Abstract
Prostaglandin (PG) E2, a potent mediator produced in inflamed tissues, can substantially influence mast cell responses including adhesion to basement membrane proteins, chemotaxis, and chemokine production. However, the signaling pathways by which PGE2 induces mast cell chemotaxis and chemokine production remains undefined. In this study, we identified the downstream target of phosphatidylinositol 3-kinase, mammalian target of rapamycin (mTOR), as a key regulator of these responses. In mouse bone marrow-derived mast cells, PGE2 was found to induce activation of mTORC1 (mTOR complexed to raptor) as indicated by increased p70S6K and 4E-BP1 phosphorylation, and activation of mTORC2 (mTOR complexed to rictor), as indicated by increased phosphorylation of AKT at position Ser473. Selective inhibition of the mTORC1 cascade by rapamycin or by the use of raptor-targeted shRNA failed to decrease PGE2-mediated chemotaxis or chemokine generation. However, inhibition of the mTORC2 cascade through the dual mTORC1/mTORC2 inhibitor Torin, or through rictor-targeted shRNA, resulted in a significant attenuation in PGE2-mediated chemotaxis, which was associated with a comparable decrease in actin polymerization. Furthermore, mTORC2 down-regulation decreased PGE2-induced production of the chemokine monocyte chemoattractant protein-1 (CCL2), which was linked to a significant reduction in ROS production. These findings are consistent with the conclusion that activation of mTORC2, downstream of PI3K, represents a critical signaling locus for chemotaxis and chemokine release from PGE2-activated mast cells.
Keywords: Chemotaxis, Mast Cell, mTOR, mTOR Complex (mTORC), Prostaglandins, Signal Transduction
Introduction
Mast cells contribute to innate and adaptive host defense mechanisms through the release of an arsenal of inflammatory mediators upon activation induced by ligation of cell surface receptors (1–3). Inappropriate or exaggerated activation of mast cells in this manner, however, contributes to atopy, anaphylaxis, and other allergic disorders (4–6). The high affinity IgE receptor (FcϵRI),3 when aggregated through binding of a specific antigen to receptor-bound IgE, is the major receptor involved in such reactions (7, 8). However, other receptors expressed on mast cells have the capacity to significantly enhance antigen/IgE-mediated mast cell degranulation, eicosanoid production, and cytokine production, or to induce the release of these mediators by themselves (9, 10). Hence, activation of an additional receptor(s) may well contribute to mast cell-related disease states under specific conditions.
Mast cells are derived from CD13+/CD34+/CD117 (KIT)+ bone marrow progenitor cells (11) that migrate into peripheral tissues, during which time they undergo differentiation and maturation; processes controlled by chemotactic and growth factors present in the circulation and at sites of mast cells residency (12). Migration of mast cells into sites of inflammation is also a feature of chronic atopic disease and certain helminth and bacterial infections (13, 14) and may also contribute to inflammation associated with amyotrophic lateral sclerosis (15). Multiple receptors are expressed on mast cells that may contribute to the migration of mast cells, particularly to inflamed tissues. These include surface receptors for KIT ligand (stem cell factor (SCF)), chemokines such as CCL2, and agonists of various G protein-coupled receptors including prostaglandin (PG) E2. All of these receptors induce chemotaxis of mouse bone marrow-derived mast cells (BMMCs) under experimental conditions (16, 17). We have focused on SCF and PGE2 in particular, because both are generated in inflamed tissues and may thus participate in the homing of mast cells to these sites. SCF is generated from fibroblasts associated with chronically inflamed tissues (18), whereas PGE2 is produced in smooth muscle cells, respiratory cells, fibroblasts, macrophages, and mast cells, and is one of the major eicosanoids generated during inflammatory responses (19–21). In addition, PGE2 can function as a chemotactic factor for immature and mature BMMCs in vitro and in vivo (17).
We recently reported that chemotaxis of mouse BMMCs induced by SCF and PGE2 is dramatically enhanced upon co-stimulation with antigen/IgE (22). This enhancement is dependent on phosphoinositide 3-kinase (PI3K) and, in turn, Bruton's tyrosine kinase (Btk), leading to enhanced Rac- and calcium-dependent actin reorganization. Although the chemotactic responses to SCF and antigen alone were similarly regulated by PI3K and Btk, chemotaxis induced by PGE2 alone, and indeed the other GPCR agonists examined, was observed to be mediated by a PI3K-dependent, but Btk-independent, mechanism. However, the identity of the critical signaling element(s), downstream of PI3K, remains unknown.
PI3K regulates multiple downstream signaling pathways through its production of phosphatidylinositol 3,4,5-trisphosphate from phosphatidylinositol 4,5-bisphosphate and subsequent recruitment of pleckstrin homology domain-containing signaling proteins (21) such as Btk, phosphoinositide-dependent kinase-1, AKT, and phospholipase Cγ to the plasma membrane (23). Because PGE2 neither activates mast cell Btk (22) nor phospholipase Cγ (24), we hypothesized that a signaling module downstream of the phosphoinositide-dependent kinase-1/AKT axis may participate in the signaling processes regulating PGE2-mediated chemotaxis. A possible candidate is the serine threonine kinase, mammalian target of rapamycin (mTOR), which is activated through the AKT-dependent phosphorylation and consequential down-regulation of the negative inhibitor of mTOR signaling, tuberin (25, 26). Two distinct pathways are regulated by mTOR as a result of its binding to specific regulators, raptor and rictor to form, respectively, mTORC1 and mTORC2 complexes in association with other binding partners (27). The mTORC1 complex, through the phosphorylation of p76S6 kinase and 4E-BP1, primarily controls translational regulation (28), whereas mTORC2 promotes other cellular responses through the feedback phosphorylation of AKT (Ser473) (29). With respect to mast cells, the mTORC1 pathway is activated via FcϵRI and KIT and has been implicated in the regulation of KIT-mediated cytokine production and chemotaxis (30); however, a role for mTORC2 has yet to be defined. In view of the above, we have now investigated whether mTOR-regulated pathways are activated by PGE2 and might account for the observed PI3K-dependent, Btk-independent regulation of chemotaxis induced by PGE2.
Here we report that both mTORC1- and mTORC2-mediated signaling cascades are activated downstream of PI3K in mouse bone marrow-derived mast cells, following challenge with PGE2. Through the use of targeted gene knockdown and inhibition approaches, we demonstrate that the mTORC2 cascade is selectively utilized for the regulation of PGE2-mediated mast cell chemotaxis. Furthermore, mTORC2 also contributed to the PGE2-mediated production of monocyte chemoattractant protein-1 (CCL2) and PGD2. Taken together, these results show that mTORC2, but not mTORC1, is an important signaling intermediary in PGE2-mediated mast cell chemotaxis and mast cell mediator release.
EXPERIMENTAL PROCEDURES
Cell Isolation and Sensitization
Mouse BMMCs were obtained by flushing bone marrow cells from the femurs of C57BL/6 mice (The Jackson Laboratory) and then culturing the cells for 4–6 weeks in RPMI 1640 containing IL-3 (30 ng/ml) (Peprotech) as described (24, 31). BMMCs were cytokine-starved in cytokine-free medium for 16 h before experiments.
Cell Adhesion
BMMCs were cultured overnight in cytokine-free medium and stained with Calcein-AM (3 μg/ml) (Invitrogen) for 30 min in HEPES buffer (10 mm HEPES, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 0.4 mm Na2HPO4·7H2O, 5.6 mm glucose, 1.8 mm CaCl2·2H2O, and 1.3 mm MgSO4·7H2O) containing 0.04% BSA (Sigma-Aldrich). Multiwell tissue culture plates (96 wells; BD Bioscience) were precoated overnight with 5 mg/ml fibronectin (Sigma). The plates were washed three times with PBS, blocked with 5% BSA for 1 h, and then washed three times with PBS before the addition of BMMCs (2.5 × 104/well). The BMMCs were challenged with PGE2 (100 nm) for 1 h, and nonadherent cells were removed by washing the plates three times with PBS. Measurement of residual fluorescence, indicating attached cells, was accomplished using a GENios fluorescent plate reader (ReTiSoft, Mississauga, Canada) with an excitation wavelength of 492 nm and emission wavelength of 535 nm. The adherent cells (percentage of total added) were calculated as absorbance of sample/absorbance of total cell lysates × 100.
