Abstract
Proteases are a ubiquitous group of enzymes that play key roles in the life cycle of parasites, in the host-parasite relationship, and in the pathogenesis of parasitic diseases. Furthermore, proteases are targets for the development of new anti-parasitic therapy. Protozoan parasites like Leishmania predominantly express Clan CA cysteine proteases for key life cycle functions. It was therefore unexpected to find a high level of serine protease activity expressed by Leishmania donovani. Purification of this activity followed by mass spectrometry identified oligopeptidase B (OPB; Clan SC, family S9A) as the responsible enzyme. This was confirmed by gene knock-out of OPB, which resulted in the disappearance of the detected serine protease activity of Leishmania extracts. To delineate the specific role of OPB in parasite physiology, proteomic analysis was carried out on OPB(−/−) versus wild type parasites. Four protein species were significantly elevated in OPB(−/−) parasites, and all four were identified by mass spectrometry as enolase. This increased enolase was enzymatically inactive and associated with the parasite membrane. Aside from its classic role in carbohydrate metabolism, enolase was recently found to localize to membranes, where it binds host plasminogen and functions as a virulence factor for several pathogens. As expected, there was a striking alteration in macrophage responses to Leishmania when OPB was deleted. Whereas wild type parasites elicited little, if any, response from infected macrophages, OPB(−/−) parasites induced a massive up-regulation in gene transcription. Additionally, these OPB(−/−) parasites displayed decreased virulence in the murine footpad infection model.
Keywords: Parasite, Parasitology, Protease, Serine Protease, Trypanosome, Enolase, Leishmania, Virulence, donovani, major
Introduction
Proteases are a ubiquitous group of enzymes functioning in nearly all biological phenomena. In protozoan parasites, proteases play key roles in life cycle transition, host invasion, parasite immune evasion, and the pathogenesis of parasitic diseases (1, 2). At least 50 cysteine proteases and twenty serine proteases are known to exist in the Leishmania genome (3–5). The molecular evolution of proteases shows a striking dichotomy between the predominant Clan CA cysteine proteases of protozoa and primitive metazoa and the dominant trypsin family (S1A) serine proteases of vertebrates (6). It was therefore unexpected to find that Leishmania donovani, Leishmania major, and Leishmania mexicana (but not the related kinetoplastid parasite, Trypanosoma cruzi) express high levels of serine protease activity, relative to cysteine protease activity in parasite extracts.
To delineate the character of this serine protease activity, it was purified from Leishmania extracts and identified as Clan SC, family S9A oligopeptidase B by mass spectrometry. Oligopeptidase B (OPB)2 has been identified in other protozoa, including T. cruzi, Trypanosoma brucei, and Trypanosoma evansi (7–9). These enzymes represent a subfamily of potential chemotherapeutic targets (10) because their inhibitors have been shown to be trypanocidal (4, 11, 12). OPB of T. cruzi is involved in the invasion of host cells through the generation of an unknown signaling ligand that interacts with host cells in order to recruit lysosomes to their surface (7). A related T. cruzi oligopeptidase has also been implicated in tissue invasion by degrading components of the extracellular matrix (13).
In this study, we describe the identification and characterization of Leishmania OPB. Native and recombinant enzyme was purified and biochemically characterized. The single copy oligopeptidase B gene was deleted by targeted gene replacement. The role of oligopeptidase B was then analyzed both in in vitro culture and in the murine model of infection. To delineate a specific role for oligopeptidase B in the Leishmania life cycle, two-dimensional gel electrophoresis was used to compare the proteomes of wild type and OPB(−/−) parasites. Enolase was identified as a major target of oligopeptidase B, and a putative role for enolase in Leishmania-macrophage interactions was explored. Enolase is present on the cell surface of several parasites, including Leishmania, where it has been shown to bind host plasminogen (14). It may therefore have a role in macrophage entry via plasminogen receptors and parasite dissemination from the sand fly bite site (15, 16).
EXPERIMENTAL PROCEDURES
Animals and Parasite Strains
Commercially bred, 6–8-week-old, female BALB/c mice (Mus musculus) were used for the murine footpad infection model. Microarray analysis was performed on bone marrow-derived macrophages from female C57BL/6 mice (Charles River Laboratories International, Inc., Davis, CA). Leishmania donovani donovani MHOMISD/OO/lS-2D (Ldd IS C12) was used for enzyme isolation and knock-out studies. L. major LV39 MRHO/SU/59/P was also used for knock-out studies as well as animal infections. Serine protease activity assays were also performed on lysates from L. mexicana MNYC/BZ/62/M379 and T. cruzi M/HOM/AR/74/CA-I CL72. Leishmania promastigotes were cultured at 27 °C in M199 (Sigma) liquid medium as described previously (17). Axenic amastigotes of L. donovani were cultured at 37 °C in 100% fetal bovine serum (Omega Scientific Inc., Tarzana, CA) as described previously (18).
Parasite Lysates and Protease Determination
Cultures of Leishmania promastigotes, axenic amastigotes, and T. cruzi amastigotes and epimastigotes were pelleted and resuspended three times with phosphate-buffered saline. The resultant washed pellets were then lysed in 50 mm Tris-HCl, pH 8.0, with 0.25% Triton X-100 (Sigma) at 4 °C for 1 h. These solutions were then centrifuged, and the supernatants were isolated and sterilized with 0.2-μm syringe filters (Nalgene Nunc, Rochester, NY). Protein concentrations were determined by NanoDrop 1000 (Thermo Fisher Scientific Inc., Waltham, MA). Protease activity was determined by measuring the hydrolysis of the fluorogenic peptide substrate benzyloxycarbonyl-prolyl-arginyl-7-amido-4-methylcoumarin (Z-PR-AMC) (Peptides International, Inc., Louisville, KY) in 50 mm Tris-HCl, pH 8.0 and 5.5, with and without 10 μm N-[N-(l-3-trans-carboxyoxirane-2-carbonyl)-l-leucyl]-agmatine) (E-64) or 2 mm PEFABLOC (4-(2-aminoethyl)benzenesulfonylfluoride, HCl (AEBSF)) (EMD Chemicals, Gibbstown, NJ) (see procedures for the protease activity assay below).
Purification and Identification of Serine Protease Activity (OPB) from Leishmania Lysate by Mass Spectrometry
Filtered lysate from 109 L. donovani promastigotes, prepared described above, was loaded onto a HiTrap Q-Sepharose Fast Flow column using an ÄKTA purifier (GE Healthcare). A 50 mm Tris-HCl, pH 8.0, buffer was used for column equilibration, sample loading, and eluting at a 1 ml/min flow rate. Protein was eluted using a linear gradient of 0–1 m NaCl over 20 column volumes while collecting 0.25-ml fractions. OPB-containing fractions were identified by Z-PR-AMC hydrolysis at pH 8.0. Peak OPB-containing fractions were run on a Criterion 10–20% Tris-HCl SDS-polyacrylamide gel and stained by Coomassie (Bio-Rad). Protein bands were excised, and tryptic digests were performed. Samples were analyzed by LC-MS/MS with a QStar XL (Applied Biosystems, Foster City, CA) equipped with a Protana nanospray ion source (Protana, Inc., Toronto, Canada). A peptide mass accuracy tolerance of 100 ppm and a fragment ion mass accuracy tolerance of 0.15 Da were used. A database search was conducted using Mascot (Matrix Science Inc., Boston, MA) on the Sanger Institute L. major GeneDB.
OPB Cloning and Sequencing
Genomic DNA from L. donovani was isolated as described previously (19). The full-length OPB gene was then amplified from the genomic DNA by PCR using the Expand High Fidelity PCR System (Roche Applied Science). The forward (5′-CTC AGA TCT CCA CCC CCA CCA GCC CGG-3′) and reverse (5′-CTC AGA TCT CCT AGA GTT AAT GAT CAC GAC TC-3′) primers (BglII sites, in boldface type, were included on the primers for cloning) were designed based on the non-coding sequences flanking the Leishmania infantum OPB open reading frame (L. infantum GeneDB: LinJ09_V3.0820). The resultant 2.2-kb PCR product was cloned into pCR2.1-TOPO using TOPO TA cloning and sequenced using the included sequencing primers (Invitrogen). The amino acid sequence of L. donovani OPB was aligned with homologous sequences from L. infantum, L. major, Leishmania braziliensis, T. cruzi, T. brucei, Leishmania amazonensis, and T. evansi (respectively, GeneDB: LinJ09_V3.0820, LmjF09.0770, LbrM09_V2.0850, Tc00.1047053511557.10, and Tb11.52.0003; GenBankTM: ABQ23350 and AAS55050) using the ClustalW algorithm from MegAlign (DNASTAR, Madison, WI). The Pichia pastoris transformation construct was generated by PCR amplification of OPB, adding a 5′ XhoI site (boldface type) followed by a Kex2 cleavage site (underlined) and a 3′ XbaI site (underlined boldface type) using forward (5′-CTC CTC GAG AAA AGA ATG TTG TCG GGC AAC ACC ATC G-3′) and reverse (5′-CTC TCT AGA TTA CCT GCG AAC CAG CAG GCG-3′) primers and then cloned into pPICZα A (Invitrogen).
