Abstract
Little is known regarding how the Oct1 transcription factor regulates target gene expression. Using murine fibroblasts and two target genes, Polr2a and Ahcy, we show that Oct1 recruits the Jmjd1a/KDM3A lysine demethylase to catalyze the removal of the inhibitory histone H3K9 dimethyl mark and block repression. Using purified murine T cells and the Il2 target locus, and a colon cancer cell line and the Cdx2 target locus, we show that Oct1 recruits the NuRD chromatin-remodeling complex to promote a repressed state, but in a regulated manner can switch to a different capacity and mediate Jmjd1a recruitment to block repression. These findings indicate that Oct1 maintains repression through a mechanism involving NuRD and maintains poised gene expression states through an antirepression mechanism involving Jmjd1a. We propose that, rather than acting as a primary trigger of gene activation or repression, Oct1 is a switchable stabilizer of repressed and inducible states.
Keywords: Chromatin Immunoprecipitation (ChiP), Gene Expression, Histone Modification, Immunology, Transcription Factors, Il2, Jmjd1a, NuRD, Oct1, T Lymphocyte
Introduction
The POU2 (Pit-1, Oct1/2, Unc-86) transcription factor family includes ∼13 mammalian paralogs as well as representatives from other metazoans (1). The best known example, Oct4/POU5F1, regulates embryonic stem (ES) cell identity and is a key factor used to generate induced pluripotent stem cells from somatic cells (2–5). Oct1/POU2F1 is related to Oct4 and possesses similar in vitro DNA binding specificity (for reviewed, see Ref. 6). As with many transcription factors, these proteins are known to regulate gene expression both positively and negatively (e.g. 7, 8); however, their activity has been thought to be determined by gene context and not subject to regulation.
Loss of Oct1 inhibits oncogenic transformation in mouse embryonic fibroblasts (MEFs) and tumorigenicity in p53-deficient mice and xenograft assays, while having little effect on cell growth in culture or transformation by serial passage (9). One study indicates that Oct1 levels are increased in some human gastric cancers (10). In contrast, multiple studies have identified coordinate up-regulation of Oct1 target genes in lung and breast adenocarcinomas, leukemias, and myeloid leukemia stem cells, without concurrent up-regulation of Oct1 itself (11–14), suggesting that Oct1 activity may be deregulated in malignancy. Recent findings showing post-translational regulation of Oct1 support this possibility (15). Although Oct1 has been studied intensively, our current understanding of how it regulates gene transcription is surprisingly limited (see for example, Ref. 16).
Here, we show using three different systems (fibroblasts, primary T cells, and a colon cancer cell line) that Oct1 is a bipotential and switchable transcriptional regulator. In fibroblasts, Oct1 mediates recruitment of the Jmjd1a histone demethylase to target genes (Polr2a and Ahcy) following oxidative stress exposure. In the absence of Oct1, Jmjd1a fails to be recruited, H3K9me2 levels are elevated, and inappropriate repression is observed. In contrast, Oct1 recruits the nucleosome remodeling and histone deacetylation (NuRD/Mi-2) chromatin remodeling complex to the Il2 promoter in naïve CD4 T cells to mediate gene repression. The recruitment is regulated because upon T cell activation Oct1 loses its capacity to associate with NuRD and instead is required for recruitment of Jmjd1a. Phorbol 12-myristate 13-acetate (PMA) treatment is sufficient to switch association from NuRD to Jmjd1a, despite the fact that PMA is insufficient to activate Il2. In resting but previously stimulated T cells, Oct1 is required to maintain Jmjd1a at Il2, remove histone H3K9me2 marks and protect DNA from methylation, promoting the stronger expression associated with secondary stimulation. In DLD-1 colon adenocarcinoma cells, Oct1 bound to the Cdx2 target locus is required for mutually exclusive NuRD and Jmjd1a association. PMA treatment of DLD-1 cells results in reduced Jmjd1a association and represses Cdx2 in a manner requiring Oct1. These results show that Oct1 is a bipotential factor capable of acting through opposing mechanisms to reinforce repressed or inducible states, even at the same target gene.
EXPERIMENTAL PROCEDURES
Cell Culture
Wild-type and Oct1-deficient MEFs were cultured as described previously (17). For H2O2 treatment, fibroblasts were exposed to 4 mm H2O2 for 1 h, then incubated without H2O2 for the indicated times. EL-4 T cells were cultured as described (18). Purified primary naïve splenic T cells were isolated from wild-type C57BL/6 mice. T cells were cultured in RPMI 1640 medium (Invitrogen) with 10% heat-inactivated fetal bovine serum (FBS; Hyclone), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine (Invitrogen). T cells were stimulated in culture by 10 μg/ml plate-bound anti-CD3ϵ and 2 μg/ml anti-CD28 antibodies (eBioscience). The cells were washed and rested for 8 days without antibody stimulation and restimulated for the indicated time points with the same antibodies. PMA (Sigma) was used at the indicated concentrations for 6 h. PD98059 (Calbiochem) and SP600125 (A.G. Scientific, Inc.) were used at the indicated concentrations. T cells were preincubated with the inhibitors for 2 h and were present throughout the entire 6-h PMA stimulation. DLD-1 cells were cultured in DMEM (Invitrogen) supplemented with 10% 1:1 heat-inactivated FBS:bovine calf serum, penicillin, streptomycin, l-glutamine, and 50 μm β-mercaptoethanol (Sigma). All cells were cultured at 37 °C and 5% CO2 in a humidified atmosphere.
T Cell Purification
C57BL/6 Rag1−/− mice were irradiated and transplanted with wild-type and Oct1−/− fetal liver cells using previously published methods (19). T cells were isolated using a CD4+ T cell isolation kit (Miltenyi) with modifications. A biotin-conjugated CD44 antibody (eBioscience) was also included to eliminate CD44Hi cells. Purification was confirmed by flow cytometry. All animal procedures were approved by the institutional animal care and use committee.
Latex Nanoparticle Purification/Liquid Chromatography-Mass Spectrometry
Oct1 purification using nanoparticles was performed as described (15), except that the nanoparticles were coupled to human Il2 promoter DNA containing an Oct1 binding site (−262 to −222; CATACAGAAGGCGTTAATTGCATGAATTAGAGCTATCACC). Gel slab excision and preparation, and mass spectrometry were conducted as described (15).
