Abstract
UbcH5c, a member of the UbcH5 family of protein ubiquitin conjugase E2 enzymes, is a critical component of biological processes in human cells, being the initial ubiquitinating enzyme of substrates like IκB, TP53, and cyclin D1. We report here that the metastasis regulator protein SLUG inhibits the expression of UbcH5c directly through chromatin remodeling and thus, among other downstream effects, elevates the level of cyclin D1, thus enhancing the growth rates of breast cancer cells. Overexpression of SLUG in the SLUG-deficient breast cancer cells significantly decreased the levels of mRNA and protein of UbcH5c but only elevated the protein levels of cyclin D1. On the contrary, knockdown of SLUG in SLUG-high breast cancer cells elevated the levels of UbcH5c while decreasing the level of cyclin D1 protein. SLUG is recruited at the E2-box sequence at the UbcH5c gene promoter along with the corepressor CtBP1 and the effector HDAC1 to silence the expression of this gene. Knockdown of UbcH5c in the SLUG-deficient human breast cells elevated the level of cyclin D1 as well as the rates of proliferation and invasiveness of these cells. Whereas the growth rates of the cells are enhanced due to overexpression of SLUG or knockdown of UbcH5c in the breast cancer cells tested, ER+ cells also acquire resistance to the anti-estrogen 4-hydroxytamoxifen due to the rise of cyclin D1 levels in these cells. This study thus implicates high levels of SLUG and low levels of UbcH5c as a determinant in the progression of metastatic breast cancer.
Keywords: Breast Cancer, Chromatin Remodeling, Cyclins, Transcription Repressor, Tumor Metastases, Ubiquitin Ligase, Ubiquitination, Anti-estrogen, SLUG, UbcH5c
Introduction
The ubiquitin-proteasome system regulates protein degradation in mammalian and other eukaryotic cells to affect a wide range of biological processes, including cellular proliferation and differentiation (1–3). The covalent modification of protein substrates with ubiquitin in the form of polymeric chains, with ubiquitin moieties connected through isopeptide linkage between the ϵ-amide group of lysine and the carboxyl end of glycine, is the critical prerequisite of proteasomal degradation of proteins (4).
Proteasomal degradation of proteins is initiated by the concerted actions of three enzymes, E1, E2, and E3, which ubiquitinate the target protein (4–9). The specificity of ubiquitination is largely determined by E3 ligases, which recognize the protein substrate and facilitate ubiquitin transfer from the E2 ubiquitin-conjugating enzyme onto substrate (4–9). A single E2 enzyme can usually interact with several E3 ubiquitin ligases and thereby affect multiple targets (10). Recently, the E2 enzymes of the UbcH5 family is drawing the attention of biologists due to their perceived roles in the degradation of key regulatory molecules like IkB (11), cyclin D1 (12, 13), TP53, and MDM2 (14).
The human UbcH5 family consists of three homologs: UbcH5a, UbcH5b (also known as Ubc4), and UbcH5c (also known as UbE2D3) (15–18). The detailed mechanism of the polyubiquitination of IkBα is shown to begin with the action of the UbcH5c E2 ubiquitin-conjugating enzyme that transfers a single ubiquitin to IkBα (11). Subsequently, the Cdc34 E2 functions in the formation of polyubiquitin chains (11). Whether similar modalities involving Cdc34 are operative in the degradation of cyclin D1 or TP53 is not known. Little is also known about the potential mechanisms that may regulate the levels of UbcH5c in the breast cancer cells.
Cyclins modulate the cyclin-dependent kinases, which are the key cell cycle regulators (19–24). These molecules are often regulated by ubiquitination through different mechanisms (20–24). Binding of cyclin D1 to Cdk4 and Cdk6 leads to the phosphorylation of the retinoblastoma protein (25). Phosphorylation of retinoblastoma protein prevents it from repressing the E2F family of transcription factors and leads to the transcription of several genes required for the G1 to S phase transition, thereby promoting cellular proliferation (25). Cyclin D1 is overexpressed in ∼50% of breast cancers and is implicated as the cause of their increased rate of proliferation (26).
One of the causes for breast cancer cell proliferation and metastasis is the overexpression of the metastasis modulator transcriptional repressor protein SLUG (27–31). This protein binds to the E2-box sequence of its target gene promoters and down-regulates their expression by chromatin remodeling (28, 32). SLUG induces metastasis through the repression of several genes in breast and other cancer cells (28, 29). We report here that SLUG indirectly elevates the levels of cyclin D1 in breast cancer cells by repressing the ubiquitin conjugase enzyme UbcH5c. Cyclin D1 was shown to be a target for UbcH5c in the promyelocytic NB4 cells, where it helps proteasomal degradation of cyclin D1 (13).
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human breast cancer cells MCF7, MDA-MB-468, MDA-MB-231, and BT549 were obtained from American Type Culture Collection (Manassas, VA) and were cultured in ATCC-recommended media (32–34). Authentications of the cell lines are routinely performed in our laboratory following the instructions provided in ATCC Bulletin 8. Cell cycle distribution of SLUG-manipulated breast cancer cells were determined by FACS analysis as described (34). Mouse anti-FLAG M2 antibody was purchased from Sigma. Rabbit anti-CtBP1 (C-terminal binding protein-1) and anti-HDAC1 (histone deacetylase-1) antibodies were purchased from Upstate Millipore (Burlington, MA). Rabbit anti-cyclin D1 (H-295), goat anti-SLUG (G18), and rabbit anti-SLUG (H-140) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-cyclin D1 antibody used in immunofluorescence studies was procured from BIOSOURCE (Invitrogen). Mouse anti-UbcH5c antibody was procured from Novus Biologicals, Inc (Littleton, CO). Antibodies against AKT, phospho-AKT, GSK3β and phospho-GSK3β, cyclin D1, and phosphocyclin D1 (Thr286) were procured from Cell Signaling (Danvers, MA). Trichostatin A, 4-hydroxytamoxifen, and the proteasomal inhibitor benzyloxycarbonyl-leucyl-leucyl-leucinal (MG132) were purchased from Sigma. Wild-type human cyclin D1 (in pCMV; Addgene plasmid 19927) (35) and the T286A mutant of human cyclin D1 (C-terminally HA-tagged in pCDNA3.0; Addgene plasmid 11182) (36) as well as the N-terminally HA-tagged ubiquitin expression plasmid (in pcDNA3.1; Addgene plasmid 18712) (37) were procured from Addgene Inc. (Cambridge, MA). Human UbcH5c (C-terminally tagged with c-Myc and DDK epitopes) containing a plasmid construct in pCMV6 was procured from Origene (Rockville, MD).
Expression of Recombinant Proteins in Breast Cancer Cells
Human SLUG coding sequence was amplified (33) from RNA isolated from BT549 cells using N-terminal and C-terminal primers. The amplified cDNA (831 bp) was sequence-verified, digested with ClaI/BamHI, and cloned in ClaI/BamHI sites at the multiple cloning site of p3XFLAG-CMV-14 plasmid (Sigma). The cells were transfected with the SLUG construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 48 h, cells were plated in G418-containing medium to select a stable cell population expressing SLUG. The p3XFLAG vector- and p3XFLAG-SLUG construct-transfected cells were grown in their respective media with G418 (500 μg/ml). We generated multiple (>10) independent SLUG-transfected populations of these cells as well as the vector-transfected controls for our studies. To overexpress UbcH5c in MDA-MB-231 cells, we transfected these cells with a c-Myc-DDK-tagged ORF clone of human UbcH5c; cells were trypsinized and plated for a proliferation assay or were lysed in radioimmune precipitation assay buffer to perform the immunoblot analysis. Wild-type and T286A mutant of human cyclin D1 were overexpressed in the MDA-MB-231 cells similarly. The overexpressions of the recombinant mRNAs and proteins were evaluated by real-time RT-PCR and Western blot analysis.
