Abstract
The eight mammalian Cbx proteins are chromodomain-containing proteins involved in regulation of heterochromatin, gene expression, and developmental programs. They are evolutionarily related to the Drosophila HP1 (dHP1) and Pc (dPc) proteins that are key components of chromatin-associated complexes capable of recognizing repressive marks such as trimethylated Lys-9 and Lys-27, respectively, on histone H3. However, the binding specificity and function of the human homologs, Cbx1–8, remain unclear. To this end we employed structural, biophysical, and mutagenic approaches to characterize the molecular determinants of sequence contextual methyllysine binding to human Cbx1–8 proteins. Although all three human HP1 homologs (Cbx1, -3, -5) replicate the structural and binding features of their dHP counterparts, the five Pc homologs (Cbx2, -4, -6, -7, -8) bind with lower affinity to H3K9me3 or H3K27me3 peptides and are unable to distinguish between these two marks. Additionally, peptide permutation arrays revealed a greater sequence tolerance within the Pc family and suggest alternative nonhistone sequences as potential binding targets for this class of chromodomains. Our structures explain the divergence of peptide binding selectivity in the Pc subfamily and highlight previously unrecognized features of the chromodomain that influence binding and specificity.
Keywords: Chromatin Histone Modification, Chromatin Regulation, Histone Methylation, NMR, X-ray Crystallography, Cbx, Chromodomain
Introduction
The heterochromatin protein HP1 and Polycomb proteins are two distinct regulators of chromatin structure in Drosophila melanogaster involved in epigenetic repression of gene expression. The chromodomains of dHP13 and dPc direct the localization of their respective complexes via specific recognition of distinct but sequence-related repressive marks on histone H3 trimethylated at Lys-9 (H3K9me3) and Lys-27 (H3K27me3), respectively (1, 2). Both of these marks reside within the identical sequence motif, ARKS. These properties in the fly have served as an important paradigm for understanding chromatin dynamics and gene regulation through development (3, 4) as well as fundamental structure-function and specificity properties of the chromodomains in general (1, 2, 5–7).
In mammals the dPc and dHP1 homologs have expanded to five (Cbx2, -4, -6, -7, -8) and three (Cbx1, -3, -5) proteins, respectively. Each human/mouse Cbx contains one chromodomain with ∼55–65% sequence identity to dHP1 or dPc (8). A characteristic feature of the chromodomain is the positioning of the mono-, di-, and trimethylammonium moiety within a pocket lined with aromatic residues, often supplemented by one or more acidic side chains (9). Despite the similarity in overall structure, the peptide binding grooves of the dHP1 and dPc chromodomains show distinct features. Each protein reveals clear discrimination for its cognate site via recognition of different residues upstream of each ARKS motif in H3 that are complementary to differences in the binding grooves of the chromodomains (1).
Although the exact cellular role of each Cbx protein has not been defined, many of the Cbx proteins play important roles in human development and disease. Cbx1, -3, and -5 are important for formation of, and gene repression in heterochromatin. Cbx2, -4, -6, -7, and -8 are components of Polycomb repressive complex 1 (PRC1), a key regulator of developmental genes. Mutation of Cbx2 was shown to affect human sexual development (10). Finally, Cbx7 has been implicated in the development of leukemia (11), and Cbx7 and -8 contribute to repression of the INK4A locus (12–14).
The chromodomains of Cbx proteins are thought to, at least in part, localize the proteins and their respective complexes to appropriately marked sites of the epigenome via recognition of histone H3 trimethylated at either Lys-9 or Lys-27. A number of studies have been conducted to decipher the specificity within the Cbx family of proteins. Bernstein et al. (15) have investigated five mouse dPc homologs (Cbx2, -4, -6, -7, -8) and showed that despite a high degree of conservation, the chromodomains display significant differences in histone peptide binding preferences. Not all dPc homologs bind preferentially to H3K27me3 as might be expected based on dPc specificity (15). Cbx2 and Cbx7 recognized both H3K9me3 and H3K27me3, whereas Cbx4 preferred H3K9me3. Recently, Vincenz and Kerppola (16) studied the Pc family of Cbx proteins in ES cells and fibroblasts and observed that the ability of various Cbx proteins to bind chromatin was mediated by non-conserved regions of the protein. Using bimolecular fluorescence complementation experiments, they demonstrated that neither the chromodomains nor H3K27me3 was required for targeting of Cbx proteins to chromatin in vivo. More recently, Yap et al. (17) showed that Cbx7 employs overlapping yet distinct regions within its chromodomain for binding H3K27me3 and RNA. Collectively, these studies show that related chromodomain binding motifs have differences in binding characteristics and highlight the need to carefully analyze the differences between mammalian and fly chromodomain-containing proteins as well as differences within mammalian chromodomain subfamilies.
