Abstract
Expression of the gene encoding neurotensin/neuromedin N (NT/N) is mostly limited to the brain and specialized enteroendocrine N cells in the distal small intestine. We have identified key regulatory elements in the promoter region that are involved in human NT/N (hNT/N) gene expression in the novel human endocrine cell line, BON, which resembles intestinal N cells in several important aspects including NT/N precursor protein processing, ratios of different NT/N mRNA isoforms, and high levels of constitutive expression of the NT/N gene. In this study, we demonstrated multiple cis-regulatory elements including a proximal region containing a cAMP-responsive element (CRE)/AP-1-like element that binds both the AP-1 and CRE-binding protein (CREB)/ATF proteins (c-Jun, ATF-1, ATF-2, JunD, and CREB). Similar to the rat NT/N gene, this region is critical for constitutive hNT/N gene expression. Moreover, we identified a novel region that binds the orphan hormone receptor, NR2F2. We have demonstrated that the C terminus of NR2F2 strongly represses hNT/N transcription, whereas an N-terminal domain antagonizes this repressive effect. Regulation of NT/N expression by NR2F2 may have important consequences for lipid metabolism. We speculate that a complex interplay between the proximal CRE/AP-1-like motif and NR2F2 binding region exists to regulate hNT/N expression, which is critical for the high level of constitutive expression of NT/N in enteroendocrine cells. Finally, the BON cell line provides a unique model to characterize the factors regulating expression of the hNT/N gene and to better understand the mechanisms responsible for terminal differentiation of the N cell lineage in the gut.
Keywords: AP-1 Transcription Factor, CREB, DNA-binding Protein, Gene Expression, Transcription Promoter, ARP-1, ATF, COUP-TFII, Neuromedin, Neurotensin
Introduction
Neurotensin (NT),2 a tridecapeptide predominantly localized to specialized enteroendocrine N cells with greatest abundance in the jejenum and ileum of the gastrointestinal (GI) tract, is released by intraluminal fats (1–3). It serves several functions in the GI tract including stimulation of pancreatic and biliary secretion (4–6), inhibition of gastric and small bowel motility (7, 8), and facilitation of fatty acid translocation from the intestinal lumen (9–12). Furthermore, NT stimulates the growth of various GI tissues including the pancreas (13), colon (14), and small bowel (15–18), as well as certain colonic, pancreatic, prostatic, and lung cancers that possess NT receptors (1). The primary sequences of both the rat and human genes encoding NT and the structurally related hexapeptide, neuromedin N, have been elucidated (19, 20). In the GI tract, the NT/neuromedin N (NT/N) gene is developmentally regulated in the human and rat intestines in a temporospatial manner (21–23). The promoter proximal 5′-flanking region of the rat NT/N gene has previously been shown to integrate multiple environmental stimuli in neuroendocrine PC12 cells and to be sufficient for cell-specific expression in enteroendocrine BON cells (24, 25). These results indicated that an array of distinct cis-regulatory motifs localized in the proximal 120 bp of the 5′-flanking sequence are required for both inducible and high level, constitutive NT/N expression, depending on the cell type. A proximal promoter cAMP-responsive element (CRE)/AP-1 site and a purine-rich imperfect direct repeat sequence were particularly important for expression in BON cells.
The regulation of gene expression in eukaryotic cells involves the specific interaction of DNA-binding proteins with cis-acting DNA sequences located in the 5′-flanking region upstream from the transcriptional start site. The trans-acting factors that interact with these DNA elements either activate or repress gene transcription, and depending on the cell type, these factors, such as AP-1 and CRE-binding protein (CREB)/ATF (activating transcription factor), may be ubiquitous. The nuclear factor AP-1 superfamily consists of the jun (c-Jun, JunB, and JunD) and fos (c-Fos, Fra-1, and Fra-2) gene subfamilies that bind the AP-1 site (TGAGTCA) as either homo- or heterodimers (26, 27). Another large family of transcription factors, comprising at least 10 related proteins that bind the CRE site (TGACGTCA), includes CREB (CRE binding), ATF, and CREM (CRE modulator) (28). Moreover, members of the steroid hormone receptor superfamily also regulate transcription of a large variety of genes and play a crucial role in developmental processes (29, 30). In addition to the classical steroid-thyroid hormone receptors, a large number of genes in this superfamily have been cloned and classified as “orphan” receptors, as the ligands for these receptors have yet to be described. The novel chicken ovalbumin upstream promoter-transcription factors, now denoted as nuclear receptor subfamily 2, group F (NR2F), are among the better characterized orphan receptors and include NR2F1 (or EAR-3) and NR2F2 (or ARP-1) (31). Studies in several organisms have indicated that NR2Fs play important roles in biological processes such as cell differentiation, cell fate determination, cell cycle regulation, and metabolic homeostasis (30–32). Other closely related members of this superfamily include hepatocyte nuclear factor-4 (HNF-4) and retinoic acid receptor (RAR). The NR2Fs are capable of repressing the basal promoter activity of several target genes and have been implicated in organogenesis and the determination of cell fate (29–33). In addition, the NR2Fs and HNF-4 are important in the regulation of certain intestinal and liver-specific genes. For instance, it has been demonstrated that NR2F2 binds to the intestinal fatty acid-binding protein promoter and increases its expression (34). In contrast, NR2F2 acts as a repressor for other genes by several different mechanisms including competition with HNF-4 for binding to common promoter elements (35).
In our present study, we have utilized the novel human endocrine BON cell line to delineate the regulatory elements involved in cell-specific expression of the human NT/N (hNT/N) gene. BON cells, like the terminally differentiated N cells of the small bowel, express high levels of NT/N mRNA, synthesize and secrete NT peptide, and process the NT/N precursor protein in a fashion identical to that of the normal intestine, with neuromedin N present in its large molecular form and NT present as the small peptide (36, 37). Here, we have identified two critical cis-regulatory elements in the human NT/N promoter that serve to modulate basal NT/N expression in endocrine cells. Similar to the rat, a proximal CRE/AP-1-like motif binds both AP-1 and CREB/ATF factors and functions as a crucial element for high level basal NT/N expression. Interestingly, in contrast to the rat, a region just distal to the CRE/AP-1-like motif binds NR2F2 and functions as a repressor of NT/N expression, predominantly through the C-terminal region of NR2F2. These findings are important to better delineate the mechanisms that modulate intestinal endocrine gene expression that, currently, are poorly understood.
