Abstract
Mitochondria undergo continuous cycles of homotypic fusion and fission, which play an important role in controlling organelle morphology, copy number, and mitochondrial DNA maintenance. Because mitochondria cannot be generated de novo, the motility and distribution of these organelles are essential for their inheritance by daughter cells during division. Mitochondrial Rho (Miro) GTPases are outer mitochondrial membrane proteins with two GTPase domains and two EF-hand motifs, which act as receptors to regulate mitochondrial motility and inheritance. Here we report that although all of these domains are biochemically active, only the GTPase domains are required for the mitochondrial inheritance function of Gem1p (the yeast Miro ortholog). Mutations in either of the Gem1p GTPase domains completely abrogated mitochondrial inheritance, although the mutant proteins retained half the GTPase activity of the wild-type protein. Although mitochondrial inheritance was not dependent upon Ca2+ binding by the two EF-hands of Gem1p, a functional N-terminal EF-hand I motif was critical for stable expression of Gem1p in vivo. Our results suggest that basic features of Miro protein function are conserved from yeast to humans, despite differences in the cellular machinery mediating mitochondrial distribution in these organisms.
Keywords: Calcium-binding Proteins, Enzyme Catalysis, Fluorescence Resonance Energy Transfer (FRET), Membrane Proteins, Mitochondria, Mitochondrial Transport, Protein Domains, GTPase
Introduction
In addition to serving as the powerhouses of eukaryotic cells, mitochondria play central roles in programmed cell death and apoptosis (1), aging (2), calcium homeostasis (3), and innate immune response to viral infection (4–6). Mitochondria in many cell types are tubular and undergo cycles of homotypic fusion and fission, opposing processes that control organelle shape, copy number, and mitochondrial DNA maintenance (7, 8). Optimal cell function also relies on pathways that control mitochondrial motility and distribution. Abnormalities in mitochondrial motility and distribution can cause severe defects in highly polarized cells, like motor neurons, where mitochondria delivered to synapses maintain local ATP and calcium levels (9, 10).
Mitochondrial motility and distribution mechanisms are particularly critical during cell division because mitochondria in daughter cells cannot be generated de novo and instead arise by fission and inheritance of preexisting mitochondria from the mother cell. Movement of mitochondria is mediated by a set of conserved proteins in multicellular eukaryotes. In flies and mammals, mitochondria associate with mitochondria-specific kinesin motors (11–13) via an adaptor protein, called Milton (12, 14), which binds in turn to a tail-anchored mitochondrial outer membrane receptor, mitochondrial Rho (Miro)2 GTPase (15, 16). Recently, Miro-independent targeting of human Milton to mitochondria has also been observed (17). In budding yeast, where most organelle movement occurs along actin filaments rather than microtubules, mitochondrial transport requires a type-V myosin motor (Myo2p) (18), two Myo2p-associating proteins (Mmr1p and Ypt11p) (18, 19), and the single Miro ortholog, Gem1p (20, 21). Although these molecular machineries differ with respect to the types of cytoskeletal tracks, motors, and accessory proteins employed, they converge at the point of the Miro/Gem1p receptor on the mitochondrial surface, underscoring the importance of this receptor in mitochondrial movement.
Members of the Miro family, including Gem1p, contain two GTPase domains (GTPase I and II) that flank two bipartite Ca2+-binding EF-hand motifs (EF-I and -II) (15, 20) (Fig. 1, A and B). Because the C termini of these proteins are tail-anchored in the outer mitochondrial membrane, all four domains are exposed to the cytoplasm (Fig. 1C). Genetic studies indicate that these domains are important for the function of Drosophila Miro (16), mammalian Miro (15, 22–24), and yeast Gem1p (20). However, the predicted activities of these domains have not been experimentally established, and it is not known whether the biochemical activities of the individual domains are interdependent. In addition, it is unknown whether all four domains must be active in a single molecule for mitochondrial inheritance. In this study, we performed a structure/function analysis of the yeast Miro GTPase, Gem1p, and established that both GTPase domains are essential for mitochondrial inheritance. Conversely, Ca2+ binding by the EF-hand motifs is not required for Gem1p function. Instead, a mutation that abolishes Ca2+ binding by the N-terminal EF-I motif severely compromises protein stability.
FIGURE 1.
Domain structure of yeast Miro GTPase, Gem1p. A, a schematic view of yeast Miro GTPase, Gem1p, showing the location of the two putative GTPase domains (GTPase I and II), two EF-hand motifs (EF-hand I and II), and the transmembrane segment (TM). The amino acid positions are indicated above the structure. B, sequences of predicted Gem1p GTP-binding sites (G1 motif) and EF-hand motifs aligned with Miro homologues. Shaded characters indicate residues mutated in this study. hMiro, human Miro; dMiro, Drosophila Miro. C, schematic representation of Gem1p on the mitochondrial outer membrane (20).
EXPERIMENTAL PROCEDURES
Materials
GDP, GTP, and ATP were purchased from Sigma-Aldrich. 2′,3′-O-(N-Methylanthraniloyl)-substituted guanine nucleotides (mant-GDP and -GTPγS) were obtained from Jena Bioscience (Jena, Germany). A α-32P-labeled GTP (3,000 Ci/mmol) and 45CaCl2 (10 Ci/g) were supplied by Izotop (Budapest, Hungary) and PerkinElmer Life Sciences, respectively. Oligonucleotide DNA primers were synthesized by Genenet (Fukuoka, Japan) or the University of Utah Health Sciences Center DNA/peptide synthesis facility. All other reagents were of biochemical research grade.
Yeast Strains
The gem1Δ and gem1Δmmr1Δ yeast strains were constructed in the W303 background as described (20, 21). Standard methods were used to manipulate yeast (25, 26) and Escherichia coli (27). All mutations, disruptions, and constructs were confirmed by PCR, DNA sequencing, and Western blotting.