Chemotaxis Assay
Chemotaxis assays were performed using Transwell® permeable support chambers with 5.0 μm pore polycarbonate membranes on 6.5-mm inserts (Costar) placed within 24 well polystyrene plates essentially as described (22). Briefly, cytokine-deprived BMMCs (3 × 105 cells/100 μl in HEPES buffer containing 0.5% BSA) were placed in the upper support chamber. The upper chambers were then replaced in the lower chamber containing PGE2 (100 nm). After 4 h of incubation at 37 °C, the cells migrating to the lower chambers in response to PGE2 (100 nm) were collected and counted by microscopy. For the inhibitor studies, BMMCs were preincubated with the indicated inhibitors in the upper chamber and placed in 600 μl of HEPES buffer containing 0.5% BSA and indicated inhibitors for 30 min, and then the upper chambers were replaced in the lower chamber containing PGE2 (100 nm).
Measurement of Chemokine Release
For chemokine release studies, the cells (1 × 106 cells/ml) were incubated in cytokine-free RPMI medium for 16 h before exposure to PGE2 (100 nm) for 6 h in the same medium. CCL2 released into the medium was measured by mouse CCL2 Quantikine ELISA kits (R & D Systems).
PGD2 Measurements
The release of PGD2 from PGE2-stimulated cells was measured as described (32). Briefly, cytokine-deprived BMMCs were stimulated with PGE2 (100 nm) for 20 min, and then cell-free supernatants were analyzed for PGD2 by competitive enzyme immunoassay (Cayman Chemicals), according to the manufacturer's instructions. The results are presented as ng/ml released from 100,000 cells into 100 μl.
Intracellular ROS Detection
Intracellular ROS were measured as described (32, 33). Briefly, cytokine-deprived BMMCs were preincubated with or without the indicated inhibitors for 10–20 min. In specific experiments, the cells were treated with targeted shRNA constructs as described below, prior to conducting the assay. After centrifugation, the cells were incubated with dichlorofluorescein-diacetate (20 μm) (EMD Biosciences) in cytokine-free cell culture medium for 20 min at 4 °C. The cells were then washed with HEPES buffer containing 0.04% BSA and seeded at 2 × 105 cells/well in a black opaque 96-well microplate in the presence or absence of PGE2 (100 nm). 2′,7′-dichlorofluorescein (DCF) fluorescence was monitored at 37 °C in 1-min intervals for 30 min using GENios fluorescent plate reader (ReTiSoft) with an excitation wavelength of 492 nm and an emission wavelength of 535 nm. The results are expressed as relative fluorescent units.
Western Blot
Cytokine-deprived BMMCs were stimulated with PGE2 (100 nm) or the selective EP2 and EP3 PGE2 receptor agonists sulprostone (100 nm) and butaprost (100 nm) (Cayman Chemicals) for the indicated times, and the lysates were prepared as described (34). For the studies conducted with the mTOR inhibitors rapamycin (Calbiochem) and Torin (kind gift from Dr. Nathanael S. Gray, Harvard Medical School), and the PI3K inhibitors wortmannin (EMD), AS252424, and IC87114 (kind gift from Dr. Bart Vanhaesebroeck, Barts and the London School of Medicine and Dentistry), BMMCs were preincubated with the indicated inhibitors for 15–20 min and then stimulated with PGE2 (100 nm) for the indicated times. In the case of pertussis toxin (PTX), the cells were preincubated with PTX (1 μg/ml) for 4 h and then stimulated with PGE2 (100 nm). The proteins were separated by electrophoresis on 4–12% NuPAGE BisTris gels (Invitrogen). Following membrane transfer, the proteins were probed using the following antibodies: anti-phospho-AKT (Ser(P)473), anti-phospho-AKT (Thr(P)308), anti-phospho-mTOR (Ser(P)2448, anti-phospho-p70S6K (Thr(P)389), anti-phospho-4E-BP1 (Thr(P)37/46), anti-phospho-ERK1/2 (Thr(P)202/Tyr(P)204), anti-phospho-JNK (Thr(P)183/Tyr(P)185), anti-phospho-p38 (Thr(P)180/Tyr(P)182) (Cell Signaling), anti-EP3, anti-Syk (Santa Cruz), and anti-COX1 (Cayman). To normalize protein loading, the membranes were stripped and probed for Syk or alternatively, identically loaded gels were probed for Syk.
Intracellular Ca2+ Determination
Changes in cytosolic Ca2+ levels were determined in cytokine-deprived BMMCs following loading of the cells with Fura-2 AM ester (Molecular Probes) as described (31). Briefly, the cells were loaded with Fura-2 AM (2 μm) for 30 min at 37 °C, rinsed, resuspended in HEPES buffer containing 0.04% BSA and sulfinpyrazone (0.3 mm) (Sigma-Aldrich), and then placed in a 96-well black culture plate (2 × 104 cells/well) (CulturPlat-96 F; PerkinElmer Life Sciences). Fluorescence was measured at two excitation wavelengths (340 and 380 nm) and an emission wavelength of 510 nm. The ratio of the fluorescence readings was calculated following subtraction of the fluorescence of the cells that had not been loaded with Fura-2 AM.
Measurement of F-actin (Polymeric, Filamentous Actin) Content by Flow Cytometry
Cytokine-deprived BMMCs (1 × 106 cells/sample) were washed and then challenged with PGE2 (100 nm) for 2 min as indicated. For the inhibitor studies, the cells were preincubated with the indicated inhibitors for 15–20 min prior to challenging the cells with PGE2 (100 nm) for 2 min. The cells were then fixed at room temperature by the addition of 1 ml of 4% paraformaldehyde for 15 min and then permeabilzed with 0.1% saponin, PBS for 5 min and stained with 2 μg/ml of FITC-labeled phalloidin (Sigma) in 1% BSA, 0.1% saponin, PBS for 1 h in the dark at room temperature. After washing three times with PBS, cellular F-actin content was determined using FACScan flow cytometer by gating on 10,000 living cells.
Fluorescence Microscopy
For imaging, cytokine-starved BMMCs (50,000/sample) were stimulated with PGE2 (100 nm) for 2 min. After stimulation, the cells were immediately attached to glass slides using a cytospin centrifuge (450 rpm, 3 min, room temperature). The cells were then fixed and permeablized and stained with FITC-labeled phalloidin as previously described under “Measurement of F-actin (Polymeric, Filamentous Actin) Content by Flow Cytometry.” After washing the slides carefully with PBS, imaging was obtained using fluorescence microscopy.
Lentivirus shRNA Transfection of 293T Cells and Transduction of BMMCs
The following raptor or rictor-targeted shRNAs were purchased from Sigma (Sigma-Aldrich): CGGGCCCGAGTCTGTGAATGTAATCTCGAGATTACATTCACAGACTCGGGCTTTTTG (TRCN0000077472, raptor), CGGGCCATCTGAATAACTTCACTACTCGAGTAGTGAAGTTATTCAGATGGCTTTTTG (TRCN0000123397, rictor), CCGGGCCGCTATTGATAATGATGTTCTCGAGAACATCATTATCAATAGCGGCTTTTT (TRCN0000026617, EP3), CCGGGTGAGCTACTATACTCGCATTCTCGAGAATGCGAGTATAGTAGCTCACTTTTTG (TRCN0000067899, Cox1), and CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT (SHC002, control nontarget control vector).
The packaging vector (MissionTM lentiviral packaging mix; Sigma) and the pLKO1 transfer vectors with raptor, rictor, or control shRNA (8 μg) were co-transfected into 293T cells (15 × 106 cells) with FuGENE 6 transfection reagent (48 μl) (Roche Applied Science) and lentivirus packaging mix (78 μl) (Sigma) in antibiotic-free DMEM (545 μl). The transfected 293T cells were grown in DMEM containing FBS (10%) and l-glutamine (4 mm). Following 16–19 h of transfection, medium was removed and replaced with fresh DMEM including penicillin (100 units/ml) and streptomycin (100 μg/ml). After 48–72 h transfection, the viruses were collected by centrifugation (25,000 rpm, 1 h 40 min, 4 °C), and then the resulting pellet was resuspended in 3 ml of BMMC complete medium including SCF (30 ng/ml) and IL-3 (30 ng/ml) rather than IL-3 alone to improve transduction efficiency. Transduction of BMMCs (3–4 weeks old) was conducted by transferring the 3 ml of the resuspended virus with hexadimerthrine bromide (10 μg/ml) to a T75 culture flask containing 1 × 107 BMMCs in 10 ml of complete BMMC medium. After 24 h, the medium was changed to virus-free complete BMMC medium, and puromycin selection was initiated (1.2 μg/ml (Sigma)). The experiments were conducted on days 3 and 4 (for shRictor and shRaptor), on days 5–7 (for shCOX1), or on days 7–10 (for shEP3) after antibiotic selection.