EXPERIMENTAL PROCEDURES
Materials
Restriction, ligation, and other DNA-modifying enzymes were purchased from Promega (Madison, WI) or Stratagene (La Jolla, CA). DNase I (DPRF) was purchased from Worthington Biochemical Corp. Nucleotide and poly(dI·dC) were purchased from Pharmacia LKB Biotechnology Inc. Radioactive compounds were obtained from PerkinElmer Life Sciences. Tissue culture media and reagents were obtained from Invitrogen. The luciferase assay system was obtained from Promega. All other reagents were of molecular biology grade and were obtained from either Sigma or Amresco (Solon, OH). Elutip-d columns were purchased from Schleicher & Schuell. Specific antibodies for chromatin immunoprecipitation (ChIP) assays against NR2F2, CREB1, CREB2, ATF1, ATF2, JunD, and c-Jun were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Antisera against HNF-4, NR2F2, and NR2F1 were obtained from Drs. Frances M. Sladek (University of California, Riverside, CA), Sotirios K. Karathanasis (Pfizer Global Research and Development, Ann Arbor, MI), and Ming-Yu Tsai (Baylor College of Medicine, Houston, TX), respectively. The ChIP-IT Express Enzymatic Kit was purchased from Active Motif Inc. (Carlsbad, CA). NR2F2 and control siRNA and DharmaFECT Duo transfection reagent were purchased from Dharmacon, Inc. (Lafayette, CO). NR2F2 and the nontargeting control (NTC) shRNA lentiviral particles were from Sigma MISSION®.
Cell Culture and Generation of Stable NR2F2 Knockdown BON Cell Lines
The human carcinoid cell line BON, which was established in our laboratory (38), was cultured in Dulbecco's modified Eagle's medium (DMEM) and F12K in a 1:1 ratio supplemented with 5% fetal calf serum (FCS). COS-1 cells were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM with 10% FCS. NCI-H727 cells (lung carcinoid tumor cell line) were purchased from ATCC. QGP-1 (pancreatic endocrine cell line) was purchased from Japan Health Sciences Foundation (Osaka, Japan). NCI-H727 and QGP-1 cells were maintained in ATCC-formulated RPMI 1640 medium with 10% FBS. For generating stable NR2F2 knockdown cell lines in BON cells, BON cells were infected with NR2F2 lentiviral particles in growth medium in the presence of Polybrene (5 μg/ml) for 24 h; the cells were then incubated with growth medium for an additional 24 h. The infected cells were subcultured in 100-mm dishes in fresh medium containing puromycin (2.5 μg/ml). Puromycin-resistant cell pools were collected, and effective knockdown was monitored by Western blot. All cells were maintained in a humidified atmosphere of 95% air and 5% CO2 at 37 °C.
Plasmid Constructions and Cloning
A subcloned 0.9-kb EcoRI fragment containing the promoter region of the human NT/N gene (19) was digested with SacI and PvuII and the resulting hNT/N promoter fragment (−373 to +26) was cloned upstream of the luciferase reporter gene in SacI/SmaI-digested pXP1 (19). Deletion mutants for the hNT/N promoter were created using a PCR-based approach. A common antisense (i.e. upstream/5′) primer (5′-CACTCACTTTCAAAGCCAGGGGTCGAC-3′) was designed using sequences derived from the 5′-nontranslated region (exon 1) of the NT/N gene. A series of 3′ (or downstream) primers (Table 1) was designed such that the amplified target DNAs represented a sequential removal of 50- or 25-bp segments of the 5′-flanking sequence. The 5′-primer introduces a unique SalI restriction site into the amplified DNA, whereas the 3′-primers contain a unique SstI site. DNA fragments for each promoter deletion mutant were isolated (SstI/SmaI) and inserted into a luciferase reporter plasmid (pXP1) digested with SstI and SalI. Plasmid DNA was purified by CsCl density gradient centrifugation prior to use in transient transfection assays.
TABLE 1.
Oligonucleotides used as primers in PCR-based deletion and site-directed mutagenesis
| Oligonucleotide | Sequence |
|---|---|
| −321 | 5′-GAGCTCAGAAGTGGGAAAAGGATGATACTGGG-3′ |
| −271 | 5′-GAGCTCTATGCTGTATGTCAGTGCAGTTGAATG-3′ |
| −221 | 5′-GAGCTCCAAAGCAGCAGCAGCAGCAATTAG-3′ |
| −171 | 5′-GAGCTCTCAGAAATGGGGGAGGAGAGCAGG-3′ |
| −146 | 5′-GAGCTCGGACAAAGGAAAAGGGGAGGAGAAA-3′ |
| −122 | 5′-GAGCTCAGCAGGGCAAAGAGGGGAGGGAT-3′ |
| −95 | 5′-GAGCTCGGTGAAGATAGGGCACATCCTGCAA-3′ |
| −71 | 5′-GAGCTCAGATAATGTCTGTACAATCAATGACATCA-3′ |
| −42 | 5′-GAGCTCTCCTCCTGCTTATATATATAGGGAATG-3′ |
| −20 | 5′-GAGCTCGGAATGGCCAGAGCACCTCTCATA-3′ |
| M1 | 5′-GCAGGAGGATAATGTAATTGATTGTACAG-3′ |
| M2 | 5′-GATGTCATTGATTATAAAGACATTATCTTTGC-3′ |
| M3 | 5′-AGGAGGATGACGTCATTGATTGTACAG-3′ |
| M4 | 5′-CAGGAGGATGA_GTCATTGATTGTACAG-3′ |
| M5 | 5′-CCTATCTTCACCTCGATGCCTCCCCTC-3′ |
| M6 | 5′-CCTATCTTCACGTGCATCCCTCCCCTC-3′ |
| M7 | 5′-GTGCCCTATCTTGAGCTCCATCCCTCC-3′ |
| M8 | 5′-AGGATGTGCCGTATGTTCACCTCCATCC-3′ |
| M9 | 5′-TTTGCAGGATGTGGGCTATCTTCACCTC-3′ |
Mutagenesis
Specific regulatory elements within the hNT/N promoter were defined using replacement mutagenesis. An important advantage of this type of mutagenesis is the ability to evaluate the role of a specific sequence without altering the spatial arrangement of the promoter. The creation of the replacement mutants utilized a PCR-based strategy with anchor primers derived from sequences within the 5′-flanking region (sense: −175 to −148, 5′-GAGCTCGGTTCAGAAATGGGGGAGGAGAGC-3′) and the 5′-untranslated sequence from exon 1 of the NT/N gene (antisense: +1 to +26, 5′-AAGCTTCTGGCTTTGAAAGTGAGTGAACTATG-3′). The sense anchor primer contains a unique SstI restriction site, and the antisense primer encodes a unique HindIII site. Each replacement mutant was created using a second set of primers that targeted a specific 10-bp sequence within the NT/N promoter. The replacement sequence (5′-GCXGTCGACGC-3′) contains a unique SalI site to facilitate assembly of the mutant promoter. PCR amplification using the appropriate anchor and replacement primers was used to generate the two target DNAs that were then joined using the common SalI site from the replacement sequence. Each promoter mutant was confirmed by DNA sequence analysis prior to insertion into a luciferase reporter plasmid (pXP1). Plasmid DNA was purified by CsCl density gradient centrifugation prior to use in transient transfection assays.
