Abstract
Huntington disease (HD) is a neurodegenerative disorder caused by the expansion of a polyglutamine tract in the huntingtin (htt) protein. To uncover candidate therapeutic targets and networks involved in pathogenesis, we integrated gene expression profiling and functional genetic screening to identify genes critical for mutant htt toxicity in yeast. Using mRNA profiling, we have identified genes differentially expressed in wild-type yeast in response to mutant htt toxicity as well as in three toxicity suppressor strains: bna4Δ, mbf1Δ, and ume1Δ. BNA4 encodes the yeast homolog of kynurenine 3-monooxygenase, a promising drug target for HD. Intriguingly, despite playing diverse cellular roles, these three suppressors share common differentially expressed genes involved in stress response, translation elongation, and mitochondrial transport. We then systematically tested the ability of the differentially expressed genes to suppress mutant htt toxicity when overexpressed and have thereby identified 12 novel suppressors, including genes that play a role in stress response, Golgi to endosome transport, and rRNA processing. Integrating the mRNA profiling data and the genetic screening data, we have generated a robust network that shows enrichment in genes involved in rRNA processing and ribosome biogenesis. Strikingly, these observations implicate dysfunction of translation in the pathology of HD. Recent work has shown that regulation of translation is critical for life span extension in Drosophila and that manipulation of this process is protective in Parkinson disease models. In total, these observations suggest that pharmacological manipulation of translation may have therapeutic value in HD.
Keywords: Gene Expression, Huntington Disease, Microarray, Translation Regulation, Yeast, Huntingtin
Introduction
The fatal neurodegenerative disorder Huntington disease (HD)7 is characterized by involuntary movements, psychological abnormalities, and cognitive dysfunction. A central pathological hallmark of the disease is the selective loss of medium spiny neurons in the striatum of HD patients. HD is a gain-of-function disease caused by the expansion of a CAG repeat in the IT-15 gene, which encodes a polyglutamine (poly(Q)) tract in the huntingtin (htt) protein (1). The CAG repeat number is polymorphic in the general population with repeat length ranging from 6 to 35, whereas individuals affected by HD have a repeat length of greater than 35. The length of the poly(Q) expansion in htt correlates directly with kinetics of its aggregation in vitro and with severity of the disease in HD patients and indirectly with age of onset (2). Although increased size of the triplet repeat expansion correlates to an earlier age of onset, there is great variability in the age of onset of HD, even when controlling for repeat length. Indeed, a study by the United States-Venezuela Collaborative Research Project with HD kindreds containing over 18,000 individuals has found that ∼40% of variation in age of onset at controlled repeat lengths is due to genetic modifiers (3), suggesting that many therapeutic targets may be available for treating progression of this devastating disorder.
Since the cloning of the HD disease gene in 1993, several transgenic models of HD have been generated in a variety of organisms, including yeast, Caenorhabditis elegans, Drosophila, and mice. These models have allowed researchers to explore the underlying mechanisms of HD pathogenesis as well as to perform genetic screens and to test candidate therapeutic compounds. Yeast models have proven to be particularly powerful and facile for high throughput approaches as well as for molecular genetic manipulations (4). Although not all aspects of pathogenesis can be studied in a single-cell organism like yeast, expression of a mutant htt fragment in yeast produces several HD-relevant phenotypes, such as formation of mutant htt-containing aggregates, transcriptional dysregulation, cellular toxicity, perturbations in kynurenine pathway metabolites, increased levels of reactive oxygen species (ROS), mitochondrial dysfunction, defects in endocytosis, and apoptotic events (5).
In a genome-wide screen, we identified 28 gene deletions that suppress toxicity of a mutant htt fragment (Htt103Q) in yeast (6). We focus on three of these suppressor genes in this study: BNA4, MBF1, and UME1. BNA4 encodes the yeast homolog of the mammalian enzyme kynurenine 3-mononygenase (KMO), which catalyzes the hydroxylation of kynurenine in the kynurenine pathway of tryptophan degradation (7). Increased levels of two neurotoxic kynurenine pathway metabolites downstream of KMO have been implicated in the pathophysiology of HD: 3-hydroxykynurenine (3-HK) and quinolinic acid (8). The kynurenine pathway metabolites and enzymes are well conserved between yeast and humans, and the genetics of the pathway have been extensively characterized in yeast (7). We have dissected this pathway in yeast with regard to its influence on mutant htt toxicity and found that, much like in HD patients, the levels of 3-HK and quinolinic acid are increased in cells expressing a toxic mutant htt fragment (6, 9). Importantly, we found that lowering levels of these metabolites in yeast by genetic or pharmacological inhibition of Bna4 ameliorates disease-relevant phenotypes.
Ume1 is a component of the Rpd3 histone deacetylase (HDAC) complex in yeast. Several studies in fly and mouse models of HD have shown that inhibition of HDAC function either pharmacologically or genetically ameliorates HD-relevant phenotypes (10). In addition, we have found that HDAC inhibitors decrease levels of 3-HK and KMO activity in R6/2 HD model mice and in primary microglia cultured from these animals (8). Ume1 is required for full transcriptional repression of a subset of genes in yeast, in a mechanism requiring Rpd3 and Sin3 (11), suggesting that genetic inhibition of the yeast Rpd3 HDAC complex relieves poly(Q) toxicity in a mechanism similar to that observed in fly and mouse poly(Q) disease models. We have previously found that in ume1Δ cells expressing Htt103Q, both kynurenine pathway genes (BNA1, BNA2, BNA4, and BNA5) and kynurenine pathway metabolites (3-HK and quinolinic acid) are down-regulated as compared with wild-type cells expressing the same construct (8). Interestingly, we also found that the genes down-regulated in wild-type cells expressing Htt103Q cells are enriched for Rpd3 target genes. These observations directly link transcriptional dysregulation and perturbations in the kynurenine pathway in HD (8).
MBF1 encodes a transcriptional coactivator conserved from yeast to humans that bridges the DNA-binding region of transcriptional activator Gcn4 and TATA-binding protein (TBP) Spt15, a general transcription factor required for transcription by the three nuclear RNA polymerases (I, II, and III) (12, 13). Interestingly, a poly(Q) expansion in TBP in humans leads to spinocerebellar ataxia 17, which in many patients has phenotypes indistinguishable from HD (14). Gcn4 is considered to be the master regulator of amino acid metabolism in yeast. It is a member of the AP-1 family of transcription factors and regulates the expression of genes involved in 19 of 20 amino acid biosynthetic pathways, purine biosynthesis, autophagy (APG1, APG13, APG14), and multiple stress responses (15). In addition, it has been observed that ∼90 RPL (ribosomal protein, large subunit) and RPS (ribosomal protein, small subunit) genes, which encode ribosomal proteins, are repressed by activation of Gcn4 under stress conditions (15).
