Abstract
In higher plants, [2Fe-2S] ferredoxin (Fd) proteins are the unique electron acceptors from photosystem I (PSI). Fds are soluble, and distribute electrons to many enzymes, including Fd:NADP(H) reductase (FNR), for the photoreduction of NADP+. In addition to well studied [2Fe-2S] Fd proteins, higher plants also possess genes for significantly different, as yet uncharacterized Fd proteins, with extended C termini (FdCs). Whether these FdC proteins function as photosynthetic electron transfer proteins is not known. We examined whether these proteins play a role as alternative electron acceptors at PSI, using quantitative RT-PCR to follow how their expression changes in response to acceptor limitation at PSI, in mutant Arabidopsis plants lacking 90–95% of photosynthetic [2Fe-2S] Fd. Expression of the gene encoding one FdC protein, FdC1, was identified as being strongly up-regulated. We confirmed that this protein was chloroplast localized and increased in abundance on PSI acceptor limitation. We purified the recombinant FdC1 protein, which exhibited a UV-visible spectrum consistent with a [2Fe-2S] cluster, confirmed by EPR analysis. Measurements of electron transfer show that FdC1 is capable of accepting electrons from PSI, but cannot support photoreduction of NADP+. Whereas FdC1 was capable of electron transfer with FNR, redox potentiometry showed that it had a more positive redox potential than photosynthetic Fds by around 220 mV. These results indicate that FdC1 electron donation to FNR is prevented because it is thermodynamically unfavorable. Based on our data, we speculate that FdC1 has a specific function in conditions of acceptor limitation at PSI, and channels electrons away from NADP+ photoreduction.
Keywords: Chloroplast, Electron Paramagnetic Resonance (EPR), Electron Transfer, Iron-Sulfur Protein, Photosynthesis, Ferredoxin, Ferredoxin:NADP(H) Reductase, High Light Stress, Redox Potential
Introduction
Ferredoxins (Fds)3 are small soluble electron carrier proteins. In the final reaction of photosynthetic electron transfer (PET), photosystem I (PSI) donates electrons to Fd (1), which acts as the soluble electron donor to various acceptors in the chloroplast stroma and can also return electrons to the thylakoid in cyclic electron flow (CET) (2). The electron cascade to supply carbon fixation requires photoreduction of NADP+ by Fd, catalyzed by Fd-NADP(H) oxidoreductase (FNR) (3). Many other plastid enzymes accept electrons directly from Fd for metabolic processes. These include, but are not limited to, nitrite reductase and sulfite reductase, which are essential for assimilation of inorganic nitrogen and sulfur, respectively, and Fd-dependent glutamine oxoglutarate aminotransferase and fatty acid desaturase, which catalyze key steps in amino acid and fatty acid metabolism, respectively (4). In addition, Fd donation to thioredoxin via the Fd:thioredoxin reductase translates the redox state of the electron transfer chain into a regulatory signal controlling the activity of many enzymes (5). Fds are also capable of accepting electrons from NADPH via FNR, in a reversal of the photosynthetic reaction (6), allowing electron donation from reduced Fd to different acceptors under non-photosynthetic conditions.
Most higher plants studied possess genes for several different Fd isoproteins (7–9). There is always an isoprotein that is more abundant in non-photosynthetic tissues and has higher affinity than photosynthetic and PetF-type Fds for FNR in the non-photosynthetic (often called “root”) cascade (9, 10), where electrons are transferred from NADPH to Fd. In all plants for which we possess significant EST and cDNA information at least 2 separate photosynthetic isoproteins have been identified (7, 8). In the C4-plant maize, different functions have been identified for two of the leaf-type Fds (11). There is a higher demand for ATP (which is disproportionately produced in CET) in the bundle sheath cells of NADP+ malic enzyme type C4 plants, and maize FdI and FdII are differentially expressed in mesophyll and bundle sheath cells, respectively (12). FdII has decreased affinity for FNR (13) and demonstrates a higher activity in CET around the photosystems, whereas FdI drives linear electron flow (11). In C3 plants, this spatial distribution is not observed, but duplicate photosynthetic Fds are still present, and there is some evidence that these proteins also act differentially in linear electron flow and CET (7). In Arabidopsis, Fd2 constitutes ∼90% of leaf Fd content (9, 14), whereas Fd1 is comparatively scarce. Despite the fact that Fd1 and Fd2 have a similar affinity for FNR, they appear to perform different functions in photosynthesis, and there is evidence that Fd1 makes a specifically higher contribution to CET (7).
The Arabidopsis thaliana genome contains genes encoding four well described [2Fe-2S] Fds and also two additional genes encoding Fd proteins of unknown function (9). The coding sequences from genomic and cDNA databases suggest that both proteins (encoded by At1g32550 and At4g14890) contain significant C-terminal extensions (that of At4g14890 is shown in supplemental Fig. S1A) conserved among homologous proteins from other species, and so we have named them FdC1 and FdC2, respectively. This structural feature is intriguing because it has been demonstrated that the C terminus is critical for interaction with the C, D, and E subunits of PSI during photosynthetic electron transfer (15, 16).
In certain conditions, such as high light stress and drought, the electron transfer rate through PET can exceed the capacity of soluble stromal acceptors. When this occurs, excess electrons may reduce O2 and generate damaging reactive oxygen species (17). On acceptor limitation at PSI there is electron donation directly to O2, and the resulting superoxide radical is quickly dismutated enzymatically to form the less toxic H2O2 (18). In the classic water-water cycle, this H2O2 is reduced to H2O by the action of glutathione- and ascorbate-dependent processes (19), which are also dependent on electron donation from Fd for regeneration (20). Fd itself is also implicated in conversion of H2O2 to the highly damaging OH• radical (21). In addition to the water-water cycle, there are several other mechanisms to avoid excess reduction pressure, and therefore reactive oxygen species accumulation. These include: down-regulation of photosystem II (PSII) excitation by state transitions (22) or by the xanthophyll cycle (23), and the cycling of electrons between the cytochrome b6f complex and PSI in CET to generate a ΔpH gradient without net production of reductant (2, 24).
When Arabidopsis Fd2 is knocked out (14) or decreased by RNAi (7), Fd1 does not increase in abundance to compensate for the functional loss. In these cases, acceptor limitation at PSI results in growth retardation, but the plants grow and fix CO2 with surprising competence using only this Fd1 (around 5% of the total WT Fd content). We have investigated how transcription of the remaining Arabidopsis Fd genes respond to this PSI acceptor limitation, to try and identify possible electron sinks from PET that might alleviate this reduction pressure in the photosystems, and allow the plants to grow and reproduce. In this paper we describe how FdC1 transcripts are specifically increased in the absence of Fd2, characterize the functional properties of the purified protein, and discuss how it may act to alleviate PSI acceptor limitation, especially under high electron pressure.
