives antagonistic signals from the PKC and the p53-dependent DNA damage response pathways to control the balance of pro- and antiapoptotic Bcl-x splice variants.
Keywords: Apoptosis, DNA Damage, p53, Phosphatase, Proteasome, Protein Synthesis, RNA Splicing, Signal Transduction, Bcl-x, Oxaliplatin
Introduction
A variety of extracellular stimuli and intracellular stresses, including DNA damage, promote cell death by triggering the apoptotic pathway. Members of the Bcl-2 family of proteins play a critical role in the control of apoptosis and can display anti- or proapoptotic activity (1, 2). Overexpressing antiapoptotic members or reducing the expression of proapoptotic proteins protect cells against death stimuli. In contrast, preventing the expression of antiapoptotic effectors promotes or sensitizes cells to death stimuli. Cell death is therefore controlled via a complex and delicate balance of pro- and antiapoptotic activities.
Alternative splicing plays a major role in the control of apoptosis by dictating the production of different isoforms, often with opposite functions (3). The functional consequences of alternative splicing on apoptosis has been documented for many genes, including cell surface receptors, such as Fas; adaptor proteins and regulators, such as TRAF2 and APAF-1; mediators, such as Bcl-x, Bak, and Mcl-1; and caspases (3–5). Bcl-x is produced in two forms through the use of alternative 5′ splice sites. Bcl-xL is antiapoptotic, whereas the shorter Bcl-xS variant is proapoptotic (6). Numerous studies have reported high levels of Bcl-xL in cancer tissues, its overexpression conferring resistance to apoptotic stimuli and favoring metastasis (7, 8). In contrast, Bcl-xS can induce apoptosis and alleviate resistance to anti-cancer drugs (9, 10).
Given the critical importance of maintaining an appropriate balance of pro- and antiapoptotic splice forms, splicing regulation must be tightly attuned to environmental stresses and intracellular demands. The two principal families of RNA binding proteins involved in alternative splicing regulation, the SR and heterogeneous nuclear ribonucleoprotein families, are equipped to be on the receiving end of these pathways because they can be phosphorylated, a modification that can affect their activity and localization (11–15). However, the signaling pathways that carry information from stresses, stimuli, or drugs to kinases and phosphatases acting on splicing regulators remain poorly defined (16, 17), and links between signal transduction and the alternative splicing control of apoptotic genes have rarely been reported. In one case, Fas activation and heat shock induce the de novo synthesis of ceramide, which in turn activates protein phosphatase 1 to stimulate the production of the proapoptotic Bcl-xS isoform (18). In contrast, the phosphorylation of Sam68 by Fyn favors the antiapoptotic Bcl-xL splice variant (19). More work is required to produce a comprehensive map of intersecting signaling routes triggered by different cues to coordinate Bcl-x splicing decisions.
Recently, we showed that several anticancer drugs could modify the alternative splicing of Bcl-x (20). Among these, the genotoxic agents oxaliplatin and cisplatin displayed the broadest effect, shifting splicing in favor of the proapoptotic Bcl-xS splice variant in all cell lines tested. A recent study reported that UV irradiation increased the production of Bcl-xS, but the effect was independent of p53 (21). It was therefore of interest to investigate the pathway through which genotoxic compounds alter the balance of Bcl-x splice variants. Here we report that the increase in Bcl-xS induced by oxaliplatin requires p53 and is abrogated by inhibitors of ATM and CHK2 kinases. The splicing shift induced by the DNA damage response converges on the same regulatory element (SB1) used by the PKC signaling pathway to repress the 5′ splice site of Bcl-xS. Further, inhibiting tyrosine phosphatases antagonizes the SB1-mediated splicing shifts imposed by oxaliplatin and PKC inhibition. Thus, our results support a model in which a balance between PKC and tyrosine phosphatases activities determines the relative proportion of pro- and antiapoptotic Bcl-x splicing variants.
The convergence of signaling pathways on Bcl-x splicing control raised the possibility that phosphorylation may control the stability of a putative repressor interacting with the SB1 regulatory element. Consistent with this view, preventing proteasome-mediated protein degradation counteracted the splicing shift elicited by oxaliplatin or PKC inhibition. Finally, we observed that translation inhibition lifted the SB1-dependent repression and that proteasome inhibition rescued repression. Overall, our data suggest that PKC signaling stabilizes a low abundance splicing repressor, whereas the p53-dependent DNA damage response promotes its degradation through the proteasome pathway.
Bcl-x splicing control is therefore tightly coordinated through translation efficiency, proteasome-mediated protein degradation, PKC signaling, and the p53-mediated response to DNA damage, thereby providing a striking example of the convergence of several and sometimes antagonistic pathways to control a splicing decision critical to cell fate.
EXPERIMENTAL PROCEDURES
Cell Culture, Drugs, and Treatments
Human 293 cells (EcR-293, Invitrogen) were grown at 37 °C (5% CO2) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Oxaliplatin, cisplatin, and bortezomib were obtained from the Service Pharmaceutique du Centre de Chimiothérapie at the Centre Hospitalier de l'Université de Sherbrooke. Emetine dihydrochloride, cycloheximide, staurosporine, MG132, CHK2 inhibitor II, and pifithrin-α were from Calbiochem. Caffeine, okadaic acid, sodium orthovanadate, microcystine-LR, and cyclosporine-A were from Sigma-Aldrich.
Cells were treated with the indicated concentrations of individual drugs for 20 h unless specified otherwise. For methionine starvation treatments, cells were washed and incubated with DMEM depleted of methionine supplemented with 10% of dialyzed FBS (Wisent) for 20 h or as indicated. In the case of combinations of treatments, 293 cells were preincubated for 1 h prior to adding the second drug or application of Δ methionine treatment. For UV treatments, 293 cells were washed with phosphate-buffered saline, irradiated without medium with 20 J/m2 of UV at 254 nm. Medium was added immediately after irradiation.
Western Blot Analysis
To assess H2AX phosphorylation or the efficiency of knockdowns, whole cell extracts were prepared by lysing cells in Laemmli sample buffer and fractionating proteins on a polyacrylamide gel. Standard protocols were applied for Western blotting using an anti-phospho-H2AX (Upstate and Millipore), anti-ATM (Y170, Abcam), anti-CHK2 (a mixture of antibodies from Cell Signaling Technology recognizing phosphorylated Ser19, Ser33/35, Thr68, and Thr432), or anti-p53 antibody (Calbiochem). The same blots were decorated with an anti-β-actin antibody (A5316, Sigma-Aldrich) to evaluate total protein content in different lanes. To assess the efficiency of phosphatase inhibitors, total proteins were extracted from treated cells, and standard Western analysis was performed using an anti-diphosphorylated form of MAPK monoclonal antibody (M8159, Sigma-Aldrich) and anti-α-tubulin (4074, Abcam).
