Abstract
The synthesis of DNA in mitochondria requires the uptake of deoxynucleotides into the matrix of the organelle. We have characterized a human cDNA encoding a member of the family of mitochondrial carriers. The protein has been overexpressed in bacteria and reconstituted into phospholipid vesicles where it catalyzed the transport of all four deoxy (d) NDPs, and, less efficiently, the corresponding dNTPs, in exchange for dNDPs, ADP, or ATP. It did not transport dNMPs, NMPs, deoxynucleosides, nucleosides, purines, or pyrimidines. The physiological role of this deoxynucleotide carrier is probably to supply deoxynucleotides to the mitochondrial matrix for conversion to triphosphates and incorporation into mitochondrial DNA. The protein is expressed in all human tissues that were examined except for placenta, in accord with such a central role. The deoxynucleotide carrier also transports dideoxynucleotides efficiently. It is likely to be medically important by providing the means of uptake into mitochondria of nucleoside analogs, leading to the mitochondrial impairment that underlies the toxic side effects of such drugs in the treatment of viral illnesses, including AIDS, and in cancer therapy.
The inner membranes of mitochondria contain a family of proteins that transport various substrates and products into and out of the matrix. Family members have three tandem-repeated sequences, each of about 100 amino acids, made of two hydrophobic transmembrane α-helices joined by a large hydrophilic segment (thought to be an extramembranous loop; refs. 1–3). The tandem repeats contain conserved features. So far, 11 members of the family have been identified and sequenced. They are the uncoupling protein, and carriers for adenine nucleotides (ANC), phosphate, oxoglutarate, citrate, dicarboxylates, carnitine, ornithine, succinate-fumarate, oxaloacetate-sulfate, and oxodicarboxylates (1–7). The functions of other family members found in genome sequences are unknown. Caenorhabditis elegans encodes 38 family members, seven of them forming a subfamily related to the mammalian ANC. Three are ANC isoforms, but the rest are not. By extension of the sequence of a human expressed sequence tag (EST) encoding a protein fragment related to the subfamily, the protein sequence was completed. The protein was overexpressed in bacteria, purified, and reconstituted into phospholipid vesicles, where it transported deoxynucleoside diphosphates (or, in exchange for ADP or ATP, deoxynucleoside triphosphates less efficiently). The function of the protein is probably to act as a deoxynucleotide carrier (DNC) to supply precursors of mitochondrial DNA synthesis in the matrix. The protein also provides a potential route for uptake into the organelle of toxic antiviral nucleosides, such as 3′-azido-3′-deoxythymidine.
Materials and Methods
cDNA Sequencing.
To extend the EST sequence in 5′- and 3′-directions, PCRs were performed on adaptor-ligated double-stranded human liver cDNA (1 ng, CLONTECH; ref. 8) with primers AP1 and AP2 (CLONTECH) (NotI sites replaced by BamHI). Products were cloned into the pUC19 vector. The sequences of inserts were determined and assembled (8).
Bacterial Expression and Protein Purification.
The coding sequence was amplified from human cDNA by PCR with nucleotides 39–58 and 981–998 of the cDNA sequence (Fig. 1) as primers. The product was cloned into the pET21b vector. Transformants of Escherichia coli DH5α were selected on ampicillin (100 μg/ml) and screened by colony PCR and restriction digestion of plasmids. The sequences of inserts were verified. The encoded protein had additional C-terminal leucine and glutamate residues, followed by six histidines. The protein was overexpressed in E. coli BL21(DE3) (9). Purified inclusion bodies (9), suspended in buffer [10 mM NaCl/20 mM Pipes (pH 8.0)] were solubilized in sarkosyl [1.67% (wt/vol)] for 5 min at 0°C. This solution was diluted 20 times with buffer [0.1% sarkosyl/0.5 M NaCl/20 mM Pipes (pH 8.0)] and centrifuged (12,000 × g, 10 min, 4°C). The supernatant was put onto a Ni+-NTA-agarose column (Qiagen, Chatsworth, CA). Impurities were removed by decreasing the pH of the buffer (0.1% sarkosyl/20 mM Pipes) from 8.0 to 6.5. DNC eluted in the same buffer at pH 6.2.