Chromatin Immunoprecipitation (ChIP)
Oct1 ChIP assays used described protocols (9) and two combined anti-Oct1 antibodies (Bethyl). NuRD ChIP used antibodies against Mta2 (Abcam) or Mi-2 (Bethyl). Nuclear factor of activated T cells (NFAT) (c1+c2) antibodies were obtained from Santa Cruz Biotechnology. Antibodies against Jmjd1a and di-/trimethylated histone H3K9 were obtained from Abcam. Olignucleotide sequences were: Il2 forward, 5′-AGCATGGGAGGCAATTTATAC-3′ and reverse, 5′-TGCTTTCTGCCACACAGGTAG-3′; mouse Polr2a forward, 5′-CCATCTTGCCTGCCTTATGCATATT and reverse, 5′-TAATGAAGGCGGGGCCCTTC; Cdx2 forward, 5′-CCAATGGTTGGAGACGTCGAG-3′ and reverse, 5′-TTAGCTGTGTCAGGTCGC-TGC-3′. For real-time quantification of ChIP enrichment, crossover values for control samples were typically unobtainable. Therefore, mouse DNA enriched using the specific antibody or control (C/EBPβ) antibody (Santa Cruz Biotechnology) was amplified by PCR for both the specific target and a control nonspecific (β-actin) target. Human DNA (from DLD-1 cells) used a different nonspecific target upstream of the Lmnb2 gene. For each plate, amplification curves were compared with a standard of input controls ranging from 0.005% to 10% of input, allowing the percentage of immunoprecipitation to be calculated. -Fold enrichments were calculated as the percentage of specific DNA enriched using the specific antibody minus the percentage enriched with the control antibody, divided by the percentage of control DNA enriched with the specific antibody minus the percentage of control DNA enriched with the C/EBPβ antibody. The mouse genomic β-actin primer sequences were identical to those used for quantification of mitochondrial DNA in (9). The human control sequences were: Lmnb2-forward, 5′-TTCTATGCCAAGCCCATTCTAGGTC-3′ and reverse, 5′-GAGAGGCTCTGTCTGAGGTCACG-3′. Each displayed value represents an average of three independent experiments.
Sequential ChIP
Immune complexes were eluted at 37 °C for 30 min in 10 mm DTT. The eluted DNA was diluted 10-fold in ChIP dilution buffer followed by incubation with the indicated re-ChIP antibody. Secondary immune complexes were eluted in elution buffer. After reversal of cross-links, DNA was precipitated and analyzed by PCR. Anti-C/EBPβ antibody was used as isotype control.
qRT-PCR
RNA was isolated using TRIzol (Invitrogen), followed by RNeasy purification (Qiagen) using the RNA cleanup procedure. cDNA was synthesized using SuperScript III and random hexamers (Invitrogen). For Il2 RT-PCR, 50 ng of cDNA was used for qRT-PCR with a LightCycler 480 (Roche Applied Science). Sequences of the primers and Taqman probe for Il2 quantification were taken from Setoguchi et al. (20). ΔCt values were determined by subtracting input DNA, and ΔΔCt was determined by subtracting the ΔCt value for control primers. The ΔΔCt values were converted to -fold change using the formula -fold change = 2ΔΔCt and were averaged. Sequences for Ahcy qRT-PCR were: Ahcy forward, 5′-CGCATGCGCATCAATCCTG and Ahcy reverse, 5′-CCAGCATGTCTGATAAACTGCC. Control β-actin primers for mouse qRT-PCR were taken from Ref. 15. Sequences for Cdx2 and control Sdha qRT-PCR primers were taken from Ref. 21.
Bisulfite Sequencing
Bisulfite DNA modification was performed using a kit (Zymo Research). Primers for bisulfite sequencing were: Il2 forward, 5-GGTAGTTTTTTAGTATGGGAGG and reverse, 5-TAATTTTAACAAAAAAAACATTTTCA. PCR fragments were ligated into the TOPO TA cloning vector (Invitrogen) and sequenced.
RNAi
Oct1 and control siRNAs were obtained from Santa Cruz Biotechnology. siRNA transfection of DLD-1 cells was performed twice, 24 h apart, using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Cells were assayed 24 h after the second transfection.
RESULTS
Oct1-facilitated Recruitment of Jmjd1a Mediates Polr2a and Ahcy Antirepression in Fibroblasts
We demonstrated previously that Oct1 possesses an antirepressive function at a target gene to which it binds following stress treatment (Polr2a): the locus is expressed in normal MEFs but is inappropriately repressed in Oct1-deficient MEFs. Transient transfection of luciferase linked to the Polr2a promoter did not recapitulate this effect, suggesting that chromosomal context is important for this form of regulation (15). Oct1 binding to the Ahcy promoter, which encodes an enzyme that regulates methionine catabolism and S-adenosylmethionine levels, is also induced by treatment with H2O2 (15). Similar inappropriate repression in Oct1-deficient cells was observed at Ahcy upon exposure to H2O2, whereas wild-type cells were minimally affected (Fig. 1A).
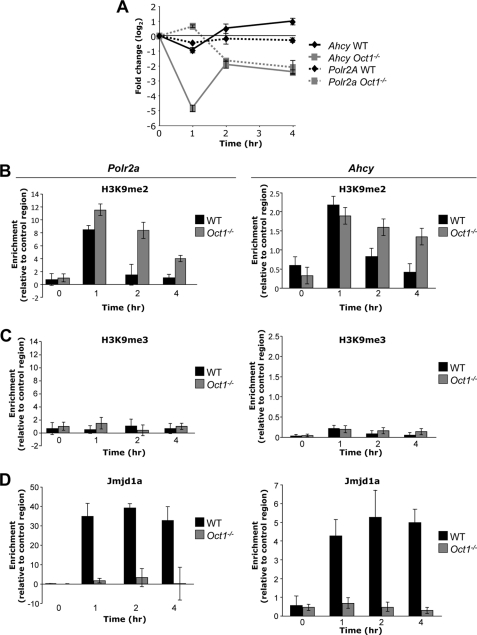
FIGURE 1.
Oct1 recruits Jmjd1a to housekeeping gene loci to maintain chromatin free of H3K9 dimethylation. A, Ahcy mRNA levels following H2O2 exposure in primary early-passage Oct1-deficient and matched littermate wild-type MEFs as measured by qRT-PCR. Measurements were made relative to β-actin. Data for Polr2a (15) are superimposed for comparison (dashed lines). Each value represents an average of three independent experiments. Error bars indicate S.D. B, ChIP-enriched enriched DNA from Oct1-deficient and matched littermate wild-type MEFs treated with H2O2 quantified by real-time PCR. PCR primers flanking the Oct1 binding site in Polr2a or Ahcy, and antibodies against histone H3K9me2 were used. Values are an average of three independent experiments. Error bars represent S.D. For details of quantification, see “Experimental Procedures.” C, similar experiment conducted using H3K9me3 antibodies. D, similar experiment using Jmjd1a antibodies.
These results suggested that in fibroblasts lacking Oct1, negative regulatory marks inappropriately accumulate resulting in repression. We reasoned that the nature of these marks would help identify the mechanism through which Oct1 functions. We studied multiple different marks, including DNA methylation. Despite significant gene repression, no DNA methylation was observed following H2O2 treatment in either wild-type or Oct1-deficient MEFs for either Ahcy or Polr2a as assessed with bisulfite sequencing (supplemental Fig. S1). These results are consistent with the housekeeping status of Polr2a and Ahcy. We also performed ChIP assays using antibodies directed against defined negative chromatin marks. We used real-time PCR amplification to quantify specific ChIP enrichment relative to a C/EBPβ control antibody. Surprisingly, at both Polr2a and Ahcy we identified a significant accumulation of histone H3K9me2 in both wild-type and Oct1-deficient MEFs following oxidative stress exposure; however, the mark was removed more rapidly in the wild-type condition, suggesting that Oct1 recruits an activity that catalyzes its removal (Fig. 1B). Whereas H3K9me2 is a marker of a transiently repressed state, H3K9me3 is a marker of stably repressed heterochromatin (22). In contrast to H3K9me2, we detected only minimal H3K9me3 (Fig. 1C). The deposition of H3K9me2 correlated with inappropriate prolonged repression in the Oct1-deficient condition (Fig. 1A).