Chromatin Immunoprecipitation (ChIP)-DNA Selection and Ligation (DSL) Analysis
Identification and evaluation of the gene promoters that bind to SLUG in C-terminal FLAG-tagged SLUG-overexpressing MCF7 and MDA-MB-468 cells (33) were done following the standard protocols, as described (38). We used the promoter chip and reagents from Aviva Systems Biology (San Diego, CA). The gene promoters showing significant binding were further analyzed by independent ChIP experiments.
siRNA Treatment
SLUG, cyclin D1, and UbcH5c siRNAs and corresponding control siRNAs were designed using the Block-IT RNAi designer software (Invitrogen) and purchased from Invitrogen. The nucleotide sequences of these siRNAs and respective control RNAs used in this study are given in supplemental Table 2S. We also used other commercially available (Santa Cruz Biotechnology, Inc.) validated pools of siRNAs against these targets for further validation of our knockdown data. Transfection of these siRNAs into the breast cells was done by lipofection using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions. Briefly, cells were transfected at ∼50% confluence using 100 pmol of siRNA in 6-well plates, and whole-cell lysates were prepared 48 h after transfection. We isolated RNA from these cells using TRIzol reagents (Invitrogen). Knockdown of the expressions of the target mRNAs by the experimental siRNA and the corresponding protein were verified by real-time RT-PCR and immunoblot analysis, respectively (32–34). To evaluate whether the effect of SLUG knockdown on the level of cyclin D1 is indeed mediated through proteasomal degradation, we treated control and SLUG siRNA-treated cells with a 5 μm concentration of the proteasomal inhibitor MG132 for 45 min in the culture medium at 37 °C. Cells were lysed, and immunoblot analysis was performed to evaluate the levels of cyclin D1, SLUG, and β-actin in these cells.
Real-time RT-PCR Analysis
Total RNA was isolated from the cultured cells using TRIzol reagent (Invitrogen). The cDNA was synthesized from 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR quantification was performed following standard protocols using SYBR Green dye (Bio-Rad). The sequences of the primers used for quantitative PCR are shown in supplemental Table 1S. RT-PCR was performed in the iCycler (Bio-Rad), as described (34). The -fold change over control samples was calculated using Ct, ΔCt, and ΔΔCt values (32, 34). β-Actin RNA was used as an endogenous control.
Immunoblot Analysis
Cells transfected with control or SLUG construct plasmids were grown in complete medium. Protein extracts were made, and Western blotting was performed as described (32–34). Cell lysates containing equal amounts of protein were resolved by 4–12% SDS-PAGE, transferred to nitrocellulose membranes, probed with the appropriate antibodies, and detected by means of enhanced chemiluminescence (32–34).
Luciferase Reporter Assay
We PCR-amplified human UbcH5c promoter (−850 to +200, NM_003340; see supplemental material for the nucleotide sequences) from total DNA isolated from MDA-MB-231 cells with specific primers (supplemental Table 1S). This promoter sequence has one E2-box at the upstream (−776 to −781) of the transcription start site (see the supplemental material for nucleotide sequence). The amplified DNA was cloned into the pCR4.0/TOPO vector (Invitrogen) and subsequently subcloned into the HindIII/PstI sites of pRL-Null vector (Promega). Cells were seeded on 24-well tissue culture plates in triplicate and allowed to grow overnight to reach 90–95% confluence. The following day, cells were transfected with pGL3-Control and pRL-UbcH5c promoter construct using Lipofectamine 2000 transfection reagent (Invitrogen). Forty-eight hours later, luciferase activities were measured using the Dual Luciferase reporter assay system (Promega) (32–34). Renilla luciferase activity was normalized to firefly luciferase activity (32–34). Overexpression of non-functional SNAG domain-deleted SLUG protein (33) in MCF7 cells failed to repress the function of the UbcH5c promoter (data not shown), suggesting that the intact repressor domain of SLUG is essential for this inhibition.
Site-directed Mutagenesis
A PCR-based site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Stratagene, La Jolla, CA) technique was used for the generation of reporter gene construct with E2-box mutation following the manufacturer's instructions. The E2-box element was mutated from 5′-CACCTG-3′ to 5′-GTTACT-3′ (sense strand). Oligonucleotides used for site-directed mutagenesis are given in the supplemental Table 1S.
ChIP Assay
ChIP assays were performed as described previously (32–34). Immunoprecipitations were performed using FLAG (for recombinant SLUG), SLUG antibody H140 (for endogenous SLUG), CtBP1, or HDAC1 antibodies. For the evaluation of acetylated histones H3 and H4, we used ChIP grade antibodies against acetylated histone H3 (Lys9 and Lys14) and acetylated histone H4 (Lys5, Lys8, Lys12, and Lys16) from Millipore. Quantitative ChIP analysis was done as described (34). UbcH5c promoter DNA was amplified from the ChIP DNA using the primers described (supplemental Table 1S).
Cell Proliferation Assay
Cells were seeded at 5 × 103 cells/well in 96-well plates and cultured in 100 μl of culture medium. The CellTiter 96 assay reagents from Promega were used for the evaluation of cell proliferation. Briefly, after 48 h, 20 μl of MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) dye solution was added to each well and mixed, and samples were then incubated at 37 °C for 4 h. The absorbance was then read at 490 nm using a 96-well plate reader. The quantity of formazan product as measured by the absorbance was directly proportional to the number of living cells in the culture (39).
Invasion Assay
Invasiveness of the breast cancer cells was evaluated following a modified Boyden chamber method using Matrigel matrix (39). Transwells (8-μm pore size, 6.5 mm in diameter) from Costar (Cambridge, MA) were coated with Matrigel and then left in an incubator for 2 h. Cells were trypsinized, washed with PBS, resuspended in serum-free medium, and then seeded in transwells (100,000 cells/transwell). Cells were allowed to grow in transwells in the presence of 10% fetal bovine serum-containing medium in the lower chamber of the transwells for 72 h. Cells remaining inside the insert were removed with cotton swabs, and the cells that had traversed to the reverse side of the insert were rinsed with PBS, fixed in 3.7% formaldehyde for 30 min at room temperature, and stained with 1% crystal violet for 1 h at room temperature. Cells were counted under a light microscope (at ×20 power), and invasive cell number was the average of those counted from five areas on each insert.
Immunohistochemistry
Immunohistochemistry was performed following the manufacturer's protocol (RayBiotech, Norcross, GA) on tissue microarray. Identifications of the spots are shown in the supplemental material (supplemental Table 3S, as provided by RayBiotech). In brief, deparaffinization was done by heating the slide in an oven (60 °C) for 20 min and then three washes in xylene and dehydration by two ethanol (100% and 95%) washes for 5 min each. The slide was washed with distilled water and then with PBS for 5 min each. Antigen unmasking was done by heating a slide in 10 mm sodium citrate buffer, pH 6.0 for 1 min at 100 °C, followed by 9 min at 95 °C. After cooling, the slide was washed with PBS three times for 5 min each, and the slide was further blocked by 10% goat serum for 1 h. The slide was incubated in primary antibodies (rabbit SLUG and mouse cyclin D1; 1:200) overnight at 4 °C, and thereafter, the slide was washed three times with PBS. It was further incubated with corresponding Alexafluor-conjugated secondary antibodies (donkey anti-rabbit R488 and donkey anti-mouse R555 from Invitrogen) for 1 h at room temperature. The slide was again washed five times with PBS, embedded in glycerol/PBS-based mounting medium, and examined using a fluorescent microscope (Nikon TE2000-E).