Cloning and Mutagenesis
To generate Gem1p bacterial expression constructs, the plasmid pRS416-GEM1 (20), which contains the complete GEM1 coding sequence, was used as a template to PCR-amplify regions encoding residues 1–616 (cytosolic domain), 1–200 (GTPase I domain), and 441–616 (GTPase II domain) with a forward primer containing a 5′ BamHI restriction site and a reverse primer containing a 3′ XhoI site, included a stop codon (TGA). The amplified fragments were cloned into a glutathione S-transferase (GST)-encoding vector, pGEX-6P-1 (GE Healthcare), to generate pGEX6P1-GEM1(1–616), pGEX6P1-GEM1(1–200), and pGEX6P1-GEM1(441–616). Mutations were introduced into pGEX6P1-GEM1(1–616) (S19N, S462N, S19N/S462N, E225K, E354K, and E225K/E354K) by site-directed mutagenesis (Stratagene).
The pRS416-MET25-GEM1 yeast plasmid contains the MET25 promoter followed by sequence encoding the GEM1 open reading frame (5′-ATG through 3′ stop codon). Plasmids harboring GEM1 domain mutations were generated by site-directed mutagenesis (Stratagene) of pRS416-GEM1 and pRS416-MET25-GEM1 (20) (this study).
Protein Expression and Purification
All proteins were expressed in E. coli strain BL21(DE3) cells. Overnight cultures (25 ml) were used to inoculate 1 liter of Luria broth (LB) medium and grown to log phase at 37 °C. Overproduction of protein was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.1 mm at 15 °C for overnight induction. The next day, cells were collected by centrifugation (6,000 rpm for 15 min), and the pellets were stored frozen (−20 °C) until purification. Bacterial pellets were resuspended in 50 mm Tris-buffered saline (pH 7.2) containing 300 mm NaCl, 5 mm MgCl2, 5 mm dithiothreitol (DTT), and 1 mm phenylmethylsulfonyl fluoride (PMSF), lysed by sonication, and centrifuged at 15,000 rpm for 15 min to obtain a soluble fraction. After clarification, the GST-tagged proteins were affinity-purified on glutathione-Sepharose 4B columns (GE Healthcare) at 4 °C. Proteins were eluted with 50 mm Tris-HCl buffer (pH 8.5) containing 300 mm NaCl, 5 mm MgCl2, and 20 mm reduced glutathione. After elution, all proteins were purified to >95% purity by gel filtration chromatography using a Sephacryl S-300 column (GE Healthcare) equilibrated with 50 mm Tris-HCl buffer (pH 7.2) containing 150 mm NaCl, 5 mm MgCl2, 5% (w/v) glycerol in the presence of 5 μm GDP.
To remove the N-terminal GST tag from the Gem1p(1–616) construct, the fusion protein was dialyzed against 50 mm Tris-HCl buffer (pH 7.2) containing 150 mm NaCl, 5 mm MgCl2, 5 μm GDP, and 1 mm DTT in the presence of GST-Precision protease (GE Healthcare) at 4 °C (18 h) and then subjected to glutathione-Sepharose 4B chromatography to remove the protease and uncleaved protein. All protein concentrations were determined by absorbance at 280 nm in 6 m guanidine hydrochloride (28).
GTP Hydrolysis Assay
The GTP hydrolysis activity of GST-Gem1p variants (final concentration of 5 μm) was assayed in 20 μl of 50 mm Tris-HCl buffer (pH 7.2) containing 100 mm KCl, 5 mm MgCl2, 1% (w/v) glycerol, 10 μm cold GTP, 5 μm GDP, 1 mm DTT in the presence of 18 nm hot GTP (α-32P-labeled) at 30 °C (without any free Ca2+ ions in the reaction buffer). At each incubation time (0, 10, 20, 30, 40, 60, 90, and 120 min), the reaction was quenched by the addition of an equal volume of a stop solution (0.5% SDS, 10 mm EDTA, and 2 mm DTT) and heating at 65 °C for 1 min. One microliter of each reaction was spotted onto a polyethyleneimine-cellulose thin layer chromatography (TLC) plate (Sigma-Aldrich) and resolved in 1 m formic acid and 0.5 m LiCl solution. The TLC plate was exposed to an imaging plate (Fuji Film, Tokyo, Japan), and the signal was detected by using an FLA 5100 phosphor imager (Fuji Film).
To determine the catalytic constant (Kcat) of GST-Gem1p variants, a 5 μm concentration of each protein was incubated with various concentrations (0, 10, 20, 50, 100, 200, 500, 750, and 1000 μm) of cold-GTP in the presence of 18 nm hot GTP. After incubation at 30 °C, the reactions were spotted onto a TLC plate and analyzed as described above. The Kcat from the GTP hydrolysis of GST-Gem1p variants was determined by Hanes-Woolf plot. In this assay, the GTP hydrolysis activity of WT Gem1p was also examined in the presence of 2 mm CaCl2 to evaluate the effect of Ca2+ binding in the enzymatic activity.
Ca2+ Binding Assay
Purified GST-Gem1p variants (10 μm) were incubated in 50 mm Tris-HCl buffer (pH 7.2) containing 150 mm NaCl, 5 mm MgCl2, and 0.15 mm 45Ca for 30 min at 25 °C. After incubation, 2 μl of each reaction was spotted onto a PVDF membrane (Millipore, Billerica, MA), washed three times with Tris-buffered saline (pH 7.2) containing 0.1% (w/v) Tween 20, exposed to an imaging plate, and analyzed using an FLA 5100 phosphor imager.
Nucleotide-binding Assay
To prepare the samples for nucleotide binding assays, the recombinant WT Gem1p(1–616) was dialyzed against 50 mm Tris-HCl buffer (pH 7.2) containing 5 mm EDTA, 150 mm NaCl, 5% (w/v) glycerol, and 1 mm DTT in order to generate nucleotide-free forms of the protein. After dialysis, 3 ml of the protein solution (1 μm) was incubated with a 5 μm concentration of either mant-GDP or -GTPγS for 15 min at 25 °C in the presence of 5 mm MgCl2. Binding of either mant-GDP or -GTPγS to the WT Gem1p(1–616) was measured using a fluorescence spectrophotometer (JASCO FP-6300, Kyoto, Japan) with the excitation wavelength (λex) at 290 nm and the emission spectra (λem) from 300 to 550 nm at 25 °C. The emission spectra were collected with Spectra Manager software (JASCO Co.).