Figure 1.
Sequence of a human cDNA and the encoded DNC. Amino acids are numbered from 1–320. An asterisk denotes the stop codon. Primers and probes are shaded. The nested primers 1F/2F and 1R/2R and probe 1P were used to confirm the EST sequence. The partial cDNA sequence was extended in 3′ and 5′ directions with primers AP1 and AP2 and nested oligonucleotides 3F/4F or 3R/4R, respectively. Primers RT 1F and RT 1R and probe RT 1P were used in reverse transcription–PCR experiments. Horizontal arrows pointing right and left indicate that primers were synthesized as shown or as the complement, respectively.
Proteins were analyzed by SDS/PAGE and stained with Coomassie blue. Their N termini were sequenced (8). The amount of pure DNC was estimated by laser densitometry of stained samples (8).
Transport Assays.
The recombinant protein in sarkosyl was reconstituted into liposomes in the presence of substrates (10). Both cardiolipin (1.14 mg/ml) and EDTA (1 mM) are essential for success. Transport was measured at 25°C with internal and external pHs at 6.8. It was started by adding [α-35S]dATP or [14C]-ADP and terminated after 2 min by addition of 100 μM of p-chloromercuribenzene sulfonate (10). Entrapped radioactivity was counted (10). The initial transport rate was calculated from the radioactivity taken up by proteoliposomes within 2 min (in the initial linear range). Other transport activities were assayed similarly (10). The amount of DNC incorporated into liposomes was measured as described (11) and varied between 15 and 20% of the protein added to the reconstitution mixture.
Expression Analysis.
Total human and mouse RNAs (2 μg) from various tissues were reverse transcribed with random hexamers or an oligo(dT)16 primer (final vol of 40 μl). Half of the reaction product was used as PCR template with forward and reverse primers RT 1F and RT 1R, respectively (Fig. 1), to amplify a cDNA fragment. The products were probed with radiolabeled oligonucleotide RT 1P (Fig. 1). As a control, a 384-bp cDNA fragment of β-actin was amplified from the rest of the reaction product with the primers 5′-GTTTGAGACCTTCAACACCC-3′ and 5′-CCAATGGTGATGACCTGGCC-3′. Mitochondria from rat tissues were solubilized in SDS. Proteins were separated by SDS/PAGE, transferred to nitrocellulose (11), and exposed to a rabbit antiserum against human DNC. Immunoconjugates were detected with a secondary antibody (horseradish peroxidase coupled to anti-rabbit Ig) and 3,3′-diaminobenzidine as peroxidase substrate.
Results
Sequence of the Human DNC.
By phylogenetic analysis of the sequences of all C. elegans mitochondrial carriers and of mammalian carriers of known function, a seven-protein subfamily related to the ANC was found. With their sequences, a human EST (THC91779) that encoded a related sequence was identified. THC91779 overlapped three other human EST clones (THC90932, THC132863, and N40412). This partial cDNA of 769 nucleotides was extended to the final sequence (Fig. 1), which encodes a protein with a molecular mass of 34,588. The assignment of the translational initiation codon is consistent with an inframe stop codon 27 base pairs upstream. The protein sequence has the characteristics of the family of mitochondrial carriers. One C. elegans clone (C42C1.10) is 39% identical to the human sequence over residues 1–364 of its 649-amino acid sequence.
Characterization of Recombinant DNC.