EXPERIMENTAL PROCEDURES
Plant Material
For all experiments shown in this work, A. thaliana plants of the ecotype Noessen (WT and fd2-knockout mutants) and Columbia (WT and fd2-RNAi lines) were used. For all measurements the plants were cultivated for 11 weeks under moderate light conditions (120 μmol quanta m−2 s−1, 20 °C) in short days as described by Becker et al. (25).
Gas Exchange and Chlorophyll Fluorescence Measurements
Gas exchange and chlorophyll fluorescence were measured simultaneously with the LI6400-XT system (LI-COR Environmental, Lincoln, NE) using the 6400-40 Leaf Chamber Fluorometer as light source and fluorometer. Leaf chamber conditions were approximately the same as the ambient growth conditions of the plants (0.04% CO2, 21% O2, 20 °C leaf temperature, and 55% relative humidity). Measurements were performed on intact leaves and calculation of the fluorescence parameters was according to Genty (26) using the following equations (nomenclature as defined by VanKooten and Snel (27)): electron transfer rate = ((Fm′ − F)/Fm′) × light intensity in μmol quanta m−2 s−1 × 0.5; qP = (Fm′ − Fs)/(Fm′ − Fo′); non-photochemical quenching = (Fm − Fm′)/Fm′; and ΦII = (Fm′ − Fs)/Fm′.
P700 Absorption Determination
For P700 absorption determination a PAM-101 fluorimeter (Walz, Effeltrich, Germany) was used. The changes in P700 absorption were detected as ΔAP700 = ΔA830 nm − ΔA860 nm (28, 29). Before every measurement plants were dark adapted for at least 10 min to ensure complete reduction of P700. The maximum P700-oxidation (ΔAP700-max) was achieved by illumination with near infrared light (NIR, λ > 700 nm), and oxidation of P700 was recorded by the PDA-100 system (Walz).
Quantitative Real Time PCR
Five μg of total RNA, isolated from leaf material was used for cDNA synthesis according to the manufacturer's instructions (Fermentas RevertAidTM First Strand cDNA Synthesis Kit, Fermentas GmbH, St. Leon-Rot, Germany). Amplification was performed in an iCycler iQTM (Bio-Rad), using specific primers for Fd1, Fd2, Fd3, FdC1, FdC2, and ubiquitin (see supplemental Table S1) in a reaction mixture containing 50 ng of cDNA and MaximaTM SYBR Green Fluorescein qPCR Mastermix (Fermentas). Plasmids for serial dilutions of copy number were created by cloning the Fd1, Fd2, Fd3, FdC1, FdC2, and ubiquitin transcripts into the pJET1.2 vector (Fermentas) according to the manufacturer's instructions (cloning primers in supplemental Table S1). Data were analyzed with the icycler iQTM Multicolor Real-time PCR Optical System Software (version 3.1) (Bio-Rad) and Fd copy number normalized for ubiquitin.
Isolation of Mesophyll Protoplasts from Arabidopsis
Leaf protoplasts were isolated from 5-week-old WT plants. Protoplast isolation was according to Seidel et al. (30), with some modifications. After harvesting, leaves were placed in water and the lower epidermis was peeled off. With the lower surface down, the leaves were placed into digesting buffer (0.4 m mannitol, 20 mm KCl, 20 mm MES, 10 mm CaCl2, 0.1% bovine serum albumin (w/v), pH 5.7) without cell wall-digesting enzymes. The isolated protoplasts were stored on ice in the dark prior to transformation.
Transient Expression of GFP-FdC1 Fusion Proteins and Fluorescence Imaging
An FdC1 full-length clone was amplified from cDNA using primers containing restriction sites XbaI (5′-ATCACTCTCTCTAGACACAAAAA-3′) and KpnI (5′-AAATCAGGTACCTGAATAGTCGT-3′), and cloned into the pGEM-T Easy (Promega, Madison, WI) plasmid. The coding region was subcloned, in-frame, into pGFP-2 (31). For transfection of protoplasts, 80–100 μg of plasmid DNA was used according to Sambrook et al. (32) in a maximum volume of 30 μl. The transfection itself was performed according to Seidel et al. (30) using polyethylene glycol. After overnight incubation in the dark at 22 °C, green fluorescence was visualized by confocal laser scanning fluorescence microscopy (cLSM 510 META, Zeiss, Göttingen, Germany) using a ×40 EC Plan Neoflur (N.A. 1.3) oil objective. FDC1-GFP fusion protein signals and autofluorescence of chlorophyll were visualized by excitation at 488 nm and emission at 500–530 and 650–710 nm, respectively.
Recombinant Expression and Purification of FdC1
The FdC1 gene was constructed and cloned into the pTRC99a vector as described for Fd1, Fd2, Fd3, and Fd4 (9). FdC1 protein was expressed and purified as described previously for Fd proteins (13) except that no acetone was used and Escherichia coli cells were instead disrupted by sonication in 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm EDTA, 1 mm MgCl2, 1 mm PMSF, and salting out was performed at 35% saturating ammonium sulfate. Native-PAGE was performed as described by Hanke et al. (9).
Detection of Protein by Western Blotting
Protein extracts for SDS-PAGE were prepared from plants, separated, and Western blotted as described previously (9). FdC1 antisera was purified as previously described (9). Rubisco and purified FdC1 primary antisera raised in rabbit were used at dilutions 1:30,000 and 1:100, respectively.
Electron Paramagnetic Resonance (EPR)
EPR spectra for oxidized and sodium dithionite-reduced FdC1 and Fd2 (∼300 μm in each case) were performed on a Bruker ELEXSYS E500 spectrometer operating at X-band and employing a Super High Q cylindrical cavity (Q factor ∼20,000) equipped with an Oxford Instruments ESR900 liquid helium cryostat linked to an ITC503 temperature controller. The microwave power was 0.2 milliwatts, with modulation frequency 100 KHz and modulation amplitude 8 G. Spectra were collected at both 15 and 70 K.
Measurements of Electron Transfer
Photoreduction of NADP+ was measured as described previously (9). The absorbance change at 340 nm was followed in a solution containing 10 μg/ml of spinach thylakoid membranes, 0.2 mm NADP+, 50 mm HEPES-NaOH, pH 7.5, 100 mm NaCl, 1 mm MgCl2, and 0.01 to 40 μm Fd, following illumination with a red light source. Reduction of Fd molecules by PSI was measured in the same way, but with substitution of NADP+ with cytochrome c, reduction of which was followed at 550 nm (Δϵ550 of reduced − oxidized cytochrome c = 18.5 mm−1 cm−1). The Fd-dependent rate was subtracted for direct electron transfer from PSI to cytochrome c. It was confirmed that electron donation occurred directly from Fd to cytochrome c, rather than via superoxide (data not shown). NADPH-dependent Fd reduction by FNR was measured as described previously (10).