Site-directed mutagenesis was performed using a procedure described previously (39). A 211-bp SstI-HindIII DNA fragment containing 175 bp of the immediate 5′-flanking sequence of the hNT/N gene was cloned into M13mp18 RF and propagated in the Escherichia coli strain CJ236. This strain is deficient in dUTPase (dut) and uracil-DNA glycosylase (ung). The M13 phage DNA produced in this strain contains 3–4 uracil residues/kilobase and served as the template for the mutagenesis reaction. The sequence of the oligonucleotide primers used in creating the mutants are listed in Table 1 with the specific base change underlined. DNA fragments (SstI-HindIII) for each mutant were cloned in the luciferase reporter plasmid (pXP1), and the sequence was confirmed using an Applied Biosystems model 394 DNA sequence analyzer.
Transient Transfection, Luciferase, and β-Galactosidase Assays
For luciferase assays, BON cells were plated at a density of 3 × 106 cells in 60-mm dishes 24 h prior to transfection. Human NT/N promoter-luciferase gene constructs (3 μg) were co-transfected with 0.2 μg of the plasmid p110 (40), which contains the β-galactosidase gene fused to the SV40 early promoter, by calcium phosphate co-precipitation as described previously (24). The β-galactosidase assays were carried out as described previously (41). Luciferase activities (mean ± S.D. after normalization to β-galactosidase) for at least three independent transfections were expressed as percentages of the wild type (either −373 to +26 or −175 to 26) hNT/N promoter fragment. NR2F2 siRNA and nontarget control siRNA were co-transfected with hNT promoter plasmids using DharmaFECT Duo transfection reagent.
Preparation of Nuclear and Whole-cell Extracts and in Vitro Footprinting
Crude nuclear extracts were prepared from BON cells according to the method described by Shapiro et al. (42). The extracts were quick-frozen, stored in aliquots at −80 °C, and used within 2 months of extraction. Whole-cell extracts from COS-1 cells transfected with vectors pMT2-HNF4 (from Dr. Frances M. Sladek, Riverside, CA), pTM2-ARP1, pMT2-ARPΔA1, pMT2-ARPΔA6, pMT2-ARPΔA7, pMT2-RXRα, pMT2-RARα, pMT2-RARβ, and pMT2-null (all kindly provided by Dr. Karathanasis, Pfizer Global Research and Development) were prepared as described previously (24). In vitro DNase I footprinting was performed using the hNT/N promoter fragment (−373 to +26) as described previously (24).
Electrophoretic Mobility Shift Assay (EMSA)
Oligonucleotides corresponding to the top and bottom strands of wild type and mutated hNT/N promoter sequences, C3P, GAGA, GATA, and HNF-1, were synthesized. The procedures for 32P oligonucleotide labeling and electrophoretic mobility shift assay were as described previously (24).
ChIP Assay
The ChIP assay was performed using the ChIP-IT Express Enzymatic Kit (Active Motif) according to the manufacturer's protocol. PCR of the hNT promoter containing the NR2F2 binding site was performed using total (input) or immunoprecipitated chromatin with the following pair of oligonucleotide primers: 5′-GGAAGATCGTCACTTTCACTCAAGGTT-3′ and 5′-GGTGCTCTGGCCATTCCCCT-3′. GAPDH primers were from Active Motif.
RNA Isolation and One-step Real-time RT-PCR
Cells were plated in 24-well plates (n = 3), and total RNA was isolated from cells using a RNeasy kit. Real-time RT-PCR was performed as described previously (43). The target gene, NT (ID: Hs00175048_m1), was analyzed. Reactions for each sample were performed in duplicate.
NT EIA for NT Secretion
To measure basal NT secretion, cells were plated in 24-well plates at a density of 2.4 × 105/well and replaced with fresh growth medium the next day. Cells were grown for 24 h and medium collected. For stimulated NT secretion, cells were incubated in serum-free medium for 30 min and then stimulated with 10 μm forskolin in fresh serum-free medium for another 30 min. The medium was collected and stored at −80 °C; NT secretion was measured by an NT EIA kit as described previously (43, 44).
Statistical Methods
One-way or two-factor analysis of variance were employed to compare mean levels of luciferase activity, real-time PCR, and NT secretion across cell culture condition groups. Tests for interaction between factors (e.g. NR2F2 and hNT/N promoter levels) and pairwise comparisons between groups were performed using contrast statements from the model. Adjusted p values of <0.05 were considered statistically significant.
RESULTS
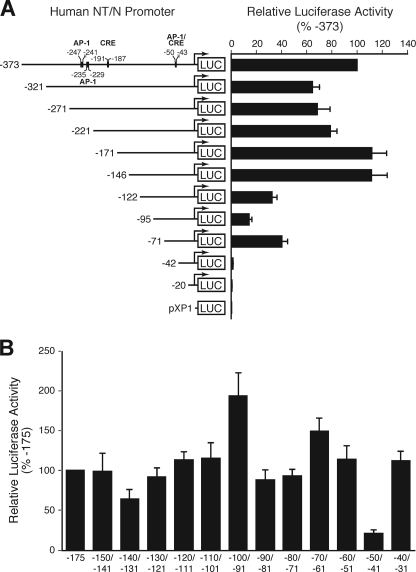
5′-Deletion and Mutational Analysis Identifies Multiple Sites in the Proximal hNT/N Promoter That Are Required for Constitutive NT/N Expression
We had shown previously that the proximal 216 bp of the rat NT/N gene is sufficient for near maximal expression in the BON human endocrine cell line, which constitutively expresses NT/N and possesses many characteristics noted in enteroendocrine cells of the intestine (24). To identify regulators of hNT/N gene expression, we performed an extensive mutational analysis of its promoter element. A series of 5′-deletions of the hNT/N promoter were generated and fused to a luciferase reporter gene. The activity of the promoter was determined by assaying luciferase activities in extracts from transfected BON cells. We found several areas that may contribute to hNT/N gene regulation, including −373 to −321, −221 to −171, −146 to −122, −122 to −95, and −71 to −42 (Fig. 1A). The decreased luciferase activity in the −373 to −321, −145 to −122, −122 to −95, and −71 to −42 deletions suggested that activating elements are located in these areas, whereas increased luciferase activity in the −221 to −171 deletions suggested that repressor elements are located in these areas. The proximal promoter region extending to −146 appeared to be the critical region for high level hNT/N expression; therefore, we next performed −10-bp replacement mutagenesis on the −150 to −31 region (spanning mutations) (Fig. 1B). Of note, mutation of the −100 to −91 region increased promoter activity, suggesting that the mutation inactivates a repressor element. In contrast, mutation of the −140 to −131 and −50 to −41 regions dramatically decreased promoter activity, suggesting that activator elements are located in this area (Fig. 1B). The latter result correlates well with the luciferase activity of the −146Luc/−122Luc and −71Luc/−42Luc constructs, respectively, as shown in Fig. 1A. Moreover, deletion analysis indicated that there is a positive element located between −122 and −95. Surprisingly, no spanning mutation in that region showed a corresponding effect, thus suggesting the presence of functionally redundant positive elements in the promoter region. Therefore, utilizing 5′-deletion and mutational analysis, we identified multiple sites in the proximal hNT/N promoter that are required for constitutive NT/N expression.
FIGURE 1.