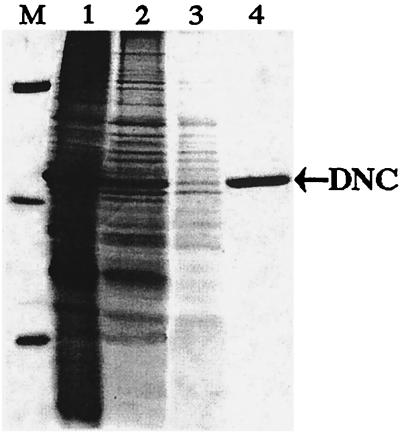
The DNC accumulated as inclusion bodies in E. coli BL21(DE3) (see Fig. 2, lane 1). The purified protein was homogeneous (Fig. 2, lane 4) with an apparent molecular mass of 36 kDa (calculated value with initiator methionine and His-tail, 36,310). Its N-terminal sequence (VGYDPKPDGR) is identical to residues 2–11 of the protein encoded in the cDNA. About 80 mg of purified protein was obtained per liter of culture.
Figure 2.
Purification of DNC by Ni+-agarose affinity chromatography. Proteins were separated by SDS/PAGE and stained with Coomassie blue. Lane M, markers (BSA, carbonic anhydrase, and cytochrome c); lane 1, sarkosyl extract of inclusion bodies; lane 2, pH 6.8 eluate; lane 3, pH 6.5 eluate; lane 4, purified DNC, eluted at pH 6.2. The position of DNC is indicated on the right by an arrow.
Transport Properties.
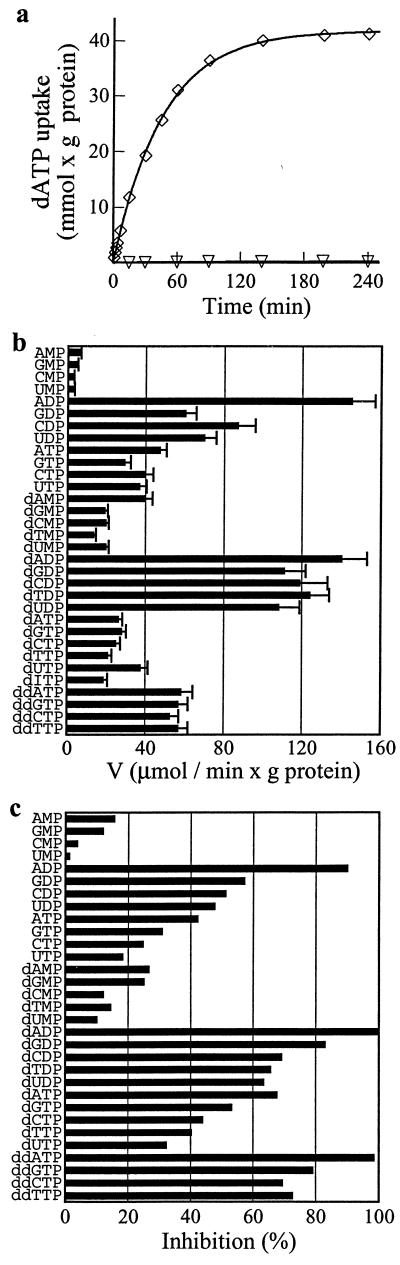
The reconstituted human DNC catalyzed the exchange of [α-35S]dATP for dADP or ADP with first-order kinetics (rate constant 0.02 min-1), isotopic equilibrium being approached exponentially (see Fig. 3a). Uptake of external substrate required internal substrate. It did not catalyze homoexchanges of malate, fumarate, oxoglutarate, carnitine, glutamate, aspartate, glutamine, or ornithine (internal concentration, 10 mM; external concentration, 1 mM). Transport under either saturating or nonsaturating concentrations of external [α-35S]dATP (1 mM and 0.02 mM, respectively) with 10 mM internal ADP, had a sharp pH optimum at 6.8.
Figure 3.