Spectroelectrochemical Redox Potentiometry
Redox titrations were performed in a glove box under anaerobic conditions in a nitrogen atmosphere (<2 ppm oxygen). All buffers and solutions were degassed by bubbling with nitrogen prior to entering the glove box to ensure removal of all traces of dioxygen. AtFdC1 and AtFd2 proteins were applied to a Bio-Rad 10-DG desalting column in the anaerobic box, pre-equilibrated with degassed 100 mm potassium phosphate, pH 7.0, with 10% (v/v) glycerol (titration buffer) to ensure removal of all traces of oxygen. The protein solutions were titrated electrochemically according to the method of Dutton (35) using sodium dithionite as the reductant and potassium ferricyanide as the oxidant, as described previously (33, 34). Titrations were performed in both reductive and oxidative directions to ensure a lack of hysteresis in the redox transitions of the heme. Dithionite and ferricyanide were delivered in ∼0.2-μl aliquots from concentrated stock solutions (typically 10–50 mm). Mediators were added to facilitate electrical communication between enzyme and electrode, prior to titration. Phenazine methosulfate (2 μm), 2-hydroxy-1,4-naphthoquinone (5 μm), methyl viologen (0.5 μm), and benzyl viologen (1 μm) were included to mediate in the range from +100 to −480 mV. The electrode was allowed to stabilize between each addition, and spectra (250–800 nm) were recorded using a Cary UV-50 Bio UV-visible scanning spectrophotometer, using a fiber optic probe running between the protein solution and the spectrophotometer external to the glove box. The electrochemical potential of the solution was measured using a Hanna pH211 meter coupled to a Pt/Calomel electrode (ThermoRussell Ltd.) at 25 ± 2 °C. The electrode was calibrated using the Fe3+/Fe2+ EDTA couple as a standard (108 mV). A factor of 244 mV was used to correct relative to the standard hydrogen electrode. Final concentrations of AtFdC1 and AtFd2 proteins used in the titrations were ∼115 and 85 μm, respectively. Absorbance changes at the major absorption feature in the visible region (at 415 nm for AtFdC1 and 424 nm for AtFd2) were plotted against the applied potential, and data were fitted to the Nernst equation to derive the midpoint potential for the [2Fe-2S] iron-sulfur cluster reduction (i.e. the [2Fe-2S]2+/1+ redox transition). Data manipulation and analysis were performed using Origin software (OriginLab, Northampton, MA). No hysteresis was observed on oxidative titration and both proteins remained stable to aggregation throughout the course of the redox titration.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: Fd1, At1g10960; Fd2, At1g60950; Fd3, At2g27510; FdC1, At4g14890; and FdC2, At1g32550.
RESULTS
Acute Acceptor Limitation in fd2 Plants under High Light
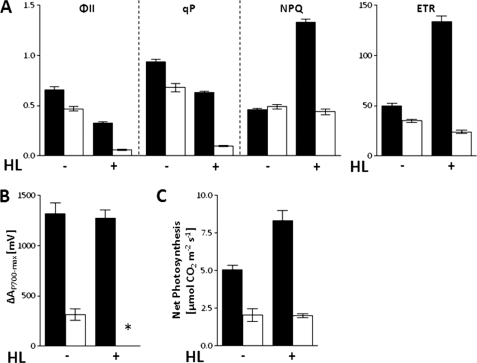
In previous work we showed that, in comparison to WT, fd2 knockout and knockdown plants contain only ∼1–10% of the total leaf Fd (7, 14). This chronic disruption to photosynthetic electron flow causes a severe phenotype (Fig. 1). Under standard growth light conditions (150 μmol quanta m−2 s−1), photosynthetic electron flow is adjusted in fd2, with decreased activity of both PSII and PSI, resulting in lower rates of photosynthetic electron transfer (Fig. 1). In combination with increased antioxidant systems (14) this allows the plant to limit problems associated with acceptor limitation at PSI, such as production of excessive free radicals. Despite this substantially reduced photosynthetic performance, the plants are capable of photoautotrophic growth, although with greatly reduced levels of CO2 fixation (Fig. 1C).
FIGURE 1.
High light induces severe PSI acceptor limitation in fd2 plants. Photosynthetic electron transport and CO2 fixation in wild type (black bars) and fd2 mutant (white bars) plants incubated at either 150 μmol quanta m−2 s−1 in growth light (HL−) or 600 μmol quanta m−2 s−1 in high light (HL+) for 2 h, then dark adapted for 15 min before measuring. A, chlorophyll fluorescence parameters for PSII capacity (ΦII), photochemical quenching (qP), non-photochemical quenching (NPQ), and electron transfer rate (ETR). B, maximum P700-oxidation in far red light (ΔAP700-max); *, no detectable activity. C, steady state CO2 assimilation rates were measured at least 2 h into the light period. Values are mean ± S.D. of at least 3 independent measurements.
On transfer to high light conditions (600 μmol quanta m−2 s−1), WT plants are capable of adjustment, increasing non-photochemical quenching to prevent damage to PSII, and increasing the electron transfer rate through the pathway (Fig. 1A). By contrast, fd2 mutants appear unable to increase non-photochemical quenching, and electron transfer rate remains constant. As a consequence, the electron transfer chain becomes loaded with electrons, and the very low photochemical quenching (qP) values indicate that PET components are predominantly reduced. Under these conditions, the capacity of PSI, measured as ΔAP700-max becomes undetectable in the fd2 plants (Fig. 1B), presumably due to a lack of electron acceptors. The fact that the plants continue to assimilate CO2 under these conditions (Fig. 1C) indicates that, although no oxidation of PSI can be measured, electrons are being transported away from PSI and used for NADPH formation. This could either be because re-reduction is so rapid that oxidation activity cannot be measured, or because this activity is catalyzed by a percentage of PSI too small for detection by ΔAP700-max. These data strongly suggest that the capacity of fd2 mutants to dissipate excess electrons from PSI is already saturated at low light concentrations.
Transcript Levels of FdC1 Are Elevated under Conditions of Acceptor Limitation
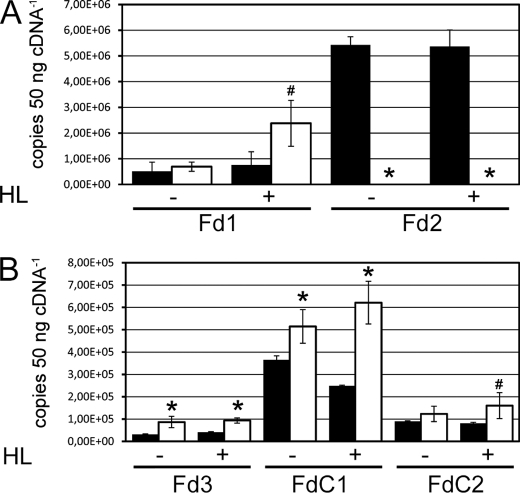
It is surprising that, in fd2 plants, only 1–10% of wild type Fd is capable of supporting photoautotrophic growth and stress alleviation pathways, such as the water-water cycle. In both fd2 knockout and knockdown plants, the alternative photosynthetic iso-protein, Fd1, shows no increase in transcript levels or protein abundance (7, 14). We therefore attempted to try and identify Fd-related proteins that might act as alternative electron acceptors from PSI, and so we examined the transcript levels of plant type and novel type Fds in the fd2 plants by quantitative real time PCR (Fig. 2). To confirm that these increases in transcript were a response to acceptor limitation, plants harvested under growth light conditions were compared with high light-treated plants.