Immunofluorescence Microscopy
Immunofluorescence staining and confocal analyses were performed as per the standard protocol (33, 34). In brief, cells were cultured in 8-well chamber slides for 24 h, rinsed with ice-cold 1× PBS, fixed with 3.7% formaldehyde for 30 min, washed three times with PBS, and fixed in ice-cold methanol for 10 min. Cells were further permeabilized with 0.2% Triton X-100 in PBS for 10 min. Thereafter, cells were washed three times with ice-cold PBS. The slides were blocked in PBS containing 5% normal goat serum and then were incubated with primary antibodies overnight at 4 °C in the blocking buffer. Slides were then washed three times with PBS, followed by incubation for 45 min with the respective Alexafluor-conjugated secondary antibody (see above). The slides were again washed five times with PBS, embedded in glycerol/PBS-based mounting medium, and examined using a fluorescent microscope (Nikon TE2000-E). Confocal images were obtained with a Nikon TE2000-UC1 laser-scanning microscope (33, 34).
Statistical Analysis
Each experiment was repeated at least three times. Results were expressed as means ± S.E. Statistical analyses were performed using GraphPad Prism software. p values were calculated using the two-sided Student's t test (paired or unpaired, as appropriate) and analysis of variance test for significance. p values of <0.05 and <0.01 were considered as significant.
RESULTS
SLUG and Cyclin D1 Levels in Human Breast Tumor Tissues and Cells Are Correlated
SLUG-overexpressing breast cancer cells are often associated with higher growth rates and aggressiveness (27–29). Because cyclin D1 levels in the cells may determine these properties, we initially evaluated whether there is a correlation between the levels of SLUG and cyclin D1 in different human breast cancer tissues and cells. SLUG and cyclin D1 levels were examined by immunohistochemistry in a human breast cancer tissue microarray (TMAH-BRC-03, RayBioTech) representing 39 metastatic breast cancer and 38 benign breast tumor/disease tissues, which are biopsies from 65 patients, as well as in different established breast cancer cell lines. Tissue spots from breast cancer patients showed strong correlation in staining of SLUG and the cyclin D1 in the tissue microarray (Fig. 1A and supplemental Fig. 1S). Tissues represented in the dots A1, A4, A5, B2, C5, D5, E2, E3, E4, I2, and J6 showed such direct correlations between SLUG and cyclin D1 expressions (see supplemental Fig. 1S and Table 3S for details). Moreover, staining intensities of SLUG and cyclin D1 in invasive cancer cell lines (MDA-MB-231 and BT549) were significantly higher compared with non-invasive breast cancer cell lines (MCF7 and MDA-MB-468) (Fig. 1B).
FIGURE 1.
Correlation between SLUG and cyclin D1 levels in breast tissues and cells. A, human breast cancer tissue microarray was examined for cyclin D1 and SLUG expressions by immunohistochemistry using mouse anti-cyclin D1 and rabbit anti-SLUG antibodies. A low magnification (×4) immunofluorescence micrograph of the array is shown in the supplemental material (supplemental Fig. 1S). We show here a higher magnification (×30) immunofluorescence micrograph for selected spots. The coordinates of the spots are marked as detailed in supplemental Table 3S. B, evaluation of the levels of cyclin D1 in normal and SLUG-expressing human breast cancer cells by immunofluorescence microscopy. 468C and MCF7C, MDA-MB-468 and MCF7 cells transfected with empty vector; 468SLUG and MCF7SLUG, cells expressing C-terminal FLAG-tagged SLUG; 231C and BT549C, normal MDA-MB-231 and BT549 cells.
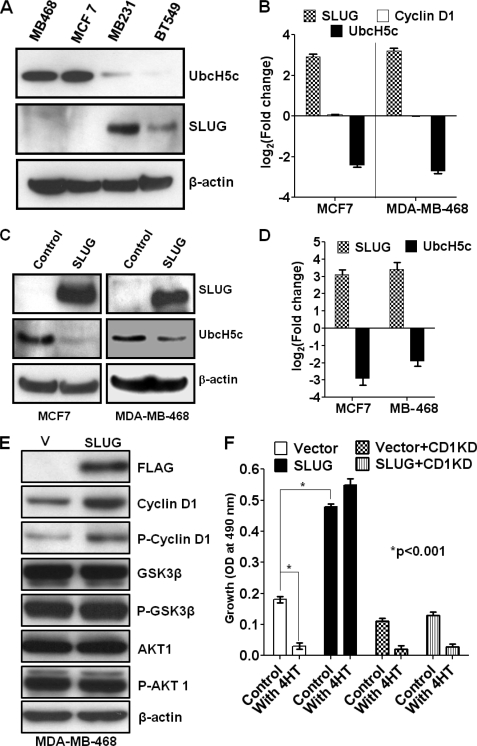
To evaluate whether SLUG expression has direct correlation with increased cyclin D1 levels in the breast cells, we overexpressed SLUG in SLUG-negative MDA-MB-468 and MCF7 cells (33) and generated multiple SLUG-transfected populations and respective vector-transfected controls. We found that stably transfected SLUG-expressing cells also have higher levels of cyclin D1 as compared with the respective control cells (Fig. 1B). There was no change in the levels of cyclin D1 mRNA in the SLUG-expressing cells (see below), suggesting that the transcriptional repressor protein SLUG perhaps stimulates the level of cyclin D1 protein by inhibiting a protein that catalyzes the turnover of cyclin D1. One of the mechanisms by which cyclin D1 is recycled is through proteasomal degradation after ubiquitination (12, 13). With multiple SLUG-transfected breast cancer cell populations, we found that the level of total cyclin D1 is increased (Fig. 2, A and B). Cell cycle distribution of MDA-MB-468 cells overexpressing SLUG and SLUG-knocked down MDA-MB-231 cells was determined by FACS analysis after propidium iodide staining. The cells were predominantly (>60%) at the S phase in the SLUG-overexpressing MDA-MB-468 cells, whereas they were predominantly in the G0/G1 phase (>65%) in the SLUG-knocked down MDA-MB-231 cells. Cyclin D1 ubiquitination is cell cycle-dependent and occurs mainly at the G1/S phase boundary (40). Because UbcH5c controls the ubiquitination of cyclin D1 (12, 13), we evaluated whether there is a correlation among SLUG, cyclin D1, and UbcH5c protein levels in four different breast cancer cell lines. Our data show that indeed there is such correlation (Fig. 2C). These results, thus, demonstrate that in breast cancer cells, SLUG, cyclin D1, and UbcH5c levels are significantly correlated.
FIGURE 2.
Effect of SLUG overexpression on the levels of cyclin D1 in MDA-MB-468 cells. A, increase in cyclin D1 levels in SLUG-expressing MDA-MB-468 cells. A Western blot shows higher levels of cyclin D1 in the recombinant cells. B, densitometric scan for cyclin D1 and SLUG levels in six independent SLUG-transfected populations and vector controls. Results are mean ± S.E. (error bars) (n = 6). The -fold changes were statistically significant (p < 0.001). C, evaluation of SLUG, cyclin D1, and UbcH5c protein levels in four different breast cancer cell lines by Western blotting. Bands were developed using IR dye-conjugated secondary antibody (LI-COR Biosciences) and visualized using the LI-COR Odyssey infrared imaging system. Quantitation and analysis of bands were performed using Odyssey software. β-Actin was used as normalization control. Levels of SLUG and cyclin D1 in the MDA-MB-231 cells and that of UbcH5c in the MCF7 cells were taken as 100 for comparison. Results are mean ± S.E. (n = 4).