Immunofluorescence Microscopy
293 cells seeded in 96-well imaging plate (BD Biosciences) pretreated with poly-l-lysine were grown until a confluence of about 40% before treating with oxaliplatin for 20 h. Cells were fixed with 3.7% paraformaldehyde in PBS for 20 min, permeabilized using 0.1% of Triton X-100 for 5 min at room temperature, washed with PBS, and then blocked with 2% bovine serum albumin in PBS for 30 min. The primary antibody, a mouse anti-phospho-H2AX (1:500) was added to cells for 1 h at room temperature. Cells were washed with 0.05% Tween and PBS and then incubated for 1 h with the secondary antibody (1:250), a goat anti-mouse IgG1 Alexa Fluor 647-conjugated (Invitrogen), and a solution of Hoechst 33342 (Invitrogen). After removal of the secondary antibody and Hoechst, cells were washed with 0.05% Tween and PBS. Controls without the primary antibody were always included. Cells were examined, and images were captured using a BD Pathway 855 High-Content Bioimager (BD Biosciences) and ×20 U-Apo 340 objective (Olympus). Sixteen images for each well were acquired and analyzed using the Attovision 1.6 software (BD Biosciences), allowing fluorescence measurements of individual cells.
Minigenes and Transfection Assays
Bcl-x inserts of plasmids X2 and X2.13 were produced by PCR amplification using plasmids CMV-X2 and CMV-X2.13 (22) as templates, the Pfu-Turbo polymerase, and primers AscI-X-Fwd (GGCGCGCCTCACTATAGGGAGACCCAAGCTGGCTAG) and X-Age-Rev (CTTACCGGTGGATCCCCCGGGCTGCAGGAATTCGAT). The derived PCR products were cleaved with AscI and AgeI and ligated into SVEDA-HIV-2 vector (a generous gift of Alberto Kornblihtt) cut with the same enzymes. Plasmid transfections were carried out with polyethyleneimide (Polysciences Inc.), and conditions for transfection of 293 cells were as described (23). For knockdowns, 293 cells were transfected with an ATM-specific siRNA (AAGCACCAGUCCAGUAUUGGCdTdT) (24), a CHK2-specific siRNA (AAGAACCUGAGGACCAAGAAC) (25), and a p53-specific siRNA (GCAUGAACCGGAGGCCCAUdTdT) (Qiagen). Drugs and other treatments or transfection of Bcl-x minigenes were applied 48 h post-treatment with the siRNAs, and cells were collected 24 h after to verify depletion through Western analysis and Bcl-x splicing profiles using RT-PCR.
RNA Extraction and RT-PCR Analysis
Total RNA was extracted from treated or transfected cells with TRIzol (Invitrogen) using the procedure described by the manufacturer. The splicing profile of Bcl-x was assessed by RT-PCR. Reverse transcription was done using the OmniScript RT kit (Qiagen) with random hexamers for endogenously derived Bcl-x mRNAs, whereas oligonucleotide RT3 (GAAGGCACAGTCGAGGCTG) was used for the plasmid-derived mRNAs. One-tenth of cDNA material was used as template for the PCR. Primers X3 (ATGGCAGCAGTAAAGCAAGCG) and X2 (TCATTTCCGACTGAAGAGTGA) were used to amplify fragments of splicing isoforms derived from endogenous Bcl-x, whereas primers X34 (AGGGAGGCAGGCGACGGCGACGAGTTT) and X-Age-Rev were used for plasmid-derived transcripts. For the conventional PCR, [α-32P]dCTP (PerkinElmer Canada Inc.) was added to PCR mixtures, and amplification products were fractionated onto a 4% native polyacrylamide gel. Gels were exposed on screens that were scanned on a STORM PhosphorImager 860 (GE Healthcare). The intensity of the bands was quantified using the ImageQuant software.
For RT-PCR data and protein synthesis assays, the two-tailed Student's t test was used to compare the means between samples and their respective controls. p values are represented by asterisks (* p < 0.05; **, p < 0.01; ***, p < 0.001). The absence of asterisks indicates that the changes relative to controls are not statistically significant.
Protein Synthesis Assays
To determine the effect of inhibitors on protein synthesis, cells were treated with individual inhibitors or methionine-depleted medium for 20 h and then washed and incubated with DMEM lacking cysteine supplemented with dialyzed FBS and 5 μCi/well [35S]cysteine (PerkinElmer Canada Inc.) and inhibitors or medium lacking methionine for an additional 4 h. After collecting cells and washing them with PBS, they were treated with 10% TCA and incubated on ice for 30 min before centrifugation at 13,000 rpm for 15 min. Pellets were then treated with 0.1 n NaOH overnight at 37 °C. Aliquots of NaOH-treated samples were transferred to scintillation vials for counting. A second aliquot of each sample was used to determine protein concentration by the Lowry method. The [35S]cysteine incorporation rate was expressed as cpm/μg of TCA-precipitated protein.
RESULTS
The Impact of Oxaliplatin on Bcl-x Splicing is p53-dependent
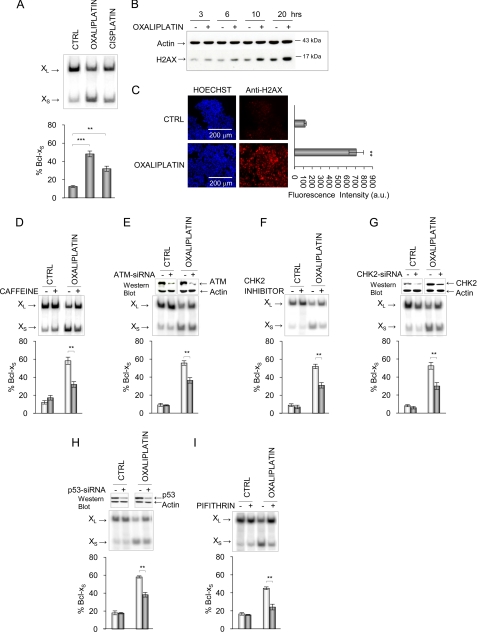
In a previous study (20), we showed that several anti-cancer agents alter the alternative splicing of Bcl-x. Among these, the genotoxic compounds oxaliplatin and cisplatin displayed the broadest spectrum of activity because they increased the production of the Bcl-xS mRNA splice variant in all cell lines that were tested, including 293 cells (Fig. 1A). Oxaliplatin probably promotes the formation of double-stranded DNA breaks because the levels of phosphorylated H2AX increased with time as judged by Western analysis (Fig. 1B) or after 24 h of treatment as detected by immunofluorescence (Fig. 1C). H2AX can be phosphorylated by the ATM and ATR kinases that sense DNA damage (26, 27), and we further observed that the Bcl-x splicing shift induced by oxaliplatin is compromised by caffeine, which inhibits the ATM/ATR kinases, and by an inhibitor of CHK2, a kinase activated by ATM/ATR (Fig. 1, D and F, respectively). Notably, the level of phosphorylated CHK2 was increased upon treatment with oxaliplatin (Fig. 1F), consistent with an induced DNA damage pathway. The role of ATM and CHK2 on Bcl-x splicing control was confirmed by RNA interference using siRNAs (Fig. 1, E and G).