FIGURE 2.
Changes in Fd iso-protein transcripts on acceptor limitation at PSI. RNA was isolated from wild type (black bars) and fd2 mutant (white bars) plants, incubated at 150 μmol quanta m−2 s−1 in growth light (HL−) or 600 μmol quanta m−2 s−1 in high light (HL+) for 2 h. The cDNA was used for real time PCR quantitation of transcript abundance, by comparison with cloned standards. A, photosynthetic Fd proteins. B, other Fd proteins. Values are mean ± S.D. of at least three independent plants. Values significantly different from the wild type are indicated by * and # for p < 0.05 and p < 0.1, respectively.
Interestingly, whereas no differences in Fd1 or Fd2 transcripts are found on high light treatment of WT plants, when fd2 plants were high light treated, the transcripts of Fd1 significantly increased (Fig. 2A). This indicates that, whereas pleiotropic mechanisms minimize acceptor limitation at PSI under growth light conditions, an increase in photosynthetic radiation can exceed this capacity, and the plant is capable of responding by increasing Fd1 expression in fd2 plants. Of the genes for other Fd proteins investigated, Fd3, FdC1, and FdC2 all showed significantly increased transcript levels in the fd2 plants. Fd3 is a specific root-type, or heterotrophic, Fd and its up-regulation is consistent with increased non-photosynthetic metabolism in fd2 plants, which has been discussed previously (14). Transcript differences were especially noticeable for FdC1, with transcripts consistently two times higher than WT levels following high light treatment. Transcript levels of FdC1 were also examined in the previously described Fd2 RNAi lines (36), and also found to be elevated (data not shown). We therefore undertook further investigation of this protein, to examine whether it could be an alternative electron acceptor at PSI, or otherwise involved in the response to acceptor limitation at PSI.
FdC1 Is Highly Conserved Among Higher Plants, and Cyanobacteria Contain Homologous Proteins
FdC1 is similar enough to well known plant-type Fds to suggest that it also possesses a [2Fe-2S] cluster (supplemental Fig. S1A). However, it is more homologous to a set of uncharacterized cyanobacterial Fd proteins than to previously studied photosynthetic or root-type Fds. This phylogenetic relationship is shown by the tree in supplemental Fig. S1B, and residues conserved among these proteins are given as black on gray in the alignment in supplemental Fig. 1A. The closely related higher plant and cyanobacterial proteins also have C-terminal extensions, and so these are also categorized here as FdC1 proteins.
These data suggest that Arabidopsis FdC1 is derived from cyanobacterial FdC1 genes acquired by the plant during the endosymbiotic origin of the chloroplast, rather than originating from gene duplication of photosynthetic or root-type Fds following endosymbiosis. FdC1 may have a conserved function between some cyanobacteria and higher plants, although it appears to have been lost from some cyanobacterial genomes.
FdC1 Is a Chloroplast Protein and Is More Abundant in fd2 Plants
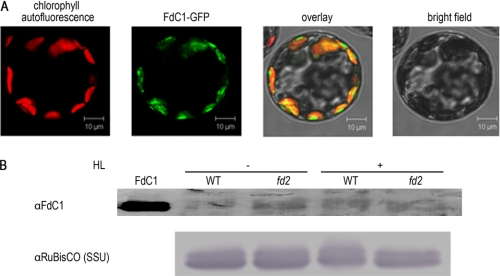
Data base information indicates that, in Arabidopsis, FdC1 is expressed predominantly in photosynthetic tissue, at a lower transcript level than photosynthetic ferredoxins (37). Homology to cyanobacterial proteins and expression in photosynthetic tissues suggests that FdC1 may be a chloroplast protein, and in comparison to cyanobacterial sequences, the higher plant FdC1s do contain N-terminal sequences that are strongly predicted to act as transit peptides by bioinformatic methods (WolfPSORT (38) and TargetP (39) predictions of 13.0 and 0.888 for the chloroplast, respectively). In addition, peptides derived from FdC1 have been identified in proteomics studies of the chloroplast FdC1 (40). To confirm that FdC1 is a chloroplast-targeted protein we constructed FdC1-GFP fusion protein vectors, and used these to transform Arabidopsis protoplasts. As can be seen from Fig. 3A, the fusion protein is clearly co-localized with chlorophyll autofluorescence when examined under the microscope, providing strong evidence that FdC1 does indeed have a chloroplast location.
FIGURE 3.
The FdC1 protein is imported into A. thaliana chloroplasts and has increased abundance on PSI acceptor limitation. A, import of FdC1 pre-protein into chloroplasts. Isolated protoplasts were transformed and after an overnight incubation in darkness, GFP expression and localization were visualized by fluorescence microscopy. Chlorophyll autofluorescence, fluorescence of GFP-FdC1 proteins, merged image of chlorophyll, and GFP fluorescence and a bright field image of the same (typical) protoplast are shown. B, increased FdC1 abundance in conditions of PSI acceptor limitation. Western blots were performed with protein extracts from WT and fd2 plants, either treated for 6 h with high light (HL+) or left in growth light conditions (HL−). Lanes were loaded with 25 μg of protein, and detection was performed using antisera raised against FdC1 (then purified against the recombinant protein). Identical blots were challenged with antisera raised against the small subunit of Rubisco (SSU) as a loading control. 10 ng of recombinant FdC1 was loaded as a control. Blots are typical of two independent experiments.
FdC1 is a good candidate as an alternative electron acceptor at PSI, due to the increased transcript in fd2 plants, and chloroplast targeting of the pre-peptide. However, it is well documented that Fd2 translation is strongly regulated (41) (referred to as FdA in the paper), and so we compared abundance of the FdC1 protein in WT and fd2 plants. We expressed the mature FdC1 protein in E. coli, purified it, and raised an antibody. Using a purified version of this antibody, we were able to specifically detect the FdC1 protein in WT and fd2 plants (Fig. 3B). The protein is present at very low abundance (around 4 pg−1 μg of protein), but is clearly increased in the leaves of fd2 plants. Interestingly, after high light treatment, WT plants also show increased FdC1 contents. This may be an indication of some translational regulation, because transcript abundance does not change significantly (Fig. 2).