In the present study we sought to understand in detail the biochemical and structural basis for histone mark recognition and discrimination among the human Cbx chromodomain protein family. We measured the binding affinities of all human Cbx chromodomains for H3K9me3 and H3K27me3 and determined the three-dimensional structures (by NMR or x-ray crystallography) of several chromodomains in complex with the modified histone peptides to elucidate the structural determinants for the specificity. The human HP1 homologs Cbx1, -3, and -5 preferentially recognize H3K9me3 in a manner similar to their dHP1 counterpart with dissociation constants in the low micromolar range. On the other hand, chromodomains from the human Pc homologs all have lower affinities for H3K9me3 and H3K27me3 peptides, do not distinguish well between these two marks, and are less sensitive to mutations within the peptide sequence. Our structural data show that the primary origin of the difference between human HP1 and Pc chromodomains lies in their respective electrostatic surfaces. Cbx1, -3, and -5 all have a large electronegative peptide binding surface that complements the basic histone peptides, whereas Cbx2, -4, -6, -7, and -8 have a much more hydrophobic surface. Further peptide array and binding studies suggest that at least some of the human Pc chromodomains bind nonhistone sequences in a methylation-dependent manner. These data suggest that Pc-class Cbx proteins may use their chromodomains to couple PRC1 complexes with novel binding partners or chromatin sites in the mammalian genome.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Chromodomains of Cbx1-(20–73), Cbx2-(9–66), Cbx3-(29–81), Cbx4-(8–65), Cbx5-(18–75), Cbx6-(8–65), Cbx7-(8–62), and Cbx8-(8–61) were overexpressed as N-terminal His6-tagged proteins at 15 °C using Escherichia coli BL21 (DE3) Codon plus RIL (Stratagene) as a host organism. For purification, cells were lysed by passing through Microfluidizer (Microfluidics Corp.) at 18,000 p.s.i. The purification procedure comprises of two chromatographic steps, an affinity chromatography on a nickel-nitrilotriacetic acid chelating column (Qiagen) and a gel filtration Superdex 75 column (26/60, GE Healthcare). The His6 tag was cleaved with tobacco etch virus protease (except for Cbx6, which has a thrombin cutting site) after the affinity column step while dialyzing against 20 mm Tris, pH 8.0, 0.5 m NaCl, and 1 mm Tris(2-carboxyethyl)phosphine.
Protein for NMR samples was prepared as follows. The bacteria were grown in M9-defined medium supplemented with [15N]ammonium chloride (0.8 g/liter) and d-[13C6]glucose (0.4 g/liter) for 13C,15N-labeled samples at room temperature. These highly expressed proteins were purified by Talon (BD Biosciences) affinity chromatography under native conditions and eluted with buffer containing 500 mm Imidazole. The proteins were treated with tobacco etch virus protease and further purified by size exclusion chromatography using a HiLoad 26/60 Superdex-75 column (GE Healthcare). The proteins were monomeric in solution as determined by size exclusion chromatography. The final NMR samples were prepared in buffer containing 20 mm sodium phosphate, pH 7.4, 200 mm NaCl, 2 mm DTT, 1 mm Tris(2-carboxyethyl)phosphine, 1 mm benzamidine, 0.5 mm PMSF for Cbx3 complexes and 10 mm sodium phosphate, pH 7.4, 300 mm NaCl, 0.5 mm Tris(2-carboxyethyl)phosphine, 1 mm benzamidine, and 0.5 mm PMSF for Cbx7 complexes. Protein-peptide complexes were prepared for NMR by titrating aliquots of unlabeled peptide into the labeled chromodomains of Cbx3 and Cbx7 in molar ratios 1:1, 1:3 (for Cbx3/K9me3), and 1:5 (for Cbx7/K27me3 and Cbx7/K9me3) until no further changes in chemical shifts were detected in the 1H,15N heteronuclear single quantum correlation spectrum.
Fluorescence Polarization
H3K9Me3 (ARTKQTARK(me3)STGGKA), H3K9Me3T6A (ARTKQAARK(me3)STGGKA), H3K27Me3 (QLATKAARK(me3)SAPATG), and H3K27Me3A24T (QLATKTARK(me3)SAPATG) were synthesized, N-terminal-labeled with fluorescein, and purified by Tufts University Core Services (Boston, MA). Binding assays were performed in a 10-μl volume at a constant labeled peptide concentration of 40 nm, and Cbx protein concentrations at saturation ranging from 800 to 1300 μm in buffer containing 20 mm Tris, pH 8.0, 250 mm NaCl, 1 mm DTT, 1 mm benzamidine, 1 mm PMSF, and 0.01% Tween 20. Fluorescence polarization assays were performed in 384-well plates using a Synergy 2 microplate reader (BioTek). The excitation wavelength of 485 nm and the emission wavelength of 528 nm were used. The data were corrected for background of the free labeled peptides. To determine Kd values, the data were fit to a hyperbolic function using Sigma Plot software (Systat Software, Inc., CA). The Kd values represent averages ± S.E. for at least three independent experiments.
Histone Peptide Array Screen
Peptides were synthesized directly on a modified cellulose membrane with a polyethylglycol linker using the peptide synthesizer MultiPep (Intavis). The degenerate screening peptide library consisted of 520 membrane-immobilized peptides corresponding to control peptides and 13-residue-long stretches of histone H3 sequences corresponding to residues 3–15 and 21–33. The lysine at positions 9 and 27 was always trimethylated, whereas flanking residues were systematically mutated to each of the 20 l-amino acids. The binding assay was performed as described previously (18). Briefly, the membrane was extensively blocked with skim milk, incubated overnight with 5 μm His6-tagged protein of interest, washed with PBS/Tween, and visualized via Western blot analysis with an anti-His antibody (Abcam).