Jmjd1a/JHDM2A/KDM3A is a jumonji C domain-containing lysine demethylase that has been shown to demethylate H3K9me1 and me2 but not H3K9me3 (23). Jmjd1a was also identified as a coactivator for the androgen receptor and shown to demethylate H3K9 at androgen receptor target genes (23). We therefore hypothesized that like androgen receptor, Oct1 uses Jmjd1a to prevent gene repression. ChIP assays using Jmjd1a antibodies indicated that it was recruited to both Polr2a and Ahcy in wild-type but not Oct1-deficient fibroblasts following oxidative stress exposure (Fig. 1D). These findings indicate that one function of Oct1 is to enable Jmjd1a recruitment to catalyze the removal of newly deposited H3K9me2.
Oct1 Promotes a Repressed State at Il2 in Naïve T Lymphocytes
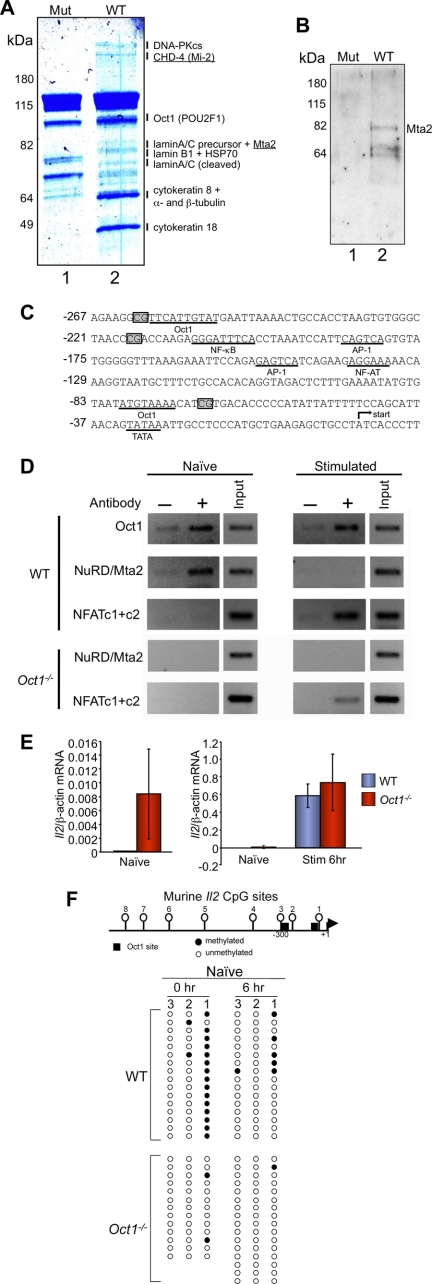
Il2 expression is confined to activated T cells, where it helps marshal the immune response (24, 25). When stimulated, naïve T cells rapidly demethylate specific CpGs in the Il2 promoter, and these sites remain demethylated after Il2 expression has ceased (26, 27). The Il2 promoter is a well known Oct1 target (28), and Oct1 occupies multiple sites in the human Il2 promoter in vivo (26, 29). However, NFAT, AP-1, and NF-κB are known to be the primary drivers of Il2 gene expression (24, 30), whereas the role of Oct1 is unclear. We previously used latex nanoparticle affinity purification to capture DNA-bound Oct1 for analysis by mass spectroscopy (15). To identify cofactors capable of associating with Oct1 bound to Il2 promoter DNA, we used nanoparticles coupled to a region of the human Il2 promoter containing a characterized Oct1 binding site (29) and HeLa nuclear extracts. Using mass spectroscopy, Oct1 itself was identified associated specifically with wild-type Il2 promoter DNA. Additionally, we identified HSP70, lamin A/C, lamin B, RecQ, α- and β-tubulin, and cytokeratin-8 as well as the DNA-dependent protein kinase catalytic subunit and two subunits of the NuRD-Mi-2 corepressor complex, CHD4-Mi-2 and Mta2 (Fig. 2A, lane 2). None of these proteins was associated with nanoparticles coupled to octamer sequences containing point mutations (lane 1). DNA-PK and lamin B have been previously described to associate with Oct1 (31–33), validating the method. NuRD also associated with Il2 promoter DNA in vitro using extracts from murine EL-4 T cells as assessed by Western blotting with anti-Mta2 antibodies (Fig. 2B, lane 2).
FIGURE 2.
Oct1 mediates Il2 repression in naïve T lymphocytes. A, Coomassie Blue-stained 10% polyacrylamide gel showing proteins from HeLa cell nuclear extracts retained on latex nanoparticles coated with human Il2 promoter DNA containing an Oct1 binding site (−262 to −222, lane 2), or an mutant octamer control (lane 1). Indicated bands showing differential retention were excised and subjected to liquid chromatography-mass spectrometry (LC/MS). Annotated bands had MASCOT scores >20 and mass errors <3 ppm. B, Western blot using Mta2 antibodies of a similar nanoparticle purification using nuclear extracts from EL-4 T cells. C, sequence of the murine Il2 proximal promoter region. CpG positions are boxed. Known transcription factor binding sites and TATA box are underlined. The transcription start site is depicted with an arrow. D, ChIP assays performed using primary splenic naïve T cells and two combined Oct1 antibodies, an antibody against the NuRD subunit Mta2, or combined antibodies against NFAT c1+c2. Amplification products using a region of the Il2 promoter encompassing Oct1 binding sites are shown. E, qRT-PCR analysis of Il2 mRNA expression levels in naïve and stimulated CD4+ splenic T cells. Error bars depict S.D. Panel on right is the averaged result of three experimental replicates. Isolated naïve cell data in left panel include six additional replicates. F, bisulfite sequencing analysis of the murine Il2 proximal promoter region. A schematic is shown at the top. Oct1 sites are shown with boxes. CpG methylation sites are numbered. Data from sequencing of clones are depicted below. Filled circles indicate the presence of a methylation event for a given clone.
NuRD is associated with transcriptional repression through DNA methylation, nucleosome remodeling, and histone deacetylation mechanisms (34–36), and NuRD associates with the Oct1 paralog Oct4 in ES cells (37–39). Mice deficient in Mta2 display normal T cell development but hyperproliferate upon activation and show increased Il2, Il4, and Ifng gene expression and lupus-like autoimmunity (40). These findings led us to test whether NuRD subunits are associated with Il2 promoter-bound Oct1 in naïve T cells, resulting in transcriptional repression. We have shown that Oct1 deficiency leads to embryonic lethality but that T cell development is grossly normal (19). A more thorough analysis of T cell subsets confirmed the prior results, although significant increases in activated (CD44hi) and decreases in memory (CD62Lhi) T cell subsets were noted (supplemental Fig. S2).
We analyzed splenic T cells from Rag-1-deficient mice repopulated with Oct1-deficient or wild-type littermate control embryonic day (E) 12.5 fetal liver hematopoietic progenitors. We focused on purified naïve CD4 helper T cells (CD8aneg, CD11bneg, CD45Rneg, DX5neg, Ter-119neg, CD44low), allowing us to work with uniform cell populations. Cells were stimulated with anti-CD3ϵ and anti-CD28 antibodies for 6 h.