Here we expand on our previous studies by using a unique combination of functional approaches and differential gene expression analysis on a genome-wide scale. In order to identify critical genes/pathways/networks involved in suppression of mutant htt toxicity, we employ oligonucleotide microarray analysis to identify genes differentially expressed in mutant htt-expressing cells compared with controls as well as in three suppressor deletion strains expressing a toxic mutant htt fragment: bna4Δ, mbf1Δ, and ume1Δ. We next functionally interrogate 380 of these differentially expressed genes (DEGs) by testing the effect of overexpression of the respective ORFs on mutant htt toxicity in yeast and thereby identify 14 DEGs that modulate mutant htt toxicity. In total, this work identifies ribosomal biogenesis and rRNA processing as critical cellular processes modulated in eukaryotic cells expressing a mutant htt fragment, which suggests that these processes are probably relevant to HD pathophysiology and therapy.
EXPERIMENTAL PROCEDURES
Yeast Strains and DNA Constructs
The strains used for microarray experiments were from the yeast gene deletion set in the MATa (BY4741) [MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0] strain background (Open Biosystems). The Y258 strain background [MATa pep4-3, his4-580, ura3-53, leu2-3,112] was used for the overexpression studies (Open Biosystems). The constructs pYES2-Htt25Q-GFP and pYES2-Htt103Q-GFP (16) were used for the microarray studies. Htt103Q is a galactose-inducible, FLAG- and GFP-tagged construct encoding the first 17 amino acids of Htt fused to a poly(Q) tract of 103 glutamines. The constructs p425GALL-Htt25Q-GFP and p425GALL-Htt103Q-GFP were used in the yeast overexpression studies and were generated by amplifying the huntingtin constructs from pYES2-Htt25Q-GFP and pYES2-Htt103Q-GFP and cloning into the SpeI and XhoI sites of p425GALL (17).
Yeast Total RNA Preparation
Synthetic complete medium lacking uracil (SC −Ura) cultures containing galactose (2%) (12 ml) were inoculated at A600 of 0.2 and incubated with shaking at 30 °C until reaching an A600 of ∼1.0. Cells were harvested and lysed with acid-washed glass beads. Total RNA was isolated with Qiagen RNeasy Midi kits, following standard protocols.
Gene Expression Analysis by DNA Oligonucleotide Arrays
Double-stranded cDNA was synthesized from total RNA, amplified as cRNA, labeled with biotin, and hybridized to Affymetrix Yeast Genome S98 Array GeneChips, which were washed and scanned at the University of Washington Center for Expression Arrays and at the J. David Gladstone Institutes Genomics Core Laboratory (University of California, San Francisco) according to the manufacturer's protocols. Images were processed with Affymetrix Microarray Suite 5.0 (MAS-5). The quality of hybridization and overall chip performance were determined from the MAS-5-generated report file.
Analysis of Microarray Data
The statistical computing language R (18) was used for quality controls, preprocessing, and analyses of the data. The quality of the microarrays was assessed by inspecting pseudoimages of the arrays, MA scatter plots of the arrays versus a pseudo-median reference chip, and histogram and boxplot of raw log intensities. Expression data are available through the Gene Expression Omnibus (GEO) data base, accession number GSE18644. The data were analyzed using the R bioconductor package affylmGUI (version 1.8.0), the graphic interface to limma (version 2.9.17) (19). Data were normalized and summarized using the GCRMA method (20). DEGs were identified by using a moderated t test (limma). To correct for multiple comparison, we have estimated the false discovery rate using the QVALUE package (21) with a typical false discovery rate threshold (q) of 10%. Gene ontology searches were performed with the DAVID Functional Annotation Tool (54). Network visualization was performed using the Osprey Network Visualization System (Version 1.2.0), which is powered by the BioGRID data base (22). Cis-regulatory elements of the DEGs were identified using the MUSA algorithm (23) in the YEASTRACT suite (24). The position weight matrix of each family was used to search for known transcription factor binding sites in the YEASTRACT data base using the Smith-Waterman local alignment algorithm (using the sum of the squared distances metric).
Real-time Quantitative PCR
The BY4741 yeast strain was transformed independently with pYES2-Htt103Q-GFP and pYES2-Htt25Q (16) by standard procedures. Yeast cells were grown overnight in SC −Ura containing glucose (2%) as a carbon source. Cultures were diluted back to optical density of 0.4 and grown in SC −Ura containing galactose (2%) to induce protein expression. Cells were harvested after growing for 25 h, pelleted, and stored at −80 °C until needed. RNA was extracted using the Qiagen RNeasy Midi kits following the manufacturer's instructions. Genomic DNA contamination was removed from RNA using Turbo DNase according to the manufacturer's protocol. 500 ng of RNA was used as a template to synthesize cDNA with the Qiagen QuantiTect® reverse transcription kit. To ensure that RNA had no genomic DNA contamination, a control reaction was included in which no reverse transcription was carried out. 17 genes were selected to analyze the mRNA expression levels by quantitative PCR, and actin-1 was chosen as a reference. Reactions were carried out in a LightCycler real-time PCR system (Roche Applied Science). cDNA was quantified using 5 μl of SYBR Green (Fermentas) and 0.3 μm forward and reverse primers. Primers were analyzed for specificity and efficiency by melting curve analysis. The efficiency was calculated at the end of each amplification reaction via relative standard curves. PCR efficiencies ranged from 0.80 to 1.0. Standard curves were calculated using the amplification of five serial dilutions of genomic DNA in triplicate. At least three independent cDNA samples were analyzed. PCRs were run in duplicate. Relative quantification was performed using LightCycler® 480 relative quantification Software. The crossing points were calculated by the second derivative method, and relative expression was calculated by the available advanced relative quantification. Further analysis and statistical tests were performed using the Relative Expression Software 2008 (REST) (25, 26). The relative quantification was corrected for PCR efficiency via both methods.
Functional Testing of DEGs
Yeast strains containing plasmids for the overexpression of selected genes were obtained from the yeast ORF collection in the Y258 strain background. The relevant yeast strains were grown overnight in 96-well plates containing 100 μl of SC −Ura supplemented with 2% glucose per well and transformed with either p425GALL-Htt25Q-GFP or p425GALL-Htt103Q-GFP using a high throughput transformation method (4). Transformants were grown to stationary phase in complete medium lacking uracil and leucine (SC −Ura−Leu) containing 2% glucose, serial diluted, and spotted onto SC −Ura−Leu medium supplemented with either 2% glucose or 2% galactose and 2% raffinose. Plates were incubated at 30 °C for 3–5 days, and yeast strains were scored for growth.
Determination of RNQ Prion Status
20-ml cultures of suppressor strains were grown to approximately an A600 of 1.0 in SC −Ura GAL/RAF medium, at which point cells were harvested by centrifugation at 3000 rpm for 5 min. Cell pellets were washed with 10 ml of water and spun as above. The cell pellets were resuspended in 200 μl of lysis buffer (100 mm Tris, pH 7.0, 200 mm NaCl, 1 mm EDTA, 5% glycerol, 0.5 mm DTT, 1× protease inhibitor mixture) and transferred to microcentrifuge tubes. The cells were lysed by the addition of ∼200 μl of acid-washed glass beads (425–600 μm; Sigma), vortexing for 1 min, the addition of 200 μl of radioimmune precipitation assay buffer (50 mm Tris pH 7.0, 200 mm NaCl, 1% Triton, 0.5% sodium deoxycholate, 0.1% SDS), and further vortexing for 10 s. Samples were then centrifuged at 3000 rpm for 15 s to pellet the glass beads and cell debris. 60 μl of supernatant was used as the “total” sample, whereas 200 μl of the supernatant was centrifuged for 30 min at 80,000 rpm in a Beckman TLA 100 Ultracentrifuge. 60 μl of the supernatant from this step was isolated and used as the “soluble” fraction, whereas the “pellet” fraction was prepared by resuspending the pellet in 100 μl of lysis buffer and 100 μl of radioimmune precipitation assay buffer, adding 67 μl of 4× protein sample buffer, and boiling for 5 min. 20 μl of 4× protein sample buffer was added to the total and soluble fractions and boiled for 5 min. The samples were resolved by SDS-PAGE and immunoblotting as described above. Curing yeast strains of endogenous prion cells was performed by growth for 5 passages on YPD supplemented with 5 mm guanidine hydrochloride (27).