5′-deletion and linker scanner (replacement) mutation analysis of hNT/N promoter activity. A, the region spanning 373 bp of the hNT/N promoter and the first 26 bp of exon 1 was progressively deleted from its 5′-end, fused to the luciferase-reporter vector pXP1, and transiently transfected into BON cells. Luciferase activities are expressed as the percentage of the −373 fusion construct and are the mean ± S.D. of four separate transfections after normalization to β-galactosidase expression. B, relative luciferase expression generated by wild type (−175 to +26) or the linker scanner (replacement) mutants of the hNT/N promoter after transfection into BON cells. Luciferase activities are expressed as a percentage of that of the wild type (−175 to +26) fusion plasmid and are the mean ± S.D. of four separate transfections after normalization to β-galactosidase expression.
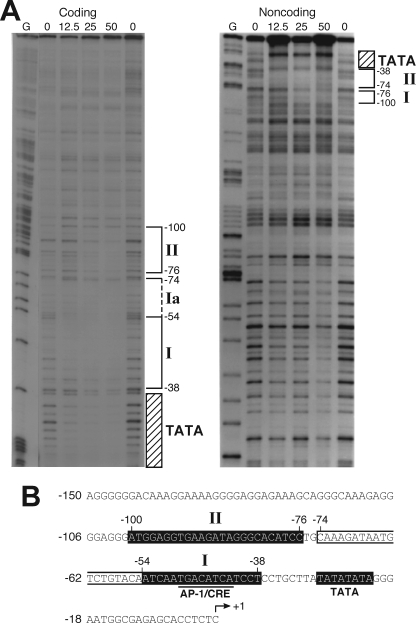
DNase I Footprinting Analysis Identifies Two Protected Regions in the Proximal hNT/N Promoter Region
To map potential binding sites in the hNT/N promoter that may contribute to the expression of the hNT/N gene, we performed DNase I footprinting assays using the full-length hNT/N gene (−373 to +26) promoter fragment. Incubation of the labeled coding strand with increasing amounts of BON cell nuclear extract resulted in the appearance of three regions of protection (I and II and the region corresponding to the TATA box) (Fig. 2A, left panel). Region I (−54 to −38) contains the CRE/AP-1-like sequence (TGACATCA) at −49 to −42. Of note, corresponding luciferase activity decreased when this protected region was deleted, indicating that protein binding to this region is crucial for constitutive hNT/N expression (Fig. 1A). Protection of region Ia (−74 to −55) required a higher concentration of BON nuclear extract, indicating that region Ia is different from region I (Fig. 2B). Region II (−100 to −76) contained a number of potential regulatory elements (e.g. GATA and GAGA binding sites) (Fig. 2B). Mutation of this region (−100 to −91) resulted in a 2-fold increase in hNT/N promoter activity (Fig. 1B), which suggested the binding of proteins that repress hNT/N promoter activity. No protection was noted between −146 and −122, despite the fact that mutation of this region resulted in a dramatic decrease in hNT/N promoter activity. Furthermore, no apparent binding was noted at the potential distal regulatory elements, which included the CRE half-site at −191 to −187 and the two potential AP-1 binding sites at −247 to −241 and −235 to −229. A similar binding pattern was confirmed using the noncoding strand as a labeled probe (Fig. 2A, right panel). Therefore, utilizing DNase I footprinting analysis, we identified two regions of protein binding (−54 to −38 and −100 to −76) in the hNT/N gene promoter.
FIGURE 2.
In vitro DNase I footprint analysis of the hNT/N promoter. A, the coding and noncoding strands of an hNT/N gene fragment, from nucleotides −373 to +26, were labeled with [32P]CTP, incubated with nuclear extracts from BON cells, and digested with DNase I. Areas protected from digestion are bracketed with the corresponding nucleotide positions, which was determined by running a DNA sequencing ladder (G reaction) in parallel. The numbers at the top of the autoradiogram are the amounts of nuclear extract (in μg of protein) used in each reaction mixture. B, sequences of the hNT/N promoter fragment and DNase I-protected regions I and II (shown as white letters on black background).
Region I, Which Contains a CRE/AP-1-like Element, Binds Both AP-1 and CREB/ATF Proteins and Is Crucial for Constitutive hNT/N Expression
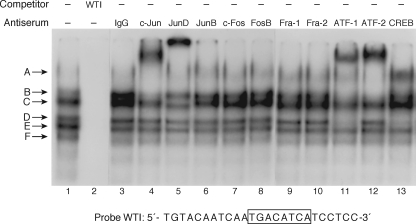
To further analyze protein binding to the promoter, the two footprinted regions were analyzed using EMSA. We first examined whether region I binds both AP-1 and CREB/ATF proteins, similar to what we had shown previously for the rat NT/N gene. As shown in Fig. 3, the addition of BON cell nuclear extract to a labeled oligonucleotide probe (−60 to −36), which includes the AP-1/CRE element at −49 to −42, produced multiple DNA-protein complexes, labeled A–F (lane 1). These complexes were effectively competed with a molar excess of unlabeled probe (Fig. 3, lane 2). The addition of antibodies specific to c-Jun (Fig. 3, lane 4), JunD (lane 5), ATF-1 (lane 11), ATF-2 (lane 12), and CREB (lane 13) produced “supershifted” complexes indicating protein binding to this region of the hNT/N gene, which is analogous to our previous findings with the rat NT/N promoter (24). Complex A was composed predominantly of c-Jun and ATF-2 proteins; complex B appeared to be composed of c-Jun, ATF-1, and ATF-2 proteins; complex C was partially supershifted by anti-JunD; and complex D was supershifted in the presence of both ATF-1 and CREB antisera. Complexes E and F were not supershifted by any of the antisera tested, and thus their composition remains uncertain. Using an oligonucleotide encompassing both regions I and Ia (−74 to −38) did not result in any additional bands (data not shown).
FIGURE 3.
Identification of the AP-1 and CREB/ATF proteins that bind to region I (−54 to −38) of the hNT/N promoter. Nuclear extracts (10 μg/lane) from BON cells were preincubated with antisera (2 μl) to various AP-1 and CREB/ATF proteins prior to the addition of labeled WT region I probe. Supershifted complexes were noted for c-Jun (lane 4), JunD (lane 5), ATF-1 (lane 11), ATF-2 (lane 12), and CREB (lane 13).