Determination of Equilibrium Dissociation Constants
Determination of equilibrium dissociation constants (Kd) of WT Gem1p(1–616) with either GDP or GTP by fluorescence measurement was performed as described previously (29) with minor modifications. Briefly, we determined the equilibrium dissociation constant of the WT Gem1p(1–616)·mant-GDP complex (KdmGDP) prior to the determination of KdGDP and KdGTP. One micromolar WT Gem1p(1–616) solution was titrated with increasing amounts of mant-GDP (0.5, 1.0, 1.5, 2.0, 3.0, 4.0, and 5.0 μm), and the apparent fluorescence intensity (Yapp) was calculated using the following equation,
 |
where FmGDP and FGem1p(1–616)·mGDP are the fluorescence intensities (per mole) of mant-GDP and WT Gem1p(1–616)·mant-GDP complex, respectively, [mGDP]total is the total concentration of mant-GDP, and [Gem1p(1–616)·mant-GDP] is the concentration of Gem1p(1–616)·mant-GDP complex. The binding of mant-GDP to WT Gem1p(1–616) was detected as fluorescence resonance energy transfer (FRET) between the mant-GDP (λem = 445 nm) and the tryptophans of WT Gem1p(1–616) (λex = 290 nm). In Equation 1, the values of [mGDP]total and [Gem1p(1–616)·mant-GDP] were used in the following equation,
 |
where [Gem1p(1–616)]total is the total concentration of WT Gem1p(1–616). The KdmGDP and FGem1p(1–616)·mGDP values were obtained by fitting the measured fluorescence data to the following equation,
 |
After estimating the KdmGDP, the equilibrium dissociation constants of GDP and GTP for WT Gem1p(1–616) were measured indirectly by competition with mant-GDP. The data fitting was performed using the program Mathematica 6 (Wolfram Research).
Protein Extraction from Yeast and Western Blotting
An alkaline extraction method (30) was used to prepare protein samples from whole yeast cells grown to log phase at 25 °C in synthetic dextrose medium. Protein extracts prepared from strains expressing plasmid-borne Gem1p WT and mutant proteins were subjected to 8% SDS-PAGE (native GEM1 promoter, 25 μl = 1.0 A600 cells/gel lane; MET25 promoter, 12.5 μl = 0.5 A600 cells/lane). Separated proteins were transferred to nitrocellulose and incubated with affinity-purified anti-Gem1p polyclonal antibody (1:500 dilution) (Shaw laboratory). Following incubation with fluorescent secondary antibodies (IRDye 680 anti-rabbit; 1:5000 Li-Cor Biosciences, Lincoln, NE), proteins were detected and quantified using an Odyssey scanner and Odyssey 3.0 analysis software (Li-Cor Biosciences). A background band recognized by the anti-Gem1p antibody was used as an internal loading control in each lane (not shown).
Analysis of Mitochondrial Inheritance
Mitochondrial inheritance was scored at 25 °C in strains expressing a matrix-targeted form of mito-GFP (pXY142-mtGFP plus) and grown to log phase (A600 0.5–1.0) in dextrose-containing medium. Data reported are the average and S.D. from three or more independent experiments (n ≥ 100).
Microscopy and Image Acquisition
Digital fluorescence and differential interference contrast microscopic images of cells were acquired using an Axioplan 2 deconvolution microscope (Carl Zeiss Microimaging, Inc.) as described previously (20). Images were processed using Zeiss Axiovision version 4.1 and assembled into figures using Adobe Photoshop CS and Adobe Illustrator CS using only linear adjustments of contrast and brightness.
RESULTS
Both Predicted Gem1p GTPase Domains Hydrolyze Nucleotide
We bacterially expressed the cytosolic domain of wild-type GST-Gem1p (designated as GST-Gem1p(1–616); Fig. 1A), which lacks the C-terminal transmembrane domain, and assayed for its ability to hydrolyze GTP in vitro. As shown in Fig. 2, A and B, time-dependent GTP hydrolysis was observed in reactions containing GST-Gem1p(1–616) but not GST alone. Introduction of S19N or S462N substitution mutations into the GTPase I or II domains of GST-Gem1p(1–616), respectively, reduced their GTPase activities to a similar extent (Fig. 2B). GTP hydrolysis activity was completely abolished when the S19N or S462N substitutions were introduced into the same molecule (Fig. 2, A and B; S19N/S462N), establishing that both domains contribute to GTP hydrolysis by Gem1p.
FIGURE 2.
GTP hydrolysis activity of Gem1p variants. A, GST-tagged WT (GST-Gem1p(1–616)) and mutant (S19N, S462N, and S19N/S462N) Gem1 proteins were incubated with α-32P-labeled GTP at 30 °C for the indicated times, and the reactants were analyzed by TLC. Equimolar GST protein alone was used as a negative control. Position of α-32P-labeled GTP and GDP are indicated by arrows. Right bottom panel, the samples used in the GTPase assay were resolved by 10% SDS-PAGE and stained with Coomassie Blue. B, the percentage of GTP hydrolysis over the indicated time course was calculated from the intensities of the α-32P-labeled GTP and GDP signals. Filled circle, WT GST-Gem1p(1–616); open square, S19N mutant; open circle, S462N mutant; open triangle, S19N/S462N; cross, GST alone. C, substrate saturation experiments were carried out by incubating GST-Gem1p(1–616) variants (5 μm) at 30 °C with increasing concentrations of cold GTP (from 0 to 1000 μm) in the presence of 18 nm α-32P-labeled GTP. The kinetic data are plotted versus the concentrations of GTP, and the symbols used are the same as in B.