NMR Spectroscopy and Data Analysis
All complex structures have been determined using the ABACUS approach (19) from NMR data collected at high resolution from nonlinearly sampled spectra and processed using multidimensional decomposition (20, 21). NMR spectra were recorded at 25 °C on Varian INOVA 500-MHz spectrometer equipped with triple resonance cold probe and Bruker Avance 600 and 800-MHz spectrometers equipped with cryoprobes. The sequence-specific assignment of 13C, 1H, and 15N resonances were assigned using the ABACUS protocol (19) from peak lists derived from manually peak picked spectra. The restraints for backbone φ and ψ torsion angles were derived from chemical shifts of backbone atoms using TALOS (22). Hydrogen bond constraints were used within regular secondary structure elements. Automated NOE assignment (23) and structure calculations were performed using CYANA (Version 2.1) according to its standard protocol. The resonances of free and bound histone peptides were assigned from two-dimensional total correlation spectroscopy (TOCSY) and NOESY experiments. Intermolecular NOEs were assigned from a three-dimensional 13C,15N-filtered, -edited NOESY spectrum (39) of 13C,15N-labeled protein bound to unlabeled peptide. A subset of unambiguous NOEs between peptide and protein were initially assigned manually and were sufficient to dock each peptide to the respective protein in CYANA calculations. Next, additional intermolecular NOEs were assigned based on knowledge of the residues expected to be in close proximity between protein and peptide (supplemental Figs. S6 and S7). The final 20 lowest-energy structures were refined within CNS (24) by a short constrained molecular dynamics simulation in explicit solvent (25) (supplemental Table S1). After refinement, 99.4 and 0.6% residues in Cbx3/K9me3, 95.7 and 4.3% residues in Cbx7/K9me3, and 94.4 and 5.6% residues in Cbx7/K27me3 were in favorable and allowed regions of the Ramachandran plot. All NOE distance and angular restraints are deposited with the atomic coordinates in the PDB. Figures were prepared using MOLMOL (26), iSee (27), and PyMOL (DeLano Scientific).
Crystallization, Data Collection, and Structure Determination
Crystals of the Cbx chromodomains were grown at 18 °C using the sitting-drop method by mixing equal volumes of the mother liquor with protein solutions. Crystallization conditions for individual structures are shown in supplemental Table S2. All diffraction data were collected at 100 K and reduced with the HKL2000 suite of programs. The crystal structures of these Cbx chromodomains and their complexes with histone peptides were solved by molecular replacement using MOLREP. COOT (28), REFMAC (29), and MOLPROBITY (30) were used for interactive model building, refinement, and validation, respectively. Crystal diffraction data and refinement statistics are displayed in supplemental Table S3.
Accession Codes
Protein Data Bank coordinates for the Cbx2/H3K27me3, Cbx5/H3K9me3, Cbx6/H3K9me3, Cbx6/H3K27me3, Cbx3/H3K9me3, Cbx7/H3K9me3, Cbx7/H3K27me3, and Cbx8/H3K9me3 have been deposited with accession codes 3H91, 3FDT, 3GV6, 3I90, 2L11, 2L12, 2L1B, and 3I91, respectively.
RESULTS
Binding Preferences of Human Cbx Proteins Diverge from Those of Drosophila
To elucidate the binding specificity for the chromodomains of human Cbx proteins, we measured the affinity of the recombinant chromodomains for both trimethylated Lys-9 and Lys-27 marks on histone H3 peptides using fluorescence polarization. Modified and mutant peptide sequences were assayed with each chromodomain (Table 1). Similar to dHP1, the human HP1 homologs (Cbx1, -3, -5) showed significant preference for H3K9me3 peptides (31) and were sensitive to mutation of Thr-6 to an alanine, confirming that these chromodomains can distinguish H3K9me3 and H3K27me3 sequences via the third residue preceding the methyllysine. In contrast, most human Pc homologs (Cbx2, -4, -6, -7, -8) had a wide range of affinities toward both marks without a distinct selectivity for one, similar to their mouse homologs (15, 31). Cbx4 and Cbx7 bound to both methylated marks but behaved more like a HP1 protein with modest binding to H3K9me3 and 2–3-fold weaker binding to H3K27me3. In fact, Cbx2 was the only Pc-like chromodomain with a clear preference for the K27me3 mark; however, the affinity was rather low. Furthermore, Cbx2, -4, and -7 were insensitive to mutation of the third residue upstream of methyllysine (Thr-6 or Ala-24). Finally, neither Cbx6 nor Cbx8 bound to K9me3 or K27me3 very weakly, with Kd values >500 μm.
TABLE 1.
Peptide binding specificity of human Cbx proteins
Dissociation constant values (Kd) for all chromodomains from human Cbx proteins with methylated peptides and their mutants are shown. Interactions were specified as no binding (N/B) when the FP data were close to background levels and are designated as >500 μm when the values were too weak to accurately estimate a Kd value. Kd values represent the averages ± S.E. for at least three independent experiments. The bold type specifies dHP1 human homologs.