Ubiquitin Conjugase Enzyme UbcH5c Expression Diminishes in Metastatic Breast Cancer Cells
Cyclin D1 level in human cells is shown to be regulated by the ubiquitin conjugase enzyme UbcH5c (12, 13). Our ChIP-DNA Selection and Ligation (DSL) analysis of the human gene promoters that bind to SLUG and are thus repressed by this transcriptional regulator revealed that UbcH5c is one such gene (supplemental Table 4S). To understand the mechanism underlying the association between SLUG and cyclin D1 in invasive breast cancer cells, we studied the expression of UbcH5c in these cell lines. Immunoblot analysis for the expression of UbcH5c protein in SLUG-deficient (MCF7 and MDA-MB-468) and SLUG-high (MDA-MB-231 and BT549) human breast cells revealed an inverse relationship between the levels of SLUG and UbcH5c in these cells (Figs. 2C and 3A). To evaluate further whether there is a causal relationship between overexpression of SLUG and a decrease in UbcH5c levels, we overexpressed C-terminally FLAG-tagged SLUG (33) in SLUG-deficient human breast cancer cells. Ectopic expression of SLUG in these SLUG-deficient cells (MCF7 and MDA-MB-468) significantly decreased the levels of UbcH5c mRNA (Fig. 3B) and protein (Fig. 3, C and D) in these cells. Although the levels of cyclin D1 protein were increased in these cells significantly (Fig. 1B), the levels of cyclin D1 mRNA remained unaltered (Fig. 3B), further suggesting potential SLUG-induced post-transcriptional regulation of cyclin D1.
FIGURE 3.
Effect of SLUG expression on UbcH5c and cyclin D1 levels in MDA-MB-468 and MCF7 cells. A, a typical immunoblot showing UbcH5c and SLUG protein levels in different human breast cancer cells. MB468, MDA-MB-468 cells; MB231, MDA-MB-231 cells. B, real-time RT-PCR analysis of the levels of SLUG, UbcH5c, and cyclin D1 mRNAs in SLUG-overexpressing MCF7 and MDA-MB-468 cells. Results are mean ± S.E. (n = 6). The differences between the experimental and control sets were statistically significant (p < 0.001). C, immunoblot analysis for SLUG and UbcH5c proteins in the control and SLUG-overexpressing (SLUG) MCF7 and MDA-MB-468 cells. D, densitometric scan for SLUG and UbcH5c levels in six independent SLUG-transfected populations and corresponding vector-transfected control cells. Results are mean ± S.E. (n = 6). The -fold changes were statistically significant (p < 0.001). E, immunoblot analysis data showing the effects of SLUG overexpression on the levels of cyclin D1, phosphocyclin D1 (at Thr286), GSK3β, phospho-GSK3β, AKT, and phospho-AKT in MDA-MB-468 cells. Control cells (V) were transfected with empty vector DNA instead of SLUG construct plasmid DNA. Recombinant SLUG was FLAG-tagged at the C-terminal end and thus was detected with anti-FLAG antibody. β-Actin was used as a loading control. F, effect of cyclin D1 knockdown (CD1KD) on the SLUG-induced increase in cell proliferation and tamoxifen (4HT; 10 μm) resistance in MCF7 cells. Control cells were transfected with empty vector DNA instead of SLUG construct plasmid DNA. Results are mean ± S.E. (n = 6). Data with cyclin D1 siRNA stealth-311 (supplemental Table 2S) are shown. Other siRNA, stealth-568, also yielded similar results (data not shown). The ability of stealth-311 to knock down cyclin D1 in MDA-MB-231 cells is shown in supplemental Fig. 3S.
Because repression of the UbcH5c gene by SLUG would decrease ubiquitination of cyclin D1 and thus its proteasomal degradation, we postulated that the stability of cyclin D1 protein is increased in SLUG-overexpressing human breast cancer cells. Cyclin D1 turnover through proteasomal degradation is mediated through its phosphorylation at Thr286 by the AKT/GSK3β pathway (40). To understand whether SLUG-induced change in ubiquitin/stability is dependent on the phosphorylation status of Thr286 in cyclin D1, we evaluated the levels of phosphocyclin D1 (at Thr286) in MDA-MB-468 cells with or without SLUG overexpression. We found that the levels of both total cyclin D1 and phosphocyclin D1 are elevated in SLUG-overexpressing cells (Fig. 3E). We then evaluated the levels of kinases in the cyclin D1 phosphorylation pathway in these cells. We did not find any significant change in the levels of GSK3β, phospho-GSK3β, AKT, and phospho-AKT in SLUG-overexpressing cells by immunoblot analysis (Fig. 3E). We postulated that although SLUG overexpression may not directly affect the levels of cyclin D1 phosphorylation pathway, SLUG-mediated knockdown of UbcH5c increased the stability of phosphocyclin D1 and thus of cyclin D1.
Expression of SLUG also increased the rate of proliferation of MCF7 cells in in vitro culture (Fig. 3F). A similar increase in the rate of proliferation was also observed with SLUG-expressing MDA-MB-468 and other SLUG-deficient breast cancer cells (e.g. T47D) (data not shown). To verify that this growth enhancement by SLUG overexpression is due to the increase in the cyclin D1 level in the cells, we knocked down cyclin D1 with two different siRNAs (see supplemental Table 2S; data for one is shown). Cyclin D1 knockdown alleviated growth-enhancing effects of SLUG in MCF7 cells (Fig. 3F).
Other biological consequences of the elevation of the level of cyclin D1 in breast cells include its direct binding to and activation of estrogen receptor (ER)2 when overexpressed (41, 42). It binds to the ER and transcriptional co-activators to stimulate ER-dependent gene expression in the absence of estrogen and even in the presence of anti-estrogen compound like 4-hydroxytamoxifen (4HT) (41, 43) (supplemental Fig. 2S). Formation of the cyclin D1-ER-4HT complex leads to the activation of ER-dependent transcription, providing one explanation for resistance of ER-positive and cyclin D1-overexpressing breast cancer cells to 4HT (44). In accordance with this molecular mechanism, clinical studies have demonstrated a notable association between 4HT resistance and overexpression of cyclin D1 in breast cancers (45–49). To evaluate whether SLUG-induced repression of UbcH5c and thus elevation of cyclin D1 renders the cells 4HT-resistant, we treated the ER-positive SLUG-overexpressing MCF7 cells to 4HT and monitored their growth. Our data show that MCF7 cells, which are otherwise sensitive to 4HT, acquire resistance to this drug when they are transfected with the SLUG-expressing construct (Fig. 3F). ER-negative MDA-MB-468 cells are naturally resistant to tamoxifen and thus did not show any change in the growth rate with or without 4HT treatment. These overexpression studies suggest that SLUG elevates the levels of biochemically functional cyclin D1 molecules through the repression of the UbcH5c gene in breast cancer cells.