FIGURE 1.
Impact of the DNA damage pathway on Bcl-x splicing. A, total RNA was extracted from 293 cells treated with 10 μm oxaliplatin or cisplatin. RT-PCR analysis was performed to amplify the Bcl-x splice products that were then fractionated in acrylamide gels (20). The Bcl-xL (456 bp) and Bcl-xS (267 bp) products are indicated. B, Western analysis of total protein with the anti-H2AX antibody (and anti-β-actin as a control) following treatment of 293 cells for various times (h) with oxaliplatin. C, immunofluorescence assays with the anti-H2AX antibody after treating 293 cells with oxaliplatin for 24 h. Hoechst staining was performed to visualize nuclei. The mean intensity of positive cells in untreated and treated cells was measured with the Pathway microscope (BD Biosciences) and is plotted in the graph on the right. D, impact on Bcl-x splicing after a pretreatment with caffeine for 1 h (7.5 μm). E, siRNA-mediated knockdown of ATM prior to the addition of oxaliplatin for 24 h in 293 cells. F, impact of the CHK2 inhibitor (10 μm). G, siRNA-mediated knockdown of CHK2 prior to the addition of oxaliplatin for 24 h in 293 cells. In H and I, 293 cells were treated with a p53-specific siRNA (catalog no. 1024849; Qiagen) or the p53 inhibitor pifithrin-α (20 μm) and then incubated with oxaliplatin. Western analysis was carried out to assess the RNAi-induced drop in ATM, CHK2, and p53 (top of G and H). Following all RT-PCR analysis in triplicate experiments, the relative abundance of the Bcl-xS splice form was plotted in graphs and expressed as mean ± S.D. (error bars). **, p < 0.01; ***, p < 0.001 compared with control or as indicated. CTRL, control.
Because p53 is a target of CHK2, we next tested the contribution of p53 in Bcl-x splicing by interfering with p53 function. The partial depletion of p53 by siRNA-mediated RNA interference promoted a significant reduction (p < 0.01) in the oxaliplatin-induced shift in 293 cells (Fig. 1H). We also tested the p53 inhibitor pifithrin-α (28) and noted a similar capacity to compromise the splicing shift elicited by oxaliplatin (Fig. 1I). The depletion or inhibition of ATM, CHK2, and p53 had no effect on Bcl-x splicing in cells that were not treated with oxaliplatin (Fig. 1, D–I). The results presented here and below have been reproduced with cisplatin, and we chose not to present the cisplatin data for reasons of space.
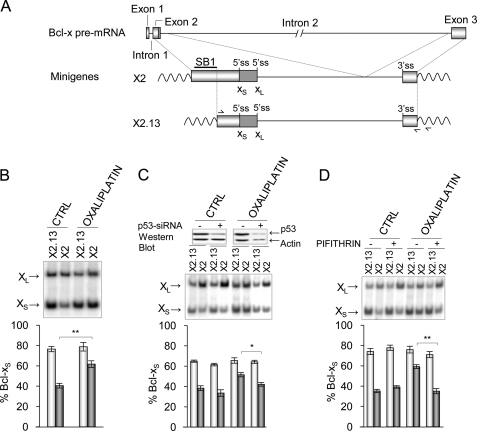
Oxaliplatin Modulates Bcl-x Splicing through the SB1 Regulatory Element
We previously reported a role for PKC in the splicing control of Bcl-x in 293 cells (23). PKC regulation of Bcl-x splicing occurs through a 361-nucleotide-long regulatory sequence (SB1) located in the first half of exon 2 (Fig. 2A). SB1 behaves as a silencer because its deletion stimulates the use of the Bcl-xS 5′ splice site and prevents further shifting upon PKC inhibition (23). To determine if oxaliplatin acts through SB1, we tested its ability to shift the splicing of transcripts derived from minigenes containing or lacking SB1 (X2 and X2.13, respectively; Fig. 2A). Notably, oxaliplatin increased the relative use of the Bcl-xS 5′ splice site only when SB1 was present (Fig. 2B). The knockdown of p53 or its inhibition by pifithrin compromised the SB1-mediated increase in the production of Bcl-xS imposed by oxaliplatin (Fig. 2, C and D). Thus, oxaliplatin alters Bcl-x splicing through a p53-dependent pathway that converges on the same regulatory element that is used by PKC.
FIGURE 2.
Oxaliplatin modulates Bcl-x splicing through the SB1 element. A, representation of the exon-intron organization of the BCL2L1 (Bcl-x) gene. The portion included in minigenes X2 and X2.13 is shown, and the SB1 region is indicated. The position of the competing 5′ splice sites and of the primers used for RT-PCR analysis of plasmid-derived transcripts is indicated. The most downstream primer shown on X2.13 hybridizes to the plasmidic portion of the sequence and was used for the reverse transcriptase step. B, 293 cells were transfected with plasmids pHIV-X2 and pHIV-X2.13. After 4 h, cells were treated for 24 h with or without oxaliplatin. C, 48 h after transfection with a p53-specific siRNA, minigenes were transfected, and 4 h later, 293 cells were treated for 24 h with or without oxaliplatin. Western analysis is shown at the top to display the relative levels of p53 and β-actin. D, 3 h after 293 cells were transfected with minigenes, they were treated for 1 h with pifithrin-α (20 μm) and then with both pifithrin and oxaliplatin for 24 h. In all cases, the Bcl-x splicing profile was measured by RT-PCR after extraction of total RNA. The percentage of Bcl-xS was plotted based on experiments performed in triplicates with S.D. values (error bars). p values are represented by asterisks (*, p < 0.05; **, p < 0.01). CTRL, control.