| Protein |
Kdpeptide |
|||
|---|---|---|---|---|
| H3K9me3 | H3K9me3T6A | H3K27me3 | H3K27me3A24T | |
| μm | ||||
| CBX1 | 5 ± 2 | 45 ± 20 | N/B | N/B |
| CBX2 | >500 | N/B | 185 ± 20 | 330 ± 50 |
| CBX3 | 15 ± 8 | N/B | N/B | N/B |
| CBX4 | 70 ± 7 | 80 ± 8 | 205 ± 20 | 250 ± 50 |
| CBX5 | 30 ± 5 | N/B | N/B | N/B |
| CBX6 | >500 | N/B | >500 | >500 |
| CBX7 | 55 ± 5 | 140 ± 15 | 110 ± 17 | 210 ± 50 |
| CBX8 | >500 | N/B | >500 | >500 |
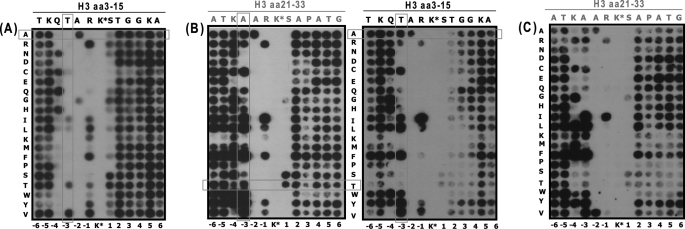
Because the human Cbx binding specificity does not correspond closely to the prototypical Drosophila specificities of HP1 and Pc proteins (2, 5, 18, 31), we sought to identify the spectrum of peptide sequences to which human Cbx chromodomains would bind using a SPOT-blot assay (18). Peptide arrays were synthesized for two sets of peptides from histone H3 covering residues 3–15 and 21–33 with trimethylated lysine (Kme3) at positions Lys-9 or -27, respectively. These two sequences were systematically mutated to each of the 20 amino acids (Fig. 1, vertical series) at each position of the peptide (Fig. 1, horizontal series). Binding was then assessed for representative HP1-like (Cbx3) and Pc-like (Cbx7 and Cbx8) chromodomains.
FIGURE 1.
SPOT-blot binding assay of chromodomain from human Cbx3 (A), Cbx7 (B), and Cbx8 (C). Two sets of peptides from histone H3 covering regions amino acids (aa) 3–15 and 21–33 with trimethylated lysine at position Lys-9 and Lys-27 (K*), respectively, were systematically mutated to each of the 20 amino acids (vertical) at every position (top) as indicated on the figure. Although Cbx7 showed binding to both peptides (see also Table 1) with an absolute dependence on ARKS motif, Cbx3 bound to H3K9me3 peptide only, and Cbx8 bound to H3K27me3. Low panel numbering corresponds H3 residues downstream (negative) or upstream (positive) to the methyl lysine.
As expected, Cbx3 chromodomain bound to H3K9me3 but not to H3K27me3-related peptides, whereas Cbx7 showed binding to both with an absolute dependence on trimethylation of Lys-9 or Lys-27 for both chromodomains (Fig. 1, A and B). Cbx8 showed binding to H3K27me3-related peptides only (Fig. 1C). Variations of the ARKS motif shared by both sequences were not tolerated for either protein except for mutations of the arginine residue to hydrophobic residues. This suggests that the hydrophobic side chain of arginine within ARKS contributes to the binding energy as opposed to its guanidino group, which points away from the interaction surface. Consistent with its similarity to dHP1 and the data in Table 1, Cbx3 binding was disrupted by most mutations of Gln-5 and Thr-6 residues upstream of the ARKS motif of H3 (5). On the other hand, many but not all mutations of Gln-5 and Thr-6 were tolerated by Cbx7, again demonstrating less sensitivity to residues in this region of the peptide. These screens confirmed that the binding event is driven primarily by the ARKS motif with little or no contribution from the C-terminal residues for either protein.
Structural Analysis of Human Cbx Chromodomain Family: Conserved Binding Features
To better understand how human Cbx chromodomains recognize their substrates, we determined the structures of Cbx2, -3, -5, -6, -7, and -8 with a combination of solution NMR and x-ray crystallography including (where possible) complexes with H3K9me3 and H3K27me3 peptides. Analysis of the structures confirmed that they have a common overall fold and bind trimethyllysine-containing peptides in a manner similar to their Drosophila homologs. In all Cbx structures, the trimethylammonium moiety is positioned within an aromatic cage consisting of three aromatic residues supplemented by one or two acidic residues and the remaining peptide binds with a surface-groove recognition mode (9). Two key elements of the peptide binding groove that are conserved among all Cbx proteins are 1) the extended β-strand conformation of the peptide forming a continuous β-sheet with the protein, and 2) a conserved binding pocket for the Ala residue at the −2 position in the peptide substrate relative to the trimethyllysine. For all Cbx histone peptide complexes, this Ala (histone residues Ala-7 or Ala-25) is buried in a small hydrophobic pocket, the size of which is sufficient to accommodate an alanine side chain but nothing larger. This explains the absolute requirement for Ala at the −2 position of the peptide as seen for the Drosophila proteins.