Expression and Purification of Recombinant OPB from P. pastoris
The pPICZα-OPB construct was electroporated into X-33 P. pastoris, and expression clones were isolated and induced for 48 h as per the manufacturer's protocol. Supernatant from induced cultures was harvested by centrifugation at 3000 × g for 10 min, followed by 0.2-μm filtration (Nalgene Nunc) and lyophilization. The crude lyophilized protein was resuspended in 20% of its original volume using 10 mm Tris-HCl, pH 8.0, with 1 m ammonium sulfate and loaded onto a HiTrap butyl-Sepharose 4FF hydrophobic interaction column (GE Healthcare). A 10 mm Tris-HCl, pH 8.0, buffer with 1 m ammonium sulfate was used for column equilibration, sample loading, and washing at a 1 ml/min flow rate. The ammonium sulfate concentration was decreased to 0.75 m for 20 column volumes to elute the OPB protein. The eluate was desalted by buffer exchanging with 10 mm Tris-HCl, pH 8.0, and then concentrated on an Amicon Ultra-4 30,000 Nominal Molecular Weight Limit filter device (Millipore, Billerica, MA). The concentrated sample was loaded onto a MonoQ HR 5/5 anion exchange column (GE Healthcare) on an ÄKTA purifier and fractionated as described above for the Q-Sepharose column. OPB-containing fractions were identified by Z-PR-AMC hydrolysis. Peak fractions were pooled, desalted, and then reconcentrated as before. OPB protein concentration was measured by NanoDrop 1000.
Protease Activity Assays and OPB Enzymatic Characterization
Protease activity was measured using peptide substrates containing C-terminal AMC or ACC (7-amino-4-carbamoyl-methylcoumarin) as the fluorogenic leaving group. Reactions contained 20 μm substrate unless otherwise noted. Enzyme samples were mixed with substrate in 50 mm Tris-HCl, pH 8.0, with 0.2% DMSO in a 150-μl total volume. Hydrolysis of the substrates was measured at 25 °C using a FlexStation microplate spectrofluorimeter (Molecular Devices, Sunnyvale, CA). Excitation/emission for AMC and ACC were 355/460 and 380/460 nm, respectively. Vmax values were calculated using the accompanying SoftMax Pro version 4.8 software. An inhibitor or salt was preincubated with OPB for 10 min prior to the addition of substrate and then read on a FlexStation. To determine the pH preference of OPB, protease activity assays were performed in triplicate using the Z-PR-AMC substrate at pH 3.0–10.5 with a 0.5-pH unit step size in 100 mm citrate phosphate buffer (pH 3.0–7.0), sodium phosphate buffer (pH 6.5–8.0), Tris-HCl buffer (pH 7.5–9.0), and glycine-NaOH buffer (pH 8.5–10.5).
Positional Scanning Synthetic Combinatorial Library Profiling and Validation
The P1–P4 substrate specificity profile for OPB was determined using a completely diverse positional scanning synthetic combinatorial library (PS-SCL) as has been described previously (20). This library is composed of substrates that are N-terminally acetylated and have an ACC fluorogenic leaving group. Protease activity assays were performed using 1 μl of substrate in a 150-μl reaction with 50 mm Tris-HCl, pH 8.0, and 0.02 ng of enzyme. Assays were performed in duplicate. To test the predictive capability of the PS-SCL substrate specificity profile, fluorogenic peptides that were predicted to be either good or bad substrates of OPB were selected for screening in protease activity assays. The following substrates were used at 20 μm: Glut-GR-AMC, Z-LR-AMC, Ac-VFRSLK-AMC, H-A-AMC, H-R-AMC, H-K-AMC, Boc-VLK-AMC, Boc-EEK-AMC, Ac-DEVD-AMC, Ac-WEHD-AMC, Z-LLE-AMC, H-L-AMC, Suc-GPLGP-AMC (Peptides International, Inc.), Ac-KQKLR-AMC (MP Biomedicals), Z-GGR-AMC, Boc-LGR-AMC, Z-GPR-AMC, Z-PR-AMC, Z-VVR-AMC, Z-RR-AMC, Boc-LRR-AMC, Ac-FR-AMC, Z-FR-AMC, Boc-RVRR-AMC, Z-VAD-AMC, Suc-AAF-AMC, H-GF-AMC, Z-GGL-AMC, Suc-LY-AMC, Suc-LLVY-AMC (Bachem California Inc., Torrance, CA), Z-AARR-AMC, Suc-PSPF-AMC, Z-AEN-AMC, Ac-ASN-AMC, and Ac-AVN-AMC. To test the PS-SCL prediction that glycine is the preferred P2 amino acid, two substrates that differ only at the P2 position, Z-GGR-AMC and Z-GPR-AMC, were compared. Protease activity assays were performed on these substrates at concentrations ranging from 80 to 0.2 μm, and the Kcat and Km values were calculated.
Constructs for Targeted Gene Deletion of OPB
Two knock-out cassettes (one for each allele) were created to delete the L. donovani and L. major OPB genes. These cassettes, cloned into the pGEM-9Zf(−) vector (Promega, Madison, WI), each contained an antibiotic resistance gene, hygr (conferring hygromycin B resistance) (21) or pacr (conferring puromycin resistance) (22), followed by 1.5 kb of the 3′-untranslated region of the L. major dhfr-ts gene (23). This untranslated region ensures high level expression during the Leishmania life cycle. To target these knock-out cassettes to the OPB locus, 5′- and 3′-targeting flanks were created and ligated upstream and downstream, respectively, of the antibiotic resistance cassettes. The targeting flanks were generated by PCR amplification of the untranslated regions directly 5′ and 3′ of the OPB open reading frame. PCR primers were designed based on the published L. infantum and L. major sequences (GeneDB): 5′-flank forward (5′-CTC ACT AGT ATC TAC GAC CAT GTG ACA CGG-3′) and reverse (5′-CTC TCT AGA CCG GGC TGG TGG GGG TGG-3′) (1.4-kb fragment) primers; 3′-flank forward (5′-CTC ACT AGT GAG TCG TGA TCA TTA ACT CTA GG-3′) and reverse (5′-CTC TCT AGA ACG CTT GTA AGC GAG CGA GG-3′) (0.9-kb fragment) primers (SpeI sites (in boldface type) and XbaI sites (underlined) were included for cloning). For targeted gene deletion, 50 μg of the targeting fragments were excised from their constructs using the flanking restriction endonucleases SpeI and XbaI (New England Biolabs, Ipswich, MA) and purified by electrophoresis on 0.8% agarose gels (24) and then purified using the QIAEX II gel extraction kit (Qiagen Inc., Valencia, CA).
Leishmania Transfections, Clone Isolation, and Southern Blot Analyses
Purified transfection fragments (as described above) were used to transfect log phase Leishmania promastigotes by electroporation (2.25 kV/cm, 500 microfarads) as described previously (23). After electroporation, the cells were grown and selected using hygromycin B and puromycin (for gene deletions) both on plates and in liquid media, and clones were isolated as described previously (25). For Southern blot analyses, genomic DNA was isolated from knock-out clones and digested with NotI (New England Biolabs), and fragments were separated by electrophoresis on 0.6% agarose gels. These were then transferred to Hybond-N+ (GE Healthcare) nylon filters by the manufacturer's instructions for alkali transfer. A knock-out probe and an OPB core probe for the Southern blots were amplified by PCR and gel-purified: knock-out probe forward (5′-TAT GCG CTG GTG ACG ATG CCG-3′) and reverse (5′-CAC GCT CAG CGG CTG CCA TG-3′) primers (1.5-kb fragment upstream of the OPB deletion locus); OPB core probe forward (5′-CGAGGTCGGCGGTGGCTTCGAC-3′) and reverse (5′-TTA CCT GCG AAC CAG CAG GCG C-3′) primers (0.9-kb fragment containing the predicted OPB catalytic core). The probes were 32P-labeled using the Rediprime II Random Prime Labeling System (GE Healthcare) as per the manufacturer's instructions. Hybridization and washing conditions were performed as described previously (26). Southern blots were scanned on a Typhoon Trio Variable Mode Imager (GE Healthcare).