RESULTS
Yeast Expressing a Mutant htt Fragment Differentially Expresses Genes Involved in Ribosome Biogenesis and rRNA Processing
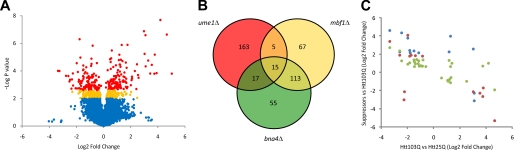
We recently performed oligonucleotide microarray hybridization assays to compare mRNA expression profiles of isogenic parental yeast (BY4741, MAT a) expressing either a wild-type or mutant htt fragment (Htt25Q or Htt103Q, respectively) (9). Here we reanalyzed the data from this experiment using the limma package implemented in R/Bioconductor (19, 28). This analysis showed that in Htt103Q-expressing cells, expression of 226 genes was up-regulated when compared with Htt25Q-expressing cells, whereas expression of 244 genes was down-regulated (q value of <0.2) (Fig. 1A). A subset of these DEGs were analyzed and confirmed via quantitative real-time PCR (supplemental Table S1).
FIGURE 1.
Identification of DEGs in wild-type and gene deletion suppressor strains expressing Htt103Q. A, volcano plot of DEGs. The log2 of the -fold change (Htt103Q versus Htt25Q) is represented on the x axis, and the negative log of p values from t test analyses is represented on the y axis. Up-regulated genes due to Htt103Q have positive -fold changes. Red, DEGs at the false discovery rate of q < 0.1; orange, 0.1 < q < 0.2; blue, q > 0.2. B, Venn diagram indicating the overlap in DEGs between bna4Δ, mbf1Δ, and ume1Δ strains expressing Htt103Q compared with the parental BY4741 strain. 15 DEGs are shared among the three deletion strains. C, inverse correlation of log -fold change (M) in DEGs in suppressors expressing Htt103Q on the y axis and in a wild-type strain expressing Htt103Q (compared with Htt25Q-expressing cells) on the x axis. Blue, bna4Δ (r = −0.77, t = −4.19, df = 12, p < 0.01); red, mbf1Δ (r = −0.74, t = −4.10, df = 14, p = 0.01); green, ume1Δ (r = −0.67, t = −4.84, df = 29, p < 0.0001).
We used the DAVID Bioinformatics Resources Functional Annotation Tool (available on the World Wide Web) to test whether the DEGs were enriched by known or predicted function using gene ontology (GO) (29). In a manner similar to our previous analysis, functional groups up-regulated significantly in the Htt103Q-expressing cells included genes involved in protein folding (p < 1.0 × 10−3) and response to stress (p < 0.01) (supplemental Table S2) (9). In addition, the new analysis found enrichment in the GO terms of response to unfolded protein (p < 1.0 × 10−5), ubiquitin cycle (p < 0.01), post-translational protein modification (p < 0.01), and vacuolar protein catabolic process (p < 0.05). We previously described that down-regulated genes in Htt103Q-expressing cells were involved in the functional groups of ribosome biogenesis (p < 1.0 × 10−39) and rRNA processing and metabolism (p < 1.0 × 10−20) (9), and we have confirmed those observations in our new analyses (supplemental Table S3). These data suggest that yeast cells expressing Htt103Q mount a response to deal with this toxic, misfolded protein, via up-regulation of proteins involved with protein misfolding, protein degradation, autophagy, and stress response. At the same time, in a manner similar to classic heat shock response, the presence of Htt103Q in yeast cells causes a dramatic reduction in expression of genes involved in rRNA metabolism and ribosome biogenesis, suggesting that general protein synthesis in these cells is significantly repressed, ultimately contributing to Htt103Q-dependent toxicity.
Common Mechanisms Underlie Mutant htt Toxicity Suppression in Gene Deletion Suppressor Strains
In order to discern if there are common mechanisms underlying toxicity suppression in mutant htt suppressor strains, we wished to next monitor gene expression perturbations in three gene deletion Htt103Q suppressor strains: bna4Δ, mbf1Δ, and ume1Δ (6). Thus, we performed microarray experiments to identify DEGs in bna4Δ, mbf1Δ, or ume1Δ yeast expressing Htt103Q versus parental wild-type cells expressing Htt103Q. We tested the 200 DEGs that showed the highest -fold change (and q < 0.1) from each deletion suppressor for enrichment of GO terms. We found that in bna4Δ cells expressing Htt103Q, there is an enrichment of several GO terms as compared with control cells, including carboxylic acid metabolism (p < 1.0 × 10−4), translation elongation (p < 1.0 × 10−3), nitrogen compound metabolism (p < 1.0 × 10−3), water-soluble vitamin biosynthesis (p < 0.01), and vesicle organization and biogenesis (p < 0.05) (supplemental Table S4). DEGs in mbf1Δ cells showed enrichment in many GO term categories, including amino acid metabolism (p < 1.0 × 10−11), nitrogen compound metabolism (p < 1.0 × 10−9), carboxylic acid metabolism (p < 1.0 × 10−7), and urea cycle intermediate metabolic process (p < 1.0 × 10−4) (supplemental Table S5). The ume1Δ suppressor strain exhibited DEGs with enrichment in GO term categories of water-soluble vitamin biosynthesis (p < 0.01), NAD biosynthesis (p < 0.01), nitrogen compound metabolism (p < 0.01), and carboxylic acid metabolism (p < 0.05), among others (supplemental Table S6). The four genes present within the NAD biosynthesis GO group are the central kynurenine pathway genes of BNA1, BNA2, BNA4, and BNA5, confirming our original analysis with this data set (8). It is critical to note that although some of the GO groups enriched in these data are specific to individual suppressors, several categories are common among these suppressors, such as nitrogen compound metabolism and carboxylic acid metabolism.