The murine Il2 promoter contains two described Oct1 binding sites, at −75 and −250 bp relative to the transcription initiation site (Fig. 2C). The two sites are too close to accurately resolve by conventional ChIP. We confirmed Oct1 binding to this region in wild-type cells (Fig. 2D, top panel). Oct1 was constitutively associated with Il2 promoter DNA, as a significant ChIP signal was detected in naïve and stimulated cells. The results are consistent with the idea that Oct1 is not a predominant trigger of Il2 expression. These results differ from a report using primary T cells in which Oct1 binding is observed only following stimulation (26). However, that study used human cord blood T cells purified using anti-CD4 antibodies, which contain populations other than naïve cells. An earlier report using a murine cell line also described constitutive Oct1 binding (29).
ChIP of the same promoter region using antibodies against the NuRD subunit Mta2 showed association in naïve but not stimulated wild-type cells. In contrast and as expected, NFAT (c1+c2), a primary target of T cell receptor stimulation, only associated in stimulated T cells. To determine whether Oct1 recruits Mta2 to Il2 in naïve but not stimulated T cells, we tested Oct1-deficient cells (Fig. 2D). NuRD failed to associate with Il2 in either naïve or stimulated Oct1-deficient cells, whereas NFAT association was unaffected. These results are consistent with a regulated recruitment mechanism in which Oct1 associates with NuRD in naïve cells to help mediate transcriptional repression and loses this capacity upon T cell activation. We tested this model using qRT-PCR to assess Il2 mRNA levels. Il2 expression was completely absent in wild-type purified naïve cells (Fig. 2E). In contrast, a very low, but statistically significant (p < 0.05) amount of Il2 mRNA expression was observed in Oct1-deficient naïve T cells. The weak expression is consistent with a lack of transcription factors such as NFAT to drive Il2 transcription, but a loss of repressive mechanisms. Following stimulation, strong mRNA expression was noted regardless of Oct1 status.
NuRD components have been associated with CpG DNA methylation (e.g. 34). We used bisulfite sequencing to assess DNA methylation status in naïve and stimulated T cells. Relative to the transcription start site, the three most proximal upstream CpGs in murine Il2 are located at positions −69 (no. 1), −217 (no. 2), and −262 (no. 3, Fig. 2F). The distal CpG is immediately adjacent to an Oct1 site and is demethylated in wild-type cells. The proximal CpG site in wild-type cells is heavily methylated but becomes partially demethylated at short time points after T cell activation (Fig. 2F). This finding differs from an earlier report using lymph node T cells (27), but is consistent with another more recent report using spleen and lymph node T cells (41). Our splenic T cells were also harvested from Rag-1-deficient hematopoietic transfer recipients, although this difference is unlikely to underlie the different observations because cells purified from wild-type C57BL/6 mice show similar patterns (supplemental Fig. S3).
In contrast to wild-type cells, we observed inappropriate and nearly complete Il2 promoter demethylation at site 1 in Oct1-deficient naïve cells (Fig. 2F). The unmethylated state was maintained following T cell stimulation. We performed similar experiments using total splenic CD4+ T cells, which include populations of naïve, regulatory, preactivated, and memory T cells. Promoter demethylation was again observed in the Oct1-deficient condition; however, this time at site 3 (supplemental Fig. S4). Site 3 is adjacent to another Oct1 binding site. Unlike CpG no. 1 and 2, CpG no. 3 is conserved to humans. These results indicate that in unstimulated T cells, Oct1 is not required fοr the initial process of Il2 gene expression, but rather helps maintain a repressed state.
Oct1 Blocks Repression at Il2 in Previously Activated T Cells
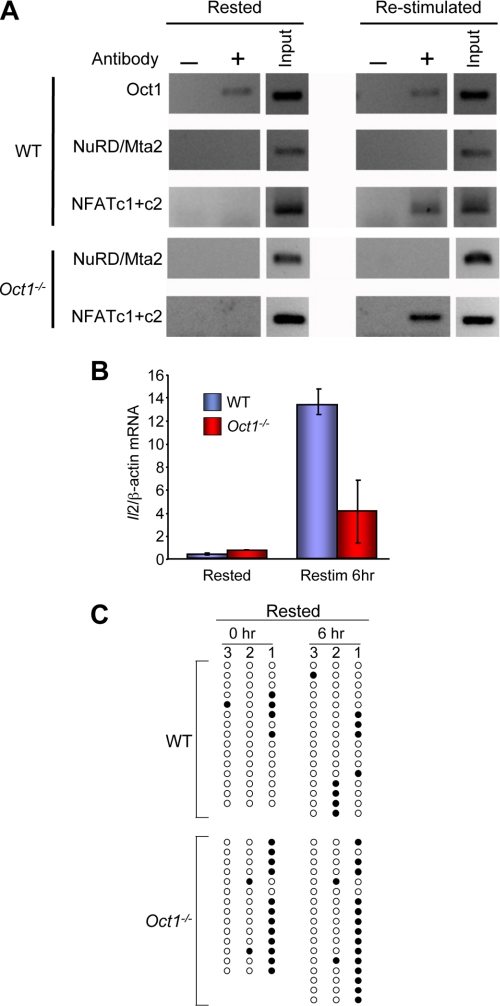
It remained to be determined whether Oct1 plays a role following T cell stimulation. Previous work has shown that T lymphocytes harbor an “epigenetic memory” at Il2 such that resting but previously stimulated cells display faster induction kinetics and higher expression levels upon secondary stimulation (26). We stimulated naïve T cells purified as before for 2 days, removed the stimulus, and rested the cells for 8 additional days, repeating the ChIP experiments before and after a second 6-h stimulation. Oct1 binding was again constitutive, showing identical characteristics in resting and restimulated cells (Fig. 3A). In contrast to naïve cells, Mta2 was not detected, suggesting that the change(s) blocking the ability of Oct1 to associate with NuRD following stimulation of naïve cells are maintained over an extended period. As expected, NFAT associated only in restimulated cells.
FIGURE 3.
Oct1 promotes expression and inhibits DNA methylation at Il2 following T cell stimulation. A, cells stimulated for 2 days and cultured for 8 additional days in the absence of stimulus. ChIP assays were performed on these rested cells or following 6-h stimulation as in Fig. 2D. B, real-time RT-PCR analysis of Il2 mRNA expression levels in resting and restimulated splenic T cells. Analysis was performed identically to Fig. 2E. C, bisulfite sequencing analysis of the murine Il2 proximal promoter region in resting and restimulated T cells. Analysis was performed similarly to Fig. 2F.
Following restimulation of rested cells with anti-CD3ϵ and anti-CD28 antibodies, strong and rapid Il2 induction was observed (Fig. 3B). Relative to the initial stimulation, restimulated wild-type cells displayed >10-fold stronger Il2 expression (compare scale in Fig. 3B with Fig. 2E). In contrast to the primary 6-h simulation in which little difference in Il2 expression was noted between Oct1-deficient and wild-type naïve cells, significant defects were observed upon restimulation of resting Oct1-deficient cells (Fig. 3B). Oct1-deficient T cells appeared microscopically normal with no evidence of cell death; however, they did not expand to the same degree as wild-type, consistent with the poor Il2 expression (data not shown). These results suggest that in rested cells, Oct1 no longer mediates repression but instead helps maintain functional memory of the previous stimulation by blocking repression.
