Abstract
Insulin-dependent glucose homeostasis is highly sensitive to the levels of insulin-responsive glucose transporter 4 (GLUT4) expression in adipocytes. The level of GLUT4 protein expression is highly dependent on the rate of GLUT4 gene transcription. GLUT4 gene transcription is decreased in a variety of physiologic states of insulin resistance including type 2 diabetes, obesity, and prolonged fasting. GLUT4 gene expression in adipocytes is differentiation-dependent, with full expression delayed until late in the differentiation program. In this paper, we have tested the hypothesis that differentiation-dependent GLUT4 gene expression in 3T3-L1 adipocytes is dependent on the nuclear concentration of a class II histone deacetylase (HDAC) protein, HDAC5. We have tested this hypothesis by reducing the levels of class II HDACs in the nuclear compartment of 3T3-L1 preadipocytes using two experimental approaches. First, preadipocytes were treated with phenylephrine, an α-adrenergic receptor agonist, to drive HDACS out of the nuclear compartment. Also, the class II HDAC concentrations were reduced using siRNA knockdown. In each case, reduction of nuclear class II HDAC concentration resulted in increased expression of endogenous GLUT4 mRNA in preadipocytes. Together, our data indicate that class II HDAC expression is the major regulatory mechanism for inhibiting GLUT4 expression in the predifferentiated state.
Keywords: Adipocyte, Differentiation, Gene Regulation, Glucose Transport, Histone Deacetylase, GLUT4
Introduction
The facilitative glucose transporter 4 (GLUT4)2 is responsible, in part, for insulin-mediated glucose uptake in skeletal muscle, heart, and adipose tissues (1). Several studies have demonstrated that insulin-dependent glucose homeostasis is highly sensitive to changes in GLUT4 protein expression (2–4). GLUT4 expression is altered in different physiological and pathological states including fasting, insulin resistance, and type 2 diabetes (5–9). Therefore, it is of great importance to understand how GLUT4 is properly regulated to understand the significant changes that result in insulin resistance, and ultimately, type 2 diabetes.
We have previously shown that the human GLUT4 promoter, when expressed in transgenic mice, is governed by two cis-acting domains: a MEF2-binding domain (10) and Domain I (11, 12). Using both transgenic and cultured cell models, we have shown that maximal GLUT4 transcriptional activation is achieved when MEF2 proteins are bound to the MEF2 domain and GLUT4 enhancer factor (GEF) is bound to Domain I (10–12). GEF proteins form a complex with MEF2 proteins that, in turn, serves as a binding site for other transcriptional mediators. It is well established that class II HDACs (HDAC4, HDAC5, HDAC7, and HDAC9) down-regulate MEF2 dependent gene transcription (13–16). GLUT4, like other MEF2-dependent genes, is regulated by recruitment of the class II histone deacetylase, HDAC5 (17). The class II HDACs possess an N-terminal domain that has been found to mediate its interactions with other proteins, including the MEF2 proteins (13, 14, 18). We have shown that HDAC5 can also form a protein complex with GEF informing us of the molecular basis by which MEF2, GEF, and HDAC5 regulate the GLUT4 promoter (17).
Several lines of evidence support the role of a class II HDAC, HDAC5, in transcriptional regulation of the GLUT4 promoter in adipocytes. First, it has been shown that constitutive localization of HDAC5 into the nucleus in cardiac tissue has resulted in a significant (∼3-fold) decrease in GLUT4 expression (19). Also, an increase in GLUT4 expression during exercise correlates with a decreased association between MEF2 and HDAC5, suggesting that HDAC5 mediates the repression of GLUT4 through MEF2 interactions (20). We have observed that the expression of HDAC5 during adipocyte differentiation is inversely correlated with GLUT4 expression (17). Using in vitro transcription assays, we have shown that HDAC5 specifically represses transcriptional activation of the GLUT4 promoter. In vivo, ChIP assays demonstrate that HDAC5 associates with the GLUT4 promoter in preadipocytes.
In the current study, we tested the hypothesis that nuclear HDAC5 levels in preadipocytes are responsible for repression of GLUT4 gene transcription prior to differentiation. Using a variety of techniques to reduce nuclear HDAC expression, we were able to induce GLUT4 mRNA expression in preadipocytes, a state in which GLUT4 is not normally expressed, confirming that HDAC-mediated repression of the GLUT4 promoter is a major mechanism responsible for regulation of differentiation-dependent GLUT4 gene expression.
MATERIALS AND METHODS
Cell Culture and Transfections
3T3-L1 cells were maintained and transfected via electroporation as described previously (21). Briefly, 10-cm plates on day 5 after differentiation 3T3-L1 adipocytes or preadipocyte fibroblasts were washed with PBS and incubated in trypsin/EDTA, PBS, and collagenase. The cells were resuspended and pelleted at 500 × g for 5 min. The cells were resuspended in DMEM containing 25 mm glucose. Five hundred microliters of cell suspension was transferred to a 0.4-cm electroporation cuvette (Bio-Rad), and 50 μg of each of the indicated plasmids were added. Empty vector (pcDNA3) was used to normalize the total DNA added in all of the experiments. The cells were electroporated using a Gene Pulser II (Bio-Rad) at 0.18 kV and 950 microfarads. The cells were allowed to recover for 10 min at room temperature. Equivalent amounts of cell suspension and fresh media were added together and plated according to the intended experiment. COS-7 cells were transiently transfected using the FuGENE 6 transfection reagent (Roche Applied Science) as described previously (17). Nuclear extracts were prepared using the NE-PER protein extraction kit (Pierce), with protease inhibitors (Roche Applied Science) added to prevent proteolysis during nuclear isolation. Whole cell extracts were prepared as described (17). The cells were initially washed in PBS and resuspended in ice-chilled whole cell extract buffer (1× PBS, 0.5% Triton X-100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1× Complete-mini EDTA-free protease inhibitors (Roche Applied Science)). The extracts were sonicated on ice, and insoluble debris was pelleted by microcentrifugation at 12,000 × g for 10 min at 4 °C. Total protein concentrations were determined with Coomassie Plus protein assay reagent (Thermo Scientific) according to the manufacturer's protocol.
siRNA Transfections
Experiments using siRNA were transfected in a similar manner as described above, with the following modifications: a final concentration of 50 μm of either scrambled negative control siRNA (#5, Ambion, AM4638) or predesigned siRNA specifically for HDAC4, HDAC5, and HDAC9 purchased from Santa Cruz Biotechnology (sc-35541, sc-35543, and sc-35551). The cells were then plated in 10-cm culture dishes and incubated for 72 h, followed by a second transfection with either empty vector (pcDNA3) or transactivating factors as indicated. RNA was harvested 24 h after the last transfection and subjected to real time (RT)-PCR as described below.