Animal Infections
Metacyclic L. major promastigotes were purified from day 4 OPB(−/−) and wild type cultures using negative selection by binding to peanut agglutinin as has been described previously (27). BALB/c mice (groups of 5) were anesthetized by isoflurane inhalation and infected subcutaneously in the left hind footpad with 5 × 106 metacyclic promastigotes in 50 μl of Hanks' balanced salt solution. Footpad swelling was measured weekly after inoculation using a Mitutoyo caliper. Parasites were recovered from infected mice by resection of the left popliteal lymph node.
Two-dimensional Gel Electrophoresis
Lysates from OPB knock-out and wild type L. donovani promastigotes (405 μg each) were prepared with the ReadyPrep 2-D Cleanup Kit and dissolved in 300 μl of rehydration/sample buffer. Using the ReadyPrep 2-D Starter Kit, samples were absorbed into 17-cm pH 4–7 isoelectric focusing strips, isoelectrically focused, and then separated on 8–16% Tris-HCl PROTEAN II Ready Gels using the manufacturer's protocols (Bio-Rad). Gels were stained using SimplyBlue SafeStain (Invitrogen) and imaged on a Typhoon Trio using the red laser (633 nm) in the fluorescence acquisition mode with no emission filter. Images taken of the gels with wild type and OPB knock-out lysates were compared. Spots that had significantly different intensities or that had shifted were excised from the gels and trypsin-digested as described above. Samples were analyzed by LC/MS/MS on an LTQ Orbitrap (Thermo Fisher Scientific Inc.) mass spectrometer. The survey scans were performed in the Orbitrap at a mass resolution of ∼60,000, whereas all of the collision-induced dissociation experiments were performed in the ion trap. Each survey scan was followed by six collision-induced dissociation experiments on computer-selected multiply charged ions. Once an ion was selected for collision-induced dissociation analysis, it was excluded from the precursor selection for ∼1 min. The reversed phase separation and data processing were performed as has been described previously (28). Peak picking was performed by PAVA, a software developed in-house. The database search was performed on the UniProt.2007.12.04 using a mass accuracy of 20 ppm for MS and 0.8 Da for fragments.
Enolase Enzymatic Activity and Differential Detergent Fractionation
Enolase activity was performed as has been described previously (14). Briefly, enolase conversion of 2-phosphoglycerate into phosphoenolpyruvate was measured spectrophotometrically at 240 nm. For differential fractionations, promastigotes from day 5 cultures were pelleted and washed twice with PBS. The washed pellet was resuspended in ice-cold Fractionation Buffer: 10 mm PIPES, pH 7.2, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 5 mm EDTA, 0.5 mm PMSF, 0.25% Protease Inhibitor Mixture III (EMD Chemicals). Cells were pelleted, resuspended in Fractionation Buffer with 0.02% digitonin, incubated for 5 min on ice, and pelleted. The supernatant represents proteins released from the partially disrupted plasma membrane. This concentration of digitonin is known to disrupt the Leishmania membrane without the bulk release of the cytoplasm (29). The pellet was resuspended in Fractionation Buffer with 1% Triton X-100, incubated for 5 min on ice, and pelleted. This supernatant represents the cytosolic fraction. The pellet was resuspended in Fractionation Buffer with 0.5% deoxycholate and 1% Tween 40, homogenized by hand, incubated for 5 min on ice, and pelleted. The supernatant from this fraction represents the nuclear fraction. This pellet, representing the cytoskeletal fraction, was resuspended in 10 mm sodium phosphate buffer with 5% SDS.
Microarray Analysis of Leishmania-infected Bone Marrow-derived Macrophages
Bone marrow-derived macrophages were isolated from the femora and tibiae of five female C57BL/6 mice. The cells were pooled and cultured in DMEM with 20% FBS and macrophage colony-stimulating factor on 10-cm non-tissue culture-treated plates at a density of 5 × 106 cells/plate. The cells were fed an additional 10 ml of medium on day 3. On day 6, macrophages were harvested from the plate using ice-cold PBS and frozen at 107 cells/ml in 90% FBS with 10% DMSO. The day prior to infecting with Leishmania, macrophages were thawed and plated on T25 flasks at 5 × 106 cells/flask in medium containing macrophage colony-stimulating factor. On the day of infection, macrophages were washed once with PBS, and their medium was replaced with DMEM with 0.5% FBS and L. donovani promastigotes, from 5-day-old cultures, infecting at a multiplicity of infection of 10. Uninfected control cells received medium without parasites. The flasks were spun down at 1000 rpm for 5 min to synchronize the infections. RNA was collected at 0, 2, 6, 12, and 24 h time points by removing the medium and lysing cells with ice-cold TRIzol (Invitrogen). RNA was isolated from the lysates using the RNeasy Mini Kit (Qiagen Inc.) and then amplified using the Amino Allyl MessageAmp II amplified RNA amplification kit (Applied Biosystems). This amplification incorporates amino allyl-UTP into the amplified RNA during in vitro transcription, which facilitates coupling to N-hydroxysuccinimide ester dyes.
Four micrograms of amplified RNA from each sample were labeled with Cy5 dye. A pooled reference (consisting of an equal mass of RNA from all of the time points within a particular infection time course) was labeled with Cy3 dye and hybridized against the Cy5-labeled samples on mouse exonic evidence-based oligonucleotide (MEEBO) arrays. These arrays were hybridized in a 63 °C water bath overnight. After hybridization, the arrays were sequentially washed in three solutions: 0.6× SSC with 0.01% SDS, 0.6× SSC, and 0.06× SSC. Arrays were scanned using a GenePix 4000B scanner with GenePix PRO version 4.1 software (Molecular Devices). The SpotReader program (Niles Scientific, Portola Valley, CA) was used for array gridding and image analysis. The resultant data files were uploaded to Acuity version 4.0 (Molecular Devices), where the raw data were log-transformed, filtered for “good quality spots” (((‘RgnR2^(635/532)’>0.6) AND (‘Flags’> = 0))AND((‘F532Mean-B532’>200) OR (‘F635Median-B635’>200))), and normalized to the 0 h control. The non-mouse control genes were removed from the data set as well as genes lacking data in at least 70% of the arrays. These data were then analyzed for statistically significant gene expression differences using the SAM (Statistical Analysis of Microarray) software version 2.23A, which works with the open source R software package (available at on the World Wide Web).
Three approaches were used in data analysis. First, pairwise comparisons were performed between cells infected with Leishmania and the uninfected control cells in order to determine the number of genes significantly affected by the infection and the relative -fold changes of these genes. To do this, two-class unpaired SAM analysis was employed using a false discovery rate cut-off of <1% and the condition that genes must have at least a 2-fold change. Each time point within a time course was treated as a replicate for the purposes of statistical analysis. Second, multiclass comparisons were performed between cells infected with wild type Leishmania, cells infected with OPB knock-out Leishmania, and uninfected cells to determine the number of genes significantly different between these groups. This was accomplished through multiclass analysis in SAM with a false discovery rate of <0.1%. Cluster software version 2.11 (available at the Eisen Lab Web site) was then utilized to make self-organizing maps and to perform average linkage clustering of both genes and arrays. Third, gene ontology and pathway analyses were performed using DAVID (Database for Annotation Visualization and Integrated Discovery; available on the World Wide Web). The list of up-regulated genes identified by the pairwise SAM analysis were input into DAVID along with a background gene list representing all of the genes used in the pairwise analysis. The pathways and molecular functions that were overrepresented in the list of up-regulated genes were identified and sorted based on -fold change and p value.
RESULTS
Leishmania Expresses High Levels of Serine Protease Activity
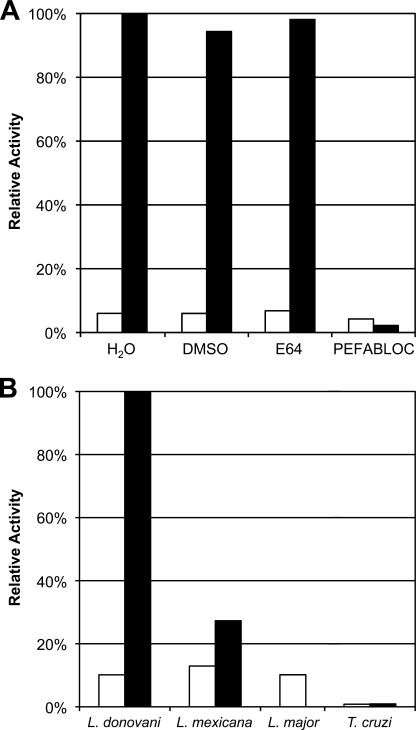
In a search for cysteine protease activity similar to that reported in L. major and L. mexicana (30), the fluorescent peptide substrate, Z-Pro-Arg-AMC, detected high levels of protease activity in extracts of L. donovani. However, this activity was inhibited by the class-specific serine protease inhibitor PEFABLOC and was resistant to the cysteine protease inhibitor E-64. Additionally, this activity was observed at pH 8.0 and was absent at pH 5.5, a profile consistent with serine protease activity (Fig. 1A). Because Clan CA cysteine proteases are the major proteolytic activity in other protozoan parasites (2, 31, 32), a comparison was made between the activity found in three Leishmania species and protease activity against the same substrate in the related intracellular kinetoplastid parasite, T. cruzi. In contrast to Leishmania, T. cruzi parasites did not express significant levels of serine protease activity in either the epimastigote or amastigote stages. Oligopeptidase B activity was detected in both the promastigote and axenic amastigote stages of Leishmania (note that L. major amastigotes cannot be cultured axenically); however, this activity was significantly elevated in the amastigote stage for both L. donovani and L. mexicana (Fig. 1B).