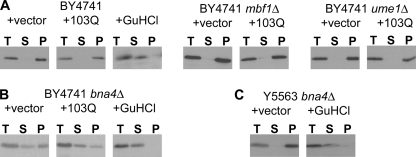
Because full levels of Htt103Q toxicity are dependent upon the presence of the Rnq1 yeast prion [RNQ+] (16), we analyzed Rnq1 prion status in the three gene deletion strains to ensure that the DEGs identified above are independent of the Rnq1 prion. To this end, we analyzed Rnq1 by sedimentation analysis in all three suppressor strains (Fig. 2, A and B). We found that in all three strains, Rnq1 was present in the pellet fraction of cells expressing Htt103Q or carrying empty vector (pYES2), indicating that Rnq1 is present in its prion form independent of Htt103Q expression. Interestingly, in the case of the bna4Δ strain, we observed that Rnq1 is also present in the soluble fraction, suggesting that this population of cells contains both prion and non-prion forms of Rnq1 (Fig. 2B). A similar phenotype has been described with other deletion strains from this library (30). Treatment with the prion curing agent guanidine hydrochloride shifted Rnq1 from the pellet fraction to the soluble fraction, providing further support that Rnq1 status is “mixed” in the bna4Δ strain (Fig. 2B). To test whether prion status was directly altered by the deletion of BNA4, we analyzed a second bna4Δ strain, and consistent with previously published work (30), we found that Rnq1 was entirely in prion form, indicating that the mixed prion phenotype is independent of BNA4 deletion (Fig. 2C). In total, these data suggest that DEGs identified in the mbf1Δ and ume1Δ strains are independent of Rnq1 prion status, whereas a subset of DEGs from the BY4741 bna4Δ strain may arise from [RNQ+]-dependent modulation of Htt103Q.
FIGURE 2.
Rnq1p is present in prion conformation in gene deletion suppressor strains. [RNQ+] prion status in wild-type yeast and deletion suppressors (BY4741 parental strain) carrying the pYES2 empty vector or expressing Htt103Q was determined by a combination of high speed centrifugation and immunoblotting. T, total extract for each yeast strain; S, supernatant fraction (soluble form of Rnq1); P, pellet fraction (prion form of Rnq1, [RNQ+]). A, immunoblotting with α-Rnq1 antibody showed that Rnq1 is found exclusively in the pellet fraction of the BY4741 wild-type strain as well as the mbf1Δ and ume1Δ strains, indicating that the protein is in the prion conformation. B, in the BY4741 bna4Δ, strain, [RNQ+] prion status is mixed, with protein present in both pellet and supernatant fractions. Treatment of BY4741 bna4Δ and parental cells carrying pYES2 with guanidine hydrochloride (GuHCL) cures [RNQ+] prion, shifting Rnq1p from the pellet fraction to the supernatant fraction. C, [RNQ+] prion status is independent of BNA4 deletion. In Y5563 bna4Δ cells, all Rnq1 is found in the pellet fraction. Treatment of Y5563 bna4Δ cells with guanidine hydrochloride cured [RNQ+] prion present in the pellet fraction of untreated cells carrying pYES2, moving Rnq1 to the supernatant fraction.
In order to filter out DEGs dependent upon modulation of Rnq1 prion status and to ascertain if common genes/mechanisms underlie suppression in the three suppressor strains, we cross-compared the three sets of DEGs identified above (Fig. 1B). Strikingly, we found that 15 annotated genes were common among these three groups, seven of which are also differentially expressed in WT cells expressing Htt103Q (Table 1). Assuming independence, the probability of finding 15 genes shared among these three groups is <1.0 × 10−13, which supports the notion that the suppressors share common mechanisms of mutant htt suppression. The genes shared among the suppressor strains function in a variety of cellular processes, including translation elongation (ANB1), stress response (DAK2), amino acid transport (DIP5), lactate metabolism (DLD3), and mitochondrial transport (YMC2). Interestingly, three of the 15 genes are predicted to encode tRNAs, all of which are down-regulated in the suppressor strains. Of these genes, two encode tRNAs for tRNAPro, and one encodes tRNALys (Table 1). In 13 of 15 cases, the genes are differentially expressed in the same direction in all three suppressor strains, reinforcing the notion that the shared DEGs represent common underlying mechanisms involved in mutant htt toxicity. Intriguingly, of these 13 genes, five are differentially expressed in the opposite direction in Htt103Q-expressing cells (AQR1, DAK2, YGR035C, YMC2, and YOR338W) (Table 1 and supplemental Table S7).
TABLE 1.
Common DEGs between bna4Δ, mbf1Δ, and ume1Δ suppressor strains expressing Htt103Q as compared with the parental BY4741 wild-type strain
Boldface type indicates genes differentially expressed in all three suppressor strains as well as in Htt103Q-expressing parental cells (in the opposite direction). Htt103Q differential expression is related to Htt25Q-expressing parental cells (q < 0.2). DEGs in the three suppressor strains expressing Htt103Q (bna4Δ, mbf1Δ, and ume1Δ) are relative to parental cells expressing Htt103Q (top 200 annotated genes, q < 0.1). NC, not changed.
| Gene | Htt103Q | bna4Δ | mbf1Δ | ume1Δ | Function |
|---|---|---|---|---|---|
| ANB1 | NC | Up | Up | Up | Translation elongation factor eIF-5A |
| AQR1 | Down | Up | Up | Up | Plasma membrane multidrug transporter |
| COS7 | Up | Up | Up | Down | Mitochondrial protein of unknown function |
| DAK2 | Up | Down | Down | Down | Dihydroxyacetone kinase |
| DIP5 | NC | Up | Up | Up | Dicarboxylic amino acid permease |
| DLD3 | NC | Up | Up | Up | d-Lactate dehydrogenase |
| LYS20 | NC | Up | Up | Up | Homocitrate synthase isozyme |
| MMP1 | NC | Up | Up | Up | High affinity S-methylmethionine permease |
| SPG4 | Up | Up | Down | Down | Protein required for survival at high temperatures |
| SUF2 | NC | Down | Down | Down | tRNAPro, 1 of 3 nuclear tRNAs; anticodon AGG |
| SUF10 | NC | Down | Down | Down | tRNAPro, 1 of 3 nuclear tRNAs; anticodon AGG |
| TK(CUU)J | NC | Down | Down | Down | tRNALys, imported into mitochondria; AAG |
| YGR035C | Down | Up | Up | Up | Protein of unknown function, Cdc28 substrate |
| YMC2 | Down | Up | Up | Up | Mitochondrial inner membrane transporter |
| YOR338W | Down | Up | Up | Up | Protein of unknown function; regulated by Azf1 |
This observation led us to compare the overlapping DEGs from the individual suppressor strains with the top 200 DEGs from Htt103Q-expressing cells to ascertain if a negative correlation exists in expression between these groups of DEGs. Strikingly, in all three comparisons, we found a significant negative correlation in differential expression of the overlapping genes (Fig. 1C). This suggests that the differential expression observed in the parental WT strain due to expression of Htt103Q is relieved in the three deletion suppressor strains. Although it is likely that these changes in expression profiles directly contribute to mutant htt toxicity, we cannot exclude the possibility that the changes are simply a downstream consequence of cellular toxicity.