Rac Activation Assay
Affinity precipitation with GST-PBD (p21-binding domain) was performed using PAK-1 PBD (Rac effector protein, p21 activated kinase-1) assay kit (Upstate) according to the manufacturer's instructions. BMMCs were preincubated with Torin or rapamycin (100 nm) for 15 min and then stimulated with PGE2 (100 nm) for 2 min, and the reaction was terminated by adding cold PBS. The cells were lysed with MLB buffer, and then BMMC lysates were mixed with 10 μl of GST-PBD bound to glutathione-agarose and incubated for 4 h at 4 °C. For a positive control, the cell lysates were incubated for 15 min at 30 °C with 100 μm GTPγS in the presence of 10 mm EDTA. For a negative control, 1 mm GDP was added. GTPγS-loaded lysates were incubated with GST-PBD for 30 min at 4 °C. Finally, precipitates were washed three times with MLB and suspended in Laemmli sample buffer. The proteins were separated on 4–12% NuPAGE BisTris gels, transferred onto nitrocellulose membrane, and blotted with anti-Rac1 antibody. To establish equal Rac content in the reactions, cell lysates from each sample were probed by immunoblot analysis before reaction with GST-PBD.
Statistical Analysis
The data are represented as the means ± S.E. The statistical analyses were performed by unpaired Student's t test. The differences were considered significant when p < 0.05. The n values represent the number of experiments from multiple preparations.
RESULTS
PGE2 Induces Activation of mTORC1 and mTORC2 in Mouse BMMCs
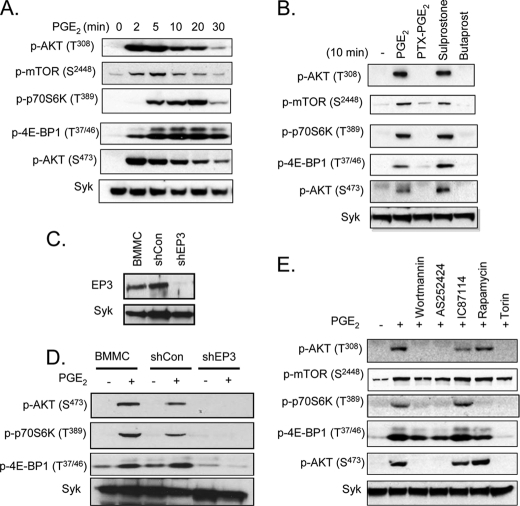
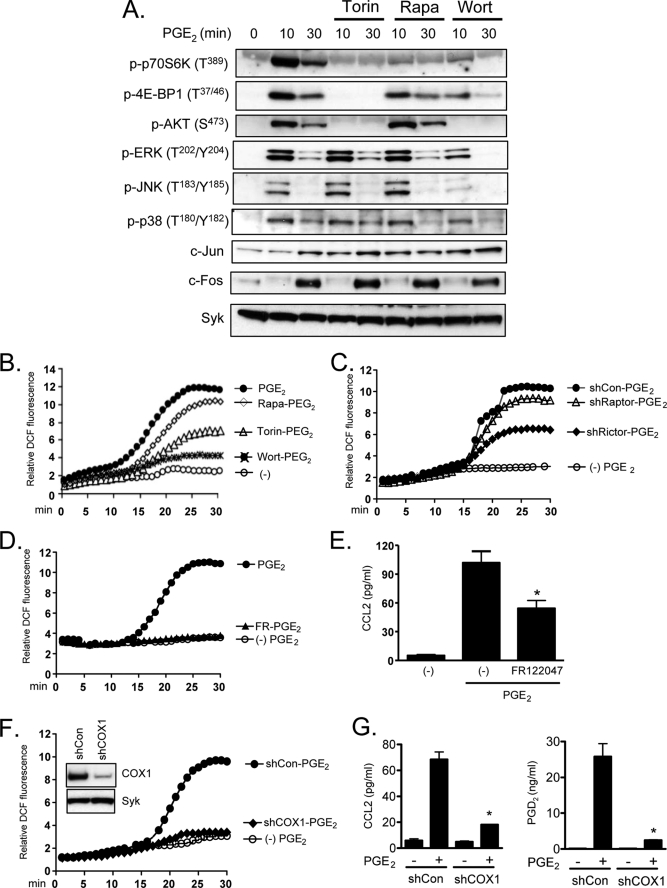
To explore the potential role of the mTOR signaling pathways in PGE2-induced chemotaxis in mast cells, we first examined whether PGE2 activates mTORC1 and mTORC2 in mouse BMMCs. The cells were stimulated for periods up to 30 min with PGE2. The lysates were then prepared and examined for phosphorylation of AKT (Thr308) and mTOR (Ser2448) as indicators of PI3K and mTOR activation, respectively. The activation of mTORC1 and mTORC2 were monitored by the phosphorylation of p70S6K (Thr389) and 4E-BP1 (Thr37/46), in the case of mTORC1, and AKT (Ser473), in the case of mTORC2. By these criteria, PI3K, mTOR, mTORC1, and mTORC2 were activated following PGE2 stimulation (Fig. 1A). The phosphorylations occurred rapidly, reaching a maximum between 2 and 5 min except for phosphorylation events related to mTORC1 activation (p-p70S6K and p-4E-BP1), which reached a maximum 10–20 min after PGE2 stimulation.
FIGURE 1.
PGE2 activates mTOR signaling via EP3. A, cytokine-deprived BMMCs were stimulated with PGE2 (100 nm) for the indicated time. B, cytokine-deprived BMMCs were stimulated with PGE2 (100 nm), sulprostone (100 nm), or butaprost (100 nm) for 10 min. In the case of PTX, the cells were preincubated with PTX (1 μg/ml) for 4 h and then stimulated with PGE2 (100 nm) for 10 min. C and D, for shRNA studies, BMMCs were transduced by lentivirus expressing shRNA for control (shCon) or shRNA for EP3 (shEP3) as described under “Experimental Procedures.” To test knockdown of EP3, total cell lysates were subjected to Western blotting with anti-EP3 antibody (C). Lentivirus-transduced BMMCs were stimulated with PGE2 (100 nm) for 10 min (D). E, for the inhibitor studies, BMMCs were preincubated with the indicated inhibitors (wortmannin (100 nm), rapamycin (100 nm), Torin (100 nm), AS252424 (3 μm), or IC87114 (3 μm)) for 20 min and then stimulated with PGE2 (100 nm) for 10 min. After stimulation, protein lysates were subjected to Western blotting. To normalize protein loading, the membranes were stripped and probed for Syk, or alternatively identically loaded samples were probed for Syk. The blots are representative of three independent experiments.
PGE2-induced Activation of mTOR in BMMCs Is Mediated by Gαi-linked EP3 Receptor and PI3K
The biological actions of PGE2 are mediated by four different GPCRs: EP1, EP2, EP3, and EP4 (35). The EP2 and EP4 receptors, which signal via by the GPCR Gαs subunit, are known to inhibit mast cell mediator release through increased cAMP production (36). In contrast, EP1 and EP3 receptors activate mast cells through Gαq and Gαi, respectively, to mobilize Ca2+ (37). Thus, EP3-induced mast cell mediator release is sensitive to PTX (35). To identify the receptor and G protein responsible for PGE2-mediated mTOR activation, we examined whether this response was mimicked by either the EP3 agonist, sulprostone, or the EP2 agonist, butaprost. Sulprostone (100 nm) induced the phosphorylation of AKT (Thr308), mTOR, p70S6K, 4E-BP1, and AKT (Ser473) to the same extent as PGE2 (Fig. 1B). The responses to PGE2 were blocked in PTX-treated cells, and butaprost failed to induce these responses. Together, these data are consistent with the conclusion that PGE2 activates the mTOR cascades via the Gαi-linked EP3 receptor. To verify this conclusion, we explored the ability of PGE2 to enhance indices of mTORC1 and mTORC2 activation in cells treated with EP3-targeted shRNA. As shown in Fig. 1C, the EP3-targeted shRNA construct markedly reduced the expression of EP3 in the BMMCs but did not alter the expression of Syk. This reduction was associated with a significant decrease in PGE2-mediated phosphorylation of p70S6K, 4E-BP1, and AKT (Ser473) (Fig. 1D), demonstrating that the ability of PGE2 to respectively enhance mTORC1 and mTORC2 activation is indeed mediated by the EP3 receptor.