Time course of dATP/ADP exchange and substrate specificity of human DNC. (a) Time course of [α-35S]dATP/ADP exchange in proteoliposomes reconstituted with the recombinant DNC. [α-35S]dATP (1 mM) was added to proteoliposomes containing 10 mM ADP (⋄) or 10 mM NaCl (▿) (b) Dependence of DNC activity on internal substrate. Proteoliposomes were preloaded internally with various substrates (concentration 10 mM). Transport was started by addition of 20 μM [α-35S]dATP and stopped after 2 min. The values are means ± SD of at least three experiments. (c) Inhibition of the rate of [α-35S]dATP uptake by external substrates. Proteoliposomes were preloaded internally with 10 mM ADP. Transport was started by adding 125 μM [α-35S]dATP and stopped after 2 min. External substrates (concentration 0.5 mM) were added together with [α-35S]dATP. The extents of inhibition (%) from a representative experiment are reported. The control value for uninhibited exchange was 0.45 mmol/min per gram of protein.
The recombinant DNC had its highest affinity for ADP and dNDPs from the internal side of the proteoliposomal membrane (Fig. 3b). The highest rates of [α-35S]dATP uptake into proteoliposomes were with internal ADP or dADP. High activities also were found with the other internal dNDPs; significant activities were found with GDP, CDP, and UDP; much lower activities with NTPs, dNTPs, dNMPs, and pyrophosphate (not shown); and virtually no activity with NMPs and (not shown) NADH, adenine, deoxyadenosine, phosphate, oxoglutarate, citrate, glycine, carnitine, adenosine, guanosine, cytidine, uridine, deoxyguanosine, deoxycytidine, deoxythymidine, deoxyuridine, guanine, cytosine, thymine, or uracil. The exchange of pyrophosphate, but not of adenine or deoxyadenosine, shows that phosphate groups are essential for transport, but as pyrophosphate exchange was 22% of ADP exchange, the nucleoside moiety is also important. Dideoxynucleoside triphosphates also exchanged with dATP at twice the rate of dNTPs (see Fig. 3b).
External nucleoside and deoxynucleoside mono-, di-, and triphosphates inhibited the [α-35S]dATP/ADP exchange (see Fig. 3c). Nucleoside diphosphates were more effective than either triphosphates or monophosphates. Deoxynucleotides were more potent than the corresponding nucleotides, and dideoxynucleoside triphosphates were as effective as dNDPs. The rate of dATP uptake was more sensitive to purine than pyrimidine nucleotides, and adenine nucleotides inhibited better than guanine nucleotides. Adenosine, guanosine, cytidine, uridine, deoxyadenosine, deoxyguanosine, deoxycytidine, deoxythymidine, deoxyuridine, adenine, guanine, cytosine, thymine, and uracil had no effect (data not shown).
The uptake of 50 μM [α-35S]dATP (internal substrate, 10 mM ADP; reaction time, 2 min) was inhibited completely by 0.1 mM p-chloromercuribenzene sulfonate and 10 mM pyridoxal 5′-phosphate (inhibitors of many mitochondrial carriers), and partly (41%) by 10 mM bathophenathroline [another strong inhibitor of several mitochondrial carriers (5–9)]. High concentrations of carboxyatractyloside (0.1 mM) and bongkrekate (0.01 mM) (inhibitors of the ANC) were partly effective on the DNC (42 and 37% inhibition, respectively). A specific inhibitor of the mitochondrial citrate carrier, 2 mM 1,2,3-benzenetricarboxylate, reduced the dATP/ADP exchange rate to 40%, possibly because this compound as the optimal substrate for DNC carries three negative charges. No significant inhibition was observed with 2 mM butylmalonate, phenylsuccinate, α-cyano-4-hydroxycinnamate and N-ethylmaleimide (inhibitors of other characterized mitochondrial carriers), and 0.033 mM cytochalasin B (inhibitor of a plasma membrane nucleoside transporter).