To determine whether the loss of functional memory in Oct1-deficient resting but previously stimulated T cells is mediated by loss of epigenetic memory, we measured DNA methylation using bisulfite sequencing. Rested wild-type cells remained largely demethylated at position 1 (Fig. 3C). In contrast, this site was largely methylated in rested Oct1-deficient T cells (86% methylation in Oct1-deficient versus 25% in wild-type). Because naïve and initially stimulated Oct1-deficient naïve cells are demethylated at this position, these data indicate that in the absence of Oct1, Il2 becomes inappropriately remethylated at position 1 during the 8-day period following naïve T cell stimulation. After 6-h restimulation of rested T cells, the methylation patterns did not substantially change. Similar remethylation at site 1 was observed using total CD4+ T cells (supplemental Fig. S4). These results indicate that in addition to maintaining a repressed transcriptional state in naïve cells, Oct1 also maintains a poised state in resting but previously stimulated cells.
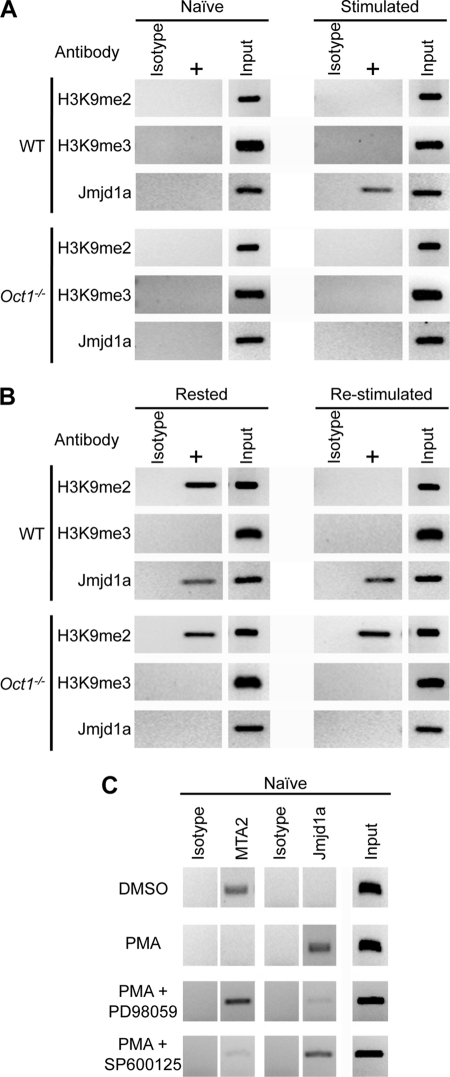
Because we had identified an Oct1 mediated anti-repression mechanism that utilized Jmjd1a in fibroblasts, we hypothesized that Oct1 antirepression in resting but previously stimulated T cells occurs through a similar mechanism. We used ChIP with antibodies directed against dimethyl and trimethyl H3K9, and Jmjd1a in naïve and rested T cells. As expected, Jmjd1a was not present at Il2 in naïve wild-type T cells but was recruited following stimulation. Jmjd1a was not recruited in Oct1-deficient T cells (Fig. 4A). As in fibroblasts, we identified a significant increase in histone H3K9me2 levels in both resting Oct1-deficient T cells and their wild-type counterparts (Fig. 4B). Following restimulation, H3K9me2 is rapidly removed (<6 h) from wild-type cells, presumably due to the action of Jmjd1a. However, the mark is inappropriately retained in Oct1-deficient T cells. No enrichment for H3K9me3, the more stably repressing mark, was observed (Fig. 4B). We infer that the function of Oct1 at the Il2 locus in rested but previously stimulated T cells is to oppose H3K9me2 (and subsequent DNA methylation at CpG position 1) through a mechanism involving Jmjd1a.
FIGURE 4.
Recruitment of Jmjd1a to Il2 by Oct1 in stimulated T cells. A, ChIP was performed on freshly isolated CD4+ naïve T cells (Naïve) and cells stimulated for 6 h (Stimulated) using antibodies specific to histone H3K9me2, K9me3, and Jmjd1a. Degree of enrichment for each indicated antibody is shown relative to control (C/EBPβ) antibodies using semiquantitative PCR. B, similar ChIP experiment performed using cell stimulated for 2 days and cultured for 8 additional days in the absence of stimulus (Rested) or the same cells stimulated for 6 h (Re-stimulated). C, PMA signaling alone sufficient to replace NuRD association with Il2 to Jmjd1a association. ChIP was performed on freshly isolated CD4+ naïve T cells treated with dimethyl sulfoxide (DMSO) carrier control or PMA for 6 h using antibodies specific to Mta2 and Jmjd1a. PMA was used at 20 ng/ml. PD98059 was used at 10 μm. SP600125 was used at 25 μm. Cells were exposed to the inhibitors for 2 h before exposure to PMA. Degree of enrichment for each indicated antibody is shown relative to isotype control (C/EBPβ) antibodies using semiquantitative PCR.
The signals emanating from the T cell receptor and CD28 are well defined and include NFAT and MAPK cascades (42). Some of these pathways can be selectively activated using specific reagents such as ionomycin to activate NFAT via Ca2+ flux, or PMA to activate MAPK signaling via PKCθ (42). We found that PMA treatment of naïve cells in isolation, although not sufficient to activate Il2 gene expression, was sufficient to induce the loss of NuRD and the association of Jmjd1a as measured by ChIP (Fig. 4C). In T cells, PMA activates PKCθ and further downstream, NF-κB and MAPK signaling. Two kinase components of the MAPK pathway are ERK1/2 and JNK. PD98059, an inhibitor of ERK1/2, but not SP600125, an inhibitor of JNK, blocked switching by PMA (Fig. 4C). These data suggest that MAPK signals impinge on Oct1 to mediate switching from repression to antirepression through a mechanism requiring ERK1/2.
Oct1 Mediates Mutually Exclusive NuRD and Jmjd1a Association with the Cdx2 Promoter in DLD-1 cells and Mediates Repression by PMA
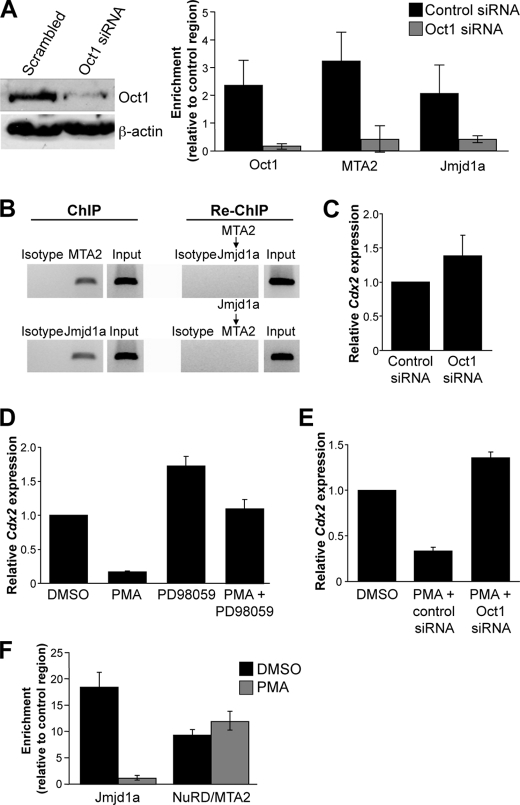
Cdx2 is a homeodomain transcription factor critical for mammalian development (43) and a known Oct1 regulatory target (44, 45). Cdx2 expression is mostly confined to the gastrointestinal tract in adults, where it acts as a tumor suppressor (46), but inappropriate expression of Cdx2 is observed in murine leukemia models and in most cases of pediatric acute myeloid leukemia and adult lymphoid leukemia (47–50), where it promotes self-renewal (49). Using DLD-1 colon adenocarcinoma cells and ChIP, we detected Oct1, NuRD, and Jmjd1a all bound to the conserved octamer site in the Cdx2 promoter (Fig. 5A). Association required Oct1, because Oct1 knockdown depleted Jmjd1a and NuRD as well as Oct1 ChIP signal (Fig. 5A). Sequential ChIP assays failed to detect any NuRD-associated DNA simultaneously associated with Jmjd1a and vice versa (Fig. 5B), suggesting that DLD-1 cells contain a heterogeneous population of Oct1 with positive and negative functionality. Consistent with this model, Oct1 siRNA knockdown produced little effect on Cdx2 gene expression (Fig. 5C).