Plasmids
Truncated HDAC5 mutant segments were created from a wild type human HDAC5 template by using PCR to delete the indicated sections. Those DNA fragments were cloned in frame into a pcDNA3.1 (Invitrogen) expression vector with an N-terminal FLAG epitope using the restriction enzyme sites KpnI/XbaI. The results were verified through enzymatic digestion and sequencing. FLAG-tagged HDAC4 and FLAG-tagged HDAC9 plasmids were gifts obtained from Dr. Johnathan R. Whetstine (Harvard Medical School).
Immunoprecipitations
Co-immunoprecipitations were carried out from whole cell extracts of COS-7 cells transiently co-transfected with either HDAC5-Δ121 or HDAC5-ΔMEF and GEF or MEF2A as indicated. Seven hundred and fifty μg of whole cell extract was incubated with 40 μl of either protein A/G beads preloaded with normal mouse IgG or EZview Red anti-FLAG M2 affinity gel beads (Sigma) prewashed in 1× PBS overnight at 4 °C gently rocking end over end. The samples were washed five times in fresh ice-chilled whole cell extract buffer, and the samples were eluted by boiling for 5 min in 2× Laemmli sample buffer.
Immunoblot Analysis
Denatured samples were separated by SDS-PAGE using 10% polyacrylamide gels for MEF2A and GEF blots and 7.5% polyacrylamide gel for HDAC5 blots. The proteins were then transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) overnight in transfer buffer (25 mm Tris, 190 mm glycine) at 0.2 Å and 4 °C. The membranes were blocked for 1 h at room temperature with blocking buffer (Rockland). The membranes were then probed with rabbit α-GEF-N sera (21), rabbit polyclonal α-MEF2A (Santa Cruz Biotechnology), rabbit polyclonal α-HDAC5 antibodies (Millipore), rabbit polyclonal α-HDAC4 antibodies (Cell Signaling), or rabbit polyclonal α-HDAC9 (Santa Cruz) antibodies overnight at 4 °C. The membranes were visualized following incubation with Alexa Fluor 680 goat anti-rabbit IgG (Molecular Probes) and quantified using the Odyssey infrared imaging system (Li-Cor Biosciences).
RNA Extraction and Quantitative Real Time PCR
RNA was extracted from three 10-cm tissue culture dishes/condition (17). Briefly, the cells were homogenized in tissue lysis buffer (5 m guanidinium isothiocyanate, 30 mm sodium acetate, pH 5.2, 10 mm EDTA, pH 8.0, 5% β-mercaptoethanol, 0.2% sarkosyl). Cesium chloride was then added to a final concentration of 3.3 m, and then the homogenate was layered over a CsCl cushion (5.7 m CsCl, 30 mm sodium acetate, 10 mm EDTA, pH 5.5). The samples were centrifuged at 120,000 × g overnight at 20 °C. After removal of the supernatant, the RNA pellet was resuspended in HES buffer (20 mm HEPES, pH 7.4, 5 mm EDTA, pH 8.0, 1% SDS) and purified by chloroform:butanol extraction. The aqueous layer containing the RNA was stored as an ethanol precipitate at −20 °C until further analysis. RNA was quantified by UV absorbance at 260, 280, and 320 nm.
Messenger RNA levels of HDAC5, HDAC4, HDAC9, mGLUT4, mPPARγ, m36B4, and actin were quantified by quantitative real time PCR. The reverse transcription-PCR conditions were 25 °C for 5 min followed with 10 min at 50 °C for the reverse transcription followed by 95 °C for 5 min and then 45 cycles at 10 s for 95 and 55 °C for 1 min. The relative copy number of actin and/or 36B4 was used for normalization. The mRNA levels were calculated using a standard curve of a mixture of representative RNA samples. Primer sequences were as follows: HDAC5, 5′-GAAGCACCTCAAGCAGCAGCAGG-3′ (forward) and 5′-CACTCTCTTTGCTCTTCTCCTTGTT-3′ (reverse); HDAC4, 5′-GGTTTGAGAGCAGGCAGAAC-3′ (forward) and 5′-CAGAGAATGAGGCCAAGGAG-3′ (reverse); HDAC9, 5′-GCTGTGAAGGTCAAGGAGGA-3′ (forward) and 5′-TTGCTGGGTGAGGTAAAACA-3′ (reverse); mGLUT4, 5′-AAAAGTGCCTGAAACCAGAG-3′ (forward) and 5′-TCACCTCCTGCTCTAAAAGG-3′ (reverse); mPPARγ, 5′-CACAATGCCATCAGGTTTGG-3′ (forward) and 5′-GCTGGTCGATATCACTGGAGATC-3′ (reverse); m36B4, 5′-CTGAGTGATGTGCAGCTGAT-3′ (forward) and 5′-AGAAGGGGGAGATGTTCAG-3′ (reverse); and actin, 5′-CCTCACTGACTACCTGATGA-3′ (forward) and 5′-AGCTCATAGCTCTTCTCCAG-3′ (reverse). All of the quantitative real time PCRs were run using a CFX96 real time PCR detection system thermal cycler (Bio-Rad).
Luciferase Assays
3T3-L1 adipocytes were differentiated and electroporated as described previously (21). Luciferase constructs −895-hGLUT4 luc and ΔΔ-hGLUT4 luc have been described previously and indicated where used (11). Luciferase assays were performed as described previously with plasmids encoding hHDAC5, hHDAC5 mutants, hHDAC4, hHDAC9, LXRα, MEF2A, and GEF (17). Luciferase assays were performed 24 h after transfection using a Dual-Glo luciferase kit (Promega).
Immunofluorescence and Imaging
3T3-L1 preadipocytes and adipocytes 5 days after differentiation were transiently transfected as described above. The cells were allowed to recover for 10 min at room temperature. Equivalent amounts of cell suspension and fresh media were added together and plated in four-chambered slides (BD Falcon) for imaging. The cells were fixed and permeablized with methanol at −20 °C for 5 min. The cells were quenched in PBS containing 0.05 m ammonium chloride for 10 min. The cells were then blocked in PBS containing 10% calf serum for 20 min and stained for HDAC5 in PBS containing 3% goat serum and 0.1% Nonidet P-40 overnight at 4 °C or for 1 h for anti-FLAG. Primary antibodies used were against FLAG (200471; Stratagene) and HDAC5 (07-040; Millipore). Primary antibodies were used at a dilution of 1:200, and secondary antibodies were used at a dilution of 1:500. Image analysis and quantification were performed using Leica LCS Lite software.