FIGURE 1.
Identification of high serine protease activity in Leishmania lysate. A, the soluble fraction of L. donovani lysate was tested for protease activity using a Z-PR-AMC substrate. Activity assays were performed at pH 5.5 (white bar) and at 8.0 (black bar), pH levels that are generally preferred by cysteine and serine proteases, respectively. The activity observed was tested for sensitivity to the cysteine protease inhibitor E-64 (10 μm) and the serine protease inhibitor PEFABLOC (2 mm). The DMSO concentration was 1%. Activities were normalized to the protease activity observed at the indicated pH without inhibitor. B, OPB activity was compared between three species of Leishmania and T. cruzi. Insect vector stages of the parasites (Leishmania promastigotes and T. cruzi epimastigotes, white bar) were compared with the vertebrate host stages (amastigotes, black bar) for L. donovani, L. mexicana, and T. cruzi. Equivalent amounts of protein from lysed parasites were measured for OPB activity at pH 8.0 using the Z-PR-AMC substrate.
Purification of the Serine Protease Activity of L. donovani Identifies Oligopeptidase B by Mass Spectrometry
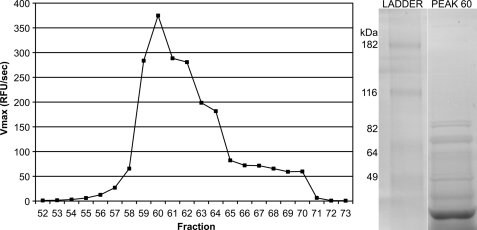
In order to identify the serine protease responsible for the observed proteolytic activity, the enzyme was purified from L. donovani utilizing Z-Pro-Arg-AMC as a substrate. The soluble fraction of L. donovani lysate was fractionated by Q-Sepharose ion exchange chromatography in a pH 8.0 Tris-HCl buffer. Activity assays were performed on eluted fractions, and one peak of activity was observed. SDS-PAGE of fraction 60, the maximal active fraction, was performed (Fig. 2). Tandem mass spectrometry was performed on bands excised from this gel. The trypsin-generated peptides from the 75–90 kDa excised band were identified by informatics analysis, consistent with Leishmania oligopeptidase B (predicted molecular mass, 83.1 kDa). This analysis yielded 23 peptides mapping to OPB with amino acid coverage of 36% and a probability-based Molecular Weight Search score of 1053.
FIGURE 2.
Purification scheme and identification of OPB from L. donovani lysate. The soluble fraction of L. donovani promastigote lysate was fractionated by Q-Sepharose ion exchange chromatography. OPB-containing fractions were identified by testing for cleavage of Z-PR-AMC at pH 8.0. Activity was plotted against fraction number, and a single peak of activity was observed. The maximally active fraction, fraction 60, was resolved on an SDS-polyacrylamide gel. Major bands were excised from the gel, and the proteins were identified by tandem mass spectrometry. The protein contained in the excised gel from 75 to 90 kDa was identified as OPB. OPB has a predicted molecular mass of 83.1 kDa.
L. donovani OPB Was Cloned, Sequenced, and Recombinantly Expressed in P. pastoris
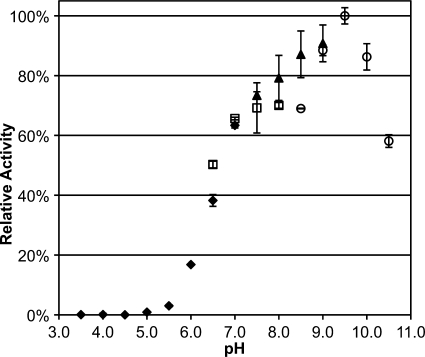
The gene encoding Leishmania donovani OPB was cloned. Sequencing yielded a 2196-bp (731-amino acid) gene. Comparisons of L. donovani OPB with other trypanosomatid OPBs are summarized in supplemental Table 1. This gene was cloned into pPICZα A and recombinantly expressed in P. pastoris. Recombinant protein was purified using a hydrophobic interaction column followed by ion-exchange chromatography. Purified OPB protein was maximally active at pH 9.5 (Fig. 3). The hydrolysis of Z-Pro-Arg-AMC by purified OPB was tested in the presence of a panel of protease inhibitors and divalent cations (Table 1). This enzymatic activity was completely abolished by antipain, PEFABLOC (AEBSF, a broadly based serine protease inhibitor), biotin-d-Phe-Pro-Arg-chloromethyl ketone (PPACK), leupeptin (reversible inhibitor of trypsin-like proteases), and TLCK (irreversible trypsin inhibitor). Benzamidine (inhibitor of trypsin-like proteases) was found to partially inhibit OPB. The catalytic domain of family S9A proteases is known to be covered by a seven-bladed β-propeller that prevents the entrance and hydrolysis of larger substrates (6). It therefore followed that the large molecular weight serine protease inhibitors α1-antitrypsin, aprotinin, and soybean trypsin inhibitor (SBTI) did not inhibit OPB. Additionally, TPCK (irreversible chymotrypsin inhibitor) and PMSF (similar to PEFABLOC) had no effect on OPB activity. As expected, inhibitors of cysteine proteases, aspartic proteases, and metalloproteases did not appreciably inhibit OPB activity. Divalent cations Ca2+, Mg2+, and Mn2+ partially reduced OPB activity, whereas Zn2+ completely blocked activity.
FIGURE 3.
Determination of the pH preference of OPB. Purified OPB was tested for pH preference by measuring the hydrolysis of Z-PR-AMC in different pH buffers. Activity was compared at pH values 3.0–10.5 with a 0.5-pH unit step size in 100 mm citrate phosphate buffer (black diamonds; pH 3.0–7.0), sodium phosphate buffer (white squares; pH 6.5–8.0), Tris-HCl buffer (black triangles; pH 7.5–9.0), and glycine-NaOH buffer (white circles; pH 8.5–10.5). Error bars, S.D.
TABLE 1.
Inhibition of OPB enzymatic activity by protease inhibitors and divalent cations
OPB was tested for sensitivity to class-specific protease inhibitors and common divalent cations. OPB activity was measured by an activity assay at pH 8.0 using the Z-PR-AMC substrate. The enzyme was preincubated with an inhibitor or cation for 10 min at the concentrations indicated. Substrate was then added, and hydrolysis was measured by AMC hydrolysis. Residual activity is the percentage of OPB activity remaining as compared with preincubation with solvent-only controls.
| Inhibitor | Concentration | Relative activity | |
|---|---|---|---|
| % | |||
| Target proteases | |||
| Serine | α1-Antitrypsin | 1 μg/ml | 100 |
| Antipain | 100 μm | 0 | |
| Aprotinin | 1 μm | 100 | |
| Benzamidine | 1 mm | 27 | |
| 3,4-DCl | 50 μm | 88 | |
| PEFABLOC | 2 mm | 0 | |
| PMSF | 1 mm | 100 | |
| PPACK | 10 μm | 0 | |
| Leupeptin | 100 μm | 0 | |
| SBTI | 10 μg/ml | 100 | |
| TLCK | 100 μm | 0 | |
| TPCK | 100 μm | 100 | |
| Threonine | Bortezomib | 10 μm | 100 |
| Cysteine | CA-074 | 1 μm | 96 |
| Cystatin | 1 μg/ml | 100 | |
| E-64 | 10 μm | 85 | |
| K11777 | 100 μm | 100 | |
| DTT | 100 μm | 100 | |
| Aspartic | Pepstatin A | 1 μm | 100 |
| Metallo | EDTA | 5 mm | 100 |
| Cations | |||
| Ca2+ | 10 mm | 69 | |
| Mg2+ | 10 mm | 73 | |
| Mn2+ | 10 mm | 73 | |
| Zn2+ | 10 mm | 0 |
Substrate Specificity Profiling of Recombinant OPB Reveals Substrate Preference
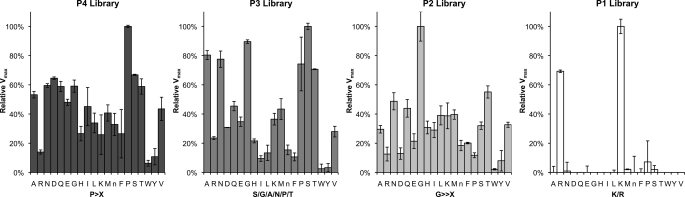
PS-SCLs have been successfully employed to determine P1–P4 subsite preferences for several proteases (20, 33). Complete libraries for each of the P1–P4 positions were used, containing 160,000 tetrapeptide substrates (Fig. 4). The PS-SCL showed that OPB has a strong preference for lysine or arginine residues at P1. OPB can accommodate several amino acids in the P2–P4 positions; however, there is a preference for glycine in P2; serine, glycine, alanine, asparagine, proline, and threonine in P3, and proline in P4. Bulky hydrophobic groups, such as tyrosine, phenylalanine, and tryptophan, are least preferred. The optimal substrate motif for OPB, as predicted by PS-SCL, is as follows: P4, Pro/X; P3, X; P2, Gly/X; P1, Lys/Arg.