Structural Basis for Cbx1, -3, and -5 Selectivity
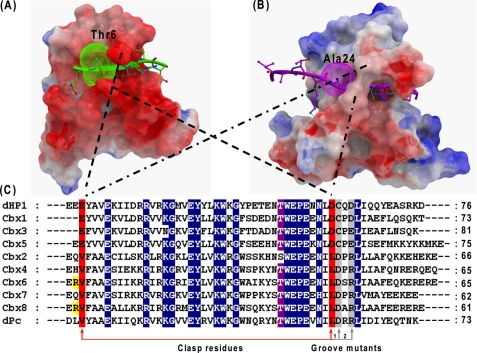
As previously reported for dHP1(5,7) and shown above for the human homologs, Thr-6 of the H3 peptide is an important determinant of the binding and selectivity of the HP1 class of chromodomains. T6V is the only mutation at peptide residue Lys-3 that is capable of interacting with Cbx3 (Fig. 1A). Comparison of the complex structures of Cbx3 and Cbx5 suggests an important role of two conserved negatively charged residues or “polar fingers.” The fingers are formed by a pair of residues Glu-29/19 (top) and Asp-68/58 (bottom) of Cbx3 and Cbx5 chromodomains, respectively, whose β and γ methylene groups are sandwiched by the side chains of Thr-6 and Arg-8 of the peptide (Figs. 2C and 3). Importantly, only valine, which is similar in size to threonine but has a methyl instead of hydroxyl group, would allow the acidic polar fingers to adopt a similar conformation. This is noteworthy because the position of the “top” finger (i.e. Glu-29/19) is important for the conformation of the subsequent residue (Phe-30/20), which contributes to the formation of the methylysine binding aromatic cage. Mutation of the polar clasp residues of Cbx3 to the corresponding residues found in the human Pc homologs (Glu-29 to Val and Asp-68 to Leu) dramatically reduced binding of Cbx3 to both H3K9me3 and H3K27me3 peptides (supplemental Table S4). Thus, a key feature of the HP1 class-peptide interactions is not a specific interaction with Thr-6 per se but, rather, the favorable polar clasp that only Thr (or Val) in this position allows.
FIGURE 2.
Recognition and specificity determinants in human Cbx protein family. Polar fingers of human dHP1 homologs (A), hydrophobic clasp of human dPc homologs (B), and sequence alignment of Drosophila and human chromodomains (C) are shown. For surface representations of the binding grooves, Cbx3/Lys-9 has a tighter groove (A), and Cbx7/Lys-27 adopt a wider groove (B) as a result of a conformational change of the turn bearing key residues. The groove contributes to peptide binding affinity but not to selectivity. Surfaces with positive electrostatic potential are blue, and negative potentials are red. The arginine residues specific to Cbx6 and Cbx8 are highlighted in yellow.
FIGURE 3.
Selectivity of human HP1 homologs is driven by polar fingers. Surface representations are shown of the polar fingers for Cbx3 (left) and Cbx5 chromodomains (right) in complex with H3K9me3 peptide. Thr-6 and Arg-8 residues of the peptide are shown in the binding cleft. The β and γ methylene groups of conserved polar fingers are sandwiched by side chains of Thr-6 and Arg-8. The right panel is rotated by 90° with respect to the left panel.
In addition, Gln-5 also seems to contribute to the binding specificity albeit to a lesser extent than Thr-6 (Fig. 1A). This residue makes main-chain interactions with Val-32 of Cbx3 and interacts with the N terminus of the chromodomain α-helix. At this position only medium sized polar residues such as Asn, Cys, Gln, and His are tolerated.
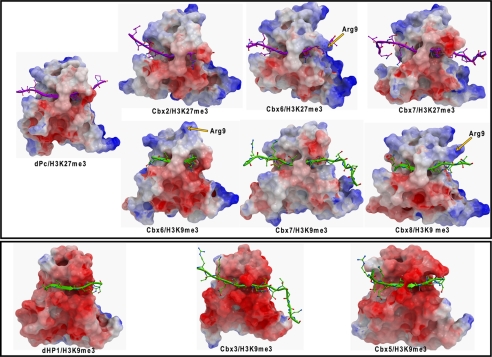
Overall, our binding data indicate that Cbx1, -3, and -5 bind with greater affinity to H3K9me3 than the Pc orthologs bind to either peptides. We attribute the stronger binding affinity of human HP1 homologs to the stark difference in electrostatic surface charge distribution as shown in Figs. 2, A and B, and 4. The HP1 class chromodomains are highly negatively charged, and this charge complimentarity with respect to the basic histone peptides is likely an important driving force for the interaction.
FIGURE 4.
Electrostatic surface depiction of the human Pc (top panel) and HP1 (bottom panel) chromodomain homologs. Surfaces with positive electrostatic potential are blue, and negative potential are red. Arginines present only in Cbx6 and Cbx8 are indicated by a yellow arrow.
Structural Basis for Loss of Selectivity for Human Pc Homologs
In contrast to the HP1-like proteins, the four human Pc-like proteins (Cbx2, -6, -7, -8) are less acidic. This very likely contributes to their lower binding affinities for basic histone peptides. Another important difference is a conserved “hydrophobic clasp” formed by Val-10 and Leu-49 (Cbx7 numbering) in place of the “polar clasp” seen in the HP1 class of chromodomains (Fig. 2B). Ala-24 of the H3K27me3 peptide interacts with this hydrophobic clasp in all four Pc-class complex structures. However, our peptide binding and array data indicate that many other amino acids including threonine and especially hydrophobic residues at this −3 position of the peptide are favorable for binding to Cbx7 (Fig. 1B). Thus, the hydrophobic clasp is an important contributor for the lack of selectivity of human dPc homologs toward the methyl lysine marks, just as the polar fingers are pivotal for selectivity within the HP1 homologs. Mutation of the hydrophobic clasp residues of Cbx7 (Val-10 and Leu-49) to the corresponding polar fingers residues in Cbx3 (Glu/Asp) abrogated the binding to both histone peptide sequences (supplemental Table S4). NMR spectra indicated that both Cbx3 and Cbx7 clasp mutants were folded but may have local or long range perturbations of their structure (supplemental Figs. S1 and S2). Thus, the “clasp” residues are not a simple element that can be swapped between HP1/Pc classes to switch binding selectivity but are context dependent within each chromodomain.