Cis-regulatory Domain Analysis of DEGs Identifies Enriched Elements
To clarify whether common regulatory mechanisms were affected in Htt103Q-expressing cells as compared with Htt25Q controls, the DEGs with a local false discovery rate of <2%, as estimated by the Rank product algorithm (n = 46), were analyzed for shared regulatory motifs using the MUSA algorithm. This analysis revealed 14 families of de novo motifs that were significantly overrepresented in these sequences (23) (Table 2). Aligning the position weight matrix of each family with the position weight matrix of known transcription factors revealed several matches. Interestingly, Gcn4 was among the transcription factors identified, suggesting a link between response to Htt103Q-dependent toxicity and this transcription factor. The list of transcription factors identified includes several zinc transcription factors, such as Hap1 (which responds to heme and oxygen levels), Azf1 (which responds to glucose), and Zap1 (which responds to zinc levels). Other candidate transcription factors include Cup2, a copper-binding transcription factor that responds to copper levels, and Ime1, which is the master regulator of meiosis and activates early meiotic genes through interactions with Ume6 (31), another transcription factor whose binding site was overrepresented in the DEGs (Table 2). Ume6 recruits the Rpd3-Sin3 HDAC complex during mitosis to repress early meiosis-specific genes via hypoacetylation of histones H3 and H4 (32). Critically, Ume6 has been shown by affinity mass spectrometry to be a component of the Rpd3-Sin3 corepressor complex along with the loss-of-function Htt103Q suppressors Ume1 and Rxt3 (6, 33). Thus, this observation further implicates transcriptional dysregulation in mutant htt toxicity and HDACs as candidate therapeutic targets.
TABLE 2.
Families of overrepresented motifs in promoter regions of genes differentitally expressed in Htt103Q versus Htt25Q yeast cells
| Family | Counta | p value | Transcription factorsb |
|---|---|---|---|
| TTTATAT | 26 | 1.77e−06 | Mig2p, Hap1p, Fzf1p, Zap1p |
| TTCTTTTC | 17 | 3.59e−06 | Azf1p, Cup2p, Zap1p, Ime1p |
| AAAAGAAA | 22 | 9.24e−06 | Azf1p, Zap1p, Cup2p, Tec1p |
| CATCGC | 22 | 3.33e−05 | Hap1p, Rfx1p, Ime1p, Rox1p, |
| GCGATG | 22 | 3.33e−05 | Hap1p, Rfx1p, Ime1p, Rox1p |
| CGCACA | 21 | 0.000104 | Crz1p, Hap1p, Aft2p, Stp2p |
| TGTGCG | 21 | 0.000104 | Crz1p, Aft2p, Stp2p, Hap1p |
| AAGAAG | 39 | 0.000146 | Tec1p, Azf1p, Zap1p, Azf1p |
| ATATTAT | 24 | 0.000166 | Arg81p, Arr1p, Mig1p,Mig3p |
| TTCTTC | 39 | 0.000257 | Tec1p, Zap1p, Abf1p, Ime1p |
| GCACGT | 18 | 0.000573 | Gcn4p, Met4p, Mig3p, Pho4p |
| ACGTGC | 18 | 0.000573 | Gcn4p, Pho4p, Met4p, Mig1p |
| GCGGCT | 16 | 0.000988 | Ume6p, Abf1p, Mig2p, Mig3p |
| AGCCGC | 16 | 0.000988 | Ume6p, Stp1p, Mig1p, Stp2p |
a Number of sequences containing at least one motif in the family (of 46).
b Only the top four matches are listed for each family.
A Unique Subset of Differential Expressed, Highly Interconnected Genes Modulate Mutant htt Toxicity
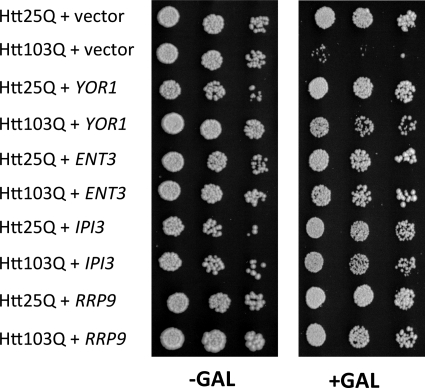
In order to ascertain the potential role of the DEGs in mutant htt toxicity, we individually tested the ability of these genes to suppress toxicity of Htt103Q when individually overexpressed in yeast. All of the genes that were available as open reading frame (ORF) constructs from the yeast ORF collection (380 of 470, 80.9%) were individually tested via growth assays for suppression of Htt103Q toxicity. We found that 12 of the DEGs suppressed toxicity of Htt103Q when overexpressed (Fig. 3 and Table 3). To eliminate the possibility that the DEGs suppressed toxicity by silencing Htt103Q expression, we analyzed expression levels by Western and dot blotting and found that Htt103Q expression was unchanged (supplemental Fig. S1, A and B). In addition, we confirmed that Rnq1 remains in its prion conformation when expression of a DEG is induced (supplemental Fig. S1C).
FIGURE 3.
Suppression of Htt103Q toxicity in yeast overexpression strains. Parental wild-type Y258 yeast containing constructs for the overexpression of the indicated yeast ORFs were transformed with p425-Htt25Q or p425-Htt103Q, and cellular viability was determined using growth assays. The expression of both the huntingtin constructs and the indicated yeast ORFs is induced by galactose. 5-Fold serial dilutions starting with equal numbers of cells of the four representative ORF suppressors are shown.
TABLE 3.
DEGs in Htt103Q-expressing cells modulate mutant htt toxicity
| Ortholog(s)a | Expressionb | Function | |
|---|---|---|---|
| Suppressor | |||
| BUD23 | + | Down | rRNA processing |
| CSE2 | − | Up | RNA polymerase II transcription |
| DBP2 | + | Down | rRNA processing |
| ENT3 | + | Up | Golgi-endosome transport |
| IPI3 | − | Down | rRNA processing |
| JJJ3 | − | Down | HSP40 chaperone |
| NSA2 | + | Down | rRNA processing |
| PRM7 | − | Down | Pheromone response |
| RAS1 | + | Down | G-protein signaling |
| RRP9 | + | Down | rRNA processing |
| UTP9 | − | Down | rRNA processing |
| YOR1 | + | Down | ABC transporter |
| Deletion suppressor | |||
| mbf1Δ | + | Up | Transcriptional coactivator |
| Deletion enhancer | |||
| apj1Δ | + | Up | HSP40 chaperone |
a Human orthologs determined via the Ensembl Genome Browser. Orthologs may be either one-to-one, one-to-many, or many-to-many. BUD23, ENT3, NSA2, RRP9, and MBF1 have one-to-one orthologs in humans that could potentially be targeted for therapeutics.
b Direction of differential expression in Htt103 versus Htt25Q cells.
Interestingly, these overexpression suppressors include genes whose expression is up-regulated as well as genes whose expression is down-regulated in cells expressing Htt103Q, indicating two different models for overexpression protection: 1) ORF overexpression mimics up-regulation of genes exerting a protective role against mutant htt toxicity, or 2) ORF overexpression rescues a depletion of a critical factor (Table 3). Excitingly, these overexpression suppressors are potential candidate therapeutic targets for HD. Of the 12 novel suppressors, seven (∼58%) have human orthologs as determined by the Ensembl Genome Browser (see the Ensembl Web site).