Knockdown of SLUG in SLUG-high MDA-MB-231 and BT549 Cells Increased the Level of UbcH5c and Decreased the Level of Cyclin D1 in These Cells
To evaluate further the correlation among SLUG, UbcH5c, and cyclin D1, we knocked down SLUG in the SLUG-positive MDA-MB-231 and BT549 cells and assessed the levels of UbcH5c and cyclin D1 proteins in these cells. SLUG was silenced in these cells by multiple “stealth” siRNAs (supplemental Table 2S). Cells with reduced SLUG expression showed significantly increased levels of UbcH5c mRNA (Fig. 4A) and protein (Fig. 4, B and C). Although the cyclin D1 mRNA levels in these cells did not change significantly (Fig. 4A), there was a considerable decrease in the levels of cyclin D1 protein (Figs. 4, B and C).
FIGURE 4.
Effect of knockdown of SLUG on cyclin D1 levels in MDA-MB-231 and BT549 cells. A, quantitative RT-PCR analysis for SLUG, UbcH5c, and cyclin D1 mRNA levels in MDA-MB-231 cells treated with different siRNAs (supplemental Table 2S). B, immunoblot analysis of UbcH5c and cyclin D1 levels in MDA-MB-231 and BT549 cells with (SLUGKD) or without (Control) knocking down SLUG (siRNA#1, stealth-21). Control cells were transfected with control siRNA. C, evaluation of SLUG, cyclin D1, and UbcH5c protein levels in MDA-MB-231 and BT549 cells with or without knockdown of SLUG. Six independent SLUG-knocked down cell populations and corresponding control siRNA-treated cells were used. Data with siRNA#1 as the reagent for the knockdown are shown. Similar results were obtained with stealth-223 (siRNA#2; data not shown). Bands were developed using IR dye-conjugated secondary antibody (LI-COR Biosciences) and visualized using the LI-COR Odyssey infrared imaging system. Quantitation and analysis of bands were performed using Odyssey software. β-Actin was used as normalization control. Results are mean ± S.E. (error bars) (n = 6). -Fold changes observed were statistically significant (p < 0.001). D, effect of knockdown of SLUG in MDA-MB-231 cells on their rate of proliferation and the role of non-degradable cyclin D1 mutant (T286A) in this process. Cells were transiently transfected with wild type or HA-tagged T286A mutant of cyclin D1 along with the siRNA (control or anti-SLUG). Results are mean ± S.E. (n = 6). The effect of SLUG knockdown with wild-type cyclin D1 was statistically significant (p < 0.0001). E, effect of the proteasome inhibitor MG132 on the decrease in cyclin D1 in SLUG-knocked down MDA-MB-231 cells. Control cells were transfected with empty vector DNA. β-Actin was used as loading control. Veh, vehicle (DMSO) for MG132 solution. F, densitometric analysis of immunoblot data as in E from three independent experiments. The upper panel (i) shows the effect on SLUG level, and the lower panel (ii) shows the effect on cyclin D1 levels. Experimental data are normalized assuming respective control as 100. Results are mean ± S.E. (n = 3); *, statistical significance in comparison with respective control (p < 0.001).
Knockdown of SLUG expression in MDA-MB-231 cells also decreased their rates of proliferation (Fig. 4D), suggesting that SLUG regulates the rate of proliferation of human breast cells through the increase in cyclin D1 levels in the cells. We hypothesize that when the SLUG level is decreased, UbcH5c levels are increased due to derepression of the UbcH5c gene. Proteasomal degradation of cyclin D1 is thus increased (12, 13), resulting in low levels of cyclin D1 with the decrease in the proliferation rates of the cells. We tested this hypothesis by expressing a non-degradable cyclin D1 mutant (T286A mutant) and evaluating whether this expression would block the changes associated with SLUG knockdown in breast cancer cells. We transiently expressed a C-terminally HA-tagged cyclin D1 T286A mutant in MDA-MB-231 cells and knocked down SLUG in these cells with siRNA. Although the expression of wild-type cyclin D1 had no significant effect, expression of the cyclin D1 mutant (T286A) in cells alleviated the effect of SLUG knockdown on the rate of proliferation of these cells (Fig. 4D). These data verify further our notion that the mechanism of SLUG-induced growth enhancement in breast cancer cells perhaps involves stabilization of cyclin D1.
To evaluate further whether the decrease in the cyclin D1 levels in SLUG-knocked down MDA-MBA-231 cells is due to enhanced proteasomal degradation of this protein, we determined whether inhibition of proteasomal activity in these cells using the proteasomal inhibitor MG132 would alleviate this effect. Control siRNA- and SLUG siRNA-treated MDA-MB-231 cells were treated with vehicle or MG132, and their cyclin D1 levels were evaluated by immunoblot analysis. Cyclin D1 levels in the SLUG-knocked down cells were indeed rescued by MG132 (Fig. 4, E and F). There was some effect of MG132 on the level of SLUG in these cells (Fig. 4, E and F), but this immunoblot analysis clearly shows that SLUG knockdown indeed enhanced proteasomal degradation of cyclin D1 in MDA-MB-231 cells.
Knockdown of UbcH5c in SLUG-deficient Breast Cancer Cells Elevated Cyclin D1 Levels as Well as Their Growth Rates, Invasiveness, and Resistance to 4HT
We postulated that SLUG-induced enhancement of cyclin D1 level in the human breast cancer cells is mediated through the repressions of UbcH5c levels in these cells. We tested this notion further by knocking down UbcH5c in the SLUG-deficient MDA-MB468 and MCF7 cells. In the UbcH5c-knocked down cells, the level of cyclin D1 was elevated significantly (Fig. 5, A–D), whereas the level of SLUG protein remained unaltered, as expected (Fig. 5, C and D). Knockdown of UbcH5c also increased the growth rates and 4HT resistance of MCF7 cells (Fig. 5E). Similar enhancement of growth rates was also observed with other SLUG-deficient UbcH5c-knocked down human breast cancer cells (data not shown). Data with stealth-1106 (supplemental Table 2S) are shown. The other siRNA against UbcH5c, stealth-1214 (supplemental Table 2S) also yielded similar observations. To verify that this growth enhancement by UbcH5c knockdown is due to the increase in the cyclin D1 level in the cells, we knocked down cyclin D1 with two different siRNAs (see supplemental Table 2S; data for one is shown). Cyclin D1 knockdown alleviated growth-enhancing effects of UbcH5c knockdown in MCF7 cells (Fig. 5E). Simultaneous knockdown of UbcH5c and cyclin D1 also brought back the sensitivity of MCF7 cells to 4HT (Fig. 5E). These data may suggest that SLUG enhances the rate of proliferation of breast cancer cells, at least in part, through the repression of the UbcH5c gene, which results in the increase in the levels of cyclin D1.
FIGURE 5.
Effect of knockdown of UbcH5c on cyclin D1 levels in MDA-MB-468 and MCF7 cells. A, immunoblot analysis showing the effect of knockdown of UbcH5c (H5cKD) on cyclin D1 level in MDA-MB-468 cells. Control cells were transfected with control siRNA. B, immunoblot analysis showing the effect of knockdown of UbcH5c on cyclin D1 level in MCF7 cells. β-Actin was used as loading control. Control cells were transfected with control siRNA. C, evaluation of cyclin D1 and UbcH5c protein levels in MDA-MB-468 cells with or without knockdown of UbcH5c. Six independent UbcH5c-knocked down cell populations and corresponding control siRNA-treated cells were used. D, evaluation of cyclin D1 and UbcH5c protein levels in MCF7 cells with or without knockdown of UbcH5c. Six independent UbcH5c-knocked down cell populations and corresponding control siRNA-treated cells were used. For the experiments in C and D, bands were developed using IR dye-conjugated secondary antibody (LI-COR Biosciences) and visualized using the LI-COR Odyssey infrared imaging system. Quantitation and analysis of bands were performed using Odyssey software. β-Actin was used as normalization control. Results are mean ± S.E. (error bars) (n = 6). Fold changes observed were statistically significant (p < 0.001). E, proliferation assays with the control and UbcH5c knockdown MCF7 cells in the absence or presence of 4HT (10 μm) and the effects of simultaneous knockdown of cyclin D1 (CD1+H5cKD) in these processes. Results are mean ± S.E. (n = 6). Data with cyclin D1 siRNA stealth-311 (supplemental Table 2S) are shown. Other siRNA, stealth-568, also yielded similar results (data not shown). F, Matrigel invasion assay with UbcH5c knocked down MCF7 and MDA-MB-468 cells. Results are mean ± S.E. (n = 6). Data with siRNA stealth-1106 (supplemental Table 2S) are shown. Other siRNA, stealth-1214, also yielded similar results (data not shown).