FdC1 Contains a [2Fe-2S] Cluster
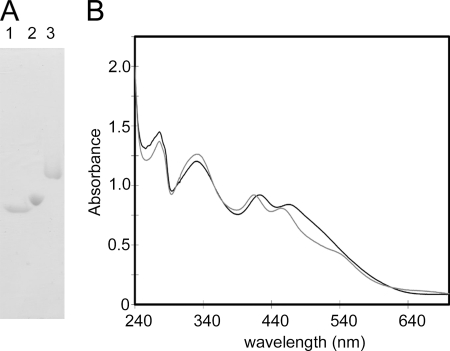
The purified FdC1 protein has absorbance peaks at around 420 and 460 nm, characteristic of the spectrum of a [2Fe-2S] cluster-containing protein (Fig. 4). However, these peaks show a significant shift to the blue when compared with many previously studied Fds, such as Fd2. This indicates FdC1 may have some differences from previously studied [2Fe-2S] Fds in the protein microenvironment around the cluster and its ligands. In addition, the purified protein shows considerably slower migration through a native PAGE gel than the photosynthetic (Fd2) and root-type (Fd3) proteins (Fig. 4A), indicating that the surface of FdC1 is less negatively charged.
FIGURE 4.
Purified, recombinant FdC1 contains a [2Fe-2S] cluster. A, native PAGE gel showing the separation of recombinant purified Arabidopsis Fd2, Fd3, and FdC1 proteins. Lanes contain 20 μg of purified protein. B, absorbance spectra of 90 μm purified recombinant Arabidopsis Fd2 (black line) and FdC1 (gray line) based on a ferredoxin ϵ420 of 10 mm−1 cm−1.
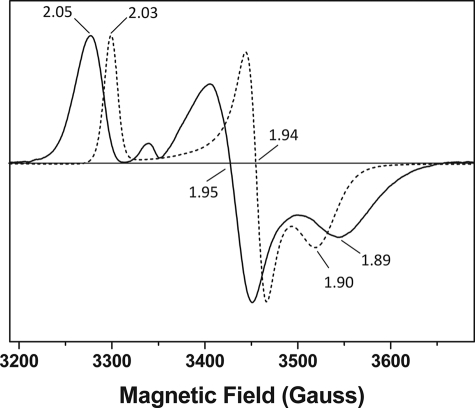
Differences between the visible spectra of Fd2 and FdC1 encouraged us to further investigate the properties of the FdC1 Fe-S cluster. Purified proteins were used for measurements of electron paramagnetic resonance (EPR), shown in Fig. 5. EPR of dithionite-reduced Fd2 and FdC1 resulted in rhombic “ferredoxin-like” spectra with g values of gz = 2.05, gy = 1.95, gx = 1.89, and gz = 2.03, gy = 1.94, gx = 1.90, respectively. No other radical signals were detected from either protein between g = 2 and g = 12 and the signal persisted even at 70 K (data not shown), indicating that only a [2Fe-2S] cluster was present in both Fd2 and FdC1. This confirmed that both purified Fd2 and FdC1 possess intact 2Fe-2S clusters, and no other Fe-S cluster type is present. Fd2 displays g values typical for those of plant-like photosynthetic Fds, and similar to that of Spinacia oleracea (spinach) Fd1 (g values at 2.040, 1.950, and 1.882) (42) and Pisum sativum (pea) Fd (g values at 2.03, 1.96, and 1.90) (43). The FdC1 [2Fe-2S] cluster elicits signals with a narrower line width and smaller anisotropy than Fd2, which are more similar to those observed for the S-1 [2Fe-2S] cluster in the Bacillus subtilis succinate dehydrogenase (g values at 2.035, 1.940, and 1.890) (44). The differences observed between the EPR spectra for A. thaliana Fd2 and FdC1 are indicative of alterations in the environment of the cluster (e.g. Fe-S bond distortions due to proximal amino acid side chains) or to charged side chains surrounding the Fe-S clusters.
FIGURE 5.
EPR spectroscopy demonstrates distinctive [2Fe-2S] cluster organization in Fd2 and FdC1. Electron paramagnetic resonance (EPR) spectra of dithionite-reduced Fd2 (solid line) and FdC1 (dashed line) showing 2Fe-2S g values of gz = 2.05, gy = 1.95, gx = 1.89, and gz = 2.03, gy = 1.94, gx = 1.90, respectively. No significant radical signals were observed for the oxidized proteins (not shown). The samples were run under identical conditions (as described under “Experimental Procedures”) at 15 K.
FdC1 Receives Electrons from PSI
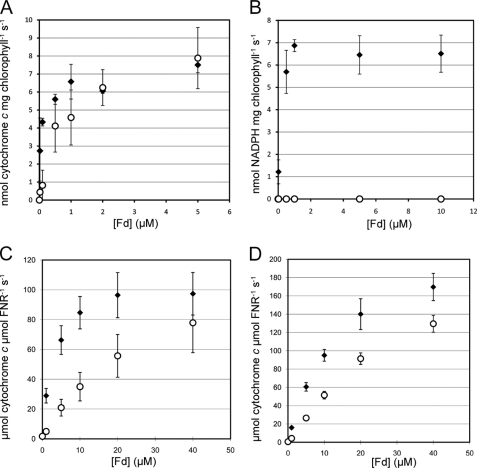
Differences in the redox center of FdC1 might confer electron transfer properties different from those previously measured for Fds. The purified FdC1 protein was therefore compared with Fd2, for its ability to act in the photosynthetic electron transport chain. Fig. 6A shows that FdC1 is capable of receiving electrons from PSI, although it has a >10-fold lower affinity (from Km values) for PSI than does Fd2 (Table 1). By contrast, FdC1 is incapable of supporting NADP+ photoreduction, being unable to transfer these PSI-derived electrons to the endogenous FNR on the thylakoid membranes (Fig. 6B).
FIGURE 6.
Electron transfer capabilities of FdC1. A comparison was made between the electron transfer abilities of Arabidopsis Fd2 (closed diamonds) and FdC1 (open circles). A, electron transfer from PSI to Fd. Electron donation was measured by illuminating spinach thylakoid membranes in the presence of the indicated Fd concentrations and cytochrome c. B, electron transfer from PSI to NADP+. Photoreduction of NADP+ was followed on illumination of spinach thylakoid membranes (with endogenous FNR) in the presence of the indicated Fd concentrations. C, electron transfer from leaf-type FNR to Fd. FNR was reduced by a glucose-6-phosphate dehydrogenase-based NADP+ re-reduction system, and electron transfer was measured at the indicated Fd concentrations. D, as for C, but with root-type FNR. Values are mean ± S.D. of at least 3 independent measurements; Km and Vmax (kcat where appropriate) values for these data are given in Table 1.
TABLE 1.