The exchange rate of internal ADP or dADP (10 mM) depended on the external concentration of [α-35S]dATP (20–1,000 μM) or [14C]ADP (8–400 μM). With both external substrates, linear functions were obtained in double-reciprocal plots. They were independent of the internal substrate and intersected the ordinate close to a common point (not shown). For ADP and dATP, the transport affinities (Km) were 42.6 ± 4.7 μM and 106 ± 15 μM (mean values of 6 and 63 experiments, respectively). The average value of Vmax was 0.85 ± 0.15 mmol/min per gram of protein. Several external substrates were competitive inhibitors of [α-35S]dATP uptake (Table 1). They increased the apparent Km without change in Vmax (not shown). These results confirm that dADP is the highest-affinity external substrate (Ki 14 μM). Furthermore, the Ki values of all of the dNDPs are two to three times and four to five times lower than those of their corresponding NDPs or dNTPs, respectively. The affinity of the DNC for ddNTPs is very high (Ki for ddATP, 25 μM) and similar to that of the dNDPs. The Ki of the antiviral drug ddCTP is 70 μM.
Table 1.
Competitive inhibition by various substrates of [α-35S]dATP uptake into proteoliposomes containing the DNC
| Substrate | Ki, μM |
| ATP | 470 ± 50 |
| ADP | 32 ± 3.4 |
| GDP | 197 ± 22 |
| CDP | 284 ± 27 |
| UDP | 380 ± 40 |
| dADP | 14 ± 1.2 |
| dGDP | 55 ± 5.9 |
| dCDP | 99 ± 9.8 |
| dTDP | 117 ± 13 |
| dUDP | 179 ± 19 |
| dGTP | 230 ± 25 |
| dCTP | 423 ± 39 |
| dTTP | 595 ± 62 |
| dUTP | 963 ± 94 |
| ddATP | 25 ± 2.3 |
| ddGTP | 60 ± 5.3 |
| ddCTP | 70 ± 7.4 |
| ddTTP | 68 ± 7.1 |
The concentration of [α-35S]dATP was 20–1,000 μM. Competing substrates were added at appropriate concentrations. For other conditions, see Fig. 3b. The Ki values were calculated from double-reciprocal plots of the rate of [α-35S]dATP uptake versus substrate concentration. The values are averages ± SD from at least three experiments.
Tissue Distribution.
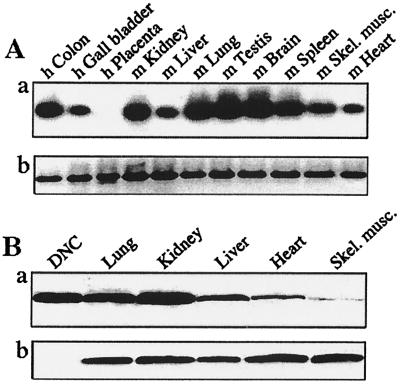
High levels of mRNA for the DNC were detected in colon, kidney, lung, testis, spleen, and brain, and lower amounts in gall bladder, liver, skeletal muscle, and heart (Fig. 4A). Similarly abundant levels of protein expression were found in rat mitochondria from kidney, lung, and liver, and lower levels in skeletal muscle and heart (Fig. 4B). The only tissue with no detectable DNC transcripts was human placenta, possibly because RNA was extracted postpartum, when biosynthetic activity is low.
Figure 4.
Expression of human DNC in various tissues. Analysis of total RNA from human (h) and mouse (m) tissues (A). (a) Hybridization of cDNA fragments for the DNC with probe RT 1P. (b) Ethidium-bromide-stained cDNA fragments for β-actin. (B) Immunodetection of the DNC in mitochondria isolated from rat tissues. In a and b, mitochondria (150 μg of protein) and human DNC (75 ng) were exposed to antisera to the DNC and subunit IV of the cytochrome c oxidase, respectively.