SLUG is also implicated in the enhancement of invasiveness of cancer cells (28, 29). We evaluated whether this enhancement of invasiveness is mediated through the repression of UbcH5c. We knocked down UbcH5c in the SLUG-negative MCF7 and MDA-MB-468 cells and evaluated the in vitro invasiveness of these cells in Matrigel using a Boyden chamber (39). We found that UbcH5c-knocked down MCF7 and MDA-MB-468 cells became significantly more invasive as compared with the corresponding scrambled siRNA-treated cells (Fig. 5F). Thus, SLUG-mediated repression of UbcH5c gene may be partially responsible for the SLUG-induced increase in the invasiveness of breast cancer cells.
Overexpression of UbcH5c in SLUG-high Breast Cancer Cells Lowered Cyclin D1 Levels as Well as Decreased Their Growth Rates
To verify further our notion that UbcH5c knockdown by SLUG is critical for SLUG-induced growth regulation in aggressive SLUG-high breast cancer cells, we overexpressed UbcH5c in the SLUG-high (thus UbcH5c-low) MDA-MB-231 cells and evaluated the level of cyclin D1 as well as their growth rates. In the UbcH5c-overexpressed cells, the level of cyclin D1 is decreased significantly (Fig. 6A). Overexpression of UbcH5c also decreased the growth rates of MDA-MB-231 cells (Fig. 6B). These data thus strengthen the idea that SLUG mediates its growth-enhancing effects in breast cancer cells in part by repression of UbcH5c gene.
FIGURE 6.
Effect of overexpression of UbcH5c on cyclin D1 levels and proliferation in SLUG-high MDA-MB-231 cells. A, immunoblot analysis showing the effect of overexpression of UbcH5c on cyclin D1 level. Control cells were transfected with empty vector DNA. UbcH5c levels were assessed by using Myc antibody for the recombinant protein and UbcH5c antibody for the total of the endogenous and recombinant protein. B, proliferation assays with the control and UbcH5c-overexpressed (UbcH5cOE) MDA-MB-231 cells. Results are mean ± S.E. (error bars) (n = 6).
SLUG Binds Directly to the Promoter of Human UbcH5c Gene in the Nucleus of Human Breast Cancer Cells
To understand further whether SLUG directly inhibits the transcription of the UbcH5c gene in human breast cancer cells, we re-evaluated the in vivo binding of SLUG to the promoter of the human UbcH5c gene. Binding of SLUG to the E2-box sequence of the target gene promoters is an essential prerequisite for SLUG-mediated repression of the gene (28, 32, 33). Analysis of the nucleotide sequences of UbcH5c promoter regions (−850 to +200) in humans (NM_003340), mice (NM_025356), and rats (NM_031237) revealed the presence of a highly conserved E2-box element (see the supplemental material for nucleotide sequences and ClustalW alignments). ChIP analysis using commercially available ChIP grade FLAG or SLUG antibody revealed that SLUG binds to the promoter of the UbcH5c gene in SLUG-expressing human breast cancer cells (Fig. 7, A and B). Overexpression of C-terminal FLAG-tagged SLUG in the SLUG-deficient breast cancer cells showed the binding of SLUG to this promoter (Fig. 7A). On the other hand, knockdown of SLUG in the SLUG-high cells abrogated the binding of this protein to the UbcH5c promoter (Fig. 7B). These data suggest that SLUG directly represses the promoter of the UbcH5c gene.
FIGURE 7.
Inhibition of the UbcH5c promoter activity in SLUG-expressing human breast cells. A, ChIP analysis for the binding of SLUG to the promoter of the UbcH5c gene in FLAG-tagged SLUG-expressing (+SLUG) MCF7 and MDA-MB-468 cells. Immunoprecipitation (IP) was done with FLAG antibody to immunoprecipitate chromatin fragments bound to FLAG-tagged SLUG. B, ChIP analysis for the binding of SLUG to the promoter of the UbcH5c gene in the control and the SLUG-knocked down MDA-MB-231 and BT549 (−SLUGKD) cells. ChIP grade SLUG antibody was used for immunoprecipitation of wild-type SLUG-bound chromatin fragments. C, repression of the UbcH5c promoter in the SLUG-expressing (SLUGOVEX) MCF7 and MDA-MB-468 cells. D, effect of E2-box mutation at the UbcH5c promoter on its activity in SLUG-expressing MCF7 cells. In C and D, the averages from six different SLUG-transfected populations and controls are shown. Results are mean ± S.E. (error bars) (n = 6). Decreases in the luciferase activities were statistically significant (p < 0.001).
SLUG Inhibits the Activity of Cloned UbcH5c Promoter in the Transfected Human Breast Cancer Cells
As compared with the control cells, SLUG expression in the recombinant cells showed significant inhibition of UbcH5c promoter activity (Fig. 7C). The inhibition of the promoter activity by SLUG is mediated through the E2-box located at the promoter, as was evidenced by the lack of inhibition of the promoter activity by SLUG when the E2-box sequence is mutated (Fig. 7D). These results suggest that SLUG down-regulates UbcH5c gene expression by binding to the E2-box sequence at this promoter.
SLUG-induced Inhibition of the UbcH5c Promoter Is Mediated through Chromatin Remodeling
We determined the binding of the co-repressor CtBP1 and the effector HDAC1 to this promoter in vivo. We used MDA-MB-468 cells expressing C-terminal FLAG-tagged SLUG for this study (33). This analysis revealed that as compared with the control vector-transfected cells, the UbcH5c promoter recruits not only SLUG but also the co-repressor CtBP1 and the histone modifier HDAC1 (Fig. 8A). The involvement of HDAC1 in the SLUG-mediated inhibition of UbcH5c gene expression is further evident from the observation that in the presence of HDAC1 inhibitor trichostatin A, SLUG failed to knock down the level of the UbcH5c protein in these cells (Fig. 8B). It is interesting to note that the apparent molecular size of the UbcH5c protein is increased slightly (∼1 kDa) in the presence of trichostatin A (Fig. 8B). The mechanism of this increase is not known. Through ChIP analysis, we find that in comparison with the control vector-transfected MDA-MB-468 cells, the SLUG-expressing cells have lower levels of the acetylated histones H3 (Lys9 and Lys14) and H4 (Lys5, Lys8, Lys12, and Lys16) (Fig. 8, C and D). These data further suggest that SLUG represses the UbcH5c promoter through chromatin remodeling (Fig. 8E).
FIGURE 8.