Statistical Analysis
The means and standard errors of the mean are reported. The differences were analyzed by analysis of co-variance or Student's t test.
RESULTS
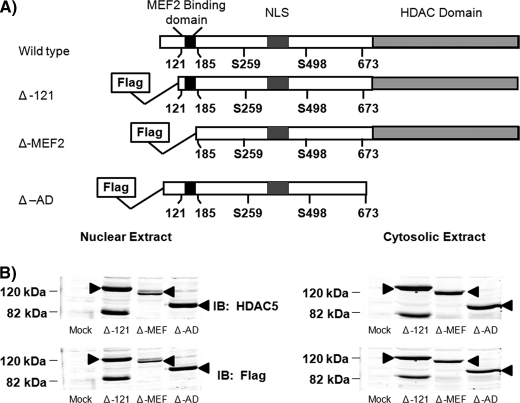
Previous studies have shown that HDAC5 and MEF2 interact both in vivo and in vitro. To determine which domains of HDAC5 were necessary for repression of GLUT4 transcription, FLAG-tagged truncated constructs were created (Fig. 1A). The consensus MEF2-binding domain is located between amino acids 121 and 185, with strong support that the first 121 amino acids do not play a role in either HDAC5 function or expression (22). Therefore, we created a mutant with the first 121 amino acids replaced with a FLAG tag to mimic wild type (Δ-121). This mutant functioned identically to wild type HDAC5 in our in vitro transcription assays (Fig. 2). A second deletion through amino acid 185 removed the consensus MEF2-binding domain (Δ-MEF). The third mutant deleted the residues C-terminal to amino acid 673, the region containing the deacetylase domain (Δ-AD). All of the constructs still contain the key serine residues (Ser-259/479) and nuclear localization signal needed for proper localization (23–26). Detection of HDAC5 mutants by Western blot analysis demonstrated that all of the proteins are efficiently expressed, appear at the expected molecular mass, and are similarly distributed between the nuclear and cytosolic compartments (Fig. 1B). We observed a band at the expected molecular mass of the Δ-121-HDAC5, but we also observed a band at 82 kDa with both the anti-HDAC5 and anti-FLAG blots. This band is similar to what has been reported previously by other groups and with other constructs (22). The band was found to be an altered form of the protein, possessing both the N-terminal tag and the C-terminal epitope. Taken together, these data suggest that some of the mRNA produced can undergo alternative splicing to create the HDAC5 variants (22). This hypothesis was confirmed by Lemercier et al. (22), yet they also noted that this variant of HDAC5 did not bind MEF2A, which is consistent with our data as well (Fig. 3).
FIGURE 1.
Expression and compartmental distribution of HDAC5 mutants in COS7 cells and 3T3-L1 adipocytes. A, schematic representation of HDAC5 wild type and FLAG-tagged truncated mutants used. B, 50 μg of nuclear or cytosolic extracts from COS-7 cells transiently overexpressing empty vector (Mock) or truncated FLAG-tagged HDAC5 were analyzed by SDS-PAGE and immunoblotted (IB) with FLAG or HDAC5 antibodies. The arrow indicates HDAC5.
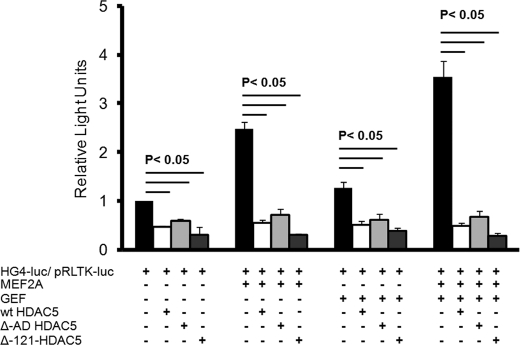
FIGURE 2.
Effect of truncation of the first 121 amino acids of HDAC5 (Δ-121) or truncation of the acetylase domain (Δ-AD) on GLUT4 promoter activity in vitro. 3T3-L1 adipocytes 6 days after differentiation were transiently transfected with luciferase reporter plasmids −895-hG4-luc (firefly luciferase, for promoter activity) and pRLTK (Renilla luciferase, for transfection efficiency), with and without plasmids encoding GEF, MEF2A, and wild type HDAC5 (wt HDAC5), Δ-121-HDAC5, or Δ-acetylase domain (AD) HDAC5. +, inclusion in transfection. The means ± S.E. from three independent experiments are shown; the data were analyzed by a one-way analysis of co-variance, and statistically significant (p < 0.05) changes are indicated.
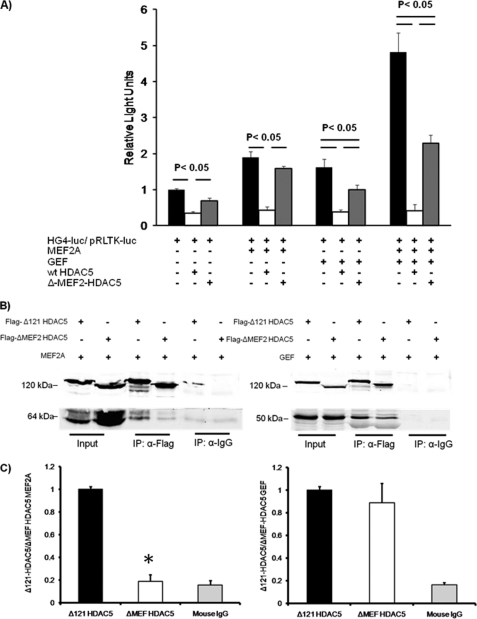
FIGURE 3.
MEF and GEF interact with HDAC5 independently. A, 3T3-L1 adipocytes 6 days after differentiation were transiently transfected with luciferase reporter plasmids −895-hG4-luc (firefly luciferase, for promoter activity) and pRLTK (Renilla luciferase, for transfection efficiency), without or with plasmids encoding GEF, MEF2A, and wild type HDAC5 (wt HDAC5) or Δ-MEF2 domain (Δ-MEF) HDAC5. +, inclusion in transfection. The data from three independent experiments are shown, and statistical significance was determined by a one-way analysis of co-variance. B, co-immunoprecipitation (IP) of HDAC5 with either MEF2A or GEF was performed in whole cell extracts from COS-7 cells transiently transfected with either MEF2A or GEF in the presence of Δ-121-HDAC5 or Δ-MEF2-HDAC5. The extracts were incubated with either preloaded normal mouse IgG-protein A/G beads or anti-FLAG beads overnight at 4 °C. The samples were then washed and analyzed by Western blot for the indicated protein. C, ratio of co-immunoprecipitated MEF2A or GEF to FLAG-tagged HDAC5 mutants. The means ± S.E. from four independent experiments are shown; statistical analysis was performed by a Student's t test.