To confirm that PGE2-mediated activation of mTORC1 and mTORC2 in mast cells was dependent on PI3K activation, we examined the ability of the PI3K inhibitor, wortmannin, to block phosphorylation of components of the mTOR signaling cascade. As predicted, wortmannin completely blocked the PGE2-induced phosphorylation of AKT at Thr308, demonstrating effective inhibition of PI3K (Fig. 1E). Similarly, phosphorylation of mTOR, p70S6K, and AKT (Ser473) was also completely blocked or, in the case of mTOR, reduced to prestimulus levels. However, as reported for antigen and SCF (30), the phosphorylation of 4E-BP1 was only partially reduced by wortmannin, suggesting that phosphorylation of 4E-BP1 may be only partially dependent on PI3K activity.
PI3K-linked responses in mast cells are described to be largely dependent on two PI3K isoforms: PI3Kδ, which is responsible for transducing signals elicited by the FcϵRI and KIT (23, 38), and PI3Kγ, which is responsible for transducing signals elicited by GPCRs (10). The PI3Kγ inhibitor, AS252424, and the PI3Kδ inhibitor, IC87114, have been demonstrated to selectively inhibit responses in mast cells elicited by these respective isoforms (39). As shown in Fig. 1E, the ability of PGE2 to induce phosphorylation of AKT (Thr308), mTOR, 4E-BP1, and AKT (Ser473) were reduced by AS252424 to the same levels as that observed with wortmannin. However, IC87114 had little effect on these parameters. Taken together, these data demonstrate that PGE2 induces mTORC1 and mTORC2 activation in mast cells through PI3Kγ downstream of the Gαi-coupled EP3 receptor.
Rapamycin and Torin Inhibition of PGE2-induced Activation of mTOR in BMMCs
Rapamycin is reported to inhibit mTORC1 activity while having a minimal effect on mTORC2 signaling following short term exposure (40). We (30) and others (40), however, have observed that rapamycin only partially inhibits 4E-BP1 phosphorylation. Recently an improved mTOR inhibitor, Torin, has been described that effectively inhibits mTORC1-mediated responses, including 4E-BP1 phosphorylation and mTORC2 activation (40). We examined the ability of both compounds to block the PGE2-induced indices of mTORC1 and mTORC2 activation in BMMCs. Rapamycin effectively inhibited mTORC1-mediated p70S6K phosphorylation and reduced the phosphorylation of 4E-BP1 to that observed in the presence of the PI3K and PI3Kγ inhibitors (Fig. 1E), while having little effect on the mTORC2 pathway. In contrast, Torin blocked both mTORC1 and mTORC2 activities (Fig. 1E). These compounds were further employed to explore the role of mTORC1 and mTORC2 in PGE2-mediated mast cell responses.
PGE2-induced Mast Cell Chemotaxis Is Mediated via mTORC2 but Not mTORC1
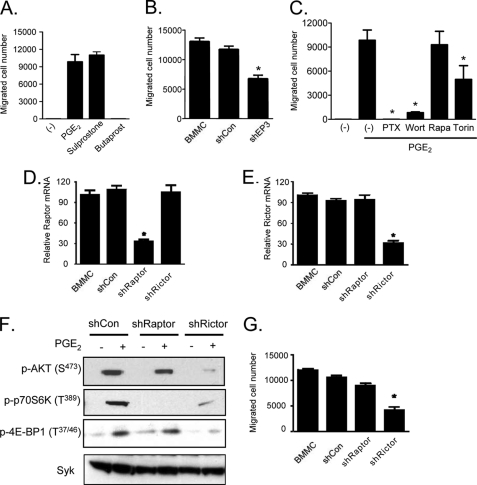
Having demonstrated that PGE2 and the EP3 agonist, sulprostone, but not the EP2 agonist, butaprost, induced mTOR activation, we next determined the relative abilities of these agonists to induce chemotaxis of mouse BMMCs. As reported (17, 22), PGE2 robustly stimulated BMMC chemotaxis (Fig. 2A). As was the case for mTOR activation, this response was mimicked by sulprostone but not by butaprost (Fig. 2A). Similarly, BMMCs treated with EP3-targeted shRNA displayed a reduced chemotactic response to PGE2 (Fig. 2B). Furthermore, as reported (17, 22), the ability of PGE2 to induce chemotaxis was also prevented by prior incubation with PTX (Fig. 2C). Thus, the Gαi-linked EP3 receptor similarly regulates both mTOR activation and chemotaxis.
FIGURE 2.
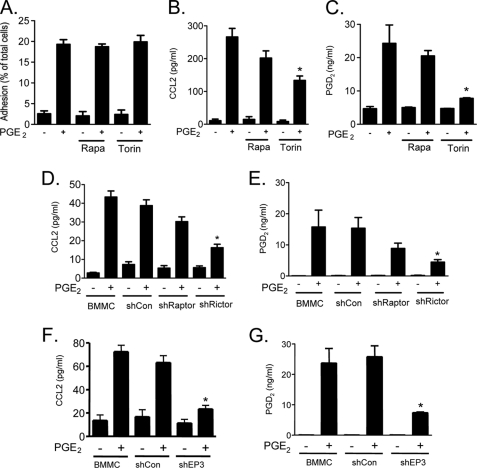
PGE2-induced chemotaxis is mediated by mTORC2. A, cytokine-deprived BMMCs were prepared, and the chemotaxis assay was performed in response to 100 nm of PGE2, sulprostone, or butaprost. B, BMMCs were transduced with lentivirus expressing shRNA for EP3, and the chemotaxis assay was performed in response to 100 nm of PGE2. C, BMMCs were preincubated with the indicated inhibitors (wortmannin (Wort), rapamycin (Rapa), or Torin (100 nm, 30 min) or PTX (1 μg/ml, 4 h)), then washed, and used for the PGE2 (100 nm)-induced chemotaxis assay as described under “Experimental Procedures.” D–G, BMMCs were transduced with lentivirus expressing control shRNA (shCon), shRNA for raptor (shRaptor), or shRNA for rictor (shRictor) as described under “Experimental Procedures.” To establish knockdown of raptor or rictor, cDNA was prepared from the cells, and real time PCR was performed using commercial TaqMan raptor or rictor probes (D and E). To confirm the knockdown, raptor or rictor's downstream molecule phosphorylation was tested. The cells were stimulated with PGE2 (100 nm) for 10 min, and protein lysates were subjected to Western blotting (F). After confirming raptor and rictor knockdown, the PGE2-induced chemotaxis assay (G) was performed. The results are the means ± S.E. of three separate experiments. *, p < 0.05 for comparison with PGE2 alone stimulation (C) or with shCon (B, D, E, and G) by Student's t test.
To investigate a potential link between PGE2-mediated mTOR activation and chemotaxis in mast cells, we examined the inhibitory activities of wortmannin, rapamycin, and Torin on the PGE2-mediated chemotactic response. Wortmannin markedly inhibited PGE2-mediated BMMC chemotaxis, as reported (22), whereas rapamycin had no effect. Torin, in contrast, significantly reduced the PGE2 response (Fig. 2C). These data thus provide evidence for a role for mTORC2, in addition to PI3K, in the regulation PGE2-mediated mast cell chemotaxis.
To verify this conclusion, shRNA technology was used to knock down rictor and raptor expression in BMMCs. Knockdown was confirmed by real time PCR, because the available antibodies against human rictor and raptor were ineffective in recognizing mouse rictor at the appropriate molecular weight. The shRNA targeting raptor (Fig. 2D) effectively knocked down raptor expression while having no effect on rictor expression (Fig. 2E), whereas shRNA targeting rictor had the opposite and intended effects on raptor and rictor expression (Fig. 2, D and E). A nontargeted control shRNA had no effect on the expression of either molecule. The ability of knockdown of raptor or rictor to respectively reduce mTORC1 and mTORC2 activities was corroborated by the inhibition of downstream signaling. As predicted and as shown in Fig. 2F, rictor-targeted shRNA markedly inhibited AKT(Ser473), and raptor-targeted shRNA significantly reduced p70S6K (Thr389) phosphorylation. Unexpectedly, although the rictor-targeted shRNA had no effect on raptor expression, it reduced both 4E-BP1 and p70S6K phosphorylation in a manner similar to that observed with the mTORC1/mTORC2 inhibitor, Torin. These data likely reflect feedback control through the regulation of AKT activation. Regardless, taken together with the real time PCR data, these data support the conclusion that raptor- and rictor-targeted shRNA constructs effectively reduce mTORC1 and mTORC2 signaling.