FIGURE 4.
Tetrapeptide substrate specificity profiling of OPB using a positional scanning synthetic combinatorial library. Subsite preference was determined using P1–P4 combinatorial libraries. All of the substrates contained an ACC fluorogenic leaving group. Assays were performed in triplicate, and the mean ± S.D. (error bars) is shown. Activity levels were normalized to the maximum mean value observed for each library. The x axis indicates the amino acid held constant at each position for a given library, designated by the one-letter code (with n representing norleucine). The determined preference for each subsite is in boldface type below the x axis (X indicates that multiple amino acids are permissible in the given subsite).
This substrate profile was verified by testing a panel of fluorogenic peptide substrates, including both good and poor predicted substrates (Table 2). OPB cleavage of each substrate is normalized to the Vmax of the Z-PR-AMC substrate. Only substrates with either a lysine or an arginine in the P1 position were cleaved. As predicted by PS-SCL analysis, the substrate Z-GGR-AMC was preferred by OPB. Kcat and Km were determined for both Z-GGR-AMC and Z-GPR-AMC. These data demonstrate that Gly-Gly-Arg is a better substrate and validate the results of the PS-SCL. Compared with Z-GGR-AMC, H-K- and H-R-AMC were poor substrates. Thus, fluorogenic peptide hydrolysis was more efficient when more than one substrate-binding site were occupied.
TABLE 2.
Substrate specificity of OPB
To test the predictive power of the PS-SCL, fluorogenic substrates were obtained that were predicted to be either good or poor matches by the PS-SCL results. Substrates were grouped and listed by decreasing initial cleavage velocities. Percentage activity is normalized to the initial velocity observed using the Z-PR-AMC substrate. All assays were performed at pH 8.0 using 20 μm substrate (except for those used to calculate Kcat and Km values). The PS-SCL predicted that the ideal P2 amino acid would be Gly. Therefore, Z-GGR-AMC and Z-GPR-AMC, which differ only in the P2 position, were compared as substrates for OPB, and the kinetic parameters were calculated. These assays were performed at substrate concentrations ranging from 80 to 0.2 μm.
| N-terminal group | P5 | P4 | P3 | P2 | P1 | Relative activity | N-terminal group | P5 | P4 | P3 | P2 | P1 | Relative activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | ||||||||||||
| Z | G | G | R | 197 | H | A | 0 | ||||||
| Boc | L | G | R | 108 | Ac | D | E | V | D | 0 | |||
| Z | G | P | R | 104 | Z | V | A | D | 0 | ||||
| Z | P | R | 100 | Ac | W | E | H | D | 0 | ||||
| Z | V | V | R | 92 | Z | L | L | E | 0 | ||||
| Glut | G | R | 91 | Suc | A | A | F | 0 | |||||
| Z | R | R | 89 | Suc | P | S | P | F | 0 | ||||
| Z | L | R | 78 | H | G | F | 0 | ||||||
| Boc | L | R | R | 69 | H | L | 0 | ||||||
| Ac | F | R | 62 | Z | G | G | L | 0 | |||||
| Ac | K | Q | K | L | R | 51 | Z | A | E | N | 0 | ||
| Z | F | R | 50 | Ac | A | S | N | 0 | |||||
| Z | A | A | R | R | 40 | Ac | A | V | N | 0 | |||
| Boc | E | K | K | 32 | Suc | G | P | L | G | P | 0 | ||
| Boc | R | V | R | R | 31 | Suc | L | Y | 0 | ||||
| Ac | F | R | S | L | K | 23 | Suc | L | L | V | Y | 0 | |
| Boc | V | R | K | 21 | TFA | Y | 0 | ||||||
| H | R | 6 | |||||||||||
| H | K | 1 |
| Kinetic parameter | Z-GPR-AMC | Z-GGR-AMC |
|---|---|---|
| Km (μm) | 7.76 | 3.08 |
| Kcat (s−1) | 4.25e4 | 6.83e4 |
| Kcat/Km (s−1 μm−1) | 0.55e4 | 2.22e4 |
OPB(−/−) Parasites Lose the Major Serine Protease Activity
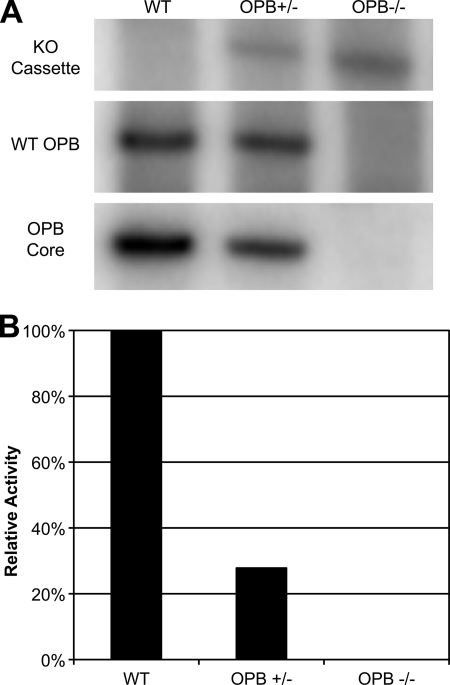
The published L. major and L. infantum genomes (GeneDB) as well as a study performed on L. amazonensis (3) indicate that Leishmania spp. contain a single copy of OPB per haploid genome. This was confirmed to be true in L. donovani and L. major by Southern blot analysis of genomic DNA (data not shown). Deletion of both alleles of the single copy gene was carried out by two rounds of targeted gene replacement and confirmed by Southern blot (Fig. 5A). To confirm that the major serine protease activity of L. donovani was attributable to oligopeptidase B, OPB(+/−) and OPB(−/−) parasites were compared with wild type (Fig. 5B). As expected, serine protease activity measured by Z-PR-AMC was completely lost in the OPB(−/−) parasites.
FIGURE 5.
Genetic and enzymatic analysis of OPB knockouts. A, successful deletion of both genomic copies of the OPB gene were determined by Southern blot analysis. Digested genomic DNA from wild type, single copy, and double copy knockouts were analyzed at the OPB locus for the presence of either the wild type locus or the knock-out cassette. The blot was stripped and reprobed for the OPB gene itself to ensure that the gene was not relocated to another site in the genome. B, lysates from wild type, single copy, and double copy knockouts were analyzed for OPB enzymatic activity using the Z-PR-AMC substrate at pH 8.0.
In culture, OPB(+/−) and OPB(−/−) parasites successfully differentiated in culture back and forth between promastigote and axenic amastigote forms. OPB(−/−) promastigotes (but not amastigotes) showed a statistically significant growth delay in culture; however, at stationary phase (day 5 post-split), these parasites attained the same density as their wild type counterparts. These knock-out parasites also efficiently infected J774 murine macrophages in culture (data not shown).