Mechanism of repression of UbcH5c promoter by SLUG in breast cancer cells. A, ChIP analysis for the co-recruitments of CtBP1 and HDAC1 with SLUG at the UbcH5c promoter. B, immunoblot analysis for the effect of trichostatin A (TSA) on SLUG-induced repression of UbcH5c levels in SLUG-expressing MDA-MB-468 cells. C, ChIP analysis showing the decrease of acetylated histones H3 and H4 at the UbcH5c promoter in SLUG-expressing MDA-MB-468 cells. D, quantitative ChIP analysis to evaluate the levels of acetylated histones H3 and H4 at the UbcH5c promoter in SLUG-expressing MDA-MB-468 cells. *, statistical significance (p < 0.001). E, model for the regulation of UbcH5c promoter by SLUG in human breast cancer cells by chromatin remodeling. BD, DNA binding domain of SLUG; RD, repressor domain for SLUG. Error bars, S.E.
DISCUSSION
The G1 to S phase transition during normal cell cycle progression is tightly controlled by the D-type and E-type cyclins (50). These cyclins are critical components of steroid- and growth factor-induced mitogenesis in breast epithelial cells (50–52). Overexpression of these genes in mammary epithelial cells leads to mammary carcinoma (26, 50).
Several interdependent and independent mechanisms may lead to the elevation of cyclin D1 levels in breast cancer cells. Included in these mechanisms are cyclin D1 gene amplification (53, 54), stimulation of cyclin D1 gene expression through the activation of ERK-MAPK or Rac-NFkB signal transduction pathways (55, 56), and inhibition of the proteasomal degradation of cyclin D1 (57, 58). In this study, we discovered a SLUG-dependent mechanism for the elevation of cyclin D1 level in human breast cancer cells. We found that in invasive breast tumor cells, SLUG is abnormally overexpressed and, among several other genes, inhibits the expression of the cyclin D1 modifier E2 enzyme UbcH5c. We hypothesize that a decrease in the level of UbcH5c decreases the turnover rate of cyclin D1. Indirect association of SLUG with cyclin D1 levels via UbcH5c is important in the understanding of the complex role played by SLUG in the etiology and metastasis of mammary carcinoma.
There could be multiple consequences of cyclin D1 level elevation in SLUG-overexpressing human breast cancer cells. The most obvious one is the increase in the rate of proliferation of these cells (51). We found a significant increase in the rates of proliferation of the breast cancer cells tested by SLUG overexpression or UbcH5c knockdown, both of which result in the elevation of cyclin D1 levels in these cells. Thus, elevation of cyclin D1 levels in the breast cancer cells through the SLUG-mediated repression of the UbcH5c gene may be one of the mechanisms by which SNAI repressors promote breast cancer aggression (28, 29). Because SLUG represses many genes in the breast cancer cells, the up-regulation of cyclin D1 in SLUG-overexpressing cells may also, at least in part, be contributed by secondary effects of the repression of other genes by SLUG in the cell.
The other consequence of cyclin D1 level increase could be the role of cyclin D1 as a transcriptional regulator (51). Cyclin D1 can form potentially functional interactions with a variety of other molecules, including cellular transcription factors (e.g. ER, androgen receptor, DMP1, STAT3, BETA2/NeuroD, and C/EBPβ) as well as both histone acetylases and deacetylases (41–43, 51). These interactions are independent of association with and activation of Cdk4 and -6 and point to a role for cyclin D1 in transcriptional regulation (51). These cyclin-dependent kinase-independent actions of cyclin D1 may lead to the invasive phenotypes found in SLUG-overexpressing and UbcH5c knockdown cells in our studies. Another consequence of estrogen-independent activation of ERα in breast cancer cells by excess cyclin D1 is the development of resistance of these cells to anti-estrogens like 4HT (44) and arzoxifen (49) (see supplemental Fig. 2S). Indeed, our data showed that elevation of SLUG or knockdown of UbcH5c in breast cancer cells made them relatively resistant to 4HT. Our study thus contributes significantly to the understanding of the multifaceted actions of overexpressed SLUG protein in human breast cancer cells.
The establishment of metastasis depends on the ability of cancer cells to acquire a migratory phenotype. In epithelial cancers, such as those of the breast, the epithelial-mesenchymal transition is associated with basal-like breast cancers, generates cells with stemlike properties, and enables cancer cell dissemination and metastasis (28, 29). SLUG is a transcriptional repressor protein that catalyzes epithelial-mesenchymal transition, inhibits apoptosis, and promotes cancer cell proliferation by inhibiting the tumor suppressor protein BRCA2 and vitamin D3 receptor (28, 29, 32, 33). Our research implies that SLUG also regulates the turnover rates of several key proteins like cyclin D1 by repressing the ubiquitin-conjugating enzyme UbcH5c. Our finding highlights the need of additional research related to the involvement of SLUG and UbcH5c in the onset and progression of breast and perhaps other epithelial cancers.
Supplementary Material
Acknowledgments
We thank Dr. Tanu Rana for valuable discussions during the development of the manuscript. We also thank Robert Smith for excellent technical assistance. We acknowledge Dr. Y. Xiong for the cyclin D1 plasmid, Dr. B. Zetter for the cyclin D1 T286A plasmid, and Dr. E. Yeh for the ubiquitin plasmid.
This work was supported in part by Department of Defense Congressionally Directed Medical Research Program IDEA Grant W81XWH-06-1-0466 and Susan G. Komen Breast Cancer Foundation Grant BCTR0707627 (to G. C.). Confocal microscopy was performed through the use of the Meharry Medical College Morphology Core, which is supported in part by National Institutes of Health Grant U54NS041071. Other core facility research was supported in part by Vanderbilt Clinical and Translational Science Award Grant UL1 RR024975 from NCRR, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1S–4S, Figs. 1S–3S, nucleotide sequences, and ClustalW alignments.
- ER
- estrogen receptor
- 4HT
- 4-hydroxytamoxifen.