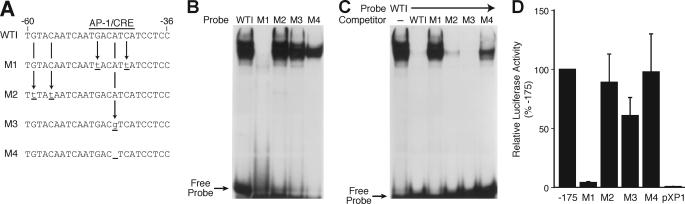
To further elucidate the importance of this region for hNT/N gene activity, site-directed mutagenesis was used to examine the role of specific elements in this region. Several oligonucleotides were synthesized with point mutations of specific bases in the −60 to −36 region (Fig. 4A). Mutation 1 (M1) consisted of point mutations of a G and a C of the CRE/AP-1 element, which have been shown previously to be important for protein binding (24). M2 mutated a G at −59 and a C at −56, which lie outside of the CRE/AP-1 element, and M3 produced a consensus CRE binding site (TGACGTCA), whereas M4 created a consensus AP-1 binding site (TGACTCA). Wild type (−60 to −36) and mutated oligonucleotides were radiolabeled and used in EMSAs with BON cell nuclear extract (Fig. 4B). Mutation of the two nucleotides in the CRE/AP-1 element (M1) completely prevented protein binding, whereas protein binding was noted with the other mutated and wild type probes. These findings were further confirmed by competition assays where the wild type (−60 to −36) oligonucleotide was used as a labeled probe with BON nuclear extract, and competition experiments were performed with unlabeled oligonucleotides (Fig. 4C). The DNA-protein complex was effectively competed with a molar excess of wild type −60 to −36, M2, and M3. The majority of the complex was competed with M4. In contrast, competition with M1 in molar excess had a minimal effect on the DNA-protein complex, thus further confirming the findings from Fig. 4B.
FIGURE 4.
Site-directed mutagenesis analysis of region I. A, schematic representation of site-directed mutagenesis of region I. B, the wild-type region I (WTI) hNT/N promoter fragment (−60 to −36) and mutant fragments (M1–M4) were radiolabeled and incubated with BON cell nuclear extracts (10 μg). and EMSA was performed as described under “Experimental Procedures.” C, competitive binding analysis by EMSA with BON nuclear extracts (10 μg) and unlabeled WTI and mutant M1–M4 probes (at 200-fold molar excess) as competitors. D, the −175 to +26 region of the hNT/N promoter and its mutations, M1–M4, were cloned into the luciferase reporter plasmid, pXP1, and transiently transfected into BON cells. Luciferase activities are expressed as a percentage of the wild type (−175 to +26) fusion plasmid and are the mean ± S.D. of four separate transfections after normalization to β-galactosidase expression.
To assess the functional consequences of these mutations on hNT/N promoter activity, site-directed mutagenesis was performed using the hNT/N promoter (−175 to +26) linked to the luciferase reporter gene, and the resulting plasmids were transiently transfected into BON cells (Fig. 4D). Mutation of the nucleotides within the CRE/AP-1 element (M1) silenced the activity of the hNT/N promoter, consistent with the EMSA analysis demonstrating absence of protein binding. Meanwhile, promoter activity remained at approximately the same level as the wild type (−175) construct with M2, M3, and M4. Interestingly, supershift analysis using BON nuclear extract with either labeled M3 or M4 oligonucleotide probes demonstrated binding of c-Jun, JunD, ATF-1, ATF-2, and CREB proteins to the consensus CRE site, similar to the wild type oligonucleotide (data not shown). Creation of a consensus AP-1 site changed the binding proteins to JunD, c-Fos, and Fra-1 (data not shown). In summary, we show that region I, which contains a CRE/AP-1-like element (−49 to −42), binds both AP-1 and CREB/ATF proteins using EMSA analysis. Moreover, mutational analysis demonstrated that this region is crucial for constitutive hNT/N gene expression.
The Orphan Hormone Receptor NR2F2 Binds to Region II of the hNT/N Promoter
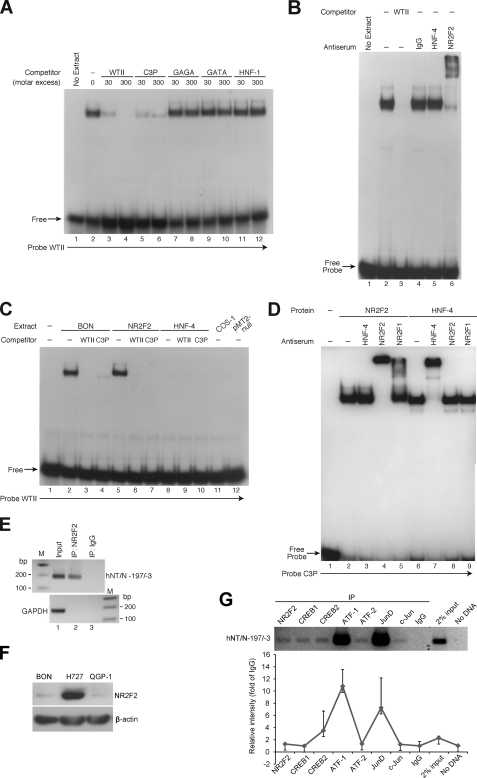
Analysis of the sequence within region II revealed homologies to known binding sites for transcription factors such as GAGA, GATA, and HNF-1. In addition, this area of the hNT/N promoter resembled an element known as C3P present in the apolipoprotein CIII (apoC III) promoter, which can bind both HNF-4 and NR2F2 (35). Accordingly, to identify the proteins binding to this region, double-stranded oligonucleotides representing consensus sequences for the binding of these transcription factors were synthesized and tested for the ability to compete for binding to the radiolabeled hNT/N oligonucleotide (−106 to −77) corresponding to this footprinted region (Fig. 5A). The addition of BON cell nuclear extract to the labeled hNT/N probe produced one major DNA-protein complex (Fig. 5A, lane 2), which was effectively competed by molar excess of unlabeled probe (lanes 3 and 4). No competition for binding was noted with consensus oligonucleotides to the GAGA, GATA, and HNF-1 sequences (Fig. 5A, lanes 7–12). In contrast, almost the entire complex was competed by the oligonucleotide corresponding to the C3P site (Fig. 5A, lanes 5 and 6). Because the C3P site binds both NR2F2 and HNF-4 in the apoC III gene, these findings suggested the binding of one or both proteins to the hNT/N gene promoter. To elucidate the specific protein binding to this region, EMSAs were repeated using antisera to HNF-4 and NR2F2 (Fig. 5B). Antisera to NR2F2 (Fig. 5B, lane 6), but not HNF-4 (lane 5), supershifted almost the entire complex with only a residual amount of the DNA-protein complex remaining, which is similar to the findings in the competition experiments (Fig. 5A, lane 6). These findings provided compelling evidence that NR2F2, but not HNF-4, binds to region II (−100 to −76) of the hNT/N promoter.
FIGURE 5.