As shown in Fig. 2G, neither control shRNA nor the raptor-targeted shRNA had any effect on PGE2-mediated mast cell chemotaxis. However, the shRNA targeting rictor significantly reduced the chemotactic response. Taken together, the inhibitor and shRNA data thus support the conclusion that Gαi-linked EP3 receptor-mediated chemotaxis is independent of mTORC1 but is at least in part regulated by mTORC2. Nevertheless, the residual activity seen in cells treated with mTORC2 inhibitors and rictor-targeted shRNA suggests that other PI3K-dependent pathways may also contribute to this response.
The Role of PI3K and Rictor in PGE2-induced Actin Polymerization
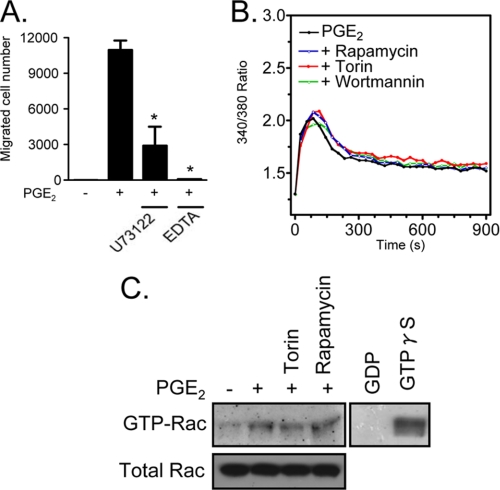
The generation of an intracellular calcium signal and actin rearrangement are both linked to chemoattractant-induced mast cell migration (22, 41). The relationship of the mTOR pathway to these two events during PGE2-induced chemotaxis was investigated. As expected from previous studies (22), inhibition of the calcium signal through treatment of the cells with the phospholipase Cγ inhibitor U73122 or through chelation of extracellular calcium with EDTA markedly attenuated PGE2-mediated BMMC chemotaxis (Fig. 3A). Nevertheless, inhibition of mTORC1 and mTORC2 with rapamycin and Torin had no effect on the PGE2-induced calcium signal (Fig. 3B).
FIGURE 3.
The role of calcium and Rac in BMMC chemotaxis. A, cytokine-deprived BMMCs were incubated with the indicated inhibitors (U73122 (1 μm) and EDTA (5 mm)) for 30 min, and then the chemotaxis assay in response to PGE2 (100 nm) was conducted. *, p < 0.05 for comparison with PGE2 alone stimulation by Student's t test. B, cytokine-deprived BMMCs were loaded with Fura-2 AM and then preincubated with the indicated inhibitors (100 nm) for 10 min. The changes in intracellular Ca2+ levels were then determined after challenge with PGE2 (100 nm). C, Rac activation was determined using a PAK-1 PBD assay kit. BMMCs were preincubated with Torin or rapamycin (100 nm, 15 min) and then stimulated with PGE2 (100 nm) for 2 min. The cell lysates were mixed with GST-PBD bound to glutathione-agarose and incubated for 4 h at 4 °C. For positive and negative controls, the cell lysates were incubated with 100 μm GTPγS or 1 mm GDP, respectively. The precipitates were washed, and the proteins were separated and blotted with an anti-Rac1 antibody. To establish equal Rac content in the reactions, cell lysates from each sample were probed by immunoblot analysis prior to reaction with GST-PBD. The results are the means ± S.E. of three separate experiments. The data from B and C are representative of three independent experiments.
In addition to the calcium signal, we have previously demonstrated (22) that antigen enhances PGE2-mediated chemotaxis through the Btk-dependent potentiation of Rac activation. However, as can be seen from Fig. 3C, although PGE2, induces Rac activation, this response was not blocked by either rapamycin or Torin, indicating that, although both calcium and Rac activation may contribute to the chemotactic response, the requirement for mTORC2 for the PGE2-mediated chemotactic response was independent of these processes.
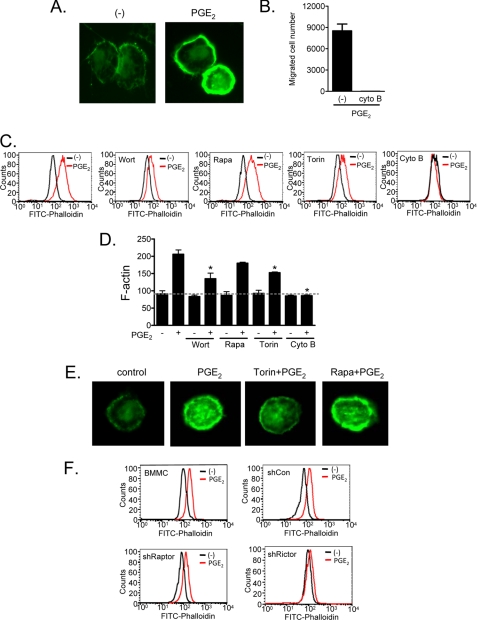
We therefore next examined whether mTORC2 contributed to actin reorganization. The necessity of cytoskeletal rearrangement, as demonstrated by actin polymerization in PGE2-stimulated cells (Fig. 4A), was confirmed by the inhibition of chemotaxis by the actin polymerization inhibitor, cytochalasin B (Fig. 4B). The effects of inhibitors on PGE2-induced actin polymerization were examined by FACS analysis of phalloidin-stained cells, which indicates polymerized F-actin but not nonpolymerized G-actin (Fig. 4, B and C). Wortmannin, Torin, and cytochalasin B suppressed PGE2-induced actin polymerization, whereas rapamycin had minimal effect (Fig. 4, C and D). Similarly, the accumulation of polymerized actin induced by PGE2 at the cell periphery, observed by fluorescence microscopy, was markedly reduced by Torin pretreatment but not by rapamycin (Fig. 4E). Consistent with these effects of Torin and rapamycin, shRNA-induced knockdown of rictor markedly reduced PGE2-dependent actin polymerization, whereas shRNA-induced knockdown of raptor minimally affected this response (Fig. 4F). From these data we conclude that mTORC2, but not TORC1, regulates PGE2-mediated chemotaxis by facilitating actin polymerization but by a mechanism independent of calcium mobilization and Rac activation.
FIGURE 4.
mTORC2 regulates PGE2-induced actin polymerization. A, cells were stimulated with PGE2 (100 nm) for 2 min then fixed and permeabilized. Actin polymerization was performed using FITC-labeled phalloidin (2 μg/ml) staining of the cells, followed by imaging by fluorescence microscopy. B, cytokine-deprived BMMCs were preincubated with the actin polymerization inhibitor cytochalasin B (10 μm) for 30 min, and then the chemotaxis assay was performed. C, BMMCs were preincubated with the indicated inhibitors (wortmannin (Wort, 100 nm, 30 min), rapamycin (Rapa, 100 nm, 30 min), Torin (100 nm, 30 min), or cytochalasin B (Cyto B, 10 μm, 30 min)), then stimulated with PGE2 (100 nm) for 2 min, fixed, and permeablized. Actin polymerization was assessed using FITC-labeled phalloidin (2 μg/ml) staining of the cells, followed by flow cytometry. D, mean values from data in C. E, BMMCs were preincubated with Torin or rapamycin for 15 min, then stimulated with PGE2 for 2 min, fixed, and permeabilized. Actin polymerization visualization was performed using FITC-labeled phalloidin (2 μg/ml) staining of the cells, followed by imaging with fluorescence microscope. F, shRNA-transduced BMMCs were stimulated with PGE2 (100 nm) for 2 min and then fixed and permeabilized. Actin polymerization was measured by flow cytometry. The results are the means ± S.E. of three separate experiments. *, p < 0.05 for comparison with PGE2 alone stimulation by Student's t test. The data in A, C, E, and F are representative from three separate experiments.
mTORC2 Is Required for Optimal PGE2-induced PGD2 and CCL2 Release
In addition to chemotaxis, PGE2 has also been documented to induce adhesion of mast cells to basement membrane proteins, as well as the production of the chemokine, CCL2, and the eicosanoid, PGD2 (42–44). We next determined whether mTORC2 played a role in these responses in addition to chemotaxis.