OPB(−/−) Parasites Accumulate Inactive Enolase Isoforms
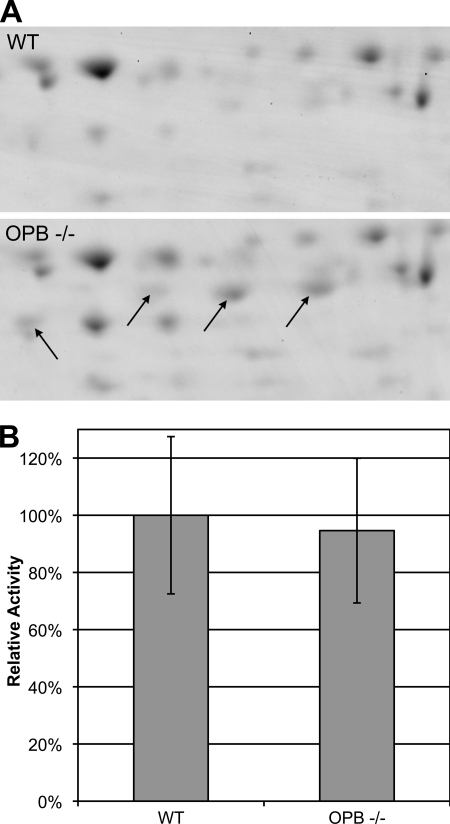
During T. cruzi infection, OPB indirectly stimulates transient Ca2+ levels in host cells. This results in the recruitment of lysosomes to the plasma membrane and allows the parasite to penetrate non-phagocytic cells (7). Leishmania does not employ this strategy and is restricted to phagocytic cells of the immune system, so we next sought to uncover the function of OPB in this genus of parasite. Two-dimensional gel electrophoresis was used to identify major differences in protein expression between wild type and OPB(−/−) parasites (Fig. 6A). Four major protein spots were significantly more intense in OPB(−/−). Mass spectrometry analysis indicated that all four were consistent with Leishmania enolase. These spots differ by their isoelectric points in a stepwise manner, suggesting that they represent different phosphorylation states of enolase. In the malaria parasite Plasmodium yoelii, different phosphorylated isoforms of enolase were localized to the parasite membrane (34). It was recently found that inactive forms of enolase are associated with the plasma membrane of L. mexicana, where they perform a function independent of the classical enzymatic activity (14). We therefore tested OPB(−/−) lysates for enolase enzymatic activity by detecting the conversion of 2-phosphoglycerate to phosphoenolpyruvate spectrophotometrically (Fig. 6B). As predicted, there was no increase in active enolase, thus the increase in enolase isoforms detected in OPB(−/−) lysates did not represent active enzyme.
FIGURE 6.
Proteomic analysis of OPB knock-out parasites. A, two-dimensional gel electrophoresis was used to compare lysates from wild type and OPB double knock-out parasites. The arrows indicate four spots that were significantly more intense in lysate from the knock-out parasites. The intensity increase was observed on replicate gels. The proteins in these spots were identified by mass spectrometry and were all found to be enolase. B, to confirm that the accumulated isoforms of enolase found in the OPB knock-out parasites did not represent increased glycolytic enolase, the glycolytic pathway function of enolase was analyzed by measuring the conversion of 2-phosphoglycerate to phosphoenolpyruvate spectrophotometrically. Error bars, S.D. of three separate experiments.
Enolase Accumulates on the Cell Surface of OPB-deficient Leishmania
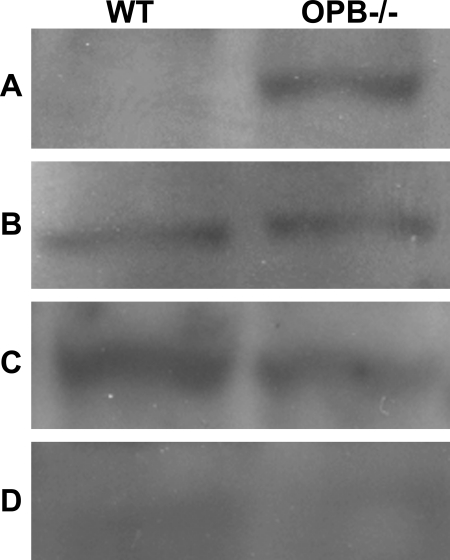
To determine whether the accumulated enolase isoforms detected by two-dimensional gel in OPB(−/−) Leishmania represent altered localization, subcellular fractionation was performed on wild type and OPB knock-out parasites. Cells were fractionated into digitonin-sensitive membrane, cytoplasmic, nuclear, and cytoskeletal fractions. These fractions were then analyzed by Western blotting using an anti-enolase antibody (Fig. 7). Enolase was detected in the cytoplasm and nucleus of both wild type and OPB(−/−) parasites. Enolase was not detected in the cytoskeletal fraction. Interestingly, enolase was detected on the cell surface of the OPB(−/−) parasites but not on wild type parasites. These data indicate that loss of OPB results in the accumulation of enolase on the parasite cell surface and that this has no effect on the level of cytoplasmic enolase.
FIGURE 7.
Differential localization of enolase in OPB knock-out versus wild type Leishmania. Wild type and OPB(−/−) Leishmania were fractionated by differential lysis. Western blots for enolase were performed on these cell extracts. Cell fractions were as follows: cell surface fraction (A), cytoplasmic fraction (B), nuclear fraction (C), and cytoskeletal fraction (D). Equivalent levels of enolase were detected in the cytoplasmic and nuclear fractions of both wild type and OBB −/− parasite, and it was not detected in the cytoskeletal fractions. Enolase was found to have accumulated on the cell surface of OPB(−/−) Leishmania but was not present on the cell surface of the wild type parasites.
OPB Is Necessary for Leishmania to Silently Infect Macrophages
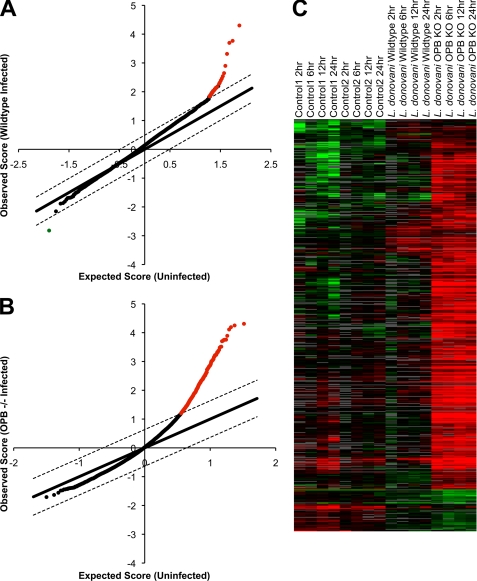
It is known that Leishmania parasites are able to evade the host immune response, at least initially, when establishing an infection (35). Macrophages infected with several species of Leishmania do not significantly alter their gene expression profiles over the first 24 h postinfection (36). To test if the accumulation of inactive surface enolase by OPB(−/−) parasites interferes with detection by macrophages, we performed microarray analysis of gene expression by bone marrow-derived macrophages infected with Leishmania. Macrophages were infected with wild type or OPB(−/−) parasites or were uninfected. RNA was collected at 0, 2, 6, 12, and 24 h postinfection. These RNAs were dye-coupled and hybridized onto MEEBO arrays. Array data were analyzed for statistically significant genes using the SAM software. -Fold change was calculated compared with uninfected cells (Fig. 8, A and B). Using a false discovery rate threshold of <1.0% and a minimum -fold change of 2 over uninfected, only 23 genes were significantly up- or down-regulated in wild type Leishmania-infected macrophages; however, a striking 495 genes were significantly up-regulated in OPB(−/−) Leishmania-infected macrophages (supplemental Table 2). Heat maps of clustered arrays illustrate the significant up-regulation of macrophage genes following infection by OPB(−/−) parasites compared with wild type-infected and uninfected controls (Fig. 8C). Families of proteins that were significantly up-regulated in the OPB(−/−) infections include proteins involved in cytokine secretion, signal transduction, and the inflammatory response (supplemental Table 3).
FIGURE 8.
Gene expression of macrophages infected by OPB knock-out parasites. Murine bone marrow-derived macrophages were infected with either wild type or OPB(−/−) parasites for 0, 2, 6, 12, and 24 h. Gene expression from these macrophages was analyzed by microarray. A and B, array data for wild type (A) and OPB(−/−) (B) infections were analyzed to find statistically significant changes in gene expression compared with uninfected macrophages using a false discovery rate threshold of <1.0% and a minimum -fold change of 2. The x axis indicates the expression level of a given gene in the uninfected control macrophages. The y axis indicates the expression level observed in the infected macrophages. Genes with a significant change in expression after Leishmania infection are colored red for up-regulation and green for down-regulation. In the wild type infections, only 23 genes were differentially expressed (false discovery rate = 0.00%) compared with uninfected cells; however, in the OPB(−/−) infections, there were 459 significantly up-regulated genes (false discovery rate = 0.67%). C, heat maps of the array data were clustered to illustrate the differences between the different time courses. The OPB knock-out-infected macrophage arrays show a significant increase in gene expression (red) compared with the uninfected and wild type Leishmania-infected macrophage arrays.