We measured the initial rate of GTP hydrolysis to determine the Kcat and Km of WT and mutant Gem1p variants. Kinetic analysis revealed that the Kcat for the S19N/S462N variant was significantly slower than that of WT (Fig. 2C and Table 1). By contrast, the individual S19N and S462N variants had more modest effects, reducing the catalytic rate of GTP hydrolysis relative to WT. Consistent with these findings, purified proteins containing only the GTPase I or II domains (Gem1p(1–200) and Gem1p(441–616)) exhibited Kcat values of ∼0.2 min−1, only slightly less than that of WT (0.24 min−1) (Table 1). Our biochemical analyses establish Gem1p as a member of the Ras GTPase superfamily, which is characterized by slower catalytic rates (31–33), rather than the Dynamin GTPase superfamily, which is characterized by faster catalytic rates (34–36).
TABLE 1.
The kinetic parameters (Kcat and Km) for Gem1p variants
| Gem1p variants | Kcat | Km |
|---|---|---|
| min−1 | μm | |
| Cytosolic domain | ||
| GST-Gem1p(1–616) (WT) | 0.24 ± 0.04 | 130.5 |
| WT + 2 mm CaCl2 | 0.26 ± 0.03 | 125.2 |
| GST-Gem1p(1–616) (S19N) | 0.16 ± 0.04 | 170.5 |
| GST-Gem1p(1–616) (S462N) | 0.11 ± 0.02 | 98.1 |
| GST-Gem1p(1–616) (S19N/S462N) | 0.02 ± 0.01 | 289.2 |
| GST-Gem1p(1–616) (E225K/E354K) | 0.23 ± 0.01 | 344.4 |
| GTPase domain | ||
| GST-Gem1p(1–200) | 0.19 ± 0.01 | 245.9 |
| GST-Gem1p(441–616) | 0.22 ± 0.02 | 91.5 |
The EF-hand Motifs of Gem1p Bind to Calcium Ions
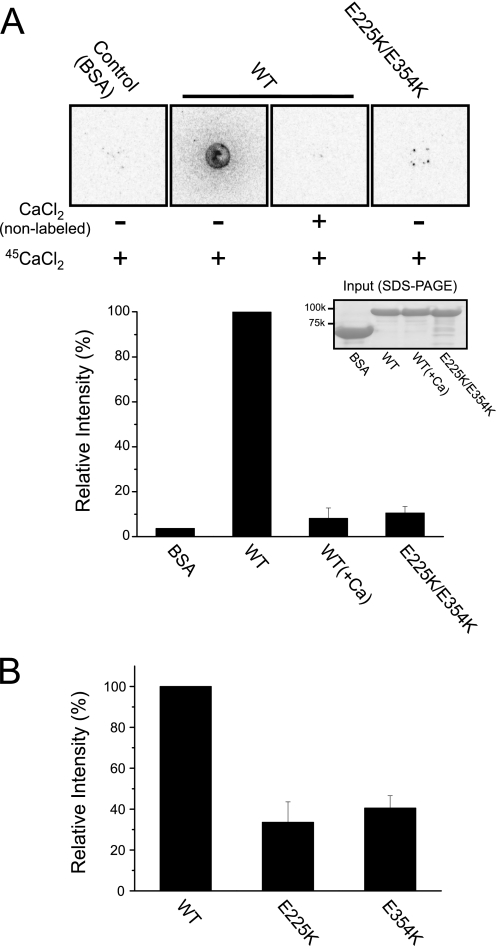
Calcium signaling mediates numerous cellular processes including mitochondrial motility (37). Although the EF-hand motifs of Miro GTPases are proposed to function as a calcium-sensitive switch (23, 24, 38), there is limited evidence that Miro GTPases physically bind calcium ions (39). To address this issue, we determined whether GST-Gem1p(1–616) containing WT or mutant (E225K or E354K) EF-hand motifs bound radioactive 45Ca ions. In this assay, the WT protein, but not the negative control (BSA) and E225K/E354K proteins (right), tightly bound to 45Ca ions (Fig. 3A). We also investigated the specificity of the protein·Ca2+ complex and found that binding of radioactive calcium ions to WT Gem1p was abolished upon competition with an excess of unlabeled competitor, verifying the specificity of the interaction. By contrast, single E225K or E354K variants exhibited reduced binding (Fig. 3B), indicating that Gem1p is a calcium-binding protein and that its calcium binding activity resides in its two EF-hand motifs. Additional experiments revealed that the hydrolysis activity of WT Gem1p was not affected by the presence of free Ca2+ ions in the reaction buffer; nor were changes observed in the GTPase activity of the E225K/E354K double mutant protein relative to that of WT protein (Table 1), establishing that nucleotide binding and hydrolysis do not require Ca2+ binding by the EF-hand motifs.
FIGURE 3.
Ca2+ binding property of Gem1p variants. A, top, 10 μm purified WT GST-Gem1p(1–616) and its E225K/E354K mutant were incubated with 0.15 mm 45Ca-labeled CaCl2 and spotted onto PVDF membrane, and binding was detected by autoradiography. The binding specificity was confirmed by addition of 200 mm unlabeled CaCl2 to the reaction. BSA was used as a negative control. Bottom, quantification of 45Ca binding to WT and EF-hand mutant Gem1 proteins, and Coomassie Blue staining of the purified proteins used in this assay. B, similar to A except showing results for the single E225K and E354K mutant proteins. Error bars, S.D.