We next compared our list of DEGs with our previously published gene deletion enhancers and suppressors of mutant htt toxicity and identified another two common genes (MBF1 and APJ1) (6, 34). Thus, in total, we identified 14 DEGs that modify mutant htt toxicity when overexpressed or deleted (Table 3). Among these 14 genes, we observed enrichment of genes within seemingly unrelated functional groups. In particular, this group was enriched for genes involved in rRNA processing (BUD23, DBP2, IPI3, NSA2, RRP9, and UTP9), stress/heat shock response (APJ1 and JJJ3), and transcription (CSE2 and MBF1) (Table 3). Interestingly, aside from rRNA processing, all of these functional groups have been extensively implicated in HD pathology.
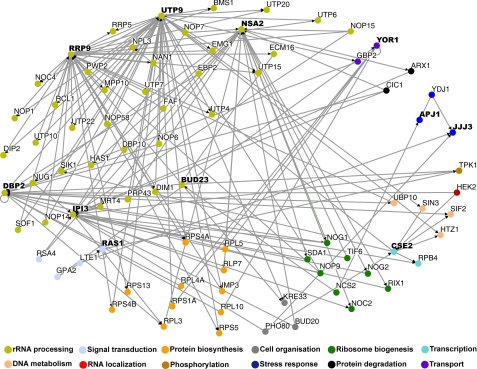
To clarify the functional connectivity among this functionally validated list of DEGs, we performed network analysis using the Osprey Network Visualization System (Version 1.2.0), which allows visualization of complex interaction networks (22). This software is powered by the BioGRID data base (available on the World Wide Web), which houses and distributes data collections of protein and genetic interactions of model organisms, including yeast, via the Saccharomyces Genome Data base (available on the World Wide Web). Via these databases, Osprey allows insertion of all known interactions for each “node” (gene of interest). The interactions types (or “edges”) include data from affinity capture experiments, two-hybrid screens, and synthetic lethality analyses, among others. Here, we have used Osprey to investigate all of the known genetic and physical interactions of the 14 functionally validated DEGs. As an initial test, we asked for all of the interactions within these 14 nodes. Interestingly, we found only one interaction among these nodes, a physical interaction between Rrp9 and Utp9, both of which are components of the small ribosomal subunit (SSU) processome involved in pre-rRNA processing, as determined by affinity capture/mass spectroscopy studies (35–37) (data not shown). This result highlights our observations from the GO analysis that showed an enrichment in genes from disparate functional groups. In order to determine if the above 14 genes function indirectly in the same network, we explored all of the interactions for these functionally validated nodes, and we found a total of 538 interactions among 464 nodes (data not shown). In order to select genes with higher level relationships, we processed the network data with an iterative minimum filter of 2, which identified all of the nodes within this group that have a minimum of two interactions with other genes within the group, and then sorted the remaining nodes by GO function (Fig. 4). This analysis uncovered a highly interconnected network of genes, which surprisingly included 11 of 14 of the original functionally validated genes. Critically, this network of 81 nodes (with 156 edges) reinforced and expanded the GO groups enriched in the mutant htt toxicity interaction network: ribosome biogenesis and assembly (71.6%, p = 1.2 × 10−49), rRNA processing (55.6%, p = 4.7 × 10−43), nuclear transport (12.3%, p = 1.5 × 10−4), and G-protein signaling (2.5%, p = 4.5 × 10−2). When a more stringent iterative minimum filter of 3 is applied, 5 of 14 modifiers are still present in the network (11 nodes; 23 edges), all of which are involved in ribosome biogenesis and assembly (p = 2.6 × 10−12), rRNA processing (p = 1.2 × 10−8), and related processes (data not shown). In total, this work suggests that although these modifiers of mutant htt toxicity play roles in disparate functional groups, they function within a highly interrelated network.
FIGURE 4.
Network analysis uncovers a high degree of interconnectivity among functionally validated DEGs. Shown is Osprey network analysis of 14 DEGs (indicated in boldface type) that suppress toxicity of Htt103Q when overexpressed. All interactions for these genes (both physical and genetic) were included in the analysis. Genes described by the same significantly enriched GO terms are color-coded and grouped together. Network data were analyzed with an iterative minimum filter of 2 (minimum of two interactions with other network genes). A total of 81 nodes and 156 edges define this network, which contains 11 of the original 14 functionally validated genes.
DISCUSSION
In this study, we utilized a novel functional approach to gene profiling experiments in order to dissect the underlying mechanisms of mutant htt toxicity in yeast. This work has not only highlighted the central role of rRNA processing and ribosome biogenesis in mutant htt toxicity in yeast but has also identified several novel suppressors of this toxicity that are candidate therapeutic targets for HD. Several gene profiling experiments with mammalian models of HD support our observations. Gene expression profiling in PC12 cells and rat striatal cells expressing mutant htt fragments has found an enrichment in genes encoding ribosomal proteins and RNA-processing proteins (38, 39). In addition, analysis of gene expression in the striatum of R6/2 HD model mice has found an enrichment in genes encoding ribosomal proteins as compared with wild-type controls (40). Recently, microarray profiling using a primary rat neuron model of HD found enrichment in genes involved in RNA splicing/RNA processing (41). Finally, an RNAi screen in Drosophila cells identified several modifiers of mutant htt aggregation that play a role in RNA processing (42). In total, these observations suggest that the results described here are not likely to be yeast-specific but may reflect cellular perturbations conserved in mammalian cells. In addition, in this study, we identified DEGs from bna4Δ, mbf1Δ, and ume1Δ strains expressing Htt103Q versus control cells to learn more about underlying mechanisms contributing to Htt103Q toxicity suppression. Analysis of prion status in these strains found that in the bna4Δ strain, Rnq1 is present in both prion and soluble forms, suggesting that a subset of these DEGs may arise from modulation of [RNQ+] status. Thus, in order to reduce [RNQ+]-dependent effects, we focused on the cross-section of these DEGs with the DEGs from the mbf1Δ and ume1Δ strains. By this approach, we identified 15 common DEGs among these three gene deletion suppressor strains. These genes include three tRNA-encoding genes as well as genes involved in a variety of cellular pathways. What is unclear in the examples above is the mechanism of these changes and how these changes contribute to mutant htt toxicity and ultimately to HD pathology. These yeast studies will serve as a strong starting point for future studies elucidating these underlying mechanisms.
A critical finding from this study is the identification of a robust network of interactions derived from 14 differentially expressed genes (nodes) that modulate toxicity of a mutant htt fragment. Despite the original nodes playing roles in diverse cellular processes, the resulting network contains 11 of these nodes within a network of 81 nodes and 156 edges and has an enrichment of genes involved in rRNA processing and ribosome biogenesis. Intriguingly, several of the functionally validated nodes appear to be highly interconnected in the network (nodes indicated in boldface type in Fig. 4), underscoring the importance of these nodes within the context of the network.