Cbx6 and Cbx8 have functional aromatic cages and hydrophobic fingers very similar to those of Cbx2, -4, and -7, but the former bind to H3K9me3 and H3K27me3 peptides with much lower affinity. We attribute this to an overall greater electropositive charge of Cbx6 and Cbx8 in the region that accommodates the RKme3S portion of the peptide. In particular, Cbx6 and Cbx8 are the only two chromodomains with an Arg residue just before residue Val-10 of the hydrophobic clasp (Fig. 2C, Arg residues are highlighted in yellow). The N-terminal residues of the chromodomain including this Arg residue is disordered in the absence of peptide, but upon complex formation it forms the upper strand of peptide-interacting β-sheet (Fig. 5). As such, the positive charge from Arg-9, present only in Cbx6 and Cbx8, may contribute to less efficient association with the basic histone peptide. Consistent with this notion, we observe a variable position or this Arg residue in the three different low affinity Cbx6 and Cbx8 complexes (Fig. 4).
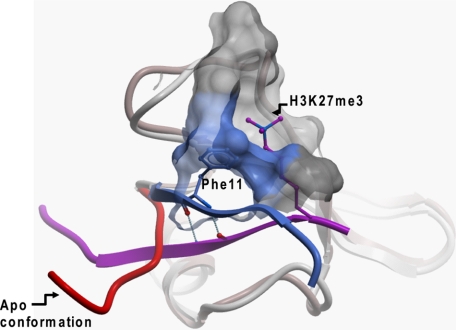
FIGURE 5.
Structural characteristics of chromodomain me-Lys binding. Superimposition of apo (red) and peptide-bound (blue) conformations of Cbx7 reveals a dramatic rearrangement of the N-terminal extremity that closes onto the histone peptide (violet). The interaction is stabilized by anti-parallel backbone hydrogen bonds and placement of a conserved aromatic residue (Phe-11 of Cbx7) in the aromatic system that reads the methyl mark (blue and gray mesh). The same trend was observed for other apo (Cbx4, PDB code 3I8Z; Cbx2, PDB code 2D9U; Cbx7, PDB code 2K1B) and peptide-bound structures within the chromodomains.
Another structural element of dPc that was noted to be important for peptide binding selectivity is an extended recognition groove that accommodates five extra histone residues (relative to dHP1) via hydrogen bonds to main-chain atoms of the peptide (1). We examined the role of residues in this groove for human Cbx2, -3, and -7 by mutagenesis. We swapped pairs of key H-bond-mediating residues along the “bottom” of this groove between the two classes of Cbx proteins (Fig. 2C, supplemental Table S4 and Fig. S3). For Cbx3 and Cbx7, these “groove” mutants had reduced binding affinities for both histone peptides. Although we did not observe a switch in peptide binding specificity, this indicates the importance of these residues for binding affinity in the context of their respective proteins. For Cbx2, the only chromodomain that has a preference for H3K27me3, the groove-2 mutant (D51C/R53D) retained the same binding affinity for H3K27me3 but had improved binding to H3K9me3. Taken together these data indicate that the extended groove likely contributes to peptide binding affinity but may not contribute to selectivity.
Human Cbx Proteins Bind to Alternative Nonhistone Methylated Sequences
Based on our peptide array data, a consensus binding motif can be derived for chromodomains of Cbx7 (A(R/I/L/F/Y/V)Kme3(S/T)) and for Cbx3 ((Q/N)(T/V)A(R/I/F/W/V)Kme3(S/T)), where the slash (/) separates alternative tolerated amino acids at each peptide residue. We hypothesized that there may be alternative or higher affinity binding partners for Cbx proteins in the human genome that include the aforementioned binding motif. We used the consensus sequence for Cbx7 to search NCBI RefSeq database for nuclear proteins that matched the array-derived consensus sequences. Numerous candidate Cbx-interacting proteins that contain at least one consensus sequence and are known or are predicted to be nuclear were identified. Then, a peptide array was designed to include these human peptide sequences from non-histone proteins that could bind to Cbx7 in a trimethylation-dependent manner (supplemental Fig. S4). Many, but not all of the sequences bound to Cbx7 chromodomain. However, if Cbx7 were to interact with one or more of these proteins in vivo it would require for Cbx7 binding partner to be trimethylated in the cell. Therefore, we searched these candidate interacting proteins for those that could potentially be methylated by virtue of their interaction with, or being a component of a known methyltransferase protein or complex.
Interestingly, SET domain bifurcated-1 (SETDB1) is a histone H3Lys-9 methyltransferase that contains three Cbx7 binding consensus sites in close proximity within the sequence that “bifurcates” the conserved SET domain. To elucidate the binding of these consensus sites for Cbx7, we measured the affinity of Cbx7 and Cbx8 chromodomains for di-, tri-, and non-methylated peptides corresponding to the three consensus sites within SETDB1 (Table 2). Cbx7/8 chromodomains bound to trimethylated SETDB1 peptides with equal or greater affinity than histone peptides and did not bind to the non-methylated peptides. Mass spectrometry of full-length human SETDB1 expressed in insect cells revealed methylation of K1170 (ALKS motif) in di- and trimethylation states (Fig. S5), suggesting that SETDB1 may be capable of automethylation. Taken together, these data show that Cbx7/8 and perhaps other Cbx proteins are likely to have an alternative non-histone binding activity.
TABLE 2.