PGE2 induced the adhesion of BMMCs to plates coated with fibronectin (Fig. 5A) but not to plates coated with other matrix proteins including vitronectin, collagen IV, and laminin (data not shown). Rapamycin had no effect on mast cell adhesion to fibronectin-coated plates (Fig. 5A) and little, if any, effect on CCL2 (Fig. 5B) and PGD2 release (Fig. 5C). Although Torin had no effect on PGE2-mediated cell adhesion, it significantly inhibited CCL2, and particularly PGD2 release, suggesting a prominent role for mTORC2, but not mTORC1, in these responses. Complementary experiments were conducted after knockdown of raptor and rictor by use of shRNAs, but it should be noted that the conditions for transfection (see “Experimental Procedures”) resulted in lower release of CCL2 and PGD2 as compared with the cells cultured under the standard protocol (compare data from nontransfected cells in Fig. 5D to Fig. 5B, and in Fig. 5E compared with Fig. 5C). Regardless, as was true for PGE2-mediated chemotaxis, only shRNA targeting rictor caused statistically significant impairment of PGE2-mediated CCL2 (Fig. 5D) and PGD2 (Fig. 5E) production. The ability of the EP3-targeted shRNA to also reduce PGE2-mediated CCL2 production (Fig. 5F) and chemotaxis (Fig. 5G) confirms the role of the EP3 receptor in these responses. These data thus support the conclusion that mTORC2 but not mTORC1 is the major contributor to the regulation of PGE2-mediated CCL2 and PGD2 production as regulated by the EP3 receptor.
FIGURE 5.
The roles of raptor and rictor in PGE2-mediated mast cell responses. A, cytokine-deprived BMMCs were preincubated with Calcein-AM (3 μg/ml) for 30 min. The cells were washed and pretreated with or indicated inhibitors (rapamycin (Rapa) or Torin (100 nm)) for 15–20 min, and then the PGE2-induced adhesion assay was performed for 1 h in fibronectin-precoated (5 μg/ml) wells. B, BMMCs were pretreated with the indicated inhibitors for 30 min and then stimulated with PGE2 (100 nm) for 6 h. Cell-free supernatants were collected, and the levels of CCL2 were determined using ELISA kit. C, BMMCs were preincubated with the indicated inhibitors for 20 min prior to stimulation with PGE2 for 20 min, and cell-free supernatants were analyzed for PGD2. D–G, BMMCs were transduced with shRNA for control, raptor, rictor, or EP3 as we described under “Experimental Procedures” and subjected to measurement of monocyte chemoattractant protein-1 (CCL2) (D and F) or PGD2 (E and G). The data are the means ± S.E. of three separate experiments. *, p < 0.05 for comparison with PGE2 alone stimulation (A–C) or with PGE2 stimulation in shCon cells (D–G) by Student's t test.
Relative Roles of MAPK and Reactive Oxygen Species in mTORC2-dependent CCL2 Production
Cytochalasin B had a minimal effect on FcϵRI-mediated degranulation and cytokine/chemokine generation (data not shown). It is therefore unlikely that mTORC2 exerts its effects on CCL2 and PGD2 release via its regulation of actin polymerization. In addition, the inability of rapamycin and Torin to block the PGE2-mediated calcium signal (Fig. 3B) and Rac activation suggests that mTORC2 regulates CCL2 and PGD2 production independently of these signals. Activation of the MAPK pathway is thought to be critical for chemokine production in a variety of cell types (45, 46). However, mTORC2 appeared to act independently of the MAPK pathways in that rapamycin and Torin had no effect on the phosphorylation of the MAPKs, ERK, p38, or JNK or the induction of their downstream targets, Fos and Jun (Fig. 6A). These results suggest that mTORC2 regulates PGE2-induced CCL2 and PGD2 generation independently of these events.
FIGURE 6.
The role of mTORC2 on activation of MAPK pathway and ROS generation. A, cytokine-deprived BMMCs were preincubated with the indicated inhibitors (wortmannin (Wort), rapamycin (Rapa), or Torin (100 nm)) for 20 min and then stimulated with PGE2 (100 nm) for 10 or 30 min. Protein lysates were then subjected to Western blotting. To normalize protein loading, the membranes were stripped and probed for Syk, or alternatively, identically loaded samples were probed for Syk. B and D, cytokine-deprived cells were preincubated with DCF-diacetate (20 μm) for 20 min. The cells were washed and incubated with inhibitors (wortmannin, rapamycin, or Torin (100 nm) or FR-122047 (300 nm)) for 20 min and then stimulated with PGE2 (100 nm). C and F, for shRNA experiment, BMMCs were transduced with shRNA for raptor, rictor, or COX1. The cells were preincubated with DCF-diacetate, and DCF fluorescence was monitored at 37 °C in 1-min intervals for 30 min following the addition of PGE2 (1000 nm). To confirm the knockdown of COX1, total cell lysates from shRNA transduced BMMCs were subjected to Western blot. E, cytokine-deprived BMMCs were preincubated with FR-122047 (300 nm) for 30 min and then stimulated with PGE2 (100 nm) for 6 h. G, shRNA transduced BMMCs (shCon and shCOX1) were stimulated with PGE2 (100 nm) for 6 h, and then cell-free supernatants were collected and performed measurement of CCL2 using an ELISA kit. shRNA transduced BMMCs (shCon and shCOX1) were stimulated with PGE2 (100 nm) for 20 min, and then cell-free supernatants were analyzed for PGD2 by competitive enzyme immunoassay. The data are the means ± S.E. of three separate experiments, and Western blot and ROS data are representative of three separate experiments. *, p < 0.05 for comparison with PGE2 alone stimulation (E) or with PGE2 stimulation in shCon cells (G) by Student's t test.
Reactive oxygen species (ROS) regulate gene expression (47) and may therefore act as important regulators of chemokine production in various cell types. We and others have demonstrated that arachidonic acid metabolism through the actions of COX-1 is one major source for generation of ROS in immune cells including mast cells (32, 33, 46). The production of PGD2, a product of COX-1 activity, in PGE2-stimulated cells would be expected to increase ROS generation in mast cells. Accordingly, PGE2 markedly increased ROS levels in BMMCs (Fig. 6B). As with PGE2-mediated PGD2 generation (Fig. 5C), this response was substantially attenuated by Torin and wortmannin but minimally so by rapamycin (Fig. 6B). Similarly, rictor-targeted but not control-targeted or raptor-targeted shRNA markedly reduced PGE2-mediated ROS production (Fig. 6C), illustrating that mTORC2 contributes to PGE2-mediated ROS generation in BMMCs.
A linkage between the mTORC2-mediated ROS and CCL2 production was examined by suppressing ROS production through inhibition of COX-1. As shown in Fig. 6 (D and E), the selective COX-1 inhibitor, FR-122047 completely ablated PGE2-mediated ROS production in the BMMCs and reduced PGE2-induced CCL2 generation to the same extent as Torin (Fig. 5B) and knockdown of rictor (Fig. 5D). To further investigate the potential link between the PGE2-mediated, mTORC2-dependent ROS generation and CCL2 production, we attenuated PGE2-mediated ROS production through knockdown of COX1 (Fig. 6F). This resulted in a significant reduction in both PGE2-mediated CCL2 and PGD2 production (Fig. 6G). These data collectively provide support for the conclusion that that the PGE2-induced, mTORC2-mediated generation of CCL2 in BMMCs is regulated through ROS generation and not through MAPK-dependent mechanisms.
DISCUSSION
In this paper we report for the first time that PGE2 activates both mTORC1 and mTORC2 in mast cells: a process mediated through the Gαi-linked EP3 receptor. Furthermore, we present evidence that supports the conclusion that mTORC2, but not mTORC1, contributes to the regulation of PGE2-mediated mast cell chemotaxis and CCL2 and PGD2 release.
PGE2 elicits multiple biological responses in mast cells in vitro, for example, adhesion to basement membrane proteins, chemotaxis (17, 42), and the production of chemokines (43) and eicosanoids (44), in addition to the up-regulation (37, 48) and down-regulation (36, 44) of antigen-mediated mast cell degranulation and cytokine production. The extent and relevance of these responses to PGE2 in a physiological setting are unclear. Nevertheless, the generation of PGE2 in inflamed tissues would, at the very least, provide a mechanism for the migration of mast cells to such tissues. Indeed, it has been shown that intradermal injection of PGE2 causes accumulation of BMMCs at the site of injection (17).