Nucleotide Binding to Gem1p
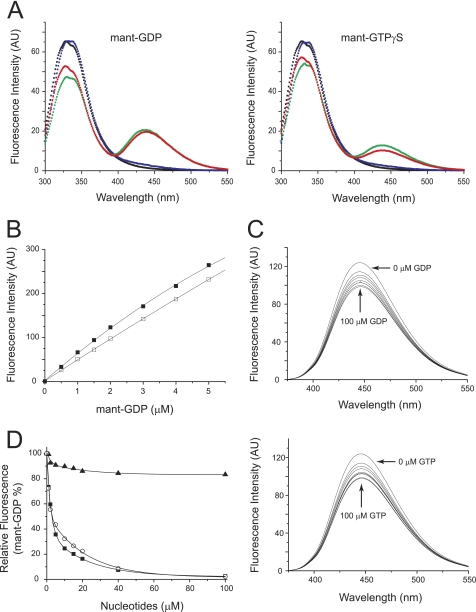
The two cytoplasmic GTPase domains in Gem1p are required for its function (20), and its GTP hydrolysis activity resides in these two domains (Fig. 2). To directly characterize their activity, we used a fluorescence assay with mant-substituted guanine nucleotides (mant-GDP and -GTPγS) to measure the affinity of nucleotide binding. These assays employed WT Gem1 protein from which GST had been proteolytically removed (designated as WT Gem1p(1–616)). In the presence of Mg2+ ions, WT Gem1p(1–616) bound to mant-GDP or mant-GTPγS (Fig. 4A, red spectra), as indicated by substantial FRET from tryptophan residues in the protein (λem = 332 nm) to the mant-nucleotides (λem = 438 nm) (black traces are negative controls showing FRET in the absence of protein). Chelating Mg2+ ions in the reaction resulted in release of the bound mant-nucleotides from WT Gem1p(1–616) and loss of FRET (blue spectra). Most importantly, reintroducing excess free Mg2+ ions resulted in decreased fluorescence intensity at 332 nm and a concomitant increase in intensity at 438 nm (green spectra), demonstrating that nucleotide binding was reversible and restored in a Mg2+-dependent manner. Titration experiments indicated that the equilibrium dissociation constant (Kd) for the mant-GDP was 0.27 μm (Fig. 4B and Table 2).
FIGURE 4.
Binding of guanine nucleotides to WT Gem1p(1–616). A, FRET from Trp residues to bound mant-GDP (left) or mant-GTPγS (right) in the WT Gem1p(1–616) complex. Fluorescence emission spectra of 1 μm WT Gem1p(1–616) in the presence (red) or absence (black) of either 5 μm mant-GDP or -GTPγS were recorded at 25 °C with 5 mm MgCl2. Blue plots represent the spectrum of the energy transfer quenched by the addition of 10 mm EDTA and subsequently restored (green) by the addition of 20 mm MgCl2 to the quenched solution, indicating that mant-nucleotides are bound to WT Gem1p(1–616). An excitation wavelength (λex) of 295 nm was used for all spectra, and fluorescence intensities are shown in arbitrary units (AU). B, titration of mant-GDP in the presence (filled squares) or absence (open squares) of 1 μm WT Gem1p(1–616). The fluorescence intensity was monitored at λem = 445 nm with excitation at 295 nm. C, competition between mant-GDP and guanine nucleotides (GDP or GTP) for binding to WT Gem1p(1–616). Emission spectra of 1 μm WT Gem1p(1–616) with 5 μm mant-GDP alone plus increasing concentrations (1, 2, 4, 5, 10, 20, 40, and 100 μm) of either GDP (top) or GTP (bottom) were monitored at 25 °C. D, the percentage of mant-GDP bound to the WT Gem1p(1–616) was plotted versus increasing concentrations of GDP (open circles), GTP (filled squares), and ATP (filled triangles).
TABLE 2.
Nucleotide binding affinities of WT Gem1p(1–616) and other GTPases by FRET
The Kd values for dynamins were derived from tetramer models.
| GTPase | Nucleotide | Kd | Reference |
|---|---|---|---|
| μm | |||
| WT Gem1p(1–616) | Mant-GDP | 0.27 ± 0.10 | This study |
| GDP | 0.78 ± 0.09 | This study | |
| GTP | 0.63 ± 0.06 | This study | |
| YihA (E. coli) | GDP | 2.7 | Ref. 29 |
| Ffh (E. coli) | GDP | 1.3 | Ref. 43 |
| GTP | 1.2 | Ref. 43 | |
| Dynamin-1 (human) | GDP | 7.4 | Ref. 44 |
| GTP | 5.4 | Ref. 44 | |
| Dynamin-2 (rat) | GDP | 7.1 | Ref. 44 |
| GTP | 13.2 | Ref. 44 |
Specificity of nucleotide binding in these experiments was established by showing that increasing concentrations of unlabeled GDP or GTP decreased the FRET signal generated by the WT Gem1p(1–616)·mant-GDP complex (Fig. 4C). Furthermore, competition for WT Gem1p(1–616) binding by added nucleotides was dependent on the guanine moiety; nucleotide exchange did not occur in the presence of ATP (Fig. 4D). Although GTP and GDP bound to WT Gem1p(1–616) with similar micromolar affinities, the protein had slightly higher affinity for GTP than for GDP (Table 2).
The EF-hand I Motif of Gem1p Is Essential for Protein Stability
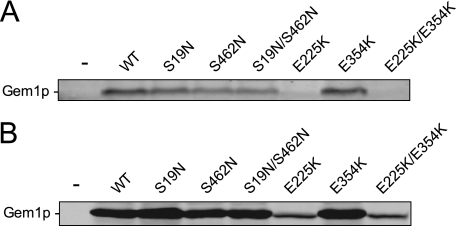
Mutation of Gem1p GTPase domains and EF-hand motifs has the potential to destabilize the mutant proteins in vivo. Using single-copy plasmids, we expressed WT and mutant Gem1p proteins from the native GEM1 promoter in a gem1Δ strain and compared steady-state protein levels by Western blotting. Although Gem1 proteins containing GTPase domain mutations (S19N, S462N, and S19N/S462N) were reproducibly detected in yeast whole cell extracts, their steady-state levels were slightly lower than WT protein (Fig. 5A). We did not detect proteins harboring the EF-I mutation E225K, although Gem1p abundance was unaffected by the EF-II mutation E354K (Fig. 5A). These data indicate that calcium binding by the N-terminal EF-hand I motif is important for Gem1p stability in vivo.
FIGURE 5.
Steady-state abundance of Gem1 WT and mutant proteins expressed in yeast. A, steady-state abundance of WT and mutant Gem1 proteins expressed from the native GEM1 promoter on a low copy (CEN) plasmid in a gem1Δ strain. B, steady-state abundance of WT and mutant Gem1 proteins expressed from the uninduced MET25 promoter on a low copy plasmid in a gem1Δ strain. Whole cell extracts separated by 8% SDS-PAGE were transferred to membrane and immunoblotted with affinity-purified anti-Gem1p polyclonal primary antibody. Protein bands were detected using a fluorescent IRDye 680-conjugated anti-rabbit secondary antibody followed by scanning on an Odyssey imaging system (Li-Cor Biosciences). The minus sign in the far left lane denotes empty vector. Extract loaded/lane in A is twice the amount loaded/lane in B.