Discussion
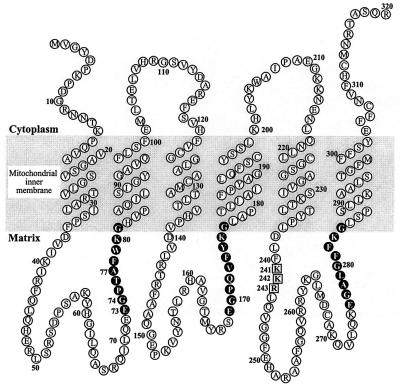
The sequence of the human DNC consists of three homologous repeats of about 100 amino acids and contains sequence motifs characteristic of the mitochondrial carrier family. Like other family members, it appears to have six transmembrane α-helices (Fig. 5). It is about 22% identical to mammalian ANCs. The ANCs contain a strictly conserved motif (RRR) in the third large hydrophilic loop thought to be involved in nucleotide binding (12) and a similar motif (KKR) is at residues 241–243 of the DNC, presumably fulfilling a similar role. The DNC also contains a sequence at residues 73–77 in the first large loop that conforms to the sequence motif EGXXA, the P-box of the DNA-binding domain of nuclear receptors (13). The same motif is also found in the loop connecting the fifth and sixth α-helices in mammalian ANCs and in the uncoupling protein (UCP1) (12, 14). In rat UCP1, the motif is thought to be involved in controlling its activity via GDP binding. Therefore, it is likely that the P-box motif in the DNC is also involved in binding nucleotide substrates or, alternatively, that it interacts directly with mtDNA.
Figure 5.
Folding of the DNC in the inner membranes of mitochondria. The topography of the six transmembrane α-helices is based on the hydrophobic profile of the sequence in Fig. 1. Each of the three tandem repeats in the sequence is folded into two transmembrane α-helices with a large intervening hydrophilic loop. The three repetitive elements are linked by shorter loops. The cytoplasmic and matrix locations of the various features are based on experimental evidence of locations of analogous features in other members of the family of mitochondrial carriers. The sequences in black are related to the DNA-binding domain of the nuclear receptor family. Residues 241–243 (in squares) correspond to the sequence RRR at residues 234–236 of the ANC from Saccharomyces cerevisiae.
Reconstituted DNC catalyzes an exchange reaction between nucleotides and deoxynucleotides. The best internal substrates are dNDPs and ADP, whereas dNDPs, dNTPs, and NDPs are the best external ones (dNDPs have the highest affinity). If the carrier is oriented in the liposomal membrane as in mitochondria, it is likely that the DNC catalyzes the uptake of dNDPs into the mitochondrial matrix. All dNDPs are transported by DNC. Once in the mitochondrion, the dNDPs will be converted to the corresponding triphosphate and incorporated into the mtDNA by the DNA polymerase-γ. Because ribonucleotide reductase is found in the cytosol of eukaryotic cells (15), the DNC appears to be essential for mtDNA synthesis. The higher Ki values for the dUDP and dUTP (Table 1), which are not incorporated into DNA, support the view that DNC is involved primarily in the mtDNA synthesis. Radioactive dNTPs are taken up by isolated mitochondria and incorporated into mtDNA (16–18) but, because they have lower affinities for the DNC than dNDPs, it is unlikely that they are its physiological substrates. The internal counterion for exchange could be ADP or ATP (Fig. 3b), but ATP is exchanged at a lower rate. In the resting state, the intramitochondrial ATP/ADP ratio is about 4 (19), and the rate of exchange of external dNDPs for internal ATP would be favored by the proton electrochemical gradient generated by electron transport. Internal GDP was exported rather poorly (Fig. 3b) and, in comparison with adenine nucleotides, is present in the mitochondrial matrix in minute amounts. It is improbable that internal GDP is the physiological counteranion for the uptake of dNDPs.