Kinetic parameters of Fd2 and FdC1 in electron transfer reactions
| Fd type | PSI to Fd |
PSI to NADP+ |
Leaf FNR to Fd |
Root FNR to Fd |
||||
|---|---|---|---|---|---|---|---|---|
| Kma | Vmax | Km | Vmax | Km | Vmax (kcat) | Km | Vmax (kcat) | |
| μm | μmol mg chlorophyll−1h−1 | μm | μmol mg chlorophyll−1h−1 | μm | μmol μmol FNR−1s−1 | μm | μmol μmol FNR−1s−1 | |
| AtFd2 | 0.06 ± 0.03 | 24.9 ± 0.5 | 0.10 ± 0.03 | 24.6 ± 0.1 | 2.8 ± 0.0 | 107 ± 16 | 13.8 ± 1.2 | 231 ± 25 |
| AtFdC1 | 0.82 ± 0.29 | 34.5 ± 3.8 | NDb | ND | 27.4 ± 1.9 | 130 ± 32 | 39.9 ± 3.2 | 261 ± 18 |
a Km and Vmax parameters were calculated from data sets used to generate the mean ± S.D. shown in Fig. 7.
b ND indicates that no activity could be detected in NADP+ photoreduction assays.
Maximum NADP+ photoreduction rates for Fd2 are lower than some published values (45, 46), but within the range of others (47). We confirmed photosynthetic competence of thylakoids by chlorophyll fluorescence (data not shown), and conclude that this variation may be due to variable leaf material and methods of thylakoid preparation.
A More Positive Redox Potential Prevents FdC1 from Supporting NADP+ Photoreduction
The absence of NADP+ photoreduction activity could either be due to the inability of FdC1 to interact with FNR, or to a difference in their relative redox potentials, which makes electron transfer thermodynamically unfavorable. To test this, we measured electron transfer between proteins in the opposite direction, i.e. from NADPH-reduced FNR to Fd (Fig. 6, C and D). Although the apparent affinity of leaf type FNR for FdC1 is 10-fold lower than that for Fd2, maximum rates of activity are equivalent within error. The apparent affinity of root-type FNR for FdC1 is only 3-fold less than for Fd2, demonstrating that FdC1 is capable of interacting with both kinds of FNR and receiving electrons from them. This suggests that the inability of FdC1 to mediate NADP+ photoreduction is due to an unfavorable (more positive) redox potential of the ferredoxin, rather than an inability to interact with FNR or PSI.
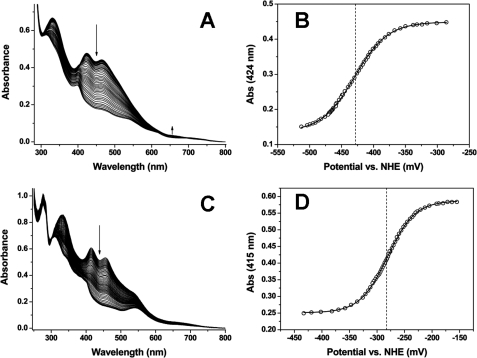
To confirm this hypothesis we compared the redox potentials of recombinant Fd2 and FdC1 proteins by redox potentiometry (Fig. 7). The major near UV-visible spectral features for the oxidized form of the FdC1 protein are at 333, 415, and 455 nm, and all are diminished in intensity on reduction of the iron-sulfur cluster (Fig. 7C). The reduced FdC1 has peaks at ∼309 and 540 nm, with a pronounced shoulder at ∼400 nm. The FdC1 absorption change at 415 nm was plotted against the applied potential and the data fitted using the Nernst equation to give a midpoint reduction potential (against the normal hydrogen electrode, NHE) of E0′ = −281 ± 3 mV (Fig. 7D). An identical value was obtained from fitting the data plotted at 455 nm. The comparable absorption features for the oxidized Fd2 are at 331, 424, and 465 nm, and again all these are decreased in intensity on iron-sulfur cluster reduction (Fig. 7A). In the region around 650 nm the reduced Fd2 has increased absorbance by comparison to that of the oxidized form (peak at ∼655 nm). Fitting of absorbance versus applied potential data for AtFd2 at 424 nm gives a value of E0′ = −429 ± 3 mV (Fig. 7B), and a data fit at 465 nm gives a value identical within error (E0′ = −428 ± 5 mV). Taken together, the electron transfer data and redox potentials suggest that FdC1 is unable to support NADP+ photoreduction because of its positive potential, making electron transfer to FNR energetically unfavorable.
FIGURE 7.
FdC1 has a more positive redox potential than photosynthetic [2Fe-2S] Fds. A, spectra accumulated during redox titration of Fd2 (∼115 μm). B, plot of Fd2 absorption data at 424 nm versus the applied potential, with the data fitted using the Nernst function. This generated a midpoint reduction potential of E0′ = −429 ± 3 mV. Arrows indicate the direction of spectral absorption change on [2Fe-2S] cluster reduction. C, spectra accumulated during redox titration of FdC1 (∼85 μm). D, plot of FdC1 absorption data at 415 nm versus the applied potential (relative to NHE), with the data fitted using the Nernst function, producing a midpoint reduction potential of E0′ = −281 ± 3 mV.
DISCUSSION
FdC1 Is a Novel Electron Acceptor at PSI
We have identified a novel Fd protein, which has a C-terminal extended by 8 amino acids in comparison to previously studied plant-type [2Fe-2S] Fd proteins from Arabidopsis, and named it FdC1. The protein has homologues among cyanobacteria (supplemental Fig. S1B), is strongly predicted to have a chloroplast transit peptide, and when expressed in protoplasts as a GFP fusion protein localizes to the chloroplast (Fig. 3). Moreover, previous proteomics studies to identify chloroplast protein contents have detected peptides from FdC1 (40). Curiously, a fragment of the putative transit peptide was also detected when the mass spectrometry data were analyzed with low stringency (40). Although the confidence levels for the presence of this part of the peptide are low, future work will have to establish that the transit peptide is truly cleaved on entry to the chloroplast.
The high conservation of FdC1 proteins among higher plants, and the presence of similar proteins in algae and cyanobacteria, suggests that FdC1 might have a conserved, unique function. The EPR spectra in Fig. 5 indicate that the A. thaliana FdC1 protein (and likely its homologues) contains a [2Fe-2S] cofactor, but that this cluster has an altered ligand environment compared with Fd2 and related plant-type photosynthetic Fds. The C-terminal extension is not wholly conserved through cyanobacteria to higher plants, but is almost identical among terrestrial plant sequences. The C-terminal of Fd has been identified as critical for efficient interaction with PSI (16), and such a dramatic difference from photosynthetic Fds would be expected to impact on the ability of the protein to accept electrons from PSI. Despite this, the Arabidopsis FdC1 was capable of accepting electrons from PSI (Fig. 6A) and we conclude that, in Arabidopsis, FdC1 is a possible alternative electron acceptor to the well described photosynthetic Fds, Fd1 and Fd2, at PSI.