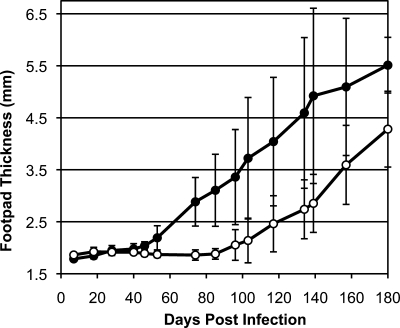
Loss of OPB Results in Delayed Lesion Formation in Mice
BALB/c mice were infected subcutaneously into their left hind footpads with either wild type or OPB(−/−) L. major parasites. Footpad swelling was measured weekly (Fig. 9). Swelling was evident in the wild type-infected mice at 46 days postinfection; however, significant swelling was not observed in mice infected with OPB(−/−) parasites until 117 days postinfection. The OPB(−/−) infections were not self-limiting and continued to increase footpad swelling, but the lesion size was consistently 7 weeks delayed compared with the wild type infections. Episomally added back OPB infections did not recapitulate wild type infections. This was not unexpected because our model of OPB function requires tight regulation of OPB expression during promastigote to amastigote differentiation and in culture.
FIGURE 9.
Mouse infections by wild type and OPB(−/−) L. major. BALB/c mice (n = 5) were infected subcutaneously in the left hind footpads with wild type (solid circle) or knock-out (empty circle) L. major. Footpad swelling was measured weekly, and the footpad thickness was plotted over time. Error bars indicate the S.D. between the five mice in each group. The mice were sacrificed at 180 days postinfection. No swelling was observed in the right hind (uninfected) footpads. Significant footpad swelling occurred at 46 days PI in wild type-infected mice (p = 0.002) and 116 days postinfection in OPB knock-out-infected mice (p = 0.036).
DISCUSSION
Proteases are key virulence factors in the pathogenesis of diseases caused by Leishmania and related kinetoplastid parasites (30). A noteworthy biochemical difference between Leishmania parasites and the closely related trypanosomatid T. cruzi is the presence, reported here, of unexpectedly high levels of serine protease activity in Leishmania. We found that three Leishmania species (L. donovani, L. major, and L. mexicana), representing the three main clinically distinct complexes of the genus, exhibited high levels of serine protease activity when compared with T. cruzi. Furthermore, this Leishmania serine protease activity increased severalfold in amastigotes, suggesting an important role for this enzyme in the vertebrate stages of the parasite for both cutaneous and visceral leishmaniasis-causing species. Whereas proteases represent a significant genomic complement of protozoa and metazoa, the Clan CA cysteine proteases predominate in invertebrates (2). The major proteolytic activity identified in Leishmania extracts was attributable to the Clan SC enzyme oligopeptidase B.
Substrate and inhibitor specificity of the Leishmania OPB revealed an enzyme that is similar yet distinct among homologues from other organisms. Previous research on OPBs from multiple trypanosomatid parasites has shown that this group of enzymes has a strong preference for basic residues in both the P1 and P2 positions (8, 9, 37) much like the OPB from E. coli (38). Alignments and homology modeling of the OPB subsites by de Matos Guedes et al. (39) revealed that both the S1 and S2 pockets from each of these organisms possess pairs of acid residues: S1 glutamic acids and S2 aspartic acids or glutamic acids for E. coli and the trypanosomes, respectively. Interestingly, the L. donovani S2 pocket contains only a single aspartic acid residue, with the second replaced with a glutamine. This single amino acid change has altered the substrate specificity of the Leishmania enzyme, giving it a preference for a P2 glycine over arginine, which makes it unique among known OPB enzymes. Leishmania OPB, like the trypanosome homologues, was found to be resistant to high molecular weight inhibitors (40), indicating that the N-terminal β-propeller domain sterically restricts access to the Leishmania OPB active site.
Previous research has not been able to identify an endogenous substrate or pathway for OPB within trypanosomatid parasites; however, OPB is known to have a role in tissue and cell invasion by T. cruzi. T. cruzi, like Leishmania, is an intracellular parasite in its vertebrate host, but unlike Leishmania, T. cruzi infects non-phagocytic cells. OPB from T. cruzi has been found to play an important role in the invasion of these non-phagocytic cells. It is known that OPB transiently increases Ca2+ levels in host cells and that this results in the recruitment of lysosomes to the host cell surface for parasite entry; however, the direct action of OPB in this process has not yet been uncovered. It is believed that the T. cruzi OPB acts through an unknown signaling ligand or parasite cell surface receptor (40). Additionally, a T. cruzi Clan SC protease has also been implicated in tissue invasion through the proteolytic cleavage of the host's extracellular matrix (13).
Our findings have elucidated the function of OPB in Leishmania and have the potential to answer some of the unresolved questions regarding the role of OPB in T. cruzi tissue and cell invasion. Proteomic analysis of wild type versus OPB(−/−) Leishmania revealed that the knock-out parasites accumulated enolase. Further investigation showed that this accumulation was due to enzymatically inactive enolase and that the accumulation was on the cell surface of these parasites. Whereas enolase functions in carbohydrate metabolism in a number of organisms, it has also recently been shown to function as a virulence factor in bacterial pathogens, such as Bacillus anthracis (41) and Streptococcus pneumoniae (42) as well as in protozoan parasites like Toxoplasma gondii (43) and Plasmodium (34), where it acts as a cell surface ligand. In fact, in Leishmania, surface-associated enolase was previously documented and shown to bind host plasminogen (44). In addition, Leishmania was found to be less infectious in plasminogen knock-out mice, supporting a role for enolase-plasminogen interactions in Leishmania virulence (16).
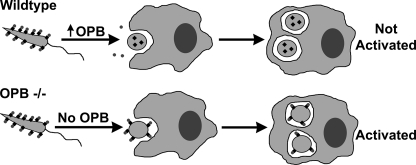
Enolase-bound plasminogen activation into plasmin or binding to plasminogen receptors may facilitate parasite entry into macrophages or prevent fibrin deposition on extracellular parasites at the site of infection. However, once in the macrophage, retention of enolase could negatively impact parasite survival by activating macrophages. As our data have shown, OPB is dramatically up-regulated upon differentiation into the amastigote form of the parasite. Clearance of surface enolase by increased OPB supports this model. Fig. 10 is a graphical representation of the proposed model for OPB function within Leishmania. Our hypothesis that OPB is needed for the silent infection of macrophages is supported by our demonstration that OPB(−/−) Leishmania caused activation of infected macrophages with expression of TLR-2, the TGF-β receptor, TNF receptors, and interferon-activated proteins (see the supplemental material). This is in stark contrast to wild type Leishmania, which elicits little, if any, macrophage reaction. A defect in the ability of Leishmania to circumvent macrophage activation during entry or to reside undetected within host macrophages would explain the significant delay in footpad swelling observed in the murine experimental model of OPB(−/−) L. major infection when compared with wild type parasites. Although not absolutely necessary for disease pathogenesis, OPB is required for full virulence.
FIGURE 10.
Model for OPB function in the Leishmania life cycle. This schematic details the proposed function of Leishmania OPB during infection of a vertebrate host. In a wild type infection, surface enolase (black diamonds) binds host plasminogen (dark gray circles) on the parasite cell surface. As the parasite begins differentiating into an amastigote, OPB is up-regulated, thereby clearing surface enolase and plasminogen. The amastigotes replicate undetected within the macrophage. In an OPB(−/−) infection, when the parasite differentiates into the amastigote stage, enolase and plasminogen are retained on the cell surface. This infection is detected by the macrophage, resulting in increased macrophage gene transcription and reduced parasite virulence.
In this study, we have directly demonstrated that Leishmania OPB is a highly active protease with unique substrate specificity when compared with related trypanosomes. OPB is responsible for regulating levels of enolase on the parasite cell surface. This moonlighting function of enolase is known to contribute to the virulence of multiple infectious agents, and its connection to OPB suggests that this is a key pathway for other trypanosomatid parasites.
Supplementary Material
Acknowledgments
We thank Juan Engel, Christopher Franklin, Elizabeth Hansell, and Judy Sakanari (University of California, San Francisco, CA) for technical assistance and P'ng Loke for providing the MEEBO arrays.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant AI35707. This work was also supported by a National Science Foundation graduate research fellowship and the Sandler Foundation. Mass spectrometry analysis was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at the University of California (San Francisco, CA) (A. L. Burlingame, Director), supported by the Biomedical Research Technology Program of the NIH National Center for Research Resources (NCRR), NIH NCRR Grant P41RR001614, and NIH NCRR Grant RR012961.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) ACV92099.