It has been well established that HDAC5 specifically inhibits MEF2-dependent gene transcription (for review see Ref. 27); however, the precise mechanism is not firmly established. To test whether GLUT4 transcriptional repression absolutely requires active deactylase activity, we employed the Δ-AD mutant (Fig. 1A) in an in vitro transcription assay with the minimal functional human GLUT4 promoter driving the expression of a firefly luciferase gene (HG4-luc), as characterized previously (17). The HG4-luc construct was transactivated without or with plasmids that encoded MEF2A, GEF, human HDAC5, the ΔAD-HDAC5 mutant, or the Δ-121-HDAC5 mutant as indicated (Fig. 2). Transfection efficiency was accounted for by normalization to a control plasmid (pRLTK-luc). As before, we observed that both MEF2A and GEF contribute to full activation of the GLUT4 promoter in an additive manner, and human HDAC5 specifically inhibits in the presence of both transcriptional activators ∼8-fold (17). The ΔAD-HDAC5 mutant expressed either alone or in the presence of either one or both transcriptional activators repressed transcriptional activity similar to wild type human HDAC5 (Fig. 2). Taken together, it appears that in vitro, the deacetylase domain of HDAC5 is not necessary for inhibition of the GLUT4 promoter. The Δ-121 mutant was also found to repress both GEF- and MEF2-dependent GLUT4 transcription (Fig. 2). The Δ-121 mutant contained a useful FLAG epitope to distinguish between endogenous HDAC5 and therefore was used as a wild type analog based on similar repression and localization compared with wild type.
We next sought to determine whether the putative MEF2-binding domain was also essential for repression of GLUT4 gene expression as seen in other MEF-dependent genes (22). To test this hypothesis, we transactivated HG4-luc without or with the Δ-MEF2 mutant (Fig. 1A) to determine whether the MEF2-binding domain was required for both MEF2- and GEF-dependent repression of the GLUT4 promoter. Removal of the MEF2-binding domain of HDAC5 completely prevented its ability to repress HG4-luc in the presence of MEF2A alone; however, it retained the ability to repress GLUT4 transcription transactivated by GEF alone (Fig. 3A). When the GLUT4 promoter was transactivated by both MEF2A and GEF, Δ-MEF2-HDAC5 mutant impaired only 50% of the transcriptional activity. These data support a model in which HDAC5 interacts with both GEF and MEF2 independently of the interaction between GEF and MEF2A.
If GEF and MEF2 bind to HDAC5 independent of the GEF and MEF2A interaction, we predicted that GEF binds HDAC5 in a domain that is distinct from the MEF2-binding domain. To test this, we transiently overexpressed MEF2A or GEF along with either the FLAG-tagged Δ-121 or Δ-MEF mutant HDACs in Cos7 cells. Whole cell detergent extracts were immunoprecipitated with either preloaded normal mouse IgG A/G beads to control for nonspecific binding or anti-FLAG beads to measure HDAC5 protein complex formation. As expected, the Δ-MEF2-HDAC5 mutant co-immunoprecipitated 80% less MEF2A protein than was co-immunoprecipitated with the Δ-121-HDAC5 mutant (Fig. 3B). The amount of MEF2A that immunoprecipitated with the Δ-MEF2-HDAC5 mutant was then due to nonspecific binding, because a similar amount was precipitated by nonimmune IgG. In contrast, there was no difference between GEF binding with the Δ-121-HDAC5 or Δ-MEF2-HDAC5 proteins (Fig. 3B). These data demonstrate that HDAC5 has distinct binding domains for MEF2A and GEF.
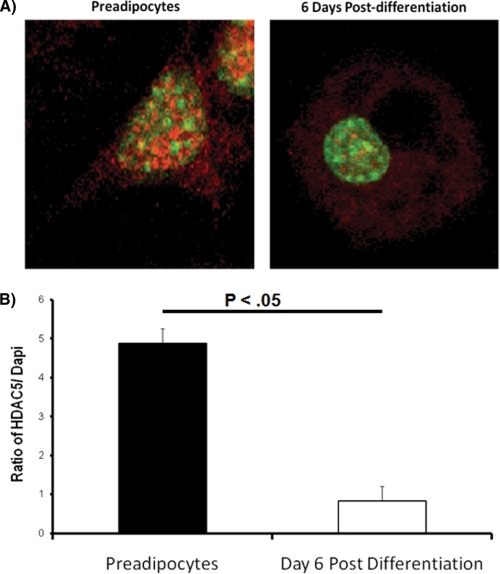
Our previous data demonstrated that GLUT4 promoter activity correlates with decreased nuclear expression of HDAC5 (17). To begin testing the role of HDAC5 as a physiologic regulator of GLUT4 transcription, we sought to determine changes in subcellular compartmentalization of HDAC5 following differentiation. Using a single cell, indirect immunofluorescence assay, we measured the level of endogenous HDAC5 in preadipocytes and fully differentiated adipocytes (6 days after differentiation). The nuclei were visualized using the DNA-specific stain, DAPI (Fig. 4A). The ratio of HDAC5 to DAPI decreased by nearly 6-fold in differentiated adipocytes compared with preadipocytes (Fig. 4B), demonstrating that HDAC5 redistributes to the cytosolic compartment as a function of differentiation.
FIGURE 4.
Distribution of endogenous HDAC5 during differentiation of adipocytes. A, 3T3-L1 preadipocytes or adipocytes 6 days after differentiation were stained with an anti-HDAC5 antibody, then stained with an Alexa 568-conjugated secondary antibody (red), and fixed in the presence of DAPI staining (green) to mark the location of the nucleus. B, a ratio of Alexa 568 signal to DAPI signal was used to determine the relative amount of HDAC5 present in the nucleus. Data from at least eight cells were analyzed and expressed as the means ± S.E. The data were analyzed by a Student's t test.