Identification of the NR2F2 protein that binds to region II (−100 to −76) of the hNT/N promoter. A, competitive binding analysis by EMSA with BON cell nuclear extracts (10 μg) and wild-type region II (WTII) as the probe. Unlabeled oligonucleotides containing consensus sites for GAGA, GATA, and HNF-1 were used as competitors at 30- and 300-fold molar excess. In addition, unlabeled WTII and oligonucleotide C3P, which contains a site that binds to both NR2F2 and HNF-4, were also used as competitors. B, nuclear extracts (10 μg/lane) from BON cells were preincubated with antisera (2 μl) to the NR2F2 and HNF-4 proteins prior to the addition of radiolabeled WTII. Supershifted complexes were noted for NR2F2 (lane 6). C, overexpressing plasmids of NR2F2 or HNF-4 and the control vector, pMT2, were transfected into COS-1 cells. Protein extracts from BON cells and transfected COS-1 cells were incubated with labeled WTII for EMSA analysis. Unlabeled WTII and oligonucleotide C3P were used as competitors at 200-fold molar excess. D, protein extracts from COS-1 cells transfected with NR2F2 or HNF-4 overexpression plasmids were preincubated with antisera to NR2F2, NR2F1, or HNF-4 prior to the addition of [32P]ATP-labeled oligonucleotide C3P. Supershifted complexes were noted for NR2F2 (lane 4), HNF-4 (lane 7), and NR2F1 (lane 5). E, BON cells were subjected to ChIP assay; soluble chromatin was prepared from BON cells transfected with NR2F2 and immunoprecipitated (IP) with NR2F2 antibody (lane 2) or IgG (lane 3). Total (Input, lane 1) and immunoprecipitated DNAs were then PCR-amplified using primer pairs covering the NR2F2 binding site within the hNT/N promoter or GAPDH gene (control). F, Western blotting analysis was performed to detect NR2F2 expression level in BON cells compared with NCI-727 and QGP-1 cells. G, a ChIP assay was performed as described in E by precipitating endogenous NR2F2, CREB1, CREB2, ATF1, ATF2, JunD, and c-Jun (top panel). Densitometric analysis composes the -fold change of PCR products to IgG from two independent PCR analyses.
To further confirm these findings, plasmids containing either the NR2F2 or HNF-4 cDNA in a pMT2 vector were transfected into COS-1 cells, and protein was extracted for analysis by EMSA (Fig. 5C). As noted previously, BON cell extract added to the labeled hNT/N probe (−106 to −77) resulted in one predominant complex, which was effectively competed with either excess unlabeled probe or the oligonucleotide containing the C3P site (Fig. 5C, lanes 2–4). The addition of protein extract from COS-1 cells after transfection with the pMT-ARP-1 (NR2F2) plasmid resulted in a protein-DNA complex with mobility identical to that of BON cell extract (Fig. 5C, lane 5). Similarly, this complex was competed with either unlabeled −106 to −77 or C3P probes (Fig. 5C, lanes 6 and 7). In contrast, no binding was noted using protein extracts from COS-1 cells alone or from those transfected with pMT2-HNF-4 or pMT2-null control plasmid (Fig. 5C, lanes 8–12). Collectively, these complementary procedures identified NR2F2, but not HNF-4, as the protein binding to region II.
To rule out a possible problem with the extract obtained from COS-1 cells transfected with pMT2-HNF-4, the C3P oligonucleotide was labeled and added to either the NR2F2 or HNF-4 extracts. Binding was noted using both extracts. The addition of HNF-4 antisera resulted in a supershifted complex within the HNF-4 extract, and NR2F2 antisera produced a supershift within the APR-1/NR2F2 extract. No cross-reactivity occurred between the two antibodies (Fig. 5D).
Occupancy of the hNT/N promoter by NR2F2 in cells was further analyzed using a ChIP assay. Cross-linked chromatin was prepared from BON cells transfected with the NR2F2 expression plasmid. The hNT/N promoter region containing the NR2F2 binding site was precipitated using either the anti-NR2F2 antibody (Santa Cruz Biotechnology) or IgG (as the negative control), and the sequence (194 bp) encompassing the NR2F2 binding site was amplified using gene-specific primers. As shown in Fig. 5E, precipitation with NR2F2 (lane 2), but not IgG (lane 3), enabled amplification. No PCR amplification of the immunoprecipitated chromatin was detected with primers for GAPDH (166 bp of PCR product, negative control), thus confirming the specificity of our results. The binding of the hNT/N promoter and NR2F2 was further confirmed by precipitating the endogenous NR2F2 protein. First, the endogenous NR2F2 expression level was detected in BON cells (Fig. 5F). Relatively low NR2F2 expression was noted in BON and QGP-1 cells (pancreatic endocrine tumor cell line) compared with NCI-H727 cells (human lung carcinoid tumor cell line). Furthermore, PCR amplification was detected from endogenous NR2F2-precipitated chromatin (Fig. 5G, top panel). In addition, PCR amplification was also noted from chromatin precipitated by endogenous CREB1, CREB2, ATF1, ATF2, JunD, and c-Jun, which further confirmed the binding of these proteins to the NT/N promoter as shown by the EMSA in Fig. 3. Densitometric analysis was performed to reflect the levels of PCR amplification (Fig. 5G, lower panel). Together, these data confirm that the hNT/N promoter is occupied by NR2F2 in BON cells within region II.
NR2F2 Represses NT/N Promoter Activity
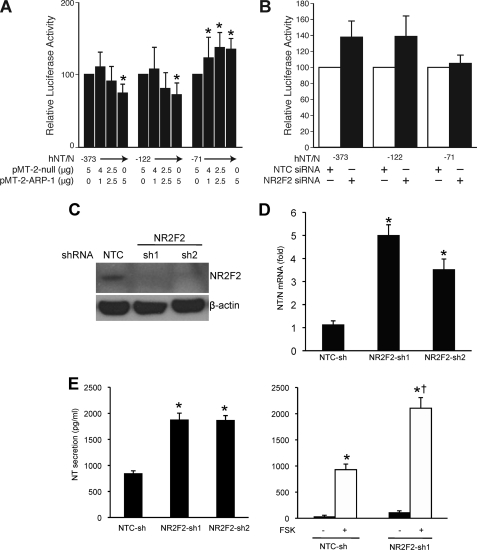
We next determined the effect of NR2F2 on the activity of the hNT/N promoter by co-transfecting the NR2F2 expression plasmid with various 5′-deletion constructs of the hNT/N promoter into BON cells (Fig. 6A). Increasing amounts of NR2F2 inhibited the activity of the hNT/N promoter in constructs that contained the NR2F2 binding site (i.e. −373 and −122). In contrast, luciferase activity increased for deletion constructs that lacked the NR2F2 site, suggesting that NR2F2 may cooperate with the transcriptional machinery to increase luciferase, as noted by others (46, 47). On the other hand, co-transfection of hNT/N promoter with NR2F2 siRNA increased the activity of the −373 and −122 hNT/N constructs (which contain the NR2F2 binding site) by more than 35%, whereas the activity for the hNT-71 plasmid (without the NR2F2 binding site) was not affected (Fig. 6B). To further confirm the physiologic regulation of NR2F2 on the NT/N gene, we established stable NR2F2 knockdown cell lines in BON cells by constitutively expressing NR2F2 shRNA. The knockdown of NR2F2 was noted in both stable knockdown cell lines compared with the NTC-sh cell line (Fig. 6C). NT/N mRNA expression was significantly increased in NR2F2 knockdown cells, as shown by real-time PCR (Fig. 6D). Moreover, NR2F2 knockdown increased basal NT peptide secretion (Fig. 6E). Previously, we had demonstrated that forskolin (FSK), a cyclic AMP activator, stimulates NT peptide secretion in BON cells (44). Therefore, we used FSK as a secretagogue to stimulate NT peptide secretion in NR2F2 knockdown cells and found that NR2F2 knockdown further increased FSK-stimulated NT peptide secretion. These data demonstrate the functional role of NR2F2 in the regulation of NT/N gene expression and NT protein secretion.