FdC1 Transcript and Protein Levels Are Related to PSI Acceptor Availability
Mutant and RNAi plants lacking Fd2 have drastically lower Fd contents, with a PSI acceptor molecule content between 1 and 10% of that seen in wild type plants (7, 14). In palisade tissue of spinach leaves, the stoichiometry of leaf Fd to PSI complex units is estimated at around 2–3:1 (48) and in potato, antisense-induced decrease of Fd to less than 40% of wild type levels is lethal (49), suggesting that Fd contents cannot viably decrease below a 1:1 ratio with PSI. However, Fd2 knockout and RNAi plants show much lower Fd contents than this, and it has been shown that in some cyanobacteria Fd protein levels are rapidly decreased on conditions of iron depletion, to levels well below this ratio (50). It is possible that the antisense approach in potato was less discriminating than that in Arabidopsis, and that in the potato experiments transcripts of multiple Fd isoforms, both Fd1, Fd2, and possibly even a FdC1 homologue, are also decreased. These data suggest that plants rely on a spectrum of acceptors at PSI, and a massive decrease in the primary photosynthetic acceptor molecule (in the case of Arabidopsis Fd2) is not as serious a defect as disruption of additional, alternative acceptors. This would explain the somewhat surprising ability of Arabidopsis Fd2 knockout plants to survive and grow photoautotrophically.
Absence of the principal acceptor does, however, lead to severe acceptor limitation at PSI in plants lacking Fd2 (Fig. 1). Under these conditions it might be expected that alternative electron acceptors are increased to compensate for the lack of Fd2, but transcript levels of the highly homologous Fd1 protein only increase significantly when plants are subjected to high light. Rather, a dramatic change is seen in increased FdC1 transcript levels (Fig. 2). This response is exaggerated in high light conditions where PSI acceptor limitation is exacerbated. It is well known that the redox state of the photosynthetic electron transport chain exerts control over the nuclear gene expression of proteins involved in PET (51) and it seems likely that this is the cause of up-regulated FdC1 expression in fd2 plants. Indeed, transcript levels of FdC1 have also been described as altered in other plants with disturbed PET. Mutants of the γ subunit of the ATPase lack a functionally assembled ATPase complex, and cannot dissipate the proton gradient formed in PET (52). Transcript analysis showed FdC1 transcripts (referred to as Fd5 in the paper) were decreased by 60% compared with the wild type.
It is interesting to speculate on what might mediate the signal from the chloroplast that regulates FdC1 expression. The best documented signal for regulating PET gene expression originates in the redox state of the plastoquinone (PQ) pool (53). However, despite having opposite expression changes in FdC1, fd2 and ATPase mutants both show extremely low photochemical quenching (qP) (7, 14, 52) indicating highly reduced PQ. This demonstrates that any signal influencing FdC1 expression probably does not originate in the PQ pool. Alternative signals, from PSI, have also been reported to influence expression of nuclear genes (54–57). It is highly unlikely that the ATPase mutant shows conditions of acceptor limitation at PSI such as that seen in fd2. It therefore seems probable that differential signals originating at PSI result in the opposite FdC1 expression patterns seen in fd2 and ATPase mutants. This is also consistent with an FdC1 expression response to acceptor limitation at PSI.
Possible Physiological Roles of FdC1
FdC1 can accept electrons from PSI, but has a much lower affinity for this reaction than does Fd2 (Fig. 6A). It has previously been reported that there is a hierarchy of electron acceptors at PSI, with donation to Fd taking precedence, and, when this capacity is saturated, donation to O2 occurs (58). Based on the results presented here, it now seems likely that FdC1 also has a place in this hierarchy, with its lower affinity (Table 1) ensuring that it never competes strongly with Fd1 or Fd2 for electrons from excited PSI. Under conditions where Fd1 and Fd2 are already reduced, such as decreased availability of stromal acceptors, or very high activity of electron donation from PSI in high light, FdC1 could then bind to PSI and channel excess electrons away from NADP+ photoreduction. Other Fd proteins incapable of driving NADP+ photoreduction have been reported in Arabidopsis (9) and Chlamydomonas (59), and it is possible that these have related functions. It is not clear whether FdC1 could donate electrons to other proteins involved in dissipating excess electron pressure, such as the as yet unidentified Fd:PQ reductase that catalyzes cyclic electron flow. Alternatively FdC1 may facilitate rapid electron transfer from PSI to O2 in the water-water cycle, or play a role in Fe-S cluster repair and assembly in photosystems under severe reduction pressure. An RNAi knockdown approach is currently being pursued in further investigations on the precise physiological role played by FdC1.
Supplementary Material
Acknowledgment
We gratefully acknowledge the excellent technical assistance of Nicolas Koenig.
This work was supported by Deutsche Forschungsgemeinschaft Grant HA5921/1-1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- Fd
- ferrodoxin
- PET
- photosynthetic electron transfer
- CET
- cyclic electron flow
- PSI
- photosystem I
- Ribisco
- ribulose-bisphosphate carboxylase/oxygenase
- PQ
- plastoquinone
- PSII
- photosystem II
- FNR
- Fd:NADP(H) reductase
- FdC
- Fd with extended C terminal.