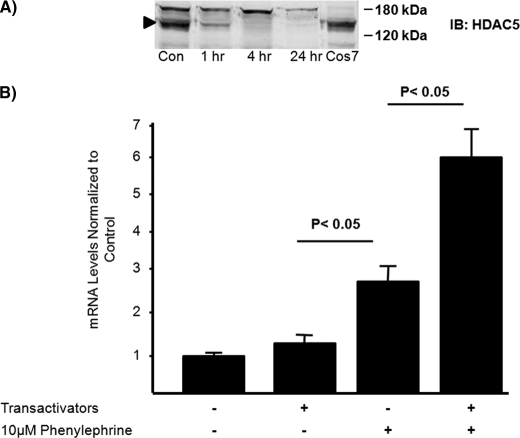
To further test the role of nuclear HDAC5 as a central regulator of GLUT4 transcription, we sought to decrease HDAC5 protein in the nuclear compartment of preadipocytes to determine whether this was sufficient for GLUT4 mRNA expression. To do this we first treated the cells with phenylephrine (PE), an α-adrenergic receptor agonist shown to induce the redistribution of class II HDACs through phosphorylation of serine 259/479 (28). To confirm the efficacy of PE in preadipocytes, we performed a time course experiment in which nuclear accumulation of HDAC5 was measured by Western blot at 0, 2, 4, and 24 h of continuous treatment with 10 μm PE. The effect was rapid, and HDAC5 was undetectable in nuclear extracts between 4 and 24 h of exposure to PE (Fig. 5A). Next, 3T3-L1 preadipocytes were transiently transfected with pcDNA3 or a combination of MEF2A, GEF, and LXRα (transactivators) and treated without or with 10 μm PE for 18 h. LXRα was added to the reaction mixture to transactivate the GLUT4 LXR response element, a site that makes a small contribution to overall GLUT4 transcriptional activity (29). The transactivators are the transcription factors known to regulate GLUT4 transcriptional activity in adipocytes (17, 29). Total RNA was isolated, and GLUT4 mRNA was quantified using RT-PCR. We found that redistribution of class II HDACs through PE treatment alone significantly increased endogenous GLUT4 mRNA in preadipocytes, whereas overexpression of the transactivators alone had no effect on GLUT4 mRNA levels (Fig. 5B). In contrast, overexpression of transactivators in combination with PE treatment further increased endogenous GLUT4 mRNA expression in preadipocytes (Fig. 5B). Given that the transfection efficiency of preadipocytes is quite low, we reasoned that the transactivators are present in only a small proportion of the cells and therefore did not expect a large increase in GLUT4 mRNA. These data support the notion that both the redistribution of the class II HDACs and the differentiation-dependent increases in activating transcription factors are required for GLUT4 mRNA expression in adipocytes.
FIGURE 5.
Phenylephrine treatment increases endogenous GLUT4 mRNA levels in preadipocytes with and without transactivating factors. A, 50 μg of COS7 whole cell extract transiently transfected with human HDAC5 or 120 μg of nuclear extracts of 3T3-L1 preadipocytes treated with or without phenylephrine for the indicated time periods and analyzed by SDS-PAGE and immunoblotting for HDAC5. The arrow indicates the expected appearance of HDAC5. Con, control; IB, immunoblot. B, 3T3-L1 preadipocytes were transiently transfected with transactivating plasmids encoding GEF, MEF2A, and LXRα. +, inclusion in transfection. The cells were treated with 10 μm phenylephrine for 24 h, and the RNA was isolated and analyzed by RT-PCR. The means ± S.E. from at least three independent experiments are shown; the data were analyzed by a Student's t test.
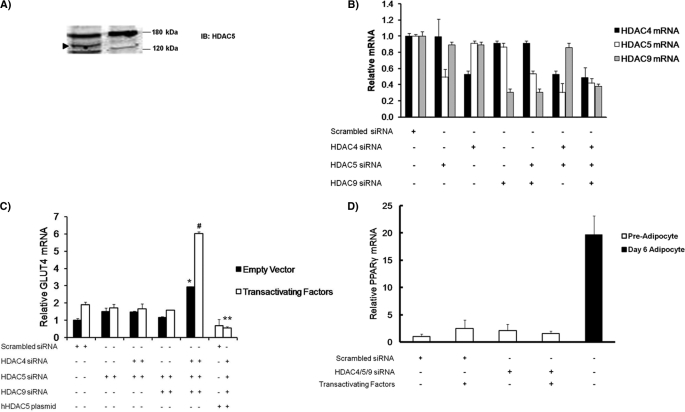
Because PE is an α-adrenergic agonist, it effects more than the distribution of class II HDACs in the cell. Therefore, we used siRNA knockdown of HDAC5 to more directly test the hypothesis that high nuclear content of class II HDACs regulates GLUT4 transcription. Other studies have demonstrated a redundant or compensatory function among class II HDACs when one is knocked down (30, 31). To control for redundancy in class II HDAC function, we established conditions to knock down HDAC5, HDAC4, and HDAC9 individually and in combination. siRNA-mediated knockdown of HDAC5 protein reduced expression below detectable levels via Western blot (Fig. 6A). This level of protein knockdown correlated with a 50% reduction in HDAC5 mRNA level (Fig. 6B). siRNA-mediated knockdown of HDAC4 and HDAC9 reduced expression of mRNA 45 and 55%, respectively. To determine the effect of class II HDAC knockdown of GLUT4 mRNA expression in preadipocytes, we transiently transfected with specific siRNAs (as indicated) and allowed the cells to incubate for 72 h. These cells were then transfected again with empty vector or the combination of transactivating factors described in Fig. 5B. After 18 h, total RNA was harvested, and GLUT4 mRNA was measured using RT-PCR. GLUT4 mRNA expression was increased only under conditions where all three class II HDACs were knocked down, confirming that these proteins have redundant function (Fig. 6C). As with PE treatment, expression of GLUT4 mRNA was further enhanced by overexpression of the GLUT4 transactivators, MEF2A, GEF, and LXR-α. To confirm that the expression of GLUT4 mRNA was not due to off target effects stemming from siRNA knockdown, we rescued the effect by overexpression of siRNA-resistant, human HDAC5 (Fig. 6C), which returned GLUT4 mRNA expression to control levels. To confirm that the HDAC siRNA knockdown was specific for GLUT4 expression and not accelerated adipocyte differentiation, we measured PPARγ mRNA levels as a marker of differentiation. PPARγ mRNA levels were not induced with HDAC knockdown, confirming that the effects were specific for GLUT4 transcription (Fig. 6D).