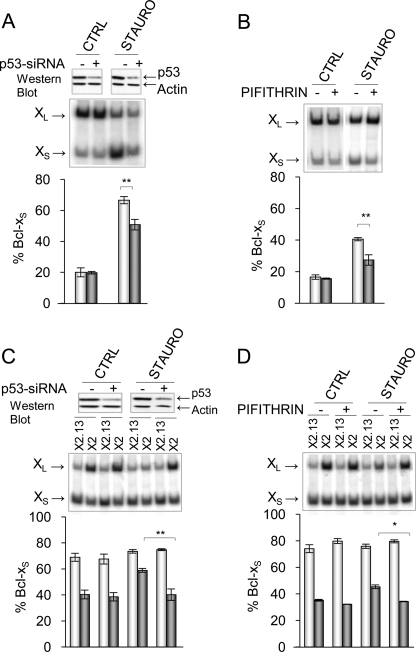
The convergence of the PKC and DNA damage response pathways was supported by showing that depleting or inhibiting p53 significantly reduced the amplitude of the splicing shift induced by staurosporine (Fig. 3, A and B) and required the SB1 element (Fig. 3, C and D). That a drop in p53 could counteract the impact of a potent PKC inhibitor indicates that the p53 pathway does not converge directly on PKC. The DNA damage and PKC pathways therefore appear distinct but may converge on a common splicing regulator.
FIGURE 3.
Convergence of the PKC and DNA damage response pathways. A and B, the impact of a p53 depletion and inhibition was tested in 293 cells treated with the PKC inhibitor staurosporine (STAURO; 60 nm). In C and D, the X2 and X2.13 minigenes were transfected into 293 cells that were treated with the siRNA against p53 or a treatment with the p53 inhibitor pifithrin. Western analysis was conducted to assess the level of p53 (top of panels). The bottom of all panels depicts labeled RT-PCR products derived from triplicate experiments plotted as described previously. p values are represented by asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars, S.D. CTRL, control.
Oxaliplatin Alters Bcl-x Splicing by Affecting the Activity of Tyrosine Phosphatases
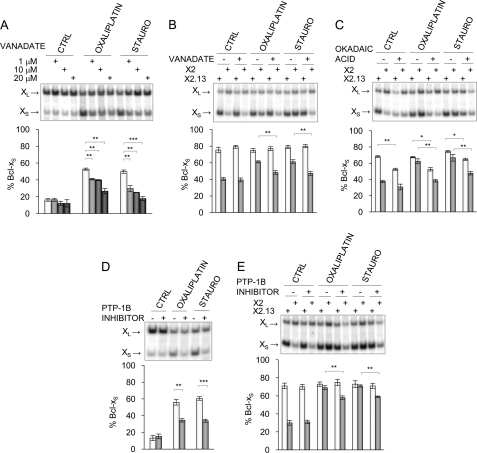
One way through which the PKC and the DNA damage response pathways may antagonistically regulate the same splicing decision is by controlling the phosphorylation of a key regulator. For example, the PKC pathway may phosphorylate a protein that represses splicing to the Bcl-xS 5′ splice site, whereas DNA damage may elicit the dephosphorylation of the same repressor. Thus, a drop in the level of a phosphorylated repressor (as when PKC is inhibited) could be compensated for by reducing the dephosphorylation rate of the repressor (as when p53 is depleted). If p53 up-regulates the activity of a phosphatase that targets this repressor, then phosphatase inhibitors should antagonize the impact of oxaliplatin and staurosporine. To test this hypothesis, we treated 293 cells with a variety of phosphatase inhibitors with the goal of identifying a class of phosphatases that control Bcl-x splicing in a manner that requires the SB1 element. Most inhibitors antagonized the splicing switch induced by oxaliplatin and staurosporine (supplemental Fig. S1). The strongest effect was obtained with the tyrosine phosphatase inhibitor orthovanadate, which nearly completely neutralized the effect of oxaliplatin and staurosporine (Fig. 4A). As observed with the inhibitors of ATM/ATR, CHK2, and p53, the phosphatase inhibitors did not affect Bcl-x splicing in cells grown in the absence of the drugs oxaliplatin and staurosporine (CTRL lanes in supplemental Fig. S1 and Fig. 4A).
FIGURE 4.
Tyrosine phosphatase inhibitors affect the SB1-mediated splicing control. A, increasing concentrations of the tyrosine phosphatase inhibitor orthovanadate (0, 1, 10, and 20 μm) were used to pretreat 293 cells for 1 h prior to the application of oxaliplatin and staurosporine (STAURO). B, 4 h post-transfection with the X2 and X2.13 minigenes, 293 cells were pretreated for 1 h with 15 μm orthovanadate before adding drugs. C, using a protocol identical to that in B, okadaic acid was used to examine the SB1-dependent response using the X2 and X2.13 minigenes. D, the PTP-1B inhibitor (50 μm) was used to pretreat 293 cells for 1 h prior to the application of oxaliplatin and staurosporine. E, 4 h post-transfection with the X2 and X2.13 minigenes, 293 cells were pretreated for 1 h with 50 μm of the PTP-1B inhibitor before adding drugs. In all cases, total RNA was extracted 24 h after the oxaliplatin and staurosporine treatments, and RT-PCR analysis was carried out as described previously. The results of triplicate experiments are plotted. p values are represented by asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars, S.D. CTRL, control.
To ask if the activity of the phosphatases occurred through the SB1 regulatory module, we tested the impact of some of the inhibitors on the splicing of transcripts derived from X2 and the SB1-lacking X2.13 minigenes. Orthovanadate antagonized the splicing shifts induced by oxaliplatin and staurosporine when Bcl-x contained the SB1 element (X2) but had no significant impact when SB1 was absent (X2.13) (Fig. 4B). In contrast, okadaic acid and the other inhibitors decreased the production of Bcl-xS on both X2 and X2.13-derived transcripts (Fig. 4C) (data not shown).
To confirm the role of protein-tyrosine phosphatases in Bcl-x splicing control, we tested a different inhibitor developed to inhibit principally PTP-1B and TC-PTP but that can also affect the activity of other tyrosine phosphatases at higher concentrations (29). The PTP-1B/TC-PTP inhibitor displayed a marked ability to counteract the Bcl-x splicing shifts induced by oxaliplatin and staurosporine (Fig. 4D). Moreover, this antagonizing activity occurred only when the SB1 regulatory element was present (Fig. 4E). Thus, our results are consistent with the view that DNA damage stimulates tyrosine phosphatases acting through the SB1 element.