REFERENCES
- 1. López-Otín C., Hunter T. (2010) Nat. Rev. Cancer 10, 278–292 [DOI] [PubMed] [Google Scholar]
- 2. Gallastegui N., Groll M. (2010) Trends Biochem. Sci. 35, 634–642 [DOI] [PubMed] [Google Scholar]
- 3. Fang Y., Fu D., Shen X. Z. (2010) Biochim. Biophys. Acta 1806, 1–6 [DOI] [PubMed] [Google Scholar]
- 4. Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 5. Baek K. H. (2006) Curr. Protein Pept. Sci. 7, 171–177 [DOI] [PubMed] [Google Scholar]
- 6. Gao M., Karin M. (2005) Mol. Cell. 19, 581–593 [DOI] [PubMed] [Google Scholar]
- 7. Sun L., Chen Z. J. (2004) Curr. Opin. Cell Biol. 16, 119–126 [DOI] [PubMed] [Google Scholar]
- 8. Conkright M. D., Wani M. A., Lingrel J. B. (2001) J. Biol. Chem. 276, 29299–29306 [DOI] [PubMed] [Google Scholar]
- 9. Li D. Q., Ohshiro K., Reddy S. D., Pakala S. B., Lee M. H., Zhang Y., Rayala S. K., Kumar R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17493–17498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X. D., Matunis M. J. (2005) Nat. Cell Biol. 7, 12–14 [DOI] [PubMed] [Google Scholar]
- 11. Wu K., Kovacev J., Pan Z. Q. (2010) Mol. Cell. 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maeda I., Ohta T., Koizumi H., Fukuda M. (2001) FEBS Lett. 494, 181–185 [DOI] [PubMed] [Google Scholar]
- 13. Hattori H., Zhang X., Jia Y., Subramanian K. K., Jo H., Loison F., Newburger P. E., Luo H. R. (2007) Blood 110, 640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saville M. K., Sparks A., Xirodimas D. P., Wardrop J., Stevenson L. F., Bourdon J. C., Woods Y. L., Lane D. P. (2004) J. Biol. Chem. 279, 42169–42181 [DOI] [PubMed] [Google Scholar]
- 15. Furukawa M., Ohta T., Xiong Y. (2002) J. Biol. Chem. 277, 15758–15765 [DOI] [PubMed] [Google Scholar]
- 16. Shembade N., Ma A., Harhaj E. W. (2010) Science 327, 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonen H., Bercovich B., Orian A., Carrano A., Takizawa C., Yamanaka K., Pagano M., Iwai K., Ciechanover A. (1999) J. Biol. Chem. 274, 14823–14830 [DOI] [PubMed] [Google Scholar]
- 18. Jensen J. P., Bates P. W., Yang M., Vierstra R. D., Weissman A. M. (1995) J. Biol. Chem. 270, 30408–30414 [DOI] [PubMed] [Google Scholar]
- 19. Helin K. (1998) Curr. Opin. Genet. Dev. 8, 28–35 [DOI] [PubMed] [Google Scholar]
- 20. Piscopo D. M., Hinds P. W. (2008) Cancer Res. 68, 5581–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Angiolella V., Donato V., Vijayakumar S., Saraf A., Florens L., Washburn M. P., Dynlacht B., Pagano M. (2010) Nature 466, 138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carrano A. C., Pagano M. (2001) J. Cell Biol. 153, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan Y., Qin L., Liu D., Wu R. C., Mussi P., Zhou S., Songyang Z., Xu J. (2007) Cancer Res. 67, 8032–8042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao F., Cheng J., Shi T., Yeh E. T. (2006) Nat. Cell Biol. 8, 1171–1177 [DOI] [PubMed] [Google Scholar]
- 25. Sherr C. J. (1995) Trends Biochem. Sci. 20, 187–190 [DOI] [PubMed] [Google Scholar]
- 26. Gillett C., Fantl V., Smith R., Fisher C., Bartek J., Dickson C., Barnes D., Peters G. (1994) Cancer Res. 54, 1812–1817 [PubMed] [Google Scholar]
- 27. Barrallo-Gimeno A., Nieto M. A. (2005) Development 132, 3151–3161 [DOI] [PubMed] [Google Scholar]
- 28. Nieto M. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 155–166 [DOI] [PubMed] [Google Scholar]
- 29. Peinado H., Olmeda D., Cano A. (2007) Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 30. Prasad C. P., Rath G., Mathur S., Bhatnagar D., Parshad R., Ralhan R. (2009) BMC Cancer 9, 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lambertini E., Franceschetti T., Torreggiani E., Penolazzi L., Pastore A., Pelucchi S., Gambari R., Piva R. (2010) BMC Mol. Biol. 11, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tripathi M. K., Misra S., Khedkar S. V., Hamilton N., Irvin-Wilson C., Sharan C., Sealy L., Chaudhuri G. (2005) J. Biol. Chem. 280, 17163–17171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mittal M. K., Myers J. N., Misra S., Bailey C. K., Chaudhuri G. (2008) Biochem. Biophys. Res. Commun. 372, 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Misra S., Sharma S., Agarwal A., Khedkar S. V., Tripathi M. K., Mittal M. K., Chaudhuri G. (2010) Mol. Cancer 9, 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Watanabe H., Pan Z. Q., Schreiber-Agus N., DePinho R. A., Hurwitz J., Xiong Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Newman R. M., Mobascher A., Mangold U., Koike C., Diah S., Schmidt M., Finley D., Zetter B. R. (2004) J. Biol. Chem. 279, 41504–41511 [DOI] [PubMed] [Google Scholar]
- 37. Kamitani T., Kito K., Nguyen H. P., Yeh E. T. (1997) J. Biol. Chem. 272, 28557–28562 [DOI] [PubMed] [Google Scholar]
- 38. Kwon Y. S., Garcia-Bassets I., Hutt K. R., Cheng C. S., Jin M., Liu D., Benner C., Wang D., Ye Z., Bibikova M., Fan J. B., Duan L., Glass C. K., Rosenfeld M. G., Fu X. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mittal M. K., Myers J. N., Bailey C. K., Misra S., Chaudhuri G. (2010) Mol. Biol. Rep. 37, 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alao J. P. (2007) Mol. Cancer 6, 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zwijsen R. M., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R. J. (1997) Cell 88, 405–415 [DOI] [PubMed] [Google Scholar]
- 42. Zwijsen R. M., Buckle R. S., Hijmans E. M., Loomans C. J., Bernards R. (1998) Genes Dev. 12, 3488–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neuman E., Ladha M. H., Lin N., Upton T. M., Miller S. J., DiRenzo J., Pestell R. G., Hinds P. W., Dowdy S. F., Brown M., Ewen M. E. (1997) Mol. Cell. Biol. 17, 5338–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ishii Y., Waxman S., Germain D. (2008) Cancer Res. 68, 852–860 [DOI] [PubMed] [Google Scholar]
- 45. Ahnström M., Nordenskjöld B., Rutqvist L. E., Skoog L., Stål O. (2005) Breast Cancer Res. Treat. 91, 145–151 [DOI] [PubMed] [Google Scholar]
- 46. Cadigan K. M., Nusse R. (1997) Genes Dev. 11, 3286–3305 [DOI] [PubMed] [Google Scholar]
- 47. Jirström K., Stendahl M., Rydén L., Kronblad A., Bendahl P. O., Stål O., Landberg G. (2005) Cancer Res. 65, 8009–8016 [DOI] [PubMed] [Google Scholar]
- 48. Stendahl M., Kronblad A., Rydén L., Emdin S., Bengtsson N. O., Landberg G. (2004) Br. J. Cancer 90, 1942–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zwart W., Rondaij M., Jalink K., Sharp Z. D., Mancini M. A., Neefjes J., Michalides R. (2009) Mol. Endocrinol. 23, 1335–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Butt A. J., Caldon C. E., McNeil C. M., Swarbrick A., Musgrove E. A., Sutherland R. L. (2008) Adv. Exp. Med. Biol. 630, 189–205 [DOI] [PubMed] [Google Scholar]
- 51. Sutherland R. L., Musgrove E. A. (2002) Breast Cancer Res. 4, 14–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sutherland R. L., Musgrove E. A. (2004) J. Mammary Gland Biol. Neoplasia 9, 95–104 [DOI] [PubMed] [Google Scholar]
- 53. Hosokawa Y., Arnold A. (1998) Genes Chromosomes Cancer 22, 66–71 [DOI] [PubMed] [Google Scholar]
- 54. Ormandy C. J., Musgrove E. A., Hui R., Daly R. J., Sutherland R. L. (2003) Breast Cancer Res. Treat. 78, 323–335 [DOI] [PubMed] [Google Scholar]
- 55. Fournier A. K., Campbell L. E., Castagnino P., Liu W. F., Chung B. M., Weaver V. M., Chen C. S., Assoian R. K. (2008) J. Cell Sci. 121, 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klein E. A., Yang C., Kazanietz M. G., Assoian R. K. (2007) Cell Cycle 6, 1115–1121 [DOI] [PubMed] [Google Scholar]
- 57. Lin D. I., Lessie M. D., Gladden A. B., Bassing C. H., Wagner K. U., Diehl J. A. (2008) Oncogene 27, 1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Masamha C. P., Benbrook D. M. (2009) Cancer Res. 69, 6565–6572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.