Several of our observations above implicate Gcn4 in mutant htt toxicity in yeast. First, expression of Htt103Q in yeast leads to down-regulation of genes involved in ribosome biogenesis, and under stress conditions, Gcn4 is known to repress transcription of ∼90 RPL and RPS genes, which encode ribosomal proteins (15). Second, MBF1, which is a deletion suppressor of Htt103Q toxicity, encodes a transcriptional coactivator that can bridge Gcn4 and TBP. Third, we found that Gcn4 binding sequences are overrepresented in the upstream regions of genes differentially expressed in Htt103Q-expressing cells. In total, this work suggests that expression of Htt103Q in yeast may down-regulate expression of ribosomal genes via induction of Gcn4 expression. Because Mbf1 expression is required for Gcn4 function, these observations collectively suggest that deletion of MBF1 may suppress mutant htt toxicity by impairing Gcn4 function. It must also be noted that induction of Gcn4 leads to up-regulation of three kynurenine pathway genes, BNA1 (3-hydroxyanthranilate 3,4-dioxygenase), BNA4 (which encodes KMO), and BNA6 (quinolinate phosphoribosyl transferase) (15). Intriguingly, deletion of either BNA1 or BNA4 suppresses toxicity of Htt103Q (6). These observations also support a hypothesis in which Gcn4 induction due to Htt103Q expression contributes to toxicity. We did not, however, see differential expression of GCN4 in our present study in Htt103Q-expressing cells (data not shown). This is not particularly surprising because GCN4, which is under strict transcriptional control, is also under translational control, via four small upstream ORFs in the 5′ leader region of the GCN4 mRNA (43). Induction of translation of the message occurs primarily under environmental stresses (44). In addition, because recruitment of TBP via Mbf1 is the rate-limiting step in Gcn4 activation (12), and MBF1 expression is up-regulated in Htt103Q-expressing yeast (Table 3), it is possible that increased levels of Mbf1 alone may be sufficient to increase Gcn4 activity, without induction of Gcn4 expression. Interestingly, life span extension in yeast due to depletion of 60 S ribosomal subunits, dietary restriction, or TOR inhibition appears to require induction of Gcn4 (45). Thus, in the case of mutant htt expression, Gcn4 induction may reflect a cellular coping mechanism gone awry. This pathway may play a similar role in humans because ATF4, the functional ortholog of Gcn4 in mammals (46), is regulated via a similar translational mechanism, and activation of Gcn4 is analogous to the mammalian integrated stress response (47).
In a related note, the yeast eIF4E-associated protein Eap1, which inhibits cap-dependent translation initiation via the TOR signaling cascade, attenuates induction of Gcn4 translation (48). Recent work has shown that overexpression of eIF4E-BP, the Eap1 functional equivalent in Drosophila, rescues Parkinsonian phenotypes in fly models of Parkinson disease by inhibiting cap-dependent translation and thereby inducing expression of genes involved in stress response (49). It has also been seen that during dietary restriction in Drosophila, 4E-BP promotes life span extension by activation of nuclear encoded mitochondrial protein translation (50).
Interestingly, in the present study, we have found that the gene encoding eIF5A, a translation elongation factor, is up-regulated in all three suppressor strains expressing Htt103Q, as compared with controls. Taken together, these data suggest that altered regulation of translation may be contributing to mutant htt toxicity and that pharmacological modulation of this process may have therapeutic relevance. Supporting this, it has been recently shown that rapamycin treatment of mouse embryonic fibroblast cells expressing mutant htt abrogates HD-relevant phenotypes by inhibition of translation, independent of effects on autophagy (51). It is also important to note that although much interest in the HD community has been focused on transcriptional dysregulation in pathogenesis and as a target for therapeutics, our study suggests that the effect of mutant htt on translational processes in the cell may also be critical.
Because most basic cellular mechanisms are conserved in yeast to higher eukaryotes, the work presented here will probably inform future studies on disease pathogenesis in HD. Due to the ease and rapidity of genetic screening in yeast, this organism is particularly amenable for integrated approaches to functional gene expression profiling. Yeast will therefore probably provide an important platform for future analyses of disease genes, further evidenced by a recent study dissecting α-synuclein toxicity in yeast (52). It is important to mention that we have identified 12 novel suppressors of mutant htt toxicity in yeast, seven of which have human orthologs as determined by the Ensembl Genome Browser. Four of these yeast genes (BUD23, ENT3, NSA2, and RRP9) have clear one-to-one orthologs in humans, which could potentially be targeted for therapy if validated in other model systems. Interestingly, ENT3 has recently been identified as a suppressor of α-synuclein toxicity in yeast (53). In summary, our study has provided new insights into the mechanisms associated with mutant htt toxicity in yeast that may be relevant to HD pathogenesis and has also identified novel candidate targets for therapeutic intervention in this disorder. Clearly, it is now critical to test the above hypotheses and to validate the candidate HD targets identified in order to ascertain how our observations are linked to mutant htt toxicity in yeast and how they may inform therapeutic strategies in HD patients. Finally, the power of using yeast to clarify mechanisms involved in HD pathogenesis and to identify candidate drug targets is underscored by our recent validation of KMO as a promising therapeutic target in HD model mice.8
Supplementary Material
Acknowledgments
We thank M. Sherman and A. Meriin for the pYES2-Htt25Q and pYES2-Htt103Q constructs.
This work was supported by Medical Research Council New Investigator Award G0700090 (to F. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S7 and Fig. S1.
D. Zwilling, S. Y. Huang, P. Guidetti, J. Lee, J. Truong, Y. Andrews-Zwilling, E. Hsieh, J. Louie, T. Wu, K. Scearce-Levie, H. Q. Wu, C. Patrick, A. Adame, F. Giorgini, S. Moussaoui, G. Laue, A. Rassoulpour, Y. Huang, J. M. Muchowski, E. Masliah, R. Schwarcz, and P. J. Muchowski, submitted for publication.
- HD
- Huntington disease
- htt
- huntingtin
- ROS
- reactive oxygen species
- KMO
- kynurenine 3-monooxygenase
- 3-HK
- 3-hydroxykynurenine
- HDAC
- histone deacetylase
- TBP
- TATA-binding protein
- DEG
- differentially expressed gene
- GO
- gene ontology
- SC
- synthetic complete.