The PKC and the DNA Damage Response Pathways Are Part of a Network Linked to Proteasome-mediated Protein Degradation
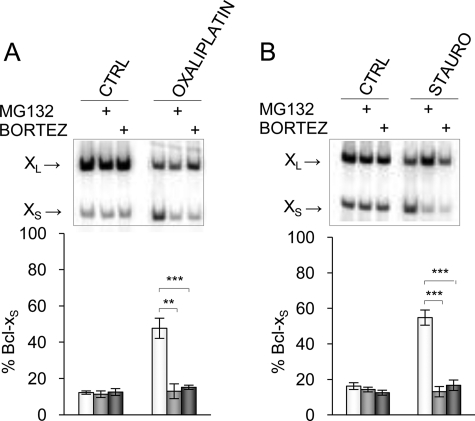
Regulating the activity of splicing modulators through phosphorylation is an emerging theme (30–34). The phosphorylation status of a protein may dictate its localization, as is the case for heterogeneous nuclear ribonucleoprotein A1 and SR proteins (31). Phosphorylation can also trigger an interaction with other proteins, as is the case for the splicing repressor SRp38 (35). Phosphorylation of the transcriptional activator SRC-3 by atypical PKC shields it from proteasomal degradation (36). Because our results support a model whereby a splicing repressor binding to SB1 exists in a non-phosphorylated and a phosphorylated state, we wanted to test if the signaling events triggered by the PKC and DNA damage pathways might be linked to protein stability. We therefore monitored the impact of 26 S subunit proteasome inhibitors on Bcl-x splicing. Although by themselves bortezomib and MG132 (37) did not affect the production of the Bcl-x splice isoforms in 293 cells, they antagonized very efficiently the Bcl-x splicing shift elicited by oxaliplatin (Fig. 5A). Bortezomib and MG132 also completely neutralized the impact of staurosporine (Fig. 5B). Inhibiting PKC or increasing the activity of tyrosine phosphatases through DNA damage may possibly shift the equilibrium toward an unphosphorylated form of the repressor that becomes a target for the proteasome, thereby lifting repression to encourage the production of Bcl-xS. In contrast, impairing proteasomal activity in these conditions would permit the accumulation of the unphosphorylated repressor, a situation that would maintain repression.
FIGURE 5.
Proteasome-mediated protein degradation affects Bcl-x splicing. 293 cells were pretreated with bortezomib or MG132 or mock-treated and then incubated with oxaliplatin (A) and staurosporine (B). Total RNA was extracted, and the Bcl-x splicing profile was measured by RT-PCR. Following quantitation, the percentage of Bcl-xS product was plotted below each panel for experiments performed in triplicates with S.D. values (error bars). p values are represented by asterisks (**, p < 0.01; ***, p < 0.001). CTRL, control.
Coupling between Translation and the SB1-mediated Control of Bcl-x Alternative Splicing
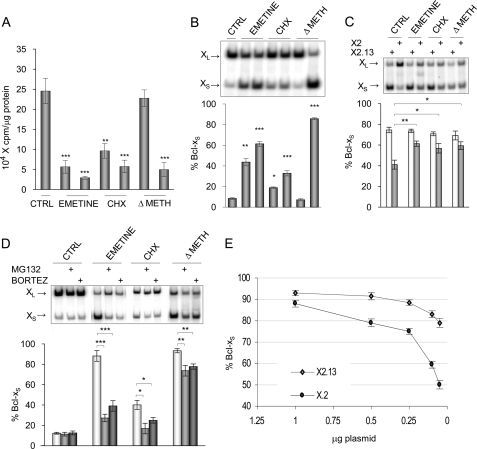
If protein degradation controls the level of a crucial splicing regulator of Bcl-x, its production may be similarly regulated. An indication that this might be the case was our previous observation that inhibiting protein synthesis with cycloheximide increased the production of Bcl-xS (23). Another group reported a similar effect on Bcl-x using emetine and cycloheximide but proposed that the shift in Bcl-x splicing might be independent of protein synthesis (38). To clarify the contribution of protein synthesis to Bcl-x splicing control, we tested the impact of a drug-free translational block. As with emetine and cycloheximide, incubating 293 cells in a medium lacking methionine compromised protein synthesis (Fig. 6A) and increased the production of Bcl-xS (Fig. 6B). Thus, a block in protein synthesis disrupts Bcl-x splicing regulation, suggesting that the availability of an important splicing regulator may be nearly limiting.
FIGURE 6.
Impact of protein synthesis on Bcl-x splicing. A, [35S]cysteine incorporation assays were carried out after applying emetine (0.2 and 0.5 μm for 20 h) or cycloheximide (CHX; 0.75 and 1.5 μg/ml for 20 h) or using a medium lacking methionine (and cysteine only for this assay) (ΔMET; 75 and 100% depleted). B, RNA from cells treated in A was extracted, and the Bcl-x splicing profile was measured by RT-PCR. C, the HIV-X2 and HIV-X2.13 Bcl-x minigenes were transfected in 293 cells. After 4 h, cells were treated for 20 h with emetine (0.5 μm), cycloheximide (1.5 μg/ml), or a medium lacking methionine. A non-treated control was included. D, 293 cells were pretreated with bortezomib or MG132 or mock-treated and then treated to inhibit protein synthesis. In all panels except A, total RNA was extracted, and the Bcl-x splicing profile was measured by RT-PCR. Following quantitation, the percentage of Bcl-xS product was plotted below each panel for experiments performed in triplicates with S.D. values (error bars). p values are represented by asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001). E, different amounts of CMV-X2 and CMV-X2.13 plasmids were transfected in 293 cells. Total RNA was extracted, and the Bcl-x splicing profile was measured by RT-PCR. CTRL, control.
Next we asked if the activity of this low abundance repressor was linked to the SB1 regulatory module. We transfected Bcl-x minigenes containing or lacking SB1 (X2 and X2.13, respectively) in 293 cells treated with medium containing emetine or cycloheximide or lacking methionine. All treatments stimulated the production of Bcl-xS but only when SB1 was present (Fig. 6C), indicating that inefficient protein synthesis counteracts the repression mediated by SB1. This result suggests that the factor existing at nearly limiting concentrations is the same one onto which converges regulation by the PKC and p53-dependent DNA damage pathways.