FIGURE 5.
Oct1 mediates NuRD and Jmjd1a recruitment at Cdx2 in DLD-1 cells and mediates cofactor switching and Cdx2 repression in response to PMA. A, ChIP enrichment quantified for Oct1, Jmjd1a, and Mta2 (NuRD) relative to C/EBPβ control antibodies in DLD-1 cells transfected with control or Oct1-specfic siRNAs. Western blot showing effect of siRNA transfection on Oct1 expression levels is shown at left. B, Jmjd1a and NuRD enrichment in sequential ChIP assays. Agarose gel images are shown from semiquantitative PCR amplification of initial ChIP material. The ChIP material was diluted, and a second sequential ChIP was performed with the indicated antibody (see “Experimental Procedures”). Amplification of the re-ChIP DNA material is shown beneath each primary ChIP. C, effect of Oct1 siRNA transfection on Cdx2 expression levels. Cdx2 mRNA was measured using qRT-PCR. Averages of three replicate experiments are shown. Error bars depict S.D. D, DLD-1 cells treated with PMA for 6 h and Cdx2 expression measured. PMA was used at 2.5 μm. Cells were also treated simultaneously with PMA and PD98059 (10 μm) to determine the effect of ERK1/2 inhibition on Cdx2 expression. Cdx2 mRNA expression was measured as in C. DMSO, dimethyl sulfoxide. E, effect of Oct1 knockdown on PMA-mediated Cdx2 repression. Cells were transfected with control and Oct1-specific siRNAs as before. 24 h after the second transfection, cells were treated with PMA and Cdx2 expression measured as before. F, effect of PMA treatment on Jmjd1a and NuRD Cdx2 promoter occupancy. Quantification of ChIP enrichment relative to C/EBPβ control antibodies is shown. PMA treatment and ChIP assays were conducted as above.
Cdx2 is repressed in colon cancer cell lines by ERK signaling (21). These findings are consistent with frequent ERK hyperactivity due to K-Ras mutation in colon cancer and the role of Cdx2 as a tumor suppressor in this tissue. However, the mechanism was not defined. 6-h PMA treatment of DLD-1 colon cancer cells silenced Cdx2 in a manner that can be blocked by inhibition of ERK activity, as expected (Fig. 5D). Oct1 but not control RNAi eliminated silencing by PMA (Fig. 5E), indicating that the ability of PMA to modulate Cdx2 in DLD-1 cells requires Oct1. We tested the ability of NuRD and Jmjd1a to interact with Cdx2 following 6-h PMA treatment. PMA substantially reduced Jmjd1a association with Cdx2, but did not significantly alter association of the NuRD subunit MTA2 (Fig. 5F). Similar results were obtained with antibodies against Mi-2, a different NuRD subunit (data not shown).
DISCUSSION
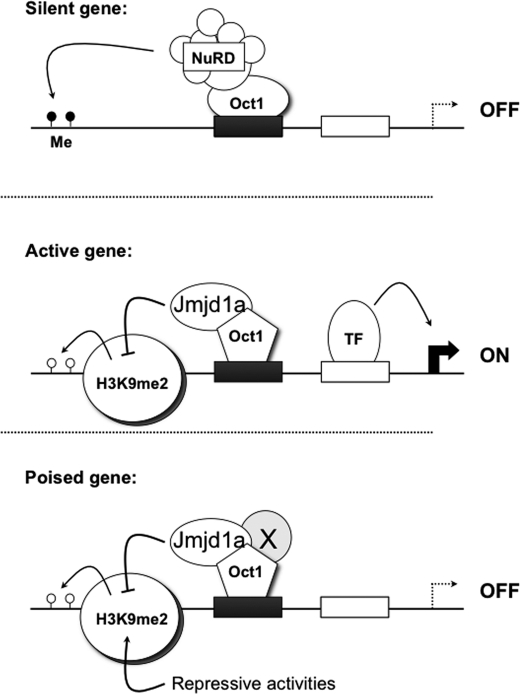
Here, we show that Oct1 is a bipotential transcription factor capable of regulated switching between repressive and antirepressive modes. Our data are consistent with the model shown in Fig. 6. Oct1 mediates antirepression at two housekeeping targets (Polr2a and Ahcy) by enabling the recruitment of Jmjd1a and maintaining chromatin free of histone H3K9me2. At the regulated Il2 target locus promoter region, Oct1 mediates recruitment of the NuRD corepressor complex in naïve T cells and recruitment of Jmjd1a in previously activated, resting T cells. When naïve cells are stimulated, Oct1 becomes modified through mechanisms involving MAPK and ERK signaling such that it no longer recruits NuRD and instead, directly or indirectly recruits Jmjd1a. We detected a biochemical interaction between Oct1 and NuRD, but have thus far failed to detect Jmjd1a. Therefore we cannot eliminate the possibility that the association with Jmjd1a is indirect. MAPK signaling is attenuated after the T cell stimulus is removed; however, the antirepressive state is retained even after 8 days in culture without stimulation. Therefore, the modification(s) induced by MAPK signals are either stable, or some other activity (e.g. the induction of Oct1 cofactors) maintains the switched state over longer times. This state allows faster Il2 induction in rested T cells relative to naïve cells and may be constitutive at Polr2a and Ahcy, allowing gene expression to recover more rapidly in wild-type fibroblasts relative to Oct1-deficient fibroblasts following stress exposure.
FIGURE 6.
Model Oct1 transcription factor mechanism. At repressed genes, Oct1 recruits NuRD and helps maintain a repressed state. At genes that are active (due in part to transcription factors labeled TF), the status of Oct1 changes such that it can no longer recruit NuRD. Instead, through either intrinsic activities and/or the recruitment of cofactors, or through indirect activities, it recruits Jmjd1a and catalyzes the removal of H9K9me2. The presence of this mark can lead to inappropriate DNA methylation in a subset of cases.