Peptide binding affinity of SETDB1 peptides
Dissociation constant values (Kd) for Cbx7 and Cbx8 chromodomains with consensus SETDB1-methylated peptides. Kd values represent the averages ± S.E. for at least two independent experiments. N/B, no binding.
| Motif | SETDB1 consensus sequence | Cbx7 Kd | Cbx8 Kd |
|---|---|---|---|
| μm | μm | ||
| VAVK(1162)S | RQVAVKme3STR | 43 ± 4 | 98 ± 15 |
| FALK(1170)S | 1165-RGFALKme3STHG | 5 ± 0.5 | 29 ± 6 |
| IAIK(1178)S | HGIAIKme3STN | 5 ± 0.8 | 37 ± 4 |
| Kme3(1162,1170,1178) | RQVAVKme3STRGFALKme3STHGIAIKme3STN | 1.2 ± 0.2 | 6 ± 2 |
| Kme2(1162,1170,1178) | RQVAVKme2STRGFALKme2STHGIAIKme2STN | 15 ± 0.5 | 180 ± 80 |
| Non-methylated | RQVAVKSTRGFALKSTHGIAIKSTN | N/B | N/B |
DISCUSSION
The present work provides a structural framework for understanding the recognition and specificity of human Cbx chromodomains. We found a key structural feature of the Cbx chromodomains that mediates interaction with the ARKme3S peptide sequence; that is, the polar fingers of Cbx1, -3, -5 and the “hydrophobic fingers” of Cbx2, -4, -6, -7, and -8. These fingers encircle the peptide, which is in an extended β-strand conformation with the Ala-7 or Ala-2 resting in a selective pocket at the bottom of the peptide binding groove and the Arg-8 or Arg-26 pointing outward and nestling against the two fingers. The peptide trimethyllysine points into a conserved aromatic cage that is formed in part by a conserved aromatic residue (Tyr or Phe) immediately adjacent to the top finger (as viewed in Figs. 3 and 5). Thus, the conformation of the finger residues is critical for both peptide sequence recognition and formation of the methyllysine binding aromatic cage. The “left” side of the fingers is adjacent to key peptide residues, Thr-6 or Ala-24, that distinguish the two important histone marks, H3K9me3 and H3K27me3, respectively.
Our structural, binding, and mutagenesis data revealed two key factors that contribute to the selectivity and lack of thereof toward histone peptides by human Cbx family. First, there is a stark difference in surface electrostatics, with the acidic HP1 class having greater affinity for basic histone peptide and the Pc class having a more hydrophobic surface resulting in lower affinities for even their most optimal histone binding peptides (Fig. 4). Second, the polar clasp of the HP1 class of chromodomains is sensitive to the residue immediately upstream from the ARKS motif, tolerating only Thr or Val with the appropriate size and shape, whereas the Pc hydrophobic clasp can tolerate most hydrophobic residues. The role of electrostatics in Cbx interactions also likely contributes to the overall weaker absolute affinities measured here for human Cbx proteins compared with those reported for mouse Cbx proteins by Bernstein et al. in a buffer with much lower salt concentration (15). Nevertheless, the overall trend in relative binding affinities among Cbx proteins is the same in both cases.
Of the eight human Cbx proteins, only Cbx2 appears to have a strong preference for the H3K27me3mark. This suggests that Cbx2 may be the functional ortholog of dPc in humans, although the binding affinity of Cbx2 for H3K27me3 peptide is rather low. We note, however, that Cbx2 is also the only Cbx protein to have a DNA binding domain; that is, an AT-hook domain (8, 32). This may increase the affinity of Cbx2 for certain H3K27me3 nucleosomes in vivo due to divalent interaction from the chromo- and AT-hook domains. Additional components of the PRC1 complex also likely contribute to added specificity and affinity in the context of nucleosomes, chromatin, and specific gene loci. For example, Yap et al. (17) recently showed that the chromodomain of Cbx7 interacts with both H3K27me3 as well as the noncoding RNA ANRIL in suppression of transcription at the INK4a locus.
Accumulating evidence suggests that many of the “readers” and “writers” of histone marks may also be active for non-histone sequences (33–35). For example, the prevalence of the QTARKS and similar sequences in the human proteome (such as those from our peptide array consensus) suggests that both H3Lys-9 methyltransferases and reader domains such as the HP1 class of chromodomains are likely to recognize non-histone proteins as well (36, 37). Our data suggest that the Pc class of Cbx proteins, which are components of PRC1 and PRC1-like complexes, also interact with nonhistone proteins (supplemental Fig. S4). Binding of Cbx2, -4, -6, -7, or -8 to epigenetic or transcription factors in a methyllysine-dependent manner would be an attractive mechanism for targeting various Cbx-containing PRC1 complexes to selective sites of the genome occupied by specific factors. This may help explain the expansion of the Pc class of Cbx proteins from insects to mammals, contributing to the greater complexity of mammalian biology.
We identified SETDB1, a H3Lys-9 methyltransferase, as a potential trimethyllysine-dependent interacting partner for Cbx7 and Cbx8. Trimethylated SETDB1 peptides have the ability to directly interact with Cbx7/8 with affinities equal or greater than histone peptides H3K27me3or H3K9me3. This is significant in light of the recent study of Bilodeau et al. (38) showing that shRNA knockdown SETDB1, Cbx7, or Cbx8 were among the most effective regulators of embryonic stem cell fate in a large scale screen of chromatin regulatory factors. This study reported that SETDB1 and H3K9me3 nucleosomes are both found at a subset of genes encoding key developmental regulators in ES cells. These SETDB1-occupied genomic sites are among those previously shown to be repressed by PcG proteins and are “bivalent,” having both H3K4me3 and H3K27me3 marks. Given the relative binding affinities observed in our experiments, it is tempting to summarize that Cbx7 and/or Cbx8 may be involved in recruiting PRC1 complexes to SETDB1 occupied sites by virtue of their chromodomain interaction with SETDB1 instead of H3K27me3.