The above model also predicts that the impact of translation defects may be delayed by inhibiting proteasome-mediated protein degradation. Indeed, bortezomib and MG132 antagonized to various extents the increase in Bcl-xS induced by different blocks in translation (Fig. 6D). Blocking protein synthesis may therefore reduce the abundance of the repressor below a threshold level required for effective repression, thereby encouraging the accumulation of Bcl-xS. On the other hand, impairing proteasome-mediated protein degradation would delay the impact of a perturbation in protein synthesis.
The nearly limiting factor subjected to regulation by phosphatases and PKC may be a regulatory protein that binds to SB1 or any upstream components of the SB1-dependent pathway. In support of the view that this critical component may be interacting with SB1 is the observation that the SB1-mediated response was abrogated when minigenes were expressed using the stronger CMV promoter and that transfecting fewer CMV-based plasmids restored the differential between X2 and the SB1-lacking X2.13 (Fig. 6E). Assuming that fewer transcripts are produced when fewer plasmids are transfected, the SB1-mediated response would therefore be sensitive to transcript levels, consistent with the existence of a nearly limiting factor that associates with the SB1-containing X2 transcript.
Overall, our results therefore support the view that the level of the SB1 repressor rapidly becomes limiting when protein synthesis is inhibited, a situation that would couple global translation efficiency with the alternative splicing of Bcl-x.
DISCUSSION
The DNA Damage Response Pathway Controls Bcl-x Splicing
Cisplatin and oxaliplatin stimulate the production of the proapoptotic Bcl-xS mRNA variant in all of the cell lines that we have tested (20). Consistent with the genotoxic mode of action of these anti-cancer drugs, the oxaliplatin-induced shift in Bcl-x splicing requires the ATM and CHK2 kinases as well as p53, a strong indication that the DNA damage response pathway regulates Bcl-x splicing. We believe that our results provide the first clear example of a link between DNA damage and alternative splicing control in mammalian cells. Previous work has shown that the DNA-damaging drug camptothecin, an inhibitor of topoisomerase I, can alter the alternative splicing of several genes in Drosophila. RNA interference assays against the ATM/ATR kinases have confirmed a role of the DNA damage response pathway in splicing control in Drosophila (39–41). In mammals, camptothecin can modify the alternative splicing of human caspase-2 (42), and a different topoisomerase I inhibitor also alters the production of splice variants for CD44, SC35, and Clk/Sty (43). Because topoisomerase I can phosphorylate SR proteins (44), it is unclear if topoisomerase I inhibitors alter splicing control directly through SR proteins or via signaling events elicited by the DNA damage response pathway. In another case, camptothecin changed the production of H-Ras splice variants in a p53-dependent manner that implicated SC35 and components of the NMD machinery (45), further challenging the connection between DNA damage and splicing control. More recently, UV irradiation was shown to alter the splicing of Bcl-x, but the process was found to be independent of p53 (21). We have confirmed the p53-independent mode of action of UV and also noted that the UV-induced change in Bcl-x splicing occurs independently of the regulatory element required for the oxaliplatin effect (supplemental Fig. S2). Ionizing radiation can also shift the Bcl-x splicing profile (data not shown), but we have not characterized the signaling pathways in this case. UV and cisplatin can alter the alternative splicing of Mdm2, but this process is also ATM/ATR- and p53-independent (46). Given that the splicing profile of Bcl-x is not affected by the depletion of Upf proteins,2 our results strongly support the view that the p53-dependent DNA damage response pathway in mammalian cells can modulate alternative splicing control.
Because p53 can become activated by a broad range of signals, including telomere shortening, hypoxia, loss of cell contact, and oncogene activation, all of these stresses would be predicted to impact apoptosis at least in part through the control of Bcl-x splicing. Because oxaliplatin and cisplatin also shifted Bcl-x splicing in cells that lack p53 (20), it is likely that the DNA damage response pathway can activate alternative routes that will ultimately control Bcl-x splicing. Nevertheless, our results suggest that restoring p53 expression in tumors may help improve their apoptotic response when platinum compounds like oxaliplatin are used as therapeutic agents.
Convergence of the PKC and DNA Damage Response Pathways
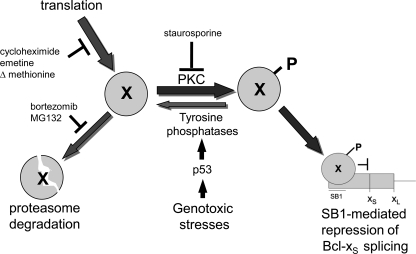
In 293 cells, the PKC pathway contributes to Bcl-x splicing regulation and acts via a 361-nucleotide region (SB1) on the Bcl-x pre-mRNA that is located ∼200 nucleotides upstream of the Bcl-xS 5′ splice site (23). Deleting SB1 stimulates the production of Bcl-xS, suggesting that SB1 recruits a factor that represses the Bcl-xS 5′ splice site (23). Notably, the impact of the DNA damage response pathway on Bcl-x splicing also requires the SB1 element, and abrogating the DNA damage response pathway by blocking p53 function compromises the ability of the cell to respond to PKC inhibition, suggesting that the PKC and the DNA damage response pathways converge on a common regulator. For this reason, we propose the existence of a repressor (X) binding to the SB1 sequence and onto which antagonistically converges the PKC and DNA damage response signaling routes (Fig. 7). In this simplistic model, the PKC pathway may activate a tyrosine kinase that produces the active form of the repressor (Xp in Fig. 7). The identity of this tyrosine kinase is unknown but unlikely to be c-Abl because c-Abl is activated by DNA damage (47, 48). The unphosphorylated form (X) could be active but would be unstable and subjected to degradation by the proteasome. In contrast, DNA damage would activate the ATM/ATR and CHK2 kinases, leading to p53 activation, which in turn would up-regulate one or several tyrosine phosphatases targeting Xp. This dephosphorylation would lead to the degradation of X to encourage the production of Bcl-xS. Consistent with this model, tyrosine phosphatase inhibitors like orthovanadate and the PTP-1B/TC-PTP inhibitor abrogated the SB1-mediated impact of oxaliplatin. Tyrosine phosphatase inhibitors neutralized the impact of the PKC inhibitor staurosporine possibly by delaying the drop in Xp. The identity of the tyrosine phosphatase(s) also remains unknown and may be revealed by targeting tyrosine phosphatases that are transcriptional targets of p53 (49). In the absence of a genotoxic stress, the PKC pathway would ensure that there is enough phosphorylated Xp to block the production of Bcl-xS in 293 cells. Abrogating p53 in the absence of a genotoxic stress would produce more Xp, but this would have no impact, given that the phosphorylated repressor is already being produced in sufficient amounts through the PKC pathway. The same reasoning applies when downstream effectors (tyrosine phosphatases) are inactivated in the absence of a genotoxic stress. The contributions of the p53 and tyrosine phosphatases therefore become apparent when the phosphorylation of the repressor is prevented by blocking PKC signaling. This putative splicing repressor would therefore sit at the receiving end of converging but antagonistic signals that dictate cell growth and apoptosis.