Human DNCs can exchange ddNTPs much more efficiently than the corresponding deoxy analogs (Fig. 3 b and c). Furthermore, the inhibition constants of external ddNTPs are close to those of dNDPs (Table 1). Therefore, ddNDPs (which are not available commercially) may well be the best substrates to be transported by the DNC. These properties suggest that the DNC is involved directly in the cytotoxicity of antiviral and anticancer nucleoside analogs such as 2′,3′-dideoxycytidine, 2′,3′-dideoxyinosine, and 3′-azido-3′-deoxythymidine. Cytoplasmic kinases convert these and other dideoxynucleosides to their mono-, di-, and triphosphate derivatives (17, 20). The latter two products would be expected to be transported into mitochondria by the DNC, there to inhibit the synthesis of mtDNA by competing with dNTPs for the active site of the DNA polymerase-γ and by chain termination (21). Clinical and laboratory findings have shown that the mechanism of toxicity of most antiviral and anticancer nucleoside analogs is to impair mitochondrial function (17, 20, 22–25). In fact, the main side effects of these drugs, myopathy, cardiomiopathy, polyneuropathy, and lactic acidosis, greatly resemble the spectrum of clinical manifestations seen in inherited mitochondrial diseases (26). Furthermore, after prolonged assumption of these drugs, histological findings commonly associated with depletion of mtDNA, such as red-ragged fibers, are observed (27). It should be noted that the antiviral nucleotide analogs strongly interfere with the action of the viral reverse transcriptases and the mtDNA polymerase-γ but have a very low affinity for the nuclear DNA polymerases (17, 28, 29). When the diphosphate and triphosphate derivatives of these drugs and other related potential antivirals become available, it should be possible to assess their potential toxicity by studying their import into proteoliposomes reconstituted with the human DNC. Such assays could help in the development of less toxic antiviral compounds.
Acknowledgments
This work was supported by the Consiglio Nazionale delle Ricerche Target Project “Biotechnology”; by the Ministero dell'Università e della Ricerca Scientifica e Tecnologica 5%-Consiglio Nazionale delle Ricerche program “Biomolecole per la Salute Umana” L. 95/95; by the Piano Biomedicina, progetto n.1 Cluster C04 L. 488/92; by an Italian National PRIN grant for Bioenergetics and Membrane Transport, by the European Social Fund, and by The Medical Research Council, Cambridge, U.K.
Abbreviations
- ANC
adenine nucleotide carrier
- DNC
deoxynucleotide carrier
- EST
expressed sequence tag
- d
deoxy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the EMBL database (accession no. AJ251857).
References
- 1.Walker J E. Curr Opin Struct Biol. 1992;2:519–526. [Google Scholar]
- 2.Palmieri F. FEBS Lett. 1994;346:48–54. doi: 10.1016/0014-5793(94)00329-7. [DOI] [PubMed] [Google Scholar]
- 3.Palmieri F, van Ommen B. In: Frontiers in Cellular Bioenergetics. Papa S, Guerrieri F, Tager J M, editors. New York: Kluwer Academic/Plenum; 1999. pp. 489–519. [Google Scholar]
- 4.Palmieri L, De Marco V, Iacobazzi V, Palmieri F, Runswick M J, Walker J E. FEBS Lett. 1997;410:447–451. doi: 10.1016/s0014-5793(97)00630-3. [DOI] [PubMed] [Google Scholar]
- 5.Palmieri L, Lasorsa F M, De Palma A, Palmieri F, Runswick M J, Walker J E. FEBS Lett. 1997;417:114–118. doi: 10.1016/s0014-5793(97)01269-6. [DOI] [PubMed] [Google Scholar]