- OPB
- oligopeptidase B
- AMC
- 7-amino-4-methylcoumarin
- ACC
- 7-amino-4-carbamoyl-methylcoumarin
- Z
- benzyloxycarbonyl
- Glut
- glutaryl
- Ac
- N-acetyl
- Boc
- N-tertbutoxycarbonyl
- Suc
- N-succinyl
- H
- free amine
- PS-SCL
- positional scanning synthetic combinatorial library
- AEBSF
- 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride
- MEEBO
- mouse exonic evidence-based oligonucleotide
- E-64
- N-[N-(l-3-trans-carboxyoxirane-2-carbonyl)-l-leucyl]-agmatine
- PPACK
- biotin-d-Phe-Pro-Arg-chloromethyl ketone
- SBTI
- soybean trypsin inhibitor
- TLCK
- Nα-p-tosyl-l-lysine chloromethyl ketone
- TPCK
- tosylphenylalanyl chloromethyl ketone.
REFERENCES
- 1. McKerrow J. H., Caffrey C., Kelly B., Loke P., Sajid M. (2006) Annu. Rev. Pathol. 1, 497–536 [DOI] [PubMed] [Google Scholar]
- 2. Sajid M., McKerrow J. H. (2002) Mol. Biochem. Parasitol. 120, 1–21 [DOI] [PubMed] [Google Scholar]
- 3. Guedes H. L., Rezende J. M., Fonseca M. A., Salles C. M., Rossi-Bergmann B., De-Simone S. G. (2007) Z. Naturforsch. C 62, 373–381 [DOI] [PubMed] [Google Scholar]
- 4. Silva-Lopez R. E., Morgado-Díaz J. A., Chávez M. A., Giovanni-De-Simone S. (2007) Parasitol. Res. 101, 1627–1635 [DOI] [PubMed] [Google Scholar]
- 5. Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J. C., Müller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'neil S., Pentony M., Pohl T. M., Price C., Purnelle B., Quail M. A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J. C., Rutter S., Saunders D., Schäfer M., Schein J., Schwartz D. C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D. F., Blackwell J. M., Stuart K. D., Barrell B., Myler P. J. (2005) Science 309, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) Nucleic Acids Res. 36, D320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burleigh B. A., Caler E. V., Webster P., Andrews N. W. (1997) J. Cell Biol. 136, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morty R. E., Lonsdale-Eccles J. D., Morehead J., Caler E. V., Mentele R., Auerswald E. A., Coetzer T. H., Andrews N. W., Burleigh B. A. (1999) J. Biol. Chem. 274, 26149–26156 [DOI] [PubMed] [Google Scholar]
- 9. Morty R. E., Pellé R., Vadász I., Uzcanga G. L., Seeger W., Bubis J. (2005) J. Biol. Chem. 280, 10925–10937 [DOI] [PubMed] [Google Scholar]
- 10. McKerrow J. H., Rosenthal P. J., Swenerton R., Doyle P. (2008) Curr. Opin. Infect. Dis. 21, 668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morty R. E., Troeberg L., Pike R. N., Jones R., Nickel P., Lonsdale-Eccles J. D., Coetzer T. H. (1998) FEBS Lett. 433, 251–256 [DOI] [PubMed] [Google Scholar]
- 12. Bal G., Van der Veken P., Antonov D., Lambeir A. M., Grellier P., Croft S. L., Augustyns K., Haemers A. (2003) Bioorg. Med. Chem. Lett. 13, 2875–2878 [DOI] [PubMed] [Google Scholar]
- 13. Yoshida N. (2006) An. Acad. Bras. Cienc. 78, 87–111 [DOI] [PubMed] [Google Scholar]
- 14. Quiñones W., Peña P., Domingo-Sananes M., Cáceres A., Michels P. A., Avilan L., Concepción J. L. (2007) Exp. Parasitol. 116, 241–251 [DOI] [PubMed] [Google Scholar]
- 15. Avilan L., Calcagno M., Figuera M., Lemus L., Puig J., Rodriguez A. M. (2000) Mol. Biochem. Parasitol. 110, 183–193 [DOI] [PubMed] [Google Scholar]
- 16. Maldonado J., Marina C., Puig J., Maizo Z., Avilan L. (2006) Exp. Mol. Pathol. 80, 289–294 [DOI] [PubMed] [Google Scholar]
- 17. Wallis A. E., McMaster W. R. (1987) J. Exp. Med. 166, 1814–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doyle P. S., Engel J. C., Pimenta P. F., da Silva P. P., Dwyer D. M. (1991) Exp. Parasitol. 73, 326–334 [DOI] [PubMed] [Google Scholar]
- 19. Medina-Acosta E., Cross G. A. (1993) Mol. Biochem. Parasitol. 59, 327–329 [DOI] [PubMed] [Google Scholar]
- 20. Choe Y., Leonetti F., Greenbaum D. C., Lecaille F., Bogyo M., Brömme D., Ellman J. A., Craik C. S. (2006) J. Biol. Chem. 281, 12824–12832 [DOI] [PubMed] [Google Scholar]
- 21. Gritz L., Davies J. (1983) Gene 25, 179–188 [DOI] [PubMed] [Google Scholar]
- 22. Lacalle R. A., Pulido D., Vara J., Zalacaín M., Jiménez A. (1989) Gene 79, 375–380 [DOI] [PubMed] [Google Scholar]
- 23. Joshi P. B., Sacks D. L., Modi G., McMaster W. R. (1998) Mol. Microbiol. 27, 519–530 [DOI] [PubMed] [Google Scholar]
- 24. Maniatis T., Fritsch E. F., Sambrook J. (1982) Molecular Cloning: A Laboratory Manual, p. 545, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25. Joshi P. B., Kelly B. L., Kamhawi S., Sacks D. L., McMaster W. R. (2002) Mol. Biochem. Parasitol. 120, 33–40 [DOI] [PubMed] [Google Scholar]
- 26. Button L. L., Russell D. G., Klein H. L., Medina-Acosta E., Karess R. E., McMaster W. R. (1989) Mol. Biochem. Parasitol. 32, 271–283 [DOI] [PubMed] [Google Scholar]
- 27. Sacks D. L., Hieny S., Sher A. (1985) J. Immunol. 135, 564–569 [PubMed] [Google Scholar]
- 28. Hansell E., Braschi S., Medzihradszky K. F., Sajid M., Debnath M., Ingram J., Lim K. C., McKerrow J. H. (2008) PLoS. Negl. Trop. Dis. 2, e262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hide M., Ritleng A. S., Brizard J. P., Monte-Allegre A., Sereno D. (2008) Parasitol. Res. 103, 989–992 [DOI] [PubMed] [Google Scholar]
- 30. Mottram J. C., Coombs G. H., Alexander J. (2004) Curr. Opin. Microbiol. 7, 375–381 [DOI] [PubMed] [Google Scholar]
- 31. Que X., Reed S. L. (2000) Clin. Microbiol. Rev. 13, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenthal P. J. (2004) Int. J. Parasitol. 34, 1489–1499 [DOI] [PubMed] [Google Scholar]
- 33. O'Brien T. C., Mackey Z. B., Fetter R. D., Choe Y., O'Donoghue A. J., Zhou M., Craik C. S., Caffrey C. R., McKerrow J. H. (2008) J. Biol. Chem. 283, 28934–28943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pal-Bhowmick I., Vora H. K., Jarori G. K. (2007) Malar. J. 6, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bogdan C., Röllinghoff M. (1998) Int. J. Parasitol. 28, 121–134 [DOI] [PubMed] [Google Scholar]
- 36. Zhang S., Kim C. C., Batra S., McKerrow J. H., Loke P. (2010) PLoS. Negl. Trop. Dis. 4, e648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morty R. E., Authié E., Troeberg L., Lonsdale-Eccles J. D., Coetzer T. H. (1999) Mol. Biochem. Parasitol. 102, 145–155 [DOI] [PubMed] [Google Scholar]
- 38. Polgár L. (1997) Proteins 28, 375–379 [PubMed] [Google Scholar]
- 39. de Matos Guedes H. L., Carneiro M. P., Gomes D. C., Rossi-Bergmanmn B., Giovanni de Simone S. (2007) Parasitol. Res. 101, 853–863 [DOI] [PubMed] [Google Scholar]
- 40. Caler E. V., Vaena de Avalos S., Haynes P. A., Andrews N. W., Burleigh B. A. (1998) EMBO J. 17, 4975–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Agarwal S., Kulshreshtha P., Bambah Mukku D., Bhatnagar R. (2008) Biochim. Biophys. Acta 1784, 986–994 [DOI] [PubMed] [Google Scholar]
- 42. Attali C., Durmort C., Vernet T., Di Guilmi A. M. (2008) Infect. Immun. 76, 5350–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferguson D. J., Parmley S. F., Tomavo S. (2002) Int. J. Parasitol. 32, 1399–1410 [DOI] [PubMed] [Google Scholar]
- 44. Vanegas G., Quiñones W., Carrasco-López C., Concepción J. L., Albericio F., Avilán L. (2007) Parasitol. Res. 101, 1511–1516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.