Supplementary Material
Acknowledgments
We are grateful to Antoine Peters for fruitful discussions. We thank Chen Chen, Brett Larsen, and Andrew James (Dr. Pawson laboratory, University of Toronto) for help with MS data acquisition and analysis. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes of Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
This work was supported in part by the Ontario Ministry of Health and Long Term Care. This work was also supported by the Canadian Cancer Society.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1–S4.
- dHP1
- Drosophila heterochromatin protein 1
- dPc
- Drosophila polycomb protein
- PRC1
- polycomb repressive complex 1
- Cbx
- chromobox proteins
- SETDB1
- SET domain bifurcated-1.
REFERENCES
- 1. Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., Khorasanizadeh S. (2003) Genes Dev. 17, 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jacobs S. A., Taverna S. D., Zhang Y., Briggs S. D., Li J., Eissenberg J. C., Allis C. D., Khorasanizadeh S. (2001) EMBO J. 20, 5232–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 4. Hublitz P., Albert M., Peters A. H. (2009) Int. J. Dev. Biol. 53, 335–354 [DOI] [PubMed] [Google Scholar]
- 5. Jacobs S. A., Khorasanizadeh S. (2002) Science 295, 2080–2083 [DOI] [PubMed] [Google Scholar]
- 6. Min J., Zhang Y., Xu R. M. (2003) Genes Dev. 17, 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., Kouzarides T., Murzin A. G., Murzina N. V., Laue E. D. (2002) Nature 416, 103–107 [DOI] [PubMed] [Google Scholar]
- 8. Whitcomb S. J., Basu A., Allis C. D., Bernstein E. (2007) Trends Genet 23, 494–502 [DOI] [PubMed] [Google Scholar]
- 9. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Biason-Lauber A., Konrad D., Meyer M., DeBeaufort C., Schoenle E. J. (2009) Am. J. Hum. Genet. 84, 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott C. L., Gil J., Hernando E., Teruya-Feldstein J., Narita M., Martínez D., Visakorpi T., Mu D., Cordon-Cardo C., Peters G., Beach D., Lowe S. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5389–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maertens G. N., El Messaoudi-Aubert S., Racek T., Stock J. K., Nicholls J., Rodriguez-Niedenführ M., Gil J., Peters G. (2009) PLoS One 4, e6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dietrich N., Bracken A. P., Trinh E., Schjerling C. K., Koseki H., Rappsilber J., Helin K., Hansen K. H. (2007) EMBO J. 26, 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernard D., Martinez-Leal J. F., Rizzo S., Martinez D., Hudson D., Visakorpi T., Peters G., Carnero A., Beach D., Gil J. (2005) Oncogene 24, 5543–5551 [DOI] [PubMed] [Google Scholar]
- 15. Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., Allis C. D. (2006) Mol. Cell. Biol. 26, 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vincenz C., Kerppola T. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16572–16577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yap K. L., Li S., Muñoz-Cabello A. M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M. J., Zhou M. M. (2010) Mol. Cell 38, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nady N., Min J., Kareta M. S., Chédin F., Arrowsmith C. H. (2008) Trends Biochem. Sci. 33, 305–313 [DOI] [PubMed] [Google Scholar]
- 19. Lemak A., Steren C. A., Arrowsmith C. H., Llinás M. (2008) J. Biomol. NMR 41, 29–41 [DOI] [PubMed] [Google Scholar]
- 20. Gutmanas A., Jarvoll P., Orekhov V. Y., Billeter M. (2002) J. Biomol. NMR 24, 191–201 [DOI] [PubMed] [Google Scholar]
- 21. Orekhov V. Y., Ibraghimov I., Billeter M. (2003) J. Biomol. NMR 27, 165–173 [DOI] [PubMed] [Google Scholar]
- 22. Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 23. Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 24. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 25. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 26. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 27. Raush E., Totrov M., Marsden B. D., Abagyan R. (2009) PLoS One 4, e7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 30. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 32. Senthilkumar R., Mishra R. K. (2009) BMC Genomics 10, 549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Couture J. F., Collazo E., Hauk G., Trievel R. C. (2006) Nat. Struct. Mol. Biol. 13, 140–146 [DOI] [PubMed] [Google Scholar]
- 34. Feng L., Lin T., Uranishi H., Gu W., Xu Y. (2005) Mol. Cell. Biol. 25, 5389–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 36. Sampath S. C., Marazzi I., Yap K. L., Sampath S. C., Krutchinsky A. N., Mecklenbräuker I., Viale A., Rudensky E., Zhou M. M., Chait B. T., Tarakhovsky A. (2007) Mol. Cell 27, 596–608 [DOI] [PubMed] [Google Scholar]
- 37. Chin H. G., Estève P. O., Pradhan M., Benner J., Patnaik D., Carey M. F., Pradhan S. (2007) Nucleic Acids Res. 35, 7313–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bilodeau S., Kagey M. H., Frampton G. M., Rahl P. B., Young R. A. (2009) Genes Dev. 23, 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zwahlen C., Pascale L., Vincent S. J. F., Greenblatt J., Konrat R., Kay L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.