REFERENCES
- 1. The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Chen S., Ferrone F. A., Wetzel R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wexler N. S., Lorimer J., Porter J., Gomez F., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S. A., Gayán J., Brocklebank D., Cherny S. S., Cardon L. R., Gray J., Dlouhy S. R., Wiktorski S., Hodes M. E., Conneally P. M., Penney J. B., Gusella J., Cha J. H., Irizarry M., Rosas D., Hersch S., Hollingsworth Z., MacDonald M., Young A. B., Andresen J. M., Housman D. E., De Young M. M., Bonilla E., Stillings T., Negrette A., Snodgrass S. R., Martinez-Jaurrieta M. D., Ramos-Arroyo M. A., Bickham J., Ramos J. S., Marshall F., Shoulson I., Rey G. J., Feigin A., Arnheim N., Acevedo-Cruz A., Acosta L., Alvir J., Fischbeck K., Thompson L. M., Young A., Dure L., O'Brien C. J., Paulsen J., Brickman A., Krch D., Peery S., Hogarth P., Higgins D. S., Jr., Landwehrmeyer B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3498–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giorgini F., Muchowski P. J. (2006) Methods Enzymol. 412, 201–222 [DOI] [PubMed] [Google Scholar]
- 5. Giorgini F., Muchowski P. J. (2009) Methods Mol. Biol. 548, 161–174 [DOI] [PubMed] [Google Scholar]
- 6. Giorgini F., Guidetti P., Nguyen Q., Bennett S. C., Muchowski P. J. (2005) Nat. Genet. 37, 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Panozzo C., Nawara M., Suski C., Kucharczyka R., Skoneczny M., Bécam A. M., Rytka J., Herbert C. J. (2002) FEBS Lett. 517, 97–102 [DOI] [PubMed] [Google Scholar]
- 8. Giorgini F. (2008) in Protein Misfolding in Biology and Disease (Outeiro T. F. ed) pp. 231–255, Transworld Research Network, Kerala, India [Google Scholar]
- 9. Giorgini F., Möller T., Kwan W., Zwilling D., Wacker J. L., Hong S., Tsai L. C., Cheah C. S., Schwarcz R., Guidetti P., Muchowski P. J. (2008) J. Biol. Chem. 283, 7390–7400 [DOI] [PubMed] [Google Scholar]
- 10. Kazantsev A. G., Thompson L. M. (2008) Nat. Rev. Drug. Discov. 7, 854–868 [DOI] [PubMed] [Google Scholar]
- 11. Mallory M. J., Strich R. (2003) J. Biol. Chem. 278, 44727–44734 [DOI] [PubMed] [Google Scholar]
- 12. Takemaru K., Harashima S., Ueda H., Hirose S. (1998) Mol. Cell. Biol. 18, 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takemaru K., Li F. Q., Ueda H., Hirose S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stevanin G., Fujigasaki H., Lebre A. S., Camuzat A., Jeannequin C., Dode C., Takahashi J., San C., Bellance R., Brice A., Durr A. (2003) Brain 126, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 15. Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., Hinnebusch A. G., Marton M. J. (2001) Mol. Cell. Biol. 21, 4347–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meriin A. B., Zhang X., He X., Newnam G. P., Chernoff Y. O., Sherman M. Y. (2002) J. Cell Biol. 157, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mumberg D., Müller R., Funk M. (1994) Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The R Development Core Team (2009) R: A Language and Environment for Statistical Computing, Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 19. Wettenhall J. M., Simpson K. M., Satterley K., Smyth G. K. (2006) Bioinformatics 22, 897–899 [DOI] [PubMed] [Google Scholar]
- 20. Wu Z., Irizarry R. A., Gentleman R., Murillo F. M., Spencer F. (2004) J. Am. Stat. Assoc. 99, 909–917 [Google Scholar]
- 21. Storey J. D., Tibshirani R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Breitkreutz B. J., Stark C., Tyers M. (2003) Genome Biol. 4, R22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mendes N. D., Casimiro A. C., Santos P. M., Sá-Correia I., Oliveira A. L., Freitas A. T. (2006) Bioinformatics 22, 2996–3002 [DOI] [PubMed] [Google Scholar]
- 24. Monteiro P. T., Mendes N. D., Teixeira M. C., d'Orey S., Tenreiro S., Mira N. P., Pais H., Francisco A. P., Carvalho A. M., Lourenço A. B., Sá-Correia I., Oliveira A. L., Freitas A. T. (2008) Nucleic Acids Res. 36, D132–D136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Nucleic Acids Res. 30, e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W. (1997) Genetics 147, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reimers M., Carey V. J. (2006) Methods Enzymol. 411, 119–134 [DOI] [PubMed] [Google Scholar]
- 29. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 30. Manogaran A. L., Fajardo V. M., Reid R. J., Rothstein R., Liebman S. W. (2010) Yeast 27, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I., Sagee S., Shenhar G. (2003) Int. Rev. Cytol. 224, 111–171 [DOI] [PubMed] [Google Scholar]
- 32. Kadosh D., Struhl K. (1997) Cell 89, 365–371 [DOI] [PubMed] [Google Scholar]
- 33. Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Biochim. Biophys. Acta 1731, 77–87; discussion 75–76 [DOI] [PubMed] [Google Scholar]
- 34. Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., Muchowski P. J. (2003) Science 302, 1769–1772 [DOI] [PubMed] [Google Scholar]
- 35. Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. (2007) Mol. Cell Proteomics 6, 439–450 [DOI] [PubMed] [Google Scholar]
- 36. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 37. Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., Gavin A. C., Hurt E. (2002) Mol. Cell 10, 105–115 [DOI] [PubMed] [Google Scholar]
- 38. Wyttenbach A., Swartz J., Kita H., Thykjaer T., Carmichael J., Bradley J., Brown R., Maxwell M., Schapira A., Orntoft T. F., Kato K., Rubinsztein D. C. (2001) Hum. Mol. Genet. 10, 1829–1845 [DOI] [PubMed] [Google Scholar]
- 39. Sipione S., Rigamonti D., Valenza M., Zuccato C., Conti L., Pritchard J., Kooperberg C., Olson J. M., Cattaneo E. (2002) Hum. Mol. Genet. 11, 1953–1965 [DOI] [PubMed] [Google Scholar]
- 40. Crocker S. F., Costain W. J., Robertson H. A. (2006) Brain Res. 1088, 176–186 [DOI] [PubMed] [Google Scholar]
- 41. Runne H., Régulier E., Kuhn A., Zala D., Gokce O., Perrin V., Sick B., Aebischer P., Déglon N., Luthi-Carter R. (2008) J. Neurosci. 28, 9723–9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doumanis J., Wada K., Kino Y., Moore A. W., Nukina N. (2009) PLoS One 4, e7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hinnebusch A. G. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hinnebusch A. G. (1997) J. Biol. Chem. 272, 21661–21664 [DOI] [PubMed] [Google Scholar]
- 45. Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., Fox L. A., Dang N., Johnston E. D., Oakes J. A., Tchao B. N., Pak D. N., Fields S., Kennedy B. K., Kaeberlein M. (2008) Cell 133, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu P. D., Harding H. P., Ron D. (2004) J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mascarenhas C., Edwards-Ingram L. C., Zeef L., Shenton D., Ashe M. P., Grant C. M. (2008) Mol. Biol. Cell 19, 2995–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsuo R., Kubota H., Obata T., Kito K., Ota K., Kitazono T., Ibayashi S., Sasaki T., Iida M., Ito T. (2005) FEBS Lett. 579, 2433–2438 [DOI] [PubMed] [Google Scholar]
- 49. Tain L. S., Mortiboys H., Tao R. N., Ziviani E., Bandmann O., Whitworth A. J. (2009) Nat. Neurosci. 12, 1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., Lu T. A., Benzer S., Kapahi P. (2009) Cell 139, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. King M. A., Hands S., Hafiz F., Mizushima N., Tolkovsky A. M., Wyttenbach A. (2008) Mol. Pharmacol. 73, 1052–1063 [DOI] [PubMed] [Google Scholar]
- 52. Yeger-Lotem E., Riva L., Su L. J., Gitler A. D., Cashikar A. G., King O. D., Auluck P. K., Geddie M. L., Valastyan J. S., Karger D. R., Lindquist S., Fraenkel E. (2009) Nat. Genet. 41, 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liang J., Clark-Dixon C., Wang S., Flower T. R., Williams-Hart T., Zweig R., Robinson L. C., Tatchell K., Witt S. N. (2008) Hum. Mol. Genet. 17, 3784–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.