FIGURE 6.
siRNA-mediated knockdown of class II HDACs increase endogenous GLUT4 mRNA in preadipocytes with and without transactivating factors. A, nuclear extracts of 3T3-L1 preadipocytes transiently transfected with scrambled siRNA or HDAC5 siRNA and analyzed by SDS-PAGE and immunoblotting for endogenous HDAC5. The arrow indicates the HDAC5 band. IB, immunoblot. B, preadipocytes were transiently transfected with either scramble siRNA or the indicated specific HDAC siRNA in combination. RNA was isolated and subjected to quantitative PCR to determine the relative effectiveness of knocking down HDAC4, HDAC5, and HDAC9. C, preadipocytes were transiently transfected with either scrambled siRNA or HDAC-specific siRNA as indicated by +. The cells were then incubated for 3 days and then transiently transfected again with either pcDNA3 (empty vector) or transactivator plasmids (MEF2A, GEF, and LXRα). RNA was isolated and analyzed by RT-PCR for endogenous GLUT4 mRNA. The data are from at least three independent experiments. The data are expressed as the means ± S.E. and analyzed by one-way analysis of co-variance. The asterisk indicates a statistically significant difference (p < 0.05) without transactivating factors. The number sign indicates a statistically significant difference from knocking down all three class II HDACs and the addition of transactivating factors (p < 0.05) over all other conditions. D, preadipocytes or adipocytes 6 days after differentiation were transiently transfected with either scrambled siRNA or HDAC-specific siRNA as indicated by +. The cells were then incubated for 3 days and then transiently transfected again with either pcDNA3 (empty vector) or transactivator plasmids (MEF2A, GEF, and LXRα). RNA was isolated and analyzed by RT-PCR for endogenous PPARγ mRNA. The data are from at least three independent experiments. The data are expressed as the means ± S.E. and analyzed by one-way analysis of co-variance. The asterisk indicates a statistically significant increase in differentiated adipocyte PPARγ compared with all other conditions (p < 0.001).
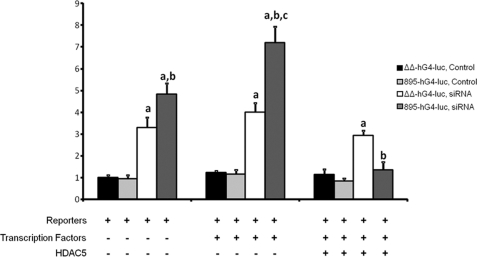
To determine whether the HDAC effects were specific to the transcription factors MEF2A and GEF, we tested activity of the GLUT4-luciferase construct carrying mutations in the MEF2A- and GEF-binding domains (ΔΔ-hG4-luc) against the fully active promoter (895-hG4-luc). MEF2A, GEF without or with HDAC5 had no effect on the mutated construct in the absence or presence of siRNA (Fig. 7). This confirmed that HDAC5 specifically inhibited the GLUT4 promoter through interaction with both MEF2A and GEF.
FIGURE 7.
The inhibition of HDAC5 is dependent on GEF and MEF2A binding to the GLUT4 promoter. 3T3-L1 preadipocytes were transiently transfected with either scrambled siRNA (control) or HDAC4/5/9-specific siRNA and incubated for 3 days and then transiently transfected again with luciferase reporter plasmids −895-hG4-luc or ΔΔ-hG4-luc (a loss of function mutation of Domain I (GEF binding site) and the MEF2-binding domain) (firefly luciferase, for promoter activity) and pRLTK (Renilla luciferase, for transfection efficiency), with and without plasmids encoding GEF, MEF2A, and wild type HDAC5 (+ indicates inclusion in transfection). The means ± S.E. from three independent experiments are shown; the data were analyzed by a one-way analysis of co-variance. Statistically significant changes are indicated as follows: a, a statistical difference between scrambled and siRNA treated similar conditions; b, a statistical difference between −895-hG4-luc and the ΔΔ-hG4-luc cells treated with siRNA; and c, a statistical difference over all other conditions (p < 0.005).
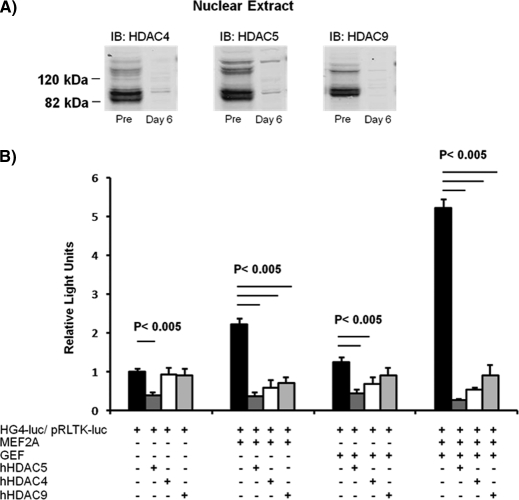
Expression GLUT4 mRNA in preadipocytes required knockdown of three class II HDACs: HDAC4, HDAC5, and HDAC9. Because only HDAC5 has been demonstrated previously to be expressed in adipose tissue (17), we determined whether HDAC4 and HDAC9 play a role in regulation of GLUT4 transcription. Abundance of HDAC4, HDAC5, and HDAC9 proteins were similar in nuclear extracts prepared from preadipocytes (Fig. 8A). The HDAC proteins were significantly diminished by 6 days after differentiation, consistent with a role in derepression of GLUT4 mRNA (Fig. 8A). Western blot analysis revealed that each of the three HDACs displayed the same splice variants, resulting in two distinct HDAC bands and sizes: one at ∼130 kDa and the other at 84 kDa. The splice variants for HDAC5 and HDAC9 have been reported previously by other labs (22, 32), consistent with our data (Fig. 1B and 8A). Although HDAC4 splicing has not been well studied, we believe that the 84-kDa species is an HDAC4 splice variant because it can be immunoprecipitated with an HDAC4 antibody (data not shown).
FIGURE 8.
Three class II HDACs are expressed in preadipocytes and can specifically regulate GLUT4 promoter activity in vitro. A, 120 μg of nuclear extracts of 3T3-L1 preadipocytes or adipocytes 6 days after differentiation were analyzed by SDS-PAGE and immunoblotted (IB) for endogenous HDAC4, HDAC5, or HDAC9 as indicated. Both the full-length protein and the lower splice variant were detected for all HDACs, but as the cells differentiated, the nuclear HDAC levels severely diminished. B, 3T3-L1 adipocytes 6 days after differentiation were transiently transfected with luciferase reporter plasmids −895-hG4-luc (firefly luciferase, for promoter activity) and pRLTK (Renilla luciferase, for transfection efficiency), with and without plasmids encoding GEF, MEF2A, wild type human HDAC4 (hHDAC4), wild type human HDAC5 (hHDAC5), or wild type human HDAC9 (hHDAC9). +, inclusion in transfection. The means ± S.E. from three independent experiments are shown; the data were analyzed by a one-way analysis of co-variance, and statistically significant (p < 0.005) changes are indicated.