REFERENCES
- 1. Arnon D. I., Tagawa K., Tsujimoto H. Y. (1963) Science 140, 378 [DOI] [PubMed] [Google Scholar]
- 2. Joliot P., Joliot A. (2006) Biochim. Biophys. Acta 1757, 362–368 [DOI] [PubMed] [Google Scholar]
- 3. Shin M., Arnon D. I. (1965) J. Biol. Chem. 240, 1405–1411 [PubMed] [Google Scholar]
- 4. Knaff D. B. (1996) in Advances in Photosynthesis. Oxygenic Photosynthesis: The Light Reactions (Ort D. R., Yocum C. F. eds) pp. 333–361, Kluwer Academic Publishers, Dordrecht [Google Scholar]
- 5. Montrichard F., Alkhalfioui F., Yano H., Vensel W. H., Hurkman W. J., Buchanan B. B. (2009) J. Proteomics 72, 452–474 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki A., Oaks A., Jacquot J. P., Vidal J., Gadal P. (1985) Plant Physiol. 78, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanke G. T., Hase T. (2008) Photochem. Photobiol. 84, 1302–1309 [DOI] [PubMed] [Google Scholar]
- 8. Hase T., Kimata Y., Yonekura K., Matsumura T., Sakakibara H. (1991) Plant Physiol. 96, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanke G. T., Kimata-Ariga Y., Taniguchi I., Hase T. (2004) Plant Physiol. 134, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Onda Y., Matsumura T., Kimata-Ariga Y., Sakakibara H., Sugiyama T., Hase T. (2000) Plant Physiol. 123, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimata-Ariga Y., Matsumura T., Kada S., Fujimoto H., Fujita Y., Endo T., Mano J., Sato F., Hase T. (2000) EMBO J. 19, 5041–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimata Y., Hase T. (1989) Plant Physiol. 89, 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsumura T., Kimata-Ariga Y., Sakakibara H., Sugiyama T., Murata H., Takao T., Shimonishi Y., Hase T. (1999) Plant Physiol. 119, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voss I., Koelmann M., Wojtera J., Holtgrefe S., Kitzmann C., Backhausen J. E., Scheibe R. (2008) Physiol. Plant 133, 584–598 [DOI] [PubMed] [Google Scholar]
- 15. Gou P., Hanke G. T., Kimata-Ariga Y., Standley D. M., Kubo A., Taniguchi I., Nakamura H., Hase T. (2006) Biochemistry 45, 14389–14396 [DOI] [PubMed] [Google Scholar]
- 16. Lelong C., Sétif P., Lagoutte B., Bottin H. (1994) J. Biol. Chem. 269, 10034–10039 [PubMed] [Google Scholar]
- 17. Asada K. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Camp W., Willekens H., Bowler C., Van Montagu M., Inzé D., Reupold-Popp P., Sandermann H., Jr., Langebartels C. (1994) Nat. Biotechnol. 12, 165–168 [Google Scholar]
- 19. Nakano Y., Asada K. (1981) Plant Cell Physiol. 22, 867–880 [Google Scholar]
- 20. Miyake C., Asada K. (1994) Plant Cell Physiol. 35, 539–549 [Google Scholar]
- 21. Snyrychová I., Pospísil P., Naus J. (2006) Photochem. Photobiol. Sci. 5, 472–476 [DOI] [PubMed] [Google Scholar]
- 22. Kanervo E., Suorsa M., Aro E. M. (2005) Photochem. Photobiol. Sci. 4, 1072–1080 [DOI] [PubMed] [Google Scholar]
- 23. Demmig B., Winter K., Krüger A., Czygan F. C. (1987) Plant Physiol. 84, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T. (2004) Nature 429, 579–582 [DOI] [PubMed] [Google Scholar]
- 25. Becker B., Holtgrefe S., Jung S., Wunrau C., Kandlbinder A., Baier M., Dietz K. J., Backhausen J. E., Scheibe R. (2006) Planta 224, 380–393 [DOI] [PubMed] [Google Scholar]
- 26. Genty B., Briantais J. M., Baker N. R. (1989) Biochim. Biophys. Acta 990, 87–92 [Google Scholar]
- 27. van Kooten O., Snel J. F. (1990) Photosynth. Res. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 28. Klughammer C., Schreiber U. (1994) Planta 192, 261–268 [Google Scholar]
- 29. Backhausen J. E., Emmerlich A., Holtgrefe S., Horton P., Nast G., Rogers J. J. M., Müller-Röber B., Scheibe R. (1998) Planta 207, 105–114 [Google Scholar]
- 30. Seidel T., Kluge C., Hanitzsch M., Ross J., Sauer M., Dietz K. J., Golldack D. (2004) J. Biotechnol. 112, 165–175 [DOI] [PubMed] [Google Scholar]
- 31. Kost B., Spielhofer P., Chua N. H. (1998) Plant J. 16, 393–401 [DOI] [PubMed] [Google Scholar]
- 32. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Habour Laboratory Press, New York [Google Scholar]
- 33. Dunford A. J., Rigby S. E., Hay S., Munro A. W., Scrutton N. S. (2007) Biochemistry 46, 5018–5029 [DOI] [PubMed] [Google Scholar]
- 34. Munro A. W., Noble M. A., Robledo L., Daff S. N., Chapman S. K. (2001) Biochemistry 40, 1956–1963 [DOI] [PubMed] [Google Scholar]
- 35. Dutton P. L. (1978) Methods Enzymol. 54, 411–435 [DOI] [PubMed] [Google Scholar]
- 36. Hanke G. T., Endo T., Satoh F., Hase T. (2008) Plant Cell Environ. 31, 1017–1028 [DOI] [PubMed] [Google Scholar]
- 37. Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004) Plant Physiol. 136, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) Nucleic Acids Res. 35, W585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 40. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) PLoS One 3, e1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vorst O., van Dam F., Weisbeek P., Smeekens S. (1993) Plant J. 3, 793–803 [DOI] [PubMed] [Google Scholar]
- 42. Aliverti A., Hagen W. R., Zanetti G. (1995) FEBS Lett. 368, 220–224 [DOI] [PubMed] [Google Scholar]
- 43. Vidakovic M., Fraczkiewicz G., Dave B. C., Czernuszewicz R. S., Germanas J. P. (1995) Biochemistry 34, 13906–13913 [DOI] [PubMed] [Google Scholar]
- 44. Hederstedt L., Maguire J. J., Waring A. J., Ohnishi T. (1985) J. Biol. Chem. 260, 5554–5562 [PubMed] [Google Scholar]
- 45. McSwain B. D., Arnon D. I. (1968) Proc. Natl. Acad. Sci. U.S.A. 61, 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hosler J. P., Yocum C. F. (1985) Biochim. Biophys. Acta 808, 21–31 [Google Scholar]
- 47. Schmitz S., Böhme H. (1995) Biochim. Biophys. Acta 1231, 335–341 [Google Scholar]
- 48. Terashima I., Yorinao I. (1985) Plant Cell Physiol. 26, 63–75 [Google Scholar]
- 49. Holtgrefe S., Bader K. P., Horton P., Scheibe R., von Schaewen A., Backhausen J. E. (2003) Plant Physiol. 133, 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pardo M. B., Gómez-Moreno C., Peleato M. L. (1990) Arch. Microbiol. 153, 528–530 [Google Scholar]
- 51. Fey V., Wagner R., Braütigam K., Wirtz M., Hell R., Dietzmann A., Leister D., Oelmüller R., Pfannschmidt T. (2005) J. Biol. Chem. 280, 5318–5328 [DOI] [PubMed] [Google Scholar]
- 52. Bosco C. D., Lezhneva L., Biehl A., Leister D., Strotmann H., Wanner G., Meurer J. (2004) J. Biol. Chem. 279, 1060–1069 [DOI] [PubMed] [Google Scholar]
- 53. Escoubas J. M., Lomas M., LaRoche J., Falkowski P. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Piippo M., Allahverdiyeva Y., Paakkarinen V., Suoranta U. M., Battchikova N., Aro E. M. (2006) Physiol. Genomics 25, 142–152 [DOI] [PubMed] [Google Scholar]
- 55. Hihara Y., Sonoike K., Kanehisa M., Ikeuchi M. (2003) J. Bacteriol. 185, 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pfannschmidt T., Schütze K., Brost M., Oelmüller R. (2001) J. Biol. Chem. 276, 36125–36130 [DOI] [PubMed] [Google Scholar]
- 57. Nakamura K., Hihara Y. (2006) J. Biol. Chem. 281, 36758–36766 [DOI] [PubMed] [Google Scholar]
- 58. Backhausen J. E., Kitzmann C., Horton P., Scheibe R. (2000) Photosynth. Res. 64, 1–13 [DOI] [PubMed] [Google Scholar]
- 59. Jacobs J., Pudollek S., Hemschemeier A., Happe T. (2009) FEBS Lett. 583, 325–329 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.