FIGURE 6.
NR2F2 represses hNT/N promoter activity. A, BON cells were co-transfected with hNT/N promoter plasmids (−373, −122, and −71) and expression plasmids for NR2F2 (pMT-2ARP-1) or with empty vector (pMT-2-null) and a β-galactosidase plasmid (internal control). Luciferase activities are expressed as a percentage of the hNT/N plasmids (−373, −122, −71) without the NR2F2 construct (*, p > 0.05 hNT/N plasmid alone). B, BON cells were co-transfected with hNT/N promoter plasmids (−373, −122, and −71) and NR2F2 siRNA or nontargeting control siRNA (Dharmacon) and with pRL-RK (Promega, internal control) using the DharmaFECT Duo transfection reagent (Dharmacon). Luciferase activities are expressed as a percentage of hNT/N promoter activity in the presence of control siRNA and are the mean ± S.D. of three separate transfections after normalization to Renilla luciferase (pRL-TK) expression. C, stable NR2F2 knockdown BON cell lines were established as shown by the Western blot. D, real-time PCR was performed to quantify NT/N mRNA levels in BON cells with stable NR2F2 knockdown (*, p > 0.05 versus NTC-sh). E, an NT EIA assay was performed to measure basal (left panel) and FSK-stimulated (right panel) NT secretion in BON cells with NR2F2 knockdown (*, p > 0.05 versus NTC-sh; *†, p > 0.05 versus NTC-sh plus FSK).
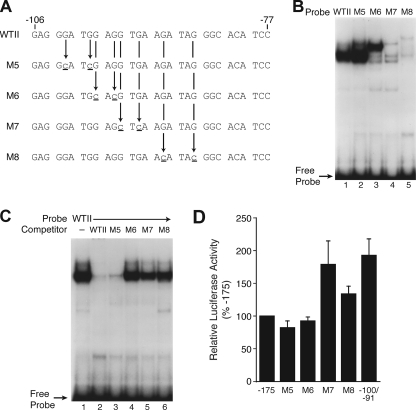
To further delineate the role of NR2F2 in the expression of hNT/N, mutational analysis was performed with two specific point mutations within region II (Fig. 7A). Similar to the analysis performed for the proximal promoter region (region I, −54 to −38), EMSAs were first performed to determine whether the site-specific mutations affected protein binding (Fig. 7B). Wild type (−106 to −77) and mutated oligonucleotides were labeled with [32P]ATP and used for EMSAs with BON cell nuclear extracts. Mutation of the two nucleotides at −102 and −99 (M5) enhanced protein binding (Fig. 7B, lane 2); no binding occurred with M7 and M8 (lanes 4 and 5), whereas a different binding pattern compared with the wild type probe was obtained with M6 (lane 3). An EMSA competition was performed to confirm these findings (Fig. 7C). The DNA-protein complex was effectively competed with a molar excess of wild type (−106 to −77) oligonucleotide and M5 (Fig. 7C, lanes 2 and 3), whereas M6, M7, and M8 had a minimal effect on DNA-protein binding (lanes 4–6).
FIGURE 7.
Site-directed mutagenesis analysis of region II. A, schematic representation of site-directed mutagenesis of region II. B, the wild type (WTII) hNT/N promoter fragment (−106 to −77) and its mutants (M5–M8) were radiolabeled and incubated with BON cell nuclear extracts (10 μg), and was performed as described under “Experimental Procedures.” C, competitive binding analysis by EMSA with BON nuclear extracts (10 μg) and unlabeled WTII and mutant probes M5–M8 (at 200-fold molar excesses) as competitors. D, the −175 to +26 region of the hNT/N promoter and its mutants (M5–M8) were cloned into the luciferase reporter plasmid, pXP1, and transiently transfected into BON cells. Luciferase activities are expressed as a percentage of the wild type (−175 to +26) fusion plasmid and are the mean ± S.D. of four separate transfections after normalization to β-galactosidase expression.
To assess the functional consequences of these mutations on hNT/N promoter activity, site-directed mutagenesis was performed using the hNT/N promoter (−175 to +26) linked to the luciferase reporter gene, and the plasmids were transiently transfected into BON cells (Fig. 7D). Mutation of the nucleotides at −102 and −99 (M5) had minimal effect on hNT/N promoter activity. Mutation of nucleotides at −95 and −93 (M7) and at −90 and −86 (M8), located in the core area of region II (−101 to −77), increased hNT/N promoter activity. These findings are consistent with the EMSAs demonstrating absence of protein binding and are similar to the −100 to −90 replacement mutagenesis results (Fig. 1B). Interestingly, mutation of the nucleotides at −98 and −96 (M6), which produced a different DNA-protein complex, had no effect on hNT/N promoter activity, indicating that NR2F2 function likely involves multiple mechanisms. Taken together, overexpression of NR2F2 decreased hNT/N promoter activity in a dose-dependent fashion; site-specific mutagenesis in the core area (−95 to −86) of region II prevented NR2F2 protein binding and increased hNT/N promoter activity, thereby indicating that the −95 to −86 area is crucial for suppression of the hNT/N promoter by NR2F2.