Similar to HDAC5, HDAC4 and HDAC9 both can specifically inhibit the GLUT4 promoter activity (Fig. 8B). Like HDAC5, HDAC4 and HDAC9 inhibited both MEF2A- and GEF-dependent GLUT4 transcription in the in vitro transcription assay.
DISCUSSION
Expression of GLUT4 is developmentally regulated in fetal development, with full expression observed after birth (33). Similarly, GLUT4 is developmentally regulated in mouse cell lines that give rise to either adipocytes or myotubes. Full expression of GLUT4 in 3T3-L1 adipocytes occurs late in the differentiation process relative to other adipocyte-specific genes (17, 34). The molecular mechanisms that underlie differentiation-dependent expression are not well established because of the paucity of cell lines expressing GLUT4 as well as numerous technical challenges encountered with the use of terminally differentiated cultured cell lines. To circumvent these problems, we have used transgenic mice mouse models to understand the mechanisms of GLUT4 gene regulation in tissues. Using transgenic mice, we have shown that regulated expression of the GLUT4 promoter in adipocytes, skeletal muscle, and cardiac muscle is largely dependent on two binding domains found within 895 bp of the major transcription initiation site (10). In adipose tissue, skeletal muscle, and cardiac muscle, these binding domains activate transcription when occupied by MEF2 proteins and GEF, respectively (12). It is possible that these binding domains are also required for differentiation-dependent expression in adipocytes and myotubes, but this mechanism of action has not been demonstrated. What confused this issue was the observation that MEF2A and GEF were expressed in preadipocytes at a time when GLUT4 expression was almost undetectable (17). Therefore, regulation of GLUT4 transcription during differentiation involves a more complex molecular mechanism. Several other transcription factors have been implicated in GLUT4 transcription including PGC1, CCAAT enhancer-binding protein, PPARγ, and FOXO (35). For example, others have reported that the expression of PPARγ is a limiting step in GLUT4 expression (36), but this may be an indirect result of PPAR-dependent changes in the rate of adipocyte differentiation. Although we cannot exclude the role of other transcription factors in GLUT4 expression during differentiation, this report provides strong evidence that the presence of transcriptional activators alone is not sufficient to activate GLUT4 gene expression. Furthermore, we tested the possibility that silencing HDACs results in an acceleration of differentiation. We found that PPARγ levels do not increase following knockdown of the three HDACs, indicating that they play no direct role in adipocyte differentiation. Here, we establish support that class II HDACs, in particular HDAC5, must be redistributed from the nucleus in addition to expression of MEF2A and GEF to support transcriptional activation of the GLUT4 gene. This work establishes HDAC5 as a key player in the differentiation-dependent regulation of GLUT4. In previous work, we showed that GEF and MEF2 proteins act as a docking site for the transcriptional co-repressor, HDAC5. In vitro, HDAC5 specifically repressed GLUT4 promoter activity. In chromatin immunoprecipitation assays, we found that HDAC5 associated with the GLUT4 promoter in preadipocytes but not fully differentiated adipocytes (17). Furthermore, we demonstrated previously that overexpression of MEF2 and GEF alone in preadipocytes was not sufficient to activate GLUT4 transcription in the in vitro GLUT4 transcription assays (17). Thus, the levels of these transcription factors are not limiting for GLUT4 gene expression. Therefore, the rate-determining factor for differentiation-dependent GLUT4 transcription resides elsewhere.
In this report, we demonstrate that class II HDACs, specifically HDAC4, HDAC5, and HDAC9, may be responsible, at least in part, for repressing GLUT4 gene expression in preadipocytes, therefore playing a significant role in the differentiation-dependent expression of GLUT4 in 3T3-L1 adipocytes. We have demonstrated that the deacetylase domain of HDAC5 does not play a role in the transcriptional repression of the GLUT4 promoter, at least from plasmid DNA (Fig. 2). Because the GLUT4 promoter/reporter plasmid DNA remains extrachromosomal, it is possible that gene expression is not repressed through chromatin remodeling but rather through formation of an inhibitory protein complex on the GLUT4 promoter. HDAC5 binding may block binding of transcriptional co-activators, as has been seen in other systems (22). HDAC5 can bind the GLUT4 promoter through associations with both GEF and MEF2 (17).
Compensatory activity between HDAC4, HDAC5, and HDAC9 was revealed in siRNA knockdown experiments (Fig. 6). Knockdown of all three isoforms was required for up-regulation of GLUT4 gene transcription. Overexpression of human HDAC5 was sufficient to reconstitute HDAC-dependent repression of the GLUT4 promoter following knockdown of HDAC4, HDAC5, and HDAC9, consistent with the notion that the class II HDACs can compensate for a single function. To date, only HDAC5 has been shown to associate with the GLUT4 promoter in vivo (17). To this end, we wanted to determine whether HDAC4 and HDAC9 proteins were present in adipocytes, and differentially regulated Western blot analysis of nuclear extracts comparing preadipocytes to day 6 after differentiation revealed high levels of all three class II HDACs (Fig. 8A). We also demonstrated that both HDAC4 and HDAC9 can specifically inhibit the GLUT4 promoter in vitro (Fig. 8B), thus further supporting a compensatory model of HDAC repression. Further work must be done to determine whether HDAC4 or HDAC9 are physiologically relevant regulators of the GLUT4 gene expression.
In conclusion, our results have provided the first evidence supporting the hypothesis that the minimal machinery needed to express GLUT4 in preadipocytes is the removal of inhibitory HDACs from the nucleus. This finding clarifies our understanding of the molecular mechanisms that underlie developmental expression of GLUT4 gene expression. We have identified a role for class II HDACs in differentiation-dependent expression of GLUT4 in 3T3-L1 adipocytes and revealed a potential mechanism for regulated expression of the GLUT4 gene by HDACs in differentiated cells. Immunofluorescence imaging of differentiated adipocytes reveals that HDAC5 is expressed but largely excluded from the nuclear compartment. Therefore, it is possible that physiologic conditions that lead to nuclear redistribution of HDAC5 may play a role in regulated expression of GLUT4 gene expression in differentiation adipocytes. For example, elevation of cAMP in neuronal cells increases HDAC5 protein levels in the nucleus (37). It is possible that increased intracellular cAMP levels may also lead to the nuclear redistribution of HDAC5 in adipocytes. Furthermore, it is well established that increased intracellular cAMP leads to down-regulation of GLUT4 transcription in adipocytes in vivo and in vitro (38, 39). Studies are currently underway to test the hypothesis that increased cAMP levels in adipocytes change nuclear distribution of HDACs in adipocytes and in turn regulate GLUT4 expression in various physiological states. Taken together, HDACs may provide a new pharmacological target in states where GLUT4 transcription is severely diminished to up-regulate GLUT4 and restore glucose homeostasis.