To increase protein expression, we cloned WT Gem1p and the mutant variants behind the MET25 promoter. When expressed from MET25 without induction, the steady-state abundance of WT, GTPase domain mutant, and E354K mutant proteins was ∼80–100-fold greater than that expressed from the native GEM1 promoter (Fig. 5B).3 Most importantly, MET25 expression produced detectable steady-state levels of E225K-containing proteins (∼20-fold overexpressed), allowing functional analysis of these Gem1p variants in vivo (Fig. 5B).3
Mitochondrial Inheritance Requires the GTPase Domains but Not Ca2+ Binding to the EF-hand Motifs of Gem1p
We previously demonstrated that Gem1p and the myosin adaptor proteins Mmr1p and Ypt11p act in independent pathways to promote mitochondrial inheritance (21). Cells lacking any two of these proteins displayed mitochondrial inheritance defects such that mother cells produced daughter cells (buds) without mitochondria. This defective mitochondrial inheritance prevented release of the affected buds from mother cells and was correlated with growth defects in the double mutant strains. Strains lacking all three proteins/pathways were essentially inviable.
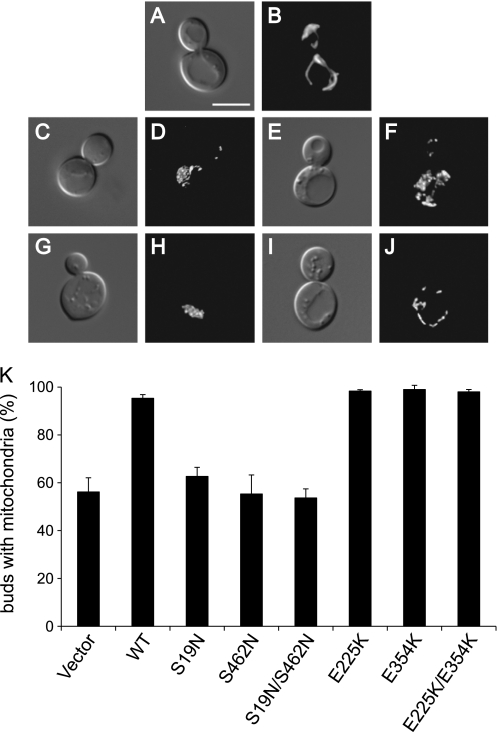
To determine the importance of Gem1p functional domains in mitochondrial inheritance, we scored the distribution of GFP-labeled mitochondria in a gem1Δmmr1Δ strain expressing WT and mutant Gem1p proteins from the uninduced MET25 promoter. In these studies, the detection of any amount of GFP-labeled mitochondria in buds was scored as successful inheritance. Cells in Fig. 6, B, D, and F, show examples of successful mitochondrial inheritance, whereas those in Fig. 6, H and J, show examples of defective mitochondrial inheritance. The gem1Δmmr1Δ double mutant displayed a strong inheritance defect in medium- and large-budded cells (only 56% of buds inherited mitochondria; Fig. 6K, vector). (The residual 56% inheritance in this strain is provided by the intact YPT11 pathway.) Expression of WT Gem1p restored mitochondrial inheritance in this strain (95% inheritance; Fig. 6K). Expression of Gem1 proteins with mutations in one or both GTPase domains (S19N, S462N, and S19N/S462N) did not rescue the gem1Δmmr1Δ defect (54–63% inheritance; Fig. 6K), consistent with the idea that GTP hydrolysis by both domains is required for Gem1p function. Significantly, mitochondrial inheritance was completely rescued by overexpressed Gem1 proteins with mutations in EF-I, EF-II, or both motifs (E225K, E354K, and E225K/E354K) (98–99% inheritance; Fig. 6K). When combined with our calcium binding studies (Fig. 3), these results provide evidence that Ca2+ binding by the two Gem1p EF-hand motifs is not essential for mitochondrial inheritance. In control experiments, scoring mitochondrial inheritance after a 4-h induction of the MET25 promoter did not alter the ability of the WT or mutant proteins to rescue gem1Δmmr1Δ inheritance defects (data not shown). Moreover, expression and overexpression of WT or mutant Gem1 proteins in a wild-type strain did not cause dominant inheritance defects or an excessive inheritance phenotype (accumulation of excess mitochondria in buds and/or depletion of mitochondria from the mother cell).
FIGURE 6.
Mitochondrial inheritance function of WT and mutant Gem1 proteins. A and B, corresponding differential interference contrast and digital fluorescence images of a wild-type strain with normal mitochondrial inheritance. C–F, gem1Δmmr1Δ cells with reduced mitochondrial inheritance. G–J, gem1Δmmr1Δ cells with defective mitochondrial inheritance. Cells are labeled with a mitochondria-targeted form of GFP (mito-GFP) and exhibit aberrant mitochondrial morphology in gem1Δmmr1Δ (21). K, quantification of mitochondrial inheritance by medium and large buds in gem1Δmmr1Δ (black bars). The presence of any mito-GFP in the bud was scored as successful inheritance. n ≥ 100. Error bars, S.D. values from three independent experiments. Bar, 5 μm.