FIGURE 7.
Regulation of Bcl-x splicing through the SB1 element. Shown is a model depicting the pathways that affect the activity of the putative unphosphorylated or phosphorylated splicing repressor (X or Xp, respectively) that binds to SB1 and down-regulates the production of Bcl-xS (for details, see “Discussion”).
Controlling the Levels of the Putative Bcl-x Splicing Repressor
As seen with many proteins, including splicing regulators, phosphorylation can modulate interactions with other proteins, in some cases shielding the protein from the degradation machinery. A similar scenario may apply to the SB1 repressor because proteasome inhibitors counteracted the SB1-dependent impact of staurosporine and oxaliplatin. Blocking proteasome-mediated protein degradation would therefore promote the accumulation of non-phosphorylated repressor, which would repress the production of Bcl-xS even when the phosphorylation of X is blocked (Fig. 7). The identity of the SB1 repressor remains for the moment unknown. T-STAR (a Sam68-related protein also known as SLM2) is the only alternative splicing factor documented to be targeted for rapid proteasome degradation (50). However, efficient down-regulation of SLM2 by RNA interference in 293 cells did not compromise the SB1-dependent splicing response (data not shown).
Another way of controlling the level of a critical regulator is through its synthesis. We already knew that the protein synthesis inhibitors cycloheximide and emetine increased the level of Bcl-xS (23, 38). Depleting methionine reproduced the pharmacologically induced splicing shifts, suggesting that cycloheximide and emetine altered Bcl-x splicing by affecting protein synthesis. The notion that splicing decisions can be altered by defects in protein synthesis has emerged recently from a study in yeast where amino acid starvation compromised the splicing efficiency of ribosomal protein-encoding genes (51). Although the rapidity of the splicing response suggests a role for signal transduction, the signaling and splicing factors involved were not identified. Alternatively, translation defects may impact splicing decisions by reducing the level of a splicing regulator below a critical threshold concentration. This situation may be occurring with Bcl-x because the splicing shift induced by blocking protein synthesis is abrogated by inhibiting proteasome-mediated protein degradation, hence probably delaying the drop in the level of the putative regulator.
Translation inhibition is known to impact apoptosis, but the mechanisms by which this connection occurs have not been clearly established (52). Our study suggests that a partial but sustained reduction in the availability of amino acids should favor the gradual production of proapoptotic Bcl-xS through alternative splicing, ultimately triggering the apoptotic cascade. Because global translation is reduced in response to most stresses, coupling protein synthesis with Bcl-x splicing may help to adjust the apoptotic response to a variety of situations in a changing environment. Relying on a splicing regulator present at a nearly limiting concentration could be advantageous because it allows cells to react rapidly, in tune with the intensity of genotoxic stresses and the magnitude of the defects in protein synthesis.
In conclusion, we have uncovered important connections between signaling routes that converge on a splicing regulatory event controlling the production of proapoptotic Bcl-xS. Our results support a model in which a delicate balance between protein synthesis and proteasome-mediated protein degradation fixes the appropriate level of a splicing repressor. The stability of this repressor would in turn be regulated positively through the PKC signaling pathway and negatively through dephosphorylation when p53 is activated by DNA damage.
Supplementary Material
Acknowledgments
We thank A. Kornblihtt for the HIV-based plasmid; N. Rivard and D. Hunting for antibodies and inhibitors; D. Garneau for help with microscopy; and S. Benchimol, J. Barbier, and J. Venables for comments on the manuscript.
Footnotes
This work was supported by Grant MOP-93791 from the Canadian Institutes of Health Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
L. Michelle and B. Chabot, unpublished data.
REFERENCES
- 1. Adams J. M. (2003) Genes Dev. 17, 2481–2495 [DOI] [PubMed] [Google Scholar]
- 2. Green D. R., Kroemer G. (2004) Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 3. Schwerk C., Schulze-Osthoff K. (2005) Mol. Cell 19, 1–13 [DOI] [PubMed] [Google Scholar]
- 4. Jiang Z. H., Wu J. Y. (1999) Proc. Soc. Exp. Biol. Med. 220, 64–72 [DOI] [PubMed] [Google Scholar]
- 5. Wu J. Y., Tang H., Havlioglu N. (2003) Prog. Mol. Subcell Biol. 31, 153–185 [DOI] [PubMed] [Google Scholar]
- 6. Boise L. H., González-García M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nuñez G., Thompson C. B. (1993) Cell 74, 597–608 [DOI] [PubMed] [Google Scholar]
- 7. Olopade O. I., Adeyanju M. O., Safa A. R., Hagos F., Mick R., Thompson C. B., Recant W. M. (1997) Cancer J. Sci. Am. 3, 230–237 [PubMed] [Google Scholar]
- 8. Du Y. C., Lewis B. C., Hanahan D., Varmus H. (2007) PLoS Biol. 5, e276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarke M. F., Apel I. J., Benedict M. A., Eipers P. G., Sumantran V., González-García M., Doedens M., Fukunaga N., Davidson B., Dick J. E., Minn A. J., Boise L. H., Thompson C. B., Wicha M., Núñez G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11024–11028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mercatante D. R., Mohler J. L., Kole R. (2002) J. Biol. Chem. 277, 49374–49382 [DOI] [PubMed] [Google Scholar]
- 11. van der Houven van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Cáceres J. F. (2000) J. Cell Biol. 149, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buxadé M., Parra J. L., Rousseau S., Shpiro N., Marquez R., Morrice N., Bain J., Espel E., Proud C. G. (2005) Immunity 23, 177–189 [DOI] [PubMed] [Google Scholar]