Oct1 also mediates recruitment of NuRD and Jmjd1a and is important for PMA-mediated gene regulation at the Cdx2 Oct1 target locus in DLD-1 cells. Also as with T cells, loss of Oct1 superficially appears to affect Cdx2 expression minimally; however, upon more careful examination Oct1 is critical for correct regulation. Several differences were also noted between the DLD-1 and T cell system. First, PMA mediates transcriptional silencing rather than transcriptional activation. Second, Oct1 is required for regulation by PMA. Third, both NuRD and Jmjd1a were detected in normally cultured cells as measured by ChIP. Sequential ChIP assays failed to detect any co-binding of the two activities, suggesting that DLD-1 cells are composed of a heterogeneous mixture of Oct1 associated with the different activities. Cumulatively, these findings suggest that Oct1 stabilizes both repressed and inducible transcriptional states. This bipotential form of transcription factor regulation has been associated with other transcription factor classes. For example, thyroid hormone receptor can switch from an activator to a repressor of the same targets in a ligand-regulated manner. This switch is mediated by exchange of coactivators and corepressors, including remodeling enzymes such as NuRD (35, 51).
Chromatin-modifying complexes are central regulators of gene expression (22, 52, 53), but the mechanisms of targeting are poorly understood. Transcription factors that maintain chromatin with the appropriate epigenetic marks through recruitment of these factors are certain to play important biological functions, for example by maintaining a silent state or priming target genes for later expression in stem/progenitor and memory cells. Our data suggest that Oct1 regulates the expression of its targets, at least in part, through spatial control of chromatin-modifying factors.
The absence of Oct1 is associated with abnormal DNA hypomethylation at a CG site proximal to the Il2 transcription start site in naïve T cells, but is also associated with abnormal hypermethylation at that site in previously stimulated, resting T cells. These changes in DNA methylation are likely an indirect readout of Oct1 regulation through NuRD and Jmjd1a. Surprisingly, both in fibroblasts following stress and in resting but previously stimulated T cells Jmjd1a was simultaneously present with H3K9me2 in wild-type cells. These results suggest that Jmjd1a is either not active in these situations (for example due to lack of cofactors) or that an opposing histone methyltransferase activity such as G9a (54) continually deposits this mark as it is being removed. We favor the latter mechanism because in wild-type T cells, the mark is present but does not accumulate or become expanded such that DNA methylation takes place, but in the absence of Oct1 it does, suggesting a finely regulated balance of activities.
The absence of Oct1 in resting T cells leads to poor activation upon restimulation relative to wild-type T cells. These findings have implications for the generation of T cell memory responses and, by extension, the ability to resist infection by certain pathogens. Oct1 is associated with the expression of multiple cytokines, including Il-3, Il-5, Il-8, Il-12/23 (55–60), and more recently Il-4 and Il-13 (61, 62). Oct1 is also associated with other loci of immunological interest (e.g. 63–65). In light of these findings, a reexamination of the role of Oct1 at these targets may shed light on their regulation. For example, a negative activity has been associated with the octamer site in the immunoglobulin heavy chain enhancer in B/T cell hybrids (66). It was postulated that a transcriptional repressor occupies this site in non-B cells. Our data suggest that the positive activity in B cells and the negative activity in non-B cells may be one and the same, and involve differential Oct1 cofactor recruitment. Rather than directly promoting expression of the aforementioned target genes, the principal role of Oct1 may be, in one mode, maintenance of repression, and in the opposite mode, prevention of transcriptional repression, i.e. poising for later expression.
Supplementary Material
Acknowledgments
We thank J. Neilson, T. Formosa, V. Planelles, and D. Stillman for critical reading of the manuscript. We thank C. Nelson and members of the University of Utah Health Sciences Center Mass Spectrometry Core for assistance with protein identification. H. Handa provided ferromagnetic latex nanoparticles for Oct1 affinity purification. J. Weis and the members of his laboratory provided expertise and reagents for isolating and culturing T lymphocytes. V. Planelles and members of his laboratory provided expertise with small molecule inhibition of MAPK signaling. D. Jones provided advice and reagents for Cdx2 gene expression analysis. We also thank D. Stillman, T. Formosa, J. Phillips, and members of their laboratories for advice and reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R21CA141009 through the NCI. This work was also supported by American Cancer Society Grant GMC-115196.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- POU
- Pit-1, Oct1/2, Unc-86
- MEF
- mouse embryonic fibroblast
- NFAT
- nuclear factor of activated T cells
- NuRD
- nucleosome remodeling and histone deacetylation
- PMA
- phorbol 12-myristate 13-acetate
- qRT-PCR
- quantitative RT-PCR
- C/EBPβ
- CCAAT-enhancer binding protein-beta.
REFERENCES
- 1. Herr W., Sturm R. A., Clerc R. G., Corcoran L. M., Baltimore D., Sharp P. A., Ingraham H. A., Rosenfeld M. G., Finney M., Ruvkun G. (1988) Genes Dev. 2, 1513–1516 [DOI] [PubMed] [Google Scholar]
- 2. Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 4. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 5. Feng B., Jiang J., Kraus P., Ng J. H., Heng J. C., Chan Y. S., Yaw L. P., Zhang W., Loh Y. H., Han J., Vega V. B., Cacheux-Rataboul V., Lim B., Lufkin T., Ng H. H. (2009) Nat. Cell Biol. 11, 197–203 [DOI] [PubMed] [Google Scholar]
- 6. Kang J., Shakya A., Tantin D. (2009) Trends Biochem. Sci. 34, 491–499 [DOI] [PubMed] [Google Scholar]
- 7. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. dela Paz N. G., Simeonidis S., Leo C., Rose D. W., Collins T. (2007) J. Biol. Chem. 282, 8424–8434 [DOI] [PubMed] [Google Scholar]
- 9. Shakya A., Cooksey R., Cox J. E., Wang V., McClain D. A., Tantin D. (2009) Nat. Cell Biol. 11, 320–327 [DOI] [PubMed] [Google Scholar]
- 10. Almeida R., Almeida J., Shoshkes M., Mendes N., Mesquita P., Silva E., Van Seuningen I., Reis C. A., Santos-Silva F., David L. (2005) J. Pathol. 207, 396–401 [DOI] [PubMed] [Google Scholar]