- 6.Palmieri L, Vozza A, Agrimi G, De Marco V, Runswick M J, Palmieri F, Walker J E. J Biol Chem. 1999;274:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- 7.Palmieri L, Agrimi G, Runswick M J, Fearnley I M, Palmieri F, Walker J E. J Biol Chem. 2001;276:1916–1922. doi: 10.1074/jbc.M004332200. [DOI] [PubMed] [Google Scholar]
- 8.Fiermonte G, Palmieri L, Dolce V, Lasorsa F M, Palmieri F, Runswick M J, Walker J E. J Biol Chem. 1998;273:24754–24759. doi: 10.1074/jbc.273.38.24754. [DOI] [PubMed] [Google Scholar]
- 9.Fiermonte G, Walker J E, Palmieri F. Biochem J. 1993;294:293–299. doi: 10.1042/bj2940293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmieri F, Indiveri C, Bisaccia F, Iacobazzi V. Methods Enzymol. 1995;260:349–369. doi: 10.1016/0076-6879(95)60150-3. [DOI] [PubMed] [Google Scholar]
- 11.Fiermonte G, Dolce V, Palmieri F. J Biol Chem. 1998;273:22782–22787. doi: 10.1074/jbc.273.35.22782. [DOI] [PubMed] [Google Scholar]
- 12.Muller V, Basset G, Nelson D R, Klingenberg M. Biochemistry. 1996;35:16132–16143. doi: 10.1021/bi960667r. [DOI] [PubMed] [Google Scholar]
- 13.Schwabe J W, Chapman L, Finch J T, Rhodes D. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Barroso M M, Fleury C, Jimenez M A, Sanz J M, Romero A, Bouillaud F, Rial E. J Mol Biol. 1999;292:137–149. doi: 10.1006/jmbi.1999.3049. [DOI] [PubMed] [Google Scholar]
- 15.Engstrom Y, Rozell B. EMBO J. 1988;7:1615–1620. doi: 10.1002/j.1460-2075.1988.tb02987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons P, Simpson M V. J Biol Chem. 1973;248:1912–1919. [PubMed] [Google Scholar]
- 17.Chang C N, Skalski V, Zhou J H, Cheng Y C. J Biol Chem. 1992;267:22414–22420. [PubMed] [Google Scholar]
- 18.Enriquez J A, Ramos J, Perez-Martos A, Lopez-Perez M J, Montoya J. Nucleic Acids Res. 1994;22:1861–1865. doi: 10.1093/nar/22.10.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letko G, Kuster U, Duszynski J, Kunz W. Biochim Biophys Acta. 1980;593:196–203. doi: 10.1016/0005-2728(80)90057-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhu C, Johansson M, Permert J, Karlsson A. Biochem Pharmacol. 1998;56:1035–1040. doi: 10.1016/s0006-2952(98)00150-6. [DOI] [PubMed] [Google Scholar]
- 21.De Clercq E. J Acquired Immune Defic Syndr. 1991;4:207–218. [PubMed] [Google Scholar]
- 22.Pedrol E, Masanes F, Fernandez-Sola J, Cofan M, Casademont J, Grau J M, Urbano-Marquez A. J Neurol Sci. 1996;138:42–48. doi: 10.1016/0022-510x(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 23.Lewis W, Dalakas M C. Nat Med. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal R P, Olivero O A. Mutat Res. 1997;390:223–231. doi: 10.1016/s1383-5718(97)00014-4. [DOI] [PubMed] [Google Scholar]
- 25.Benbrik E, Chariot P, Bonavaud S, Ammi-Said M, Frisdal E, Rey C, Gherardi R, Barlovatz-Meimon G. J Neurol Sci. 1997;149:19–25. doi: 10.1016/s0022-510x(97)05376-8. [DOI] [PubMed] [Google Scholar]
- 26.Brinkman K, ter Hofstede H J M, Burger D M, Smeitink J A, Koopmans P P. AIDS. 1998;12:1735–1744. doi: 10.1097/00002030-199814000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Dalakas M C, Illa I, Pezeshkpour G, Laukaitis J, Cohen B, Griffin J. N Engl J Med. 1990;322:1098–1105. doi: 10.1056/NEJM199004193221602. [DOI] [PubMed] [Google Scholar]
- 28.Lewis W, Simpson J F, Meyer R R. Circ Res. 1994;74:344–348. doi: 10.1161/01.res.74.2.344. [DOI] [PubMed] [Google Scholar]
- 29.Bridges E G, Abdesslem F, Sommadossi J-P. Biochem Pharmacol. 1993;45:1571–1576. doi: 10.1016/0006-2952(93)90296-9. [DOI] [PubMed] [Google Scholar]