DISCUSSION
Miro proteins are recognized as key components of the mitochondrial distribution machinery in yeast, plants, invertebrates, and mammals (7, 15, 16, 20–24, 38–40). Although genetic analyses suggest that both GTPase domains and EF-hands are required for Miro function, direct evidence for the activities of these domains is limited. Here we show that the predicted GTPase domains and EF-hand motifs in the yeast Miro, Gem1p, hydrolyze GTP and bind Ca2+ ions, respectively. By monitoring the ability of mutant proteins to rescue mitochondrial inheritance defects in vivo (Fig. 6), the nucleotide hydrolysis activities of both GTPase domains were shown to be essential for Gem1p function. Mutations that blocked Ca2+ binding to one or both EF-hand motifs did not impair mitochondrial morphology or block mitochondrial inheritance. Thus, if Ca2+ binding exerts a regulatory impact, it most likely negatively regulates the function of Gem1p in mitochondrial inheritance. We also observed that mutation of the N-terminal EF-I motif (E225K) had a dramatic effect on Gem1 protein stability (Fig. 5). Mitochondrial inheritance was unaffected in cells overexpressing WT, GTPase mutant, or EF-hand mutant Gem1 proteins, indicating that Gem1p activity is not rate-limiting in vivo and that mutant proteins do not dominantly interfere with the function of WT Gem1p or its cellular binding partners. These results address outstanding issues regarding the classification of the Gem1p GTPase domains, the roles of Ca2+ binding to Gem1p in vivo, and the conserved functions of Miro proteins.
Miro proteins were originally classified as Rho GTPases (15). A later study noted that the Gem1p N-terminal GTPase domain lacked a Rho-specific sequence insert, and the C-terminal GTPase domain sequence was not closely related to the Ras or Rho GTPase families (20). More recently, Miro proteins have been reclassified as a subfamily of the Ras GTPase superfamily (31, 38, 41). Consistent with this new classification, we showed that the rates of GTP hydrolysis by truncated Gem1 proteins containing only GTPase I or GTPase II domains are slow, with Kcat of ∼0.2 min−1 (Table 1). This poor intrinsic GTPase activity suggests that both Gem1p GTPase domains, like other members of the Ras family, utilize accessory factors in vivo to increase the rate of hydrolysis. We also observed that Gem1 proteins containing single GTPase domain mutations retained GTP hydrolysis activity (Fig. 2), indicating that the activities of the GTPase I and II domains are not interdependent. Despite this residual activity, single GTPase I and GTPase II mutations eliminated the ability of Gem1p to rescue mitochondrial inheritance defects in yeast. Additional experiments demonstrate that Gem1p with a GTPase I domain mutation could not be complemented in trans by Gem1p containing a GTPase II domain mutation.4 These combined results demonstrate that normal Gem1p function requires two active GTPase domains in a single polypeptide chain. Finally, no significant difference in GTP hydrolysis was detected when free Ca2+ ions were included in the reaction buffer or when Gem1p contained mutations in both EF-hand motifs (Table 1), suggesting that Ca2+ binding does not stimulate or inhibit the GTPase activity of one or both GTPase domains in this assay.
The EF-hand motifs of Miro proteins have never been shown to bind calcium independently; nor has the effect of Ca2+ binding on protein stability been evaluated. Here we demonstrate that point mutations in EF-I or EF-II of Gem1p reduced Ca2+ binding to a similar extent. In addition, mutation of both EF-hand motifs in a single Gem1 polypeptide chain completely abolished Ca2+ binding (Fig. 3A). Because the 45Ca binding assay does not detect low affinity Ca2+ binding (42), both EF-hands in Gem1p probably bind calcium ions with high affinity. Importantly, mutation of the EF-I motif significantly reduced the steady state abundance of Gem1p in vivo (Fig. 5). Although cellular conditions that reduce or abolish Ca2+ binding to EF-I in Gem1p have not been identified, such conditions are predicted to cause loss of protein function, presumably due to changes in protein stability and/or turnover. Consistent with this prediction, the Kcat for Gem1p GTPase activity is not altered by mutation of the EF-hands (Table 1) although the Km value is slightly increased. Gem1 proteins containing EF-hand mutations may require a higher GTP concentration to achieve a given reaction velocity because they have a less ordered structure.
Using overexpression constructs, we stably produced EF-hand mutant proteins and demonstrated that Ca2+ binding by Gem1p is not necessary for its function in mitochondrial inheritance. This result is consistent with current models for Miro protein regulation, in which Ca2+ binding to EF-hand motifs negatively regulates Miro interaction with Milton adaptor/kinesin motor complexes that promote mitochondrial movement on microtubules (24, 39). In contrast to studies performed in other systems, overexpression of WT or GTPase/EF-hand mutant forms of Gem1p had no discernable effect on mitochondrial distribution or morphology in yeast (20) (this study). Considering that yeast lack a Milton homolog and that yeast mitochondria move on actin filaments rather than microtubules, it seems likely that Miro protein function is regulated differently in distinct organisms and/or cell types. Identification of Gem1p binding partners in yeast will provide a means to study additional modes of Gem1p regulation and function.
Acknowledgments
We are grateful to members of the Shaw laboratory, especially Agnieszka Lewandowska, for helpful discussions and comments on the manuscript. We are also grateful to Takeru Nose (Kyushu University) for help with fluorescence measurements. We thank Yuko Fuchigami for technical assistance with cloning and DNA sequencing and Nasir Bashiruddin for radioisotope experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants 5T32HL007576 (to H. A. H.) and R01GM84970 (to J. M. S.). This work was also supported by the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development (P & P type D; 20301), Kanae Foundation for the Promotion of Medical Science, Astellas Foundation for Research on Metabolic Disorders, Uehara Memorial Foundation, and Takeda Science Foundation (to T. K.). Support for sequencing and oligonucleotide services at the University of Utah was provided by NCRR, National Institutes of Health, Grant M01-RR00064 (to L. Betz).
T. Koshiba, H. A. Holman, K. Kubara, K. Yasukawa, S. Kawabata, K. Okamoto, J. Macfarlane, and J. M. Shaw, unpublished data.
H. A. Holman and J. M. Shaw, unpublished data.
- Miro
- mitochondrial Rho GTPase
- mant
- 2′,3′-O-(N-methylanthraniloyl)
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- mGDP
- mant-GDP.