Although our previous studies have defined some of the signaling processes by which PGE2 influences mast cell responses (22, 24), we were unable to explain how the PI3K-dependent, Btk-independent PGE2-induced chemotaxis of BMMCs may be regulated. Our observations that mTORC1 and mTORC2 cascades are activated downstream of PI3K by PGE2 in mouse BMMCs (Fig. 1) and that down-regulation of mTORC2 activity significantly attenuates PGE2-mediated chemotaxis (Fig. 2) address this conundrum.
The role of mTOR in tumorigenesis, metabolism, and homeostasis is well established (49). However, more recent studies indicate that mTOR-regulated pathways may contribute to the regulation of other processes in non-neoplastic cells. For example, mTOR has been implicated in the modulation of innate and adaptive immune responses including dendritic cell differentiation, maturation, cytokine production, and T cell activation (50). mTOR also has been shown to regulate lipid biosynthesis, which is critical for obesity, insulin resistance, and diabetes (51).
We have previously demonstrated that mTOR signaling pathways are activated in mast cells following KIT ligation and FcϵRI aggregation as a consequence of activation of PI3Kδ (30). The activation of both mTORC1 and mTORC2 cascades by PGE2 is also clearly PI3K-dependent based on our inhibitor experiments (Figs. 1 and 2), but in contrast to KIT and FcϵRI, this activation is entirely dependent on PI3Kγ, which is normally activated by βγ subunits of trimeric G proteins. BMMCs express G protein-coupled EP1, EP3, and EP4 receptors for PGE2 (37). The Gαs-linked EP2 receptor is thought to down-regulate antigen-mediated responses, whereas the Gαi-linked EP3 receptor appears to be largely responsible for most other reported effects of PGE2 in mast cells (36, 37, 44, 48). We determined that the activation of mTOR and chemotaxis by PGE2 also occurs through the Gαi-linked EP3 receptor because of the ability of the EP3 agonist sulprostone to mimic the actions of PGE2 in activating mTOR and chemotaxis, the inability of the EP2 agonist butaprost to evoke these responses, the reversal of PGE2-mediated responses in BMMCs treated with EP3-targeted shRNA, and the suppression of these responses in cells treated with the Gαi inhibitor, PTX (Figs. 1 and 2). Of possible relevance, adenosine also activates mTORC1 and mTORC24; thus, it is possible that Gαi-linked receptors, in general, share the ability of the EP3 prostanoid receptor to activate mTOR pathways and to utilize these pathways for regulating specific mast cell responses.
We have shown that mast cell chemotaxis is dependent on calcium influx and cytoskeletal reorganization through actin polymerization/depolymerization (22). Our finding that mTORC2 also regulates chemotaxis raised the question as to whether mTORC2 is linked to calcium signaling and actin rearrangement. Although PGE2 certainly elicits a calcium signal (Fig. 3) through activation of phospholipase Cβ (24) and induces Rac activation (Fig. 3) and (22), the inability of either rapamycin or Torin to block these responses would suggest that mTOR, via TORC2, regulates PGE2-mediated chemotaxis, independently of the calcium signal and Rac activation. Instead, TORC2 appears to control chemotaxis by regulating actin polymerization, independently of these processes, as suggested by the attenuation of PGE2-induced actin polymerization by Torin and rictor-targeted shRNA (Fig. 4). Our finding is analogous to a report that mTORC2 regulates the actin cytoskeleton in serum-stimulated NIH3T3 cells in a rictor-dependent and rapamycin-insensitive manner (52). These data thus support our conclusion that PGE2-induced actin polymerization is dependent on rictor (i.e. TORC2) rather than raptor (i.e. TORC1). This scenario differs from KIT-mediated chemotaxis, which is partly controlled by TORC1 (36). Whether this difference is paradoxical or reflects differences in the way GPCRs such as EP3 or tyrosine kinase receptors such as KIT engage and utilize the TORC pathways requires studies with a more extensive repertoire of receptors.
Although our initial focus was to examine whether mTOR plays a role in PGE2-mediated chemotaxis, when examining other PGE2-mediated responses, we found that mTOR also contributed to PGD2 and CCL2 generation but had no role in cell adhesion (Fig. 5) or enhancement of antigen-mediated degranulation (data not shown). The mechanism by which mTORC2 contributes to PGD2 and CCL2 production is unlikely to be related to TORC2 regulation of actin polymerization, because cytochalasin B had no effect on CCL2 production (data not shown), although it abolished the chemotactic response to PGE2 (Fig. 4). Nor does mTORC2 act upstream of the MAPKs because neither Torin nor rapamycin blocked phosphorylation of ERK1/2, JNK, and p38 in PGE2-stimulated cells (Fig. 6), although activation of these kinases is essential for production of eicosanoids (53) and CCL2 in mast cells. Furthermore, we have previously demonstrated that PGE2 does not appear to activate PKC in BMMCs (24), indicating that, although chemokine and eicosanoid production could be potentially controlled by PKC, this would not explain how mTORC2 regulated the observed PGE2-mediated CCL2 and PGD2 generation in the BMMCs.
Our observation, however, that PGE2-mediated ROS generation was markedly attenuated following either inhibition or down-regulation of TORC2 (Fig. 6) led us to hypothesize that ROS production may link PGE2-mediated mTORC2 activation to CCL2 and PGD2 generation in the BMMCs. Our previous studies have demonstrated that antigen-mediated ROS generation in BMMCs is regulated by a PI3K/Btk pathway (33). However, although our current data show that PGE2-mediated ROS production also requires PI3K (Fig. 6), because of the lack of activation of Btk by PGE2 (22), it is unlikely that Btk contributes to this response. Thus, we propose that the PI3K/mTORC2 pathway described here represents a novel mechanism for the generation of ROS in mast cells.
There are several sources of ROS in immune cells. These include NADPH oxidases, mitochondria electron transportation, nitric-oxide synthases, and arachidonic acid metabolism. Of these, COX-1-driven arachidonic acid metabolism appears to be a major source of ROS in antigen-stimulated mast cells (32). Our data were obtained with two independent approaches: the use of a selective COX-1 inhibitor and COX-1 knockdown by targeted shRNA; the results from these approaches strongly support the conclusion that COX-1 is the major enzyme responsible for ROS production in PGE2-stimulated BMMCs (Fig. 6). Reduction of both COX-1 activation and ROS production in this manner not only significantly attenuated PGE2-mediated PGD2 production but also CCL2 generation. Thus, these data support the conclusion that mTORC2-dependent ROS production is an essential signal for PGE2-mediated CCL2 generation in addition to the requirement for COX-1 for PGE2-mediated PGD2 synthesis in BMMCs.
Previous data have supported a role for PI3K/AKT in antigen-mediated cytokine/chemokine production in mast cells (54). Thus, a similar role for mTORC2-dependent AKT activation in PGE2-mediated CCL2 may be predicted. However, the role for the mTORC2-mediated ROS production in this response is unexpected in light of the observation that antigen-mediated production of the cytokines IL-6 and TNF-α in BMMCs was unaffected by inhibition of COX-1 (32). These data indicate that alternative pathways may regulate chemokine and cytokine production in mast cells in response to different stimuli. Nevertheless, our findings do suggest that PGE2-mediated PGD2 and CCL2 production in BMMCs share a common regulatory pathway.
In summary, we have demonstrated that PGE2 activates mTORC1 and mTORC2 pathways in mast cells. Our studies further revealed that mTORC2 contributes to PGE2-induced BMMCs chemotaxis through actin rearrangement. In addition, mTORC2 is also required for optimal PGE2-mediated generation of CCL2 and PGD2 via ROS generation. Thus, mTORC2 can be considered as a key node for signaling processes downstream of PI3K for the regulation of specific PGE2-mediated responses in mast cells.
Acknowledgments
We thank Dr. Nathanael S. Gray (Dana Farber Cancer Institute, Harvard Medical School) for providing Torin and Drs. Bart Vanhaesebroeck and Khaled Ali of Barts and the London School of Medicine and Dentistry for providing AS252424 and IC87114.
Financial support was provided by the Division of Intramural Research of NIAID and NHLBI within the National Institutes of Health.
H. S. Kuehn, M.-Y. Jung, M. A. Beaven, D. D. Metcalfe, and A. M. Gilfillan, unpublished data.
- FcϵRI
- high affinity receptors for IgE
- BMMC
- bone marrow-derived mast cell
- EP
- E prostanoid receptors
- GPCR
- G protein-coupled receptor
- PG
- prostaglandin
- PI3K
- phosphoinositide 3-kinase
- PTX
- pertussis toxin
- SCF
- stem cell factor
- mTOR
- mammalian target of rapamycin
- Btk
- Bruton's tyrosine kinase
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- CCL-2
- chemokine (c-c motif) ligand 2
- DCF
- 2′,7′-dichlorofluorescein
- PDK
- phosphoinositide-dependent kinase-1
- PLC
- phospholipase C.