- 11. Reymann S., Borlak J. (2008) BMC Syst. Biol. 2, 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., Weinberg R. A. (2008) Nat. Genet. 40, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Somervaille T. C., Matheny C. J., Spencer G. J., Iwasaki M., Rinn J. L., Witten D. M., Chang H. Y., Shurtleff S. A., Downing J. R., Cleary M. L. (2009) Cell Stem Cell 4, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mattison J., Kool J., Uren A. G., de Ridder J., Wessels L., Jonkers J., Bignell G. R., Butler A., Rust A. G., Brosch M., Wilson C. H., van der Weyden L., Largaespada D. A., Stratton M. R., Futreal P. A., van Lohuizen M., Berns A., Collier L. S., Hubbard T., Adams D. J. (2010) Cancer Res. 70, 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang J., Gemberling M., Nakamura M., Whitby F. G., Handa H., Fairbrother W. G., Tantin D. (2009) Genes Dev. 23, 208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertolino E., Singh H. (2002) Mol. Cell 10, 397–407 [DOI] [PubMed] [Google Scholar]
- 17. Wang V. E., Schmidt T., Chen J., Sharp P. A., Tantin D. (2004) Mol. Cell. Biol. 24, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tantin D., Sharp P. A. (2002) Mol. Cell. Biol. 22, 1460–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang V. E., Tantin D., Chen J., Sharp P. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Setoguchi R., Hori S., Takahashi T., Sakaguchi S. (2005) J. Exp. Med. 201, 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krueger F., Madeja Z., Hemberger M., McMahon M., Cook S. J., Gaunt S. J. (2009) Cell. Signal. 21, 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jenuwein T. (2006) FEBS J. 273, 3121–3135 [DOI] [PubMed] [Google Scholar]
- 23. Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 24. Jain J., Loh C., Rao A. (1995) Curr. Opin. Immunol. 7, 333–342 [DOI] [PubMed] [Google Scholar]
- 25. Smith K. A. (1992) Curr. Opin. Immunol. 4, 271–276 [DOI] [PubMed] [Google Scholar]
- 26. Murayama A., Sakura K., Nakama M., Yasuzawa-Tanaka K., Fujita E., Tateishi Y., Wang Y., Ushijima T., Baba T., Shibuya K., Shibuya A., Kawabe Y., Yanagisawa J. (2006) EMBO J. 25, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bruniquel D., Schwartz R. H. (2003) Nat. Immunol. 4, 235–240 [DOI] [PubMed] [Google Scholar]
- 28. Ullman K. S., Flanagan W. M., Edwards C. A., Crabtree G. R. (1991) Science 254, 558–562 [DOI] [PubMed] [Google Scholar]
- 29. Garrity P. A., Chen D., Rothenberg E. V., Wold B. J. (1994) Mol. Cell. Biol. 14, 2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dienz O., Eaton S. M., Krahl T. J., Diehl S., Charland C., Dodge J., Swain S. L., Budd R. C., Haynes L., Rincon M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., van Steensel B. (2008) Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 32. Imai S., Nishibayashi S., Takao K., Tomifuji M., Fujino T., Hasegawa M., Takano T. (1997) Mol. Biol. Cell 8, 2407–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malhas A. N., Lee C. F., Vaux D. J. (2009) J. Cell Biol. 184, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Ng H. H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. (1999) Genes Dev. 13, 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xue Y., Wong J., Moreno G. T., Young M. K., Côté J., Wang W. (1998) Mol. Cell 2, 851–861 [DOI] [PubMed] [Google Scholar]
- 36. Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., Schreiber S. L. (1998) Nature 395, 917–921 [DOI] [PubMed] [Google Scholar]
- 37. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 38. Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010) Cell Stem Cell 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu X., Kovalev G. I., Chang H., Kallin E., Knudsen G., Xia L., Mishra N., Ruiz P., Li E., Su L., Zhang Y. (2008) J. Biol. Chem. 283, 13825–13833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomas R. M., Gao L., Wells A. D. (2005) J. Immunol. 174, 4639–4646 [DOI] [PubMed] [Google Scholar]
- 42. Isakov N., Altman A. (2002) Annu. Rev. Immunol. 20, 761–794 [DOI] [PubMed] [Google Scholar]
- 43. Suh E., Traber P. G. (1996) Mol. Cell. Biol. 16, 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin T., Li H. (2001) J. Biol. Chem. 276, 14752–14758 [DOI] [PubMed] [Google Scholar]
- 45. Wang P., Wang Q., Sun J., Wu J., Li H., Zhang N., Huang Y., Su B., Li R. K., Liu L., Zhang Y., Elsholtz H. P., Hu J., Gaisano H. Y., Jin T. (2009) J. Biol. Chem. 284, 26456–26465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo R. J., Suh E. R., Lynch J. P. (2004) Cancer Biol. Ther. 3, 593–601 [DOI] [PubMed] [Google Scholar]
- 47. Rawat V. P., Thoene S., Naidu V. M., Arseni N., Heilmeier B., Metzeler K., Petropoulos K., Deshpande A., Quintanilla-Martinez L., Bohlander S. K., Spiekermann K., Hiddemann W., Feuring-Buske M., Buske C. (2008) Blood 111, 309–319 [DOI] [PubMed] [Google Scholar]
- 48. Riedt T., Ebinger M., Salih H. R., Tomiuk J., Handgretinger R., Kanz L., Grünebach F., Lengerke C. (2009) Blood 113, 4049–4051 [DOI] [PubMed] [Google Scholar]
- 49. Scholl C., Bansal D., Döhner K., Eiwen K., Huntly B. J., Lee B. H., Rücker F. G., Schlenk R. F., Bullinger L., Döhner H., Gilliland D. G., Fröhling S. (2007) J. Clin. Invest. 117, 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thoene S., Rawat V. P., Heilmeier B., Hoster E., Metzeler K. H., Herold T., Hiddemann W., Gökbuget N., Hoelzer D., Bohlander S. K., Feuring-Buske M., Buske C. (2009) Leukemia 23, 649–655 [DOI] [PubMed] [Google Scholar]
- 51. Collingwood T. N., Urnov F. D., Wolffe A. P. (1999) J. Mol. Endocrinol. 23, 255–275 [DOI] [PubMed] [Google Scholar]
- 52. Denslow S. A., Wade P. A. (2007) Oncogene 26, 5433–5438 [DOI] [PubMed] [Google Scholar]
- 53. Reik W. (2007) Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 54. Tachibana M., Matsumura Y., Fukuda M., Kimura H., Shinkai Y. (2008) EMBO J. 27, 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pfeuffer I., Klein-Hessling S., Heinfling A., Chuvpilo S., Escher C., Brabletz T., Hentsch B., Schwarzenbach H., Matthias P., Serfling E. (1994) J. Immunol. 153, 5572–5585 [PubMed] [Google Scholar]
- 56. Duncliffe K. N., Bert A. G., Vadas M. A., Cockerill P. N. (1997) Immunity 6, 175–185 [DOI] [PubMed] [Google Scholar]
- 57. Kaushansky K., Shoemaker S. G., O'Rork C. A., McCarty J. M. (1994) J. Immunol. 152, 1812–1820 [PubMed] [Google Scholar]
- 58. Cron R. Q., Zhou B., Brunvand M. W., Lewis D. B. (2001) Genes Immun. 2, 464–468 [DOI] [PubMed] [Google Scholar]
- 59. Wu G. D., Lai E. J., Huang N., Wen X. (1997) J. Biol. Chem. 272, 2396–2403 [PubMed] [Google Scholar]
- 60. Zhou L., Nazarian A. A., Xu J., Tantin D., Corcoran L. M., Smale S. T. (2007) Mol. Cell. Biol. 27, 2698–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gervaziev Y. V., Olenina L. V., Krasotkina J. V., Lupatov A. Y., Mazurina S. A., Gervazieva V. B. (2010) Int. J. Immunogenet. 37, 13–20 [DOI] [PubMed] [Google Scholar]
- 62. Kiesler P., Shakya A., Tantin D., Vercelli D. (2009) Hum. Mol. Genet. 18, 4513–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thévenin C., Lucas B. P., Kozlow E. J., Kehrl J. H. (1993) J. Biol. Chem. 268, 5949–5956 [PubMed] [Google Scholar]
- 64. Zabel M. D., Wheeler W., Weis J. J., Weis J. H. (2002) J. Immunol. 168, 3341–3350 [DOI] [PubMed] [Google Scholar]
- 65. LeBowitz J. H., Kobayashi T., Staudt L., Baltimore D., Sharp P. A. (1988) Genes Dev. 2, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 66. Shen L., Lieberman S., Eckhardt L. A. (1993) Mol. Cell. Biol. 13, 3530–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.