- 13. Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A. L., Shokat K. M., Hentze M. W., Ronai Z. (2001) Nat. Cell Biol. 3, 325–330 [DOI] [PubMed] [Google Scholar]
- 14. Shin C., Feng Y., Manley J. L. (2004) Nature 427, 553–558 [DOI] [PubMed] [Google Scholar]
- 15. Shin C., Manley J. L. (2002) Cell 111, 407–417 [DOI] [PubMed] [Google Scholar]
- 16. Filippov V., Filippova M., Duerksen-Hughes P. J. (2007) Cancer Res. 67, 7621–7630 [DOI] [PubMed] [Google Scholar]
- 17. Shi J., Hu Z., Pabon K., Scotto K. W. (2008) Mol. Cell Biol. 28, 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chalfant C. E., Ogretmen B., Galadari S., Kroesen B. J., Pettus B. J., Hannun Y. A. (2001) J. Biol. Chem. 276, 44848–44855 [DOI] [PubMed] [Google Scholar]
- 19. Paronetto M. P., Achsel T., Massiello A., Chalfant C. E., Sette C. (2007) J. Cell Biol. 176, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shkreta L., Froehlich U., Paquet E. R., Toutant J., Elela S. A., Chabot B. (2008) Mol. Cancer Ther. 7, 1398–1409 [DOI] [PubMed] [Google Scholar]
- 21. Muñoz M. J., Pérez Santangelo M. S., Paronetto M. P., de la Mata M., Pelisch F., Boireau S., Glover-Cutter K., Ben-Dov C., Blaustein M., Lozano J. J., Bird G., Bentley D., Bertrand E., Kornblihtt A. R. (2009) Cell 137, 708–720 [DOI] [PubMed] [Google Scholar]
- 22. Garneau D., Revil T., Fisette J. F., Chabot B. (2005) J. Biol. Chem. 280, 22641–22650 [DOI] [PubMed] [Google Scholar]
- 23. Revil T., Toutant J., Shkreta L., Garneau D., Cloutier P., Chabot B. (2007) Mol. Cell Biol. 27, 8431–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Won J., Kim M., Kim N., Ahn J. H., Lee W. G., Kim S. S., Chang K. Y., Yi Y. W., Kim T. K. (2006) Nat. Chem. Biol. 2, 369–374 [DOI] [PubMed] [Google Scholar]
- 25. Krämer A., Mailand N., Lukas C., Syljuåsen R. G., Wilkinson C. J., Nigg E. A., Bartek J., Lukas J. (2004) Nat. Cell Biol. 6, 884–891 [DOI] [PubMed] [Google Scholar]
- 26. Ward I. M., Chen J. (2001) J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 27. Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001) J. Biol. Chem. 276, 42462–42467 [DOI] [PubMed] [Google Scholar]
- 28. Komarov P. G., Komarova E. A., Kondratov R. V., Christov-Tselkov K., Coon J. S., Chernov M. V., Gudkov A. V. (1999) Science 285, 1733–1737 [DOI] [PubMed] [Google Scholar]
- 29. Stuible M., Zhao L., Aubry I., Schmidt-Arras D., Böhmer F. D., Li C. J., Tremblay M. L. (2007) Chembiochem 8, 179–186 [DOI] [PubMed] [Google Scholar]
- 30. Novoyatleva T., Heinrich B., Tang Y., Benderska N., Butchbach M. E., Lorson C. L., Lorson M. A., Ben-Dov C., Fehlbaum P., Bracco L., Burghes A. H., Bollen M., Stamm S. (2008) Hum. Mol. Genet. 17, 52–70 [DOI] [PubMed] [Google Scholar]
- 31. Stamm S. (2008) J. Biol. Chem. 283, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 32. Blaustein M., Pelisch F., Srebrow A. (2007) Int. J. Biochem. Cell Biol. 39, 2031–2048 [DOI] [PubMed] [Google Scholar]
- 33. Tisserant A., König H. (2008) PLoS One 3, e1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong X. Y., Ding J. H., Adams J. A., Ghosh G., Fu X. D. (2009) Genes Dev. 23, 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng Y., Chen M., Manley J. L. (2008) Nat. Struct. Mol. Biol. 15, 1040–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yi P., Feng Q., Amazit L., Lonard D. M., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Mol. Cell 29, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crawford L. J., Walker B., Ovaa H., Chauhan D., Anderson K. C., Morris T. C., Irvine A. E. (2006) Cancer Res. 66, 6379–6386 [DOI] [PubMed] [Google Scholar]
- 38. Boon-Unge K., Yu Q., Zou T., Zhou A., Govitrapong P., Zhou J. (2007) Chem. Biol. 14, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katzenberger R. J., Marengo M. S., Wassarman D. A. (2006) Mol. Cell Biol. 26, 9256–9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marengo M. S., Wassarman D. A. (2008) RNA 14, 1681–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Katzenberger R. J., Marengo M. S., Wassarman D. A. (2009) J. Biol. Chem. 284, 10737–10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Solier S., Lansiaux A., Logette E., Wu J., Soret J., Tazi J., Bailly C., Desoche L., Solary E., Corcos L. (2004) Mol. Cancer Res. 2, 53–61 [PubMed] [Google Scholar]
- 43. Pilch B., Allemand E., Facompré M., Bailly C., Riou J. F., Soret J., Tazi J. (2001) Cancer Res. 61, 6876–6884 [PubMed] [Google Scholar]
- 44. Rossi F., Labourier E., Forné T., Divita G., Derancourt J., Riou J. F., Antoine E., Cathala G., Brunel C., Tazi J. (1996) Nature 381, 80–82 [DOI] [PubMed] [Google Scholar]
- 45. Barbier J., Dutertre M., Bittencourt D., Sanchez G., Gratadou L., de la Grange P., Auboeuf D. (2007) Mol. Cell Biol. 27, 7315–7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chandler D. S., Singh R. K., Caldwell L. C., Bitler J. L., Lozano G. (2006) Cancer Res. 66, 9502–9508 [DOI] [PubMed] [Google Scholar]
- 47. Yuan Z. M., Utsugisawa T., Ishiko T., Nakada S., Huang Y., Kharbanda S., Weichselbaum R., Kufe D. (1998) Oncogene 16, 1643–1648 [DOI] [PubMed] [Google Scholar]
- 48. Nehmé A., Baskaran R., Nebel S., Fink D., Howell S. B., Wang J. Y., Christen R. D. (1999) Br. J. Cancer 79, 1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Riley T., Sontag E., Chen P., Levine A. (2008) Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 50. Venables J. P., Dalgliesh C., Paronetto M. P., Skitt L., Thornton J. K., Saunders P. T., Sette C., Jones K. T., Elliott D. J. (2004) Hum. Mol. Genet. 13, 1525–1534 [DOI] [PubMed] [Google Scholar]
- 51. Pleiss J. A., Whitworth G. B., Bergkessel M., Guthrie C. (2007) Mol. Cell 27, 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Holcik M., Sonenberg N. (2005) Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.