REFERENCES
- 1. Beaven M. A. (2009) Eur. J. Immunol. 39, 11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galli S. J., Nakae S., Tsai M. (2005) Nat. Immunol. 6, 135–142 [DOI] [PubMed] [Google Scholar]
- 3. Marshall J. S. (2004) Nat. Rev. Immunol. 4, 787–799 [DOI] [PubMed] [Google Scholar]
- 4. Metcalfe D. D., Baram D., Mekori Y. A. (1997) Physiol. Rev. 77, 1033–1079 [DOI] [PubMed] [Google Scholar]
- 5. Del Prete G. (1992) Allergy 47, 450–455 [DOI] [PubMed] [Google Scholar]
- 6. Galli S. J., Tsai M., Piliponsky A. M. (2008) Nature 454, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner H., Kinet J. P. (1999) Nature 402, B24–30 [DOI] [PubMed] [Google Scholar]
- 8. Kraft S., Rana S., Jouvin M. H., Kinet J. P. (2004) Int. Arch. Allergy Immunol. 135, 62–72 [DOI] [PubMed] [Google Scholar]
- 9. Gilfillan A. M., Tkaczyk C. (2006) Nat. Rev. Immunol. 6, 218–230 [DOI] [PubMed] [Google Scholar]
- 10. Kuehn H. S., Gilfillan A. M. (2007) Immunol. Lett. 113, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirshenbaum A. S., Metcalfe D. D. (2006) Methods Mol. Biol. 315, 105–112 [DOI] [PubMed] [Google Scholar]
- 12. Okayama Y., Kawakami T. (2006) Immunol. Res. 34, 97–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madden K. B., Urban J. F., Jr., Ziltener H. J., Schrader J. W., Finkelman F. D., Katona I. M. (1991) J. Immunol. 147, 1387–1391 [PubMed] [Google Scholar]
- 14. Echtenacher B., Männel D. N., Hültner L. (1996) Nature 381, 75–77 [DOI] [PubMed] [Google Scholar]
- 15. Graves M. C., Fiala M., Dinglasan L. A., Liu N. Q., Sayre J., Chiappelli F., van Kooten C., Vinters H. V. (2004) Amyotroph. Lateral. Scler. Other Motor Neuron Disord. 5, 213–219 [DOI] [PubMed] [Google Scholar]
- 16. Meininger C. J., Yano H., Rottapel R., Bernstein A., Zsebo K. M., Zetter B. R. (1992) Blood 79, 958–963 [PubMed] [Google Scholar]
- 17. Weller C. L., Collington S. J., Hartnell A., Conroy D. M., Kaise T., Barker J. E., Wilson M. S., Taylor G. W., Jose P. J., Williams T. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11712–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hogaboam C., Kunkel S. L., Strieter R. M., Taub D. D., Lincoln P., Standiford T. J., Lukacs N. W. (1998) J. Immunol. 160, 6166–6171 [PubMed] [Google Scholar]
- 19. Humes J. L., Bonney R. J., Pelus L., Dahlgren M. E., Sadowski S. J., Kuehl F. A., Jr., Davies P. (1977) Nature 269, 149–151 [DOI] [PubMed] [Google Scholar]
- 20. Faour W. H., He Y., He Q. W., de Ladurantaye M., Quintero M., Mancini A., Di Battista J. A. (2001) J. Biol. Chem. 276, 31720–31731 [DOI] [PubMed] [Google Scholar]
- 21. Bonazzi A., Bolla M., Buccellati C., Hernandez A., Zarini S., Viganò T., Fumagalli F., Viappiani S., Ravasi S., Zannini P., Chiesa G., Folco G., Sala A. (2000) Am. J. Respir. Crit. Care Med. 162, 2272–2277 [DOI] [PubMed] [Google Scholar]
- 22. Kuehn H. S., Radinger M., Brown J. M., Ali K., Vanhaesebroeck B., Beaven M. A., Metcalfe D. D., Gilfillan A, M. (2010) J. Cell Sci. 123, 2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim M. S., Rådinger M., Gilfillan A. M. (2008) Trends Immunol. 29, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuehn H. S., Beaven M. A., Ma H. T., Kim M. S., Metcalfe D. D., Gilfillan A. M. (2008) Cell Signal. 20, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reiling J. H., Sabatini D. M. (2006) Oncogene 25, 6373–6383 [DOI] [PubMed] [Google Scholar]
- 26. Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 27. Foster K. G., Fingar D. C. (2010) J. Biol. Chem. 285, 14071–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 29. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 30. Kim M. S., Kuehn H. S., Metcalfe D. D., Gilfillan A. M. (2008) J. Immunol. 180, 4586–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tkaczyk C., Beaven M. A., Brachman S. M., Metcalfe D. D., Gilfillan A. M. (2003) J. Biol. Chem. 278, 48474–48484 [DOI] [PubMed] [Google Scholar]
- 32. Swindle E. J., Coleman J. W., DeLeo F. R., Metcalfe D. D. (2007) J. Immunol. 179, 7059–7071 [DOI] [PubMed] [Google Scholar]
- 33. Kuehn H. S., Swindle E. J., Kim M. S., Beaven M. A., Metcalfe D. D., Gilfillan A. M. (2008) J. Immunol. 181, 7706–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tkaczyk C., Metcalfe D. D., Gilfillan A. M. (2002) J. Immunol. Methods 268, 239–243 [DOI] [PubMed] [Google Scholar]
- 35. Chung K. F. (2005) Sci. STKE 2005, pe47. [DOI] [PubMed] [Google Scholar]
- 36. Kay L. J., Yeo W. W., Peachell P. T. (2006) Br. J. Pharmacol. 147, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen M., Solle M., Audoly L. P., Tilley S. L., Stock J. L., McNeish J. D., Coffman T. M., Dombrowicz D., Koller B. H. (2002) J. Immunol. 169, 4586–4593 [DOI] [PubMed] [Google Scholar]
- 38. Okkenhaug K., Ali K., Vanhaesebroeck B. (2007) Trends Immunol. 28, 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ali K., Camps M., Pearce W. P., Ji H., Rückle T., Kuehn N., Pasquali C., Chabert C., Rommel C., Vanhaesebroeck B. (2008) J. Immunol. 180, 2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Samayawardhena L. A., Kapur R., Craig A. W. (2007) Blood 109, 3679–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sakanaka M., Tanaka S., Sugimoto Y., Ichikawa A. (2008) Am. J. Physiol. Cell Physiol. 295, C1427–C1433 [DOI] [PubMed] [Google Scholar]
- 43. Nakayama T., Mutsuga N., Yao L., Tosato G. (2006) J. Leukocyte Biol. 79, 95–104 [DOI] [PubMed] [Google Scholar]
- 44. Feng C., Beller E. M., Bagga S., Boyce J. A. (2006) Blood 107, 3243–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cloutier A., Ear T., Blais-Charron E., Dubois C. M., McDonald P. P. (2007) J. Leukocyte Biol. 81, 567–577 [DOI] [PubMed] [Google Scholar]
- 46. Fialkow L., Wang Y., Downey G. P. (2007) Free Radic. Biol. Med. 42, 153–164 [DOI] [PubMed] [Google Scholar]
- 47. Schreck R., Rieber P., Baeuerle P. A. (1991) EMBO J. 10, 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gomi K., Zhu F. G., Marshall J. S. (2000) J. Immunol. 165, 6545–6552 [DOI] [PubMed] [Google Scholar]
- 49. Averous J., Proud C. G. (2006) Oncogene 25, 6423–6435 [DOI] [PubMed] [Google Scholar]
- 50. Thomson A. W., Turnquist H. R., Raimondi G. (2009) Nat. Rev. Immunol. 9, 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laplante M., Sabatini D. M. (2009) Curr. Biol. 19, R1046–R1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 53. Hirasawa N., Santini F., Beaven M. A. (1995) J. Immunol. 154, 5391–5402 [PubMed] [Google Scholar]
- 54. Kitaura J., Asai K., Maeda-Yamamoto M., Kawakami Y., Kikkawa U., Kawakami. T. (2000) J. Exp. Med. 192, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]