This work was supported, in whole or in part, by National Institutes of Health Grant DK081545.
- GLUT
- insulin-responsive glucose transporter
- MEF2
- myocyte enhancer factor 2
- GEF
- GLUT4 enhancer factor
- HDAC
- histone deacetylase
- PE
- phenylephrine
- PPAR
- peroxisome proliferator-activated receptor
- RT
- real time.
REFERENCES
- 1. Charron M. J., Katz E. B., Olson A. L. (1999) J. Biol. Chem. 274, 3253–3256 [DOI] [PubMed] [Google Scholar]
- 2. Brozinick J. T., Jr., McCoid S. C., Reynolds T. H., Nardone N. A., Hargrove D. M., Stevenson R. W., Cushman S. W., Gibbs E. M. (2001) Diabetes 50, 593–600 [DOI] [PubMed] [Google Scholar]
- 3. Christ-Roberts C. Y., Pratipanawatr T., Pratipanawatr W., Berria R., Belfort R., Kashyap S., Mandarino L. J. (2004) Metabolism 53, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 4. Gibbs E. M., Stock J. L., McCoid S. C., Stukenbrok H. A., Pessin J. E., Stevenson R. W., Milici A. J., McNeish J. D. (1995) J. Clin. Invest. 95, 1512–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berger J., Biswas C., Vicario P. P., Strout H. V., Saperstein R., Pilch P. F. (1989) Nature 340, 70–72 [DOI] [PubMed] [Google Scholar]
- 6. Garvey W. T., Huecksteadt T. P., Birnbaum M. J. (1989) Science 245, 60–63 [DOI] [PubMed] [Google Scholar]
- 7. Kahn B. B., Simpson I. A., Cushman S. W. (1988) J. Clin. Invest. 82, 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sinha M. K., Raineri-Maldonado C., Buchanan C., Pories W. J., Carter-Su C., Pilch P. F., Caro J. F. (1991) Diabetes 40, 472–477 [DOI] [PubMed] [Google Scholar]
- 9. Sivitz W. I., DeSautel S. L., Kayano T., Bell G. I., Pessin J. E. (1989) Nature 340, 72–74 [DOI] [PubMed] [Google Scholar]
- 10. Thai M. V., Guruswamy S., Cao K. T., Pessin J. E., Olson A. L. (1998) J. Biol. Chem. 273, 14285–14292 [DOI] [PubMed] [Google Scholar]
- 11. Knight J. B., Eyster C. A., Griesel B. A., Olson A. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oshel K. M., Knight J. B., Cao K. T., Thai M. V., Olson A. L. (2000) J. Biol. Chem. 275, 23666–23673 [DOI] [PubMed] [Google Scholar]
- 13. Lu J., McKinsey T. A., Nicol R. L., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4070–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miska E. A., Karlsson C., Langley E., Nielsen S. J., Pines J., Kouzarides T. (1999) EMBO J. 18, 5099–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sparrow D. B., Miska E. A., Langley E., Reynaud-Deonauth S., Kotecha S., Towers N., Spohr G., Kouzarides T., Mohun T. J. (1999) EMBO J. 18, 5085–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Youn H. D., Grozinger C. M., Liu J. O. (2000) J. Biol. Chem. 275, 22563–22567 [DOI] [PubMed] [Google Scholar]
- 17. Sparling D. P., Griesel B. A., Weems J., Olson A. L. (2008) J. Biol. Chem. 283, 7429–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grozinger C. M., Hassig C. A., Schreiber S. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czubryt M. P., McAnally J., Fishman G. I., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGee S. L., Hargreaves M. (2004) Diabetes 53, 1208–1214 [DOI] [PubMed] [Google Scholar]
- 21. Eyster C. A., Duggins Q. S., Olson A. L. (2005) J. Biol. Chem. 280, 17978–17985 [DOI] [PubMed] [Google Scholar]
- 22. Lemercier C., Verdel A., Galloo B., Curtet S., Brocard M. P., Khochbin S. (2000) J. Biol. Chem. 275, 15594–15599 [DOI] [PubMed] [Google Scholar]
- 23. Chawla S., Vanhoutte P., Arnold F. J., Huang C. L., Bading H. (2003) J. Neurochem. 85, 151–159 [DOI] [PubMed] [Google Scholar]
- 24. Davis F. J., Gupta M., Camoretti-Mercado B., Schwartz R. J., Gupta M. P. (2003) J. Biol. Chem. 278, 20047–20058 [DOI] [PubMed] [Google Scholar]
- 25. Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. (2001) J. Biol. Chem. 276, 47496–47507 [DOI] [PubMed] [Google Scholar]
- 26. McKinsey T. A., Zhang C. L., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verdin E., Dequiedt F., Kasler H. G. (2003) Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 28. Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 29. Griesel B. A., Weems J., Russell R. A., Abel E. D., Humphries K., Olson A. L. (2010) Diabetes 50, 800–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang S., McKinsey T. A., Zhang C. L., Richardson J. A., Hill J. A., Olson E. N. (2004) Mol. Cell Biol. 24, 8467–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) J. Clin. Invest. 117, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou X., Marks P. A., Rifkind R. A., Richon V. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10572–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santalucía T., Camps M., Castelló A., Muñoz P., Nuel A., Testar X., Palacin M., Zorzano A. (1992) Endocrinology 130, 837–846 [DOI] [PubMed] [Google Scholar]
- 34. Wu Z., Xie Y., Morrison R. F., Bucher N. L., Farmer S. R. (1998) J. Clin. Invest. 101, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Armoni M., Kritz N., Harel C., Bar-Yoseph F., Chen H., Quon M. J., Karnieli E. (2003) J. Biol. Chem. 278, 30614–30623 [DOI] [PubMed] [Google Scholar]
- 36. Karnieli E., Armoni M. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E38–E45 [DOI] [PubMed] [Google Scholar]
- 37. Belfield J. L., Whittaker C., Cader M. Z., Chawla S. (2006) J. Biol. Chem. 281, 27724–27732 [DOI] [PubMed] [Google Scholar]
- 38. Flores-Riveros J. R., Kaestner K. H., Thompson K. S., Lane M. D. (1993) Biochem. Biophys. Res. Commun. 194, 1148–1154 [DOI] [PubMed] [Google Scholar]
- 39. Gerrits P. M., Olson A. L., Pessin J. E. (1993) J. Biol. Chem. 268, 640–644 [PubMed] [Google Scholar]