REFERENCES
- 1. Wang X. (2001) Genes Dev. 15, 2922–2933 [PubMed] [Google Scholar]
- 2. Raha S., Robinson B. H. (2000) Trends Biochem. Sci. 25, 502–508 [DOI] [PubMed] [Google Scholar]
- 3. Rutter G. A., Rizzuto R. (2000) Trends Biochem. Sci. 25, 215–221 [DOI] [PubMed] [Google Scholar]
- 4. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 5. Moore C. B., Bergstralh D. T., Duncan J. A., Lei Y., Morrison T. E., Zimmermann A. G., Accavitti-Loper M. A., Madden V. J., Sun L., Ye Z., Lich J. D., Heise M. T., Chen Z., Ting J. P. (2008) Nature 451, 573–577 [DOI] [PubMed] [Google Scholar]
- 6. Yasukawa K., Oshiumi H., Takeda M., Ishihara N., Yanagi Y., Seya T., Kawabata S., Koshiba T. (2009) Sci. Signal. 2, ra47 [DOI] [PubMed] [Google Scholar]
- 7. Okamoto K., Shaw J. M. (2005) Annu. Rev. Genet. 39, 503–536 [DOI] [PubMed] [Google Scholar]
- 8. Chan D. C. (2006) Annu. Rev. Cell. Dev. Biol. 22, 79–99 [DOI] [PubMed] [Google Scholar]
- 9. Hollenbeck P. J., Saxton W. M. (2005) J. Cell Sci. 118, 5411–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verstreken P., Ly C. V., Venken K. J., Koh T. W., Zhou Y., Bellen H. J. (2005) Neuron 47, 365–378 [DOI] [PubMed] [Google Scholar]
- 11. Tanaka Y., Kanai Y., Okada Y., Nonaka S., Takeda S., Harada A., Hirokawa N. (1998) Cell 93, 1147–1158 [DOI] [PubMed] [Google Scholar]
- 12. Glater E. E., Megeath L. J., Stowers R. S., Schwarz T. L. (2006) J. Cell Biol. 173, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pilling A. D., Horiuchi D., Lively C. M., Saxton W. M. (2006) Mol. Biol. Cell. 17, 2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stowers R. S., Megeath L. J., Górska-Andrzejak J., Meinertzhagen I. A., Schwarz T. L. (2002) Neuron 36, 1063–1077 [DOI] [PubMed] [Google Scholar]
- 15. Fransson A., Ruusala A., Aspenström P. (2003) J. Biol. Chem. 278, 6495–6502 [DOI] [PubMed] [Google Scholar]
- 16. Guo X., Macleod G. T., Wellington A., Hu F., Panchumarthi S., Schoenfield M., Marin L., Charlton M. P., Atwood H. L., Zinsmaier K. E. (2005) Neuron 47, 379–393 [DOI] [PubMed] [Google Scholar]
- 17. Koutsopoulos O. S., Laine D., Osellame L., Chudakov D. M., Parton R. G., Frazier A. E., Ryan M. T. (2010) Biochim. Biophys. Acta 1803, 564–574 [DOI] [PubMed] [Google Scholar]
- 18. Itoh T., Toh-E A., Matsui Y. (2004) EMBO J. 23, 2520–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Itoh T., Watabe A., Toh-E A., Matsui Y. (2002) Mol. Cell. Biol. 22, 7744–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frederick R. L., McCaffery J. M., Cunningham K. W., Okamoto K., Shaw J. M. (2004) J. Cell Biol. 167, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frederick R. L., Okamoto K., Shaw J. M. (2008) Genetics 178, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fransson S., Ruusala A., Aspenström P. (2006) Biochem. Biophys. Res. Commun. 344, 500–510 [DOI] [PubMed] [Google Scholar]
- 23. Saotome M., Safiulina D., Szabadkai G., Das S., Fransson A., Aspenstrom P., Rizzuto R., Hajnóczky G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20728–20733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X., Schwarz T. L. (2009) Cell 136, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sherman F., Fink G. R., Hicks J. B. (1986) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Guthrie C., Fink G. (1991) Methods Enzymol. 194, 1–863 2005781 [Google Scholar]
- 27. Maniatis T., Fritsch E. F., Sambrook J. (1982) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 28. Edelhoch H. (1967) Biochemistry 6, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Lehoux I. E., Mazzulla M. J., Baker A., Petit C. M. (2003) Protein Expr. Purif. 30, 203–209 [DOI] [PubMed] [Google Scholar]
- 30. Kushnirov V. V. (2000) Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 31. Bourne H. R., Sanders D. A., McCormick F. (1991) Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 32. Zhang B., Wang Z. X., Zheng Y. (1997) J. Biol. Chem. 272, 21999–22007 [DOI] [PubMed] [Google Scholar]
- 33. Albert S., Will E., Gallwitz D. (1999) EMBO J. 18, 5216–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Warnock D. E., Hinshaw J. E., Schmid S. L. (1996) J. Biol. Chem. 271, 22310–22314 [DOI] [PubMed] [Google Scholar]
- 35. Fukushima N. H., Brisch E., Keegan B. R., Bleazard W., Shaw J. M. (2001) Mol. Biol. Cell. 12, 2756–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J. (2005) J. Cell Biol. 170, 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeyaraju D. V., Cisbani G., Pellegrini L. (2009) Biochim. Biophys. Acta 1787, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 38. Reis K., Fransson A., Aspenström P. (2009) FEBS Lett. 583, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 39. Macaskill A. F., Rinholm J. E., Twelvetrees A. E., Arancibia-Carcamo I. L., Muir J., Fransson A., Aspenstrom P., Attwell D., Kittler J. T. (2009) Neuron 61, 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamaoka S., Leaver C. J. (2008) Plant Cell 20, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wennerberg K., Der C. J. (2004) J. Cell Sci. 117, 1301–1312 [DOI] [PubMed] [Google Scholar]
- 42. Sienaert I., De Smedt H., Parys J. B., Missiaen L., Vanlingen S., Sipma H., Casteels R. (1996) J. Biol. Chem. 271, 27005–27012 [DOI] [PubMed] [Google Scholar]
- 43. Jagath J. R., Rodnina M. V., Lentzen G., Wintermeyer W. (1998) Biochemistry 37, 15408–15413 [DOI] [PubMed] [Google Scholar]
- 44. Solomaha E., Palfrey H. C. (2005) Biochem. J. 391, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]