The C Terminus of NR2F2 Contains the Functional Domain for hNT/N Promoter Repression
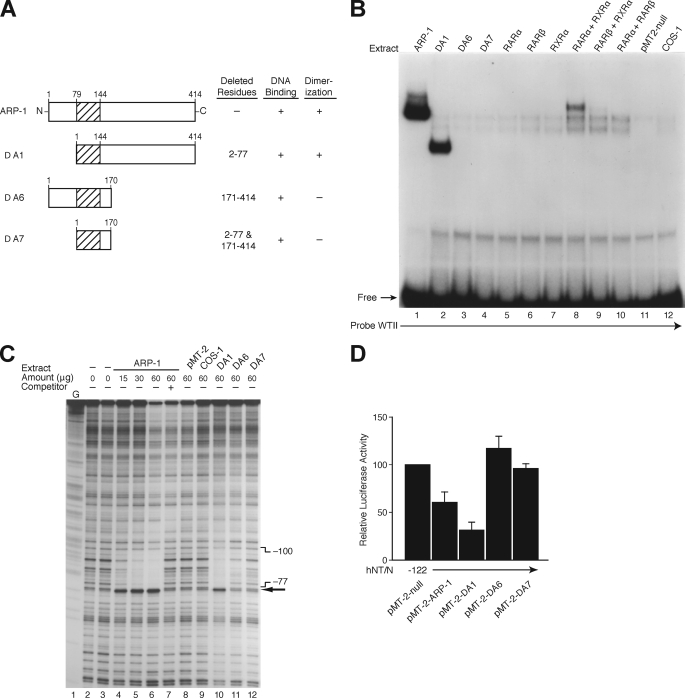
The C terminus of NR2F2 has been defined by several groups as the active repression and transrepression domain for numerous genes (31–33). To determine the mechanism for NR2F2-dependent hNT/N promoter repression, several different NR2F2 mutants (Fig. 8A) were transfected into COS-1 cells, and crude extract was made for EMSAs and the DNase I footprinting assay. For comparison, overexpression plasmids of members of the nuclear receptor superfamily, RARα, RARβ, and RXRα, were included for EMSA as it had been shown previously that NR2F2 may heterodimerize with RAR/RXR to repress promoter activity (31–33). As shown in Fig. 8A, mutant ΔA1 retains the C terminus, which contains the active repression domain. Incubation of radiolabeled wild type II probe with crude COS-1 extract containing wild type NR2F2 or the ΔA1 mutant resulted in similar patterns of protein binding (Fig. 8B, lanes 1 and 2). The binding band for ΔA1 was relatively smaller, as ΔA1 is missing 78 amino acids (∼19%) of the N terminus of wild type NR2F2. ΔA6 and ΔA7 did not effectively bind in the EMSA reactions (Fig. 8B, lanes 3 and 4). Interestingly, binding occurred for RARα combined with RXRα, RARβ combined with RXRα, and RARα combined with RARβ, thus indicating that heterodimerization is crucial for the proper functioning of these proteins. Moreover, footprinting analysis (Fig. 8C) demonstrated that incubation of the radiolabeled coding strand (−373 to +26) with crude extracts from COS-1 cells overexpressing wild type NR2F2 resulted in the appearance of a single protected region (−100 to −77) in a dose-dependent fashion (Fig. 8C, lanes 4–6); it was effectively competed by a molar excess of unlabeled probe (lane 7). These data are consistent with the footprinting results of region II shown in Fig. 2A utilizing BON cell nuclear extracts. Furthermore, the ΔA1 mutant (Fig. 8C, lane 10), but not ΔA6 and ΔA7, resulted in the same protected region as did wild type ARP-1 (lanes 4–6), which is consistent with the EMSA findings from Fig. 8B.
FIGURE 8.
Identification of the functional domains for NR2F2. A, schematic representation of the ARP-1 (NR2F2) deletion mutants used. B, expression plasmids for ARP-1 (NR2F2) and its mutants (Δ1, Δ6, and Δ7) or members of the nuclear receptor superfamily (RARα/β and RXRα/β) and the empty vector, pMT-2-null, were transfected into COS-1 cells. After 48 h, crude protein extracts (20 μg) from transfected or untransfected COS-1 cells were incubated with [32P]ATP-labeled WTII probe for EMSA analysis. C, DNase I footprinting of the NR2F2 site. The coding strand of the hNT/N promoter (−373 to +26) was labeled, incubated with crude protein from COS-1 cells transfected with or without ARP-1 (NR2F2) and its mutants (Δ1, Δ6, and Δ7), and then digested with DNase I. Areas protected from digestion are bracketed with the corresponding nucleotide positions, which were determined by running a DNA sequencing ladder (G reaction) in parallel. The numbers at the top of the autoradiogram are the amounts of nuclear extract (in μg of protein) used in each reaction mixture. A 100-fold molar excess of the oligonucleotide corresponding to the hNT/N promoter fragment −106 to −77 was used as a competitor (lane 7). D, BON cells were co-transfected with the −122 hNT/N promoter plasmid and expression plasmids for NR2F2 (pMT-2-ARP-1) or its deletion mutants (Δ1, Δ6, and Δ7) with the empty vector, pMT-2, and the β-galactosidase plasmid (internal control). Luciferase activities are expressed as the percentage of the hNT-122 plasmid without NR2F2 and are the mean ± S.D. of four separate transfections after normalization to β-galactosidase expression.
Finally, to assess the functional consequences of these NR2F2 mutations, we co-transfected wild type NR2F2 or the various mutants with the hNT/N −122 promoter plasmid (Fig. 8D). Consistent with the EMSA and footprinting assays, hNT/N −122 activity was decreased by overexpression of the wild type or ΔA1 mutant but not with the ΔA6 and ΔA7 mutants. Notably, the repressing effect of the ΔA1 mutant was stronger than wild type NR2F2. These data, taken together with the findings shown in Fig. 7D, which demonstrated that NR2F2 function is dependent on the sequences from −95 to −86, suggested that the active repression domain contained in the C terminus is crucial for hNT/N transcriptional repression and is antagonized by another domain in the N-terminal region.
DISCUSSION
The findings in this study identify crucial cis-regulatory elements of the hNT/N gene required for cell-specific transcriptional activity in the novel human endocrine BON cell line. These elements include two pivotal regions: region I (−55 to −39) that functions as an activating region and region II (−101 to −77) that binds repressor proteins. We show that the −55 to −39 region contains a CRE/AP-1-like element (−50 to −43) that binds both AP-1 and CREB/ATF proteins, including c-Jun, ATF-1, ATF-2, JunD, and CREB; this CRE/AP-1-like element is crucial for constitutive expression of the hNT/N gene. The −101 to −77 region binds to the orphan hormone receptor, NR2F2; a core area from −95 to −86 is crucial for the regulation of hNT/N promoter activity by NR2F2 as a consequence of regulation of protein binding. Specifically, the C terminus of NR2F2 functionally represses hNT/N gene transcription, whereas the N-terminal domain may antagonize this repression.
We identified a novel interaction in the protected region (region II) between nucleotides −101 and −77 of the hNT/N promoter that bind the orphan hormone receptor, NR2F2. An EMSA performed using antisera to NR2F2 supershifted almost the entire complex with only a residual amount of the DNA-protein complex remaining. Furthermore, increased expression of NR2F2 in BON cells decreased hNT/N promoter activity, whereas mutation of nucleotides in the core of region II led to increased hNT/N promoter activity, most likely due to the absence of NR2F2 protein binding. Similar to findings with other genes, we showed that the active repression domain is contained within the C terminus of NR2F2 (31–33). Interestingly, the repressive effect of the ΔA1 mutant, which lacks 78 N-terminal amino acids, was stronger than that of wild type NR2F2. We suspect that there may be another unidentified domain in the N terminus of NR2F2 that is not present in the ΔA1 mutant, which may antagonize the repressive effects of the C-terminal portion of NR2F2. Further mutational analysis of the N-terminal region will be necessary to confirm these findings.
A comparison of the human and rat NT/N proximal promoter sequences revealed a fundamental difference. We showed previously that in the rat NT/N promoter, the sequences corresponding to protected region II encode a glucocorticoid response element that is required for constitutive NT/N expression (24). However, in the human NT/N gene promoter, this region is altered and, as a result, binds NR2F2, which serves to repress NT/N transcription. There are several diverse examples of such interspecies variability in gene regulation. For instance, HNF-4α is a liver-enriched transcription factor that plays a key role in regulating liver metabolism and development. In addition to the conserved core binding sites for HNF-1α/β, HNF-6, and GATA-6 that are also found in the mouse promoter. Hatzis and Talianidis (48) identified a direct repeat sequence encoding a functional hormone response element in the human promoter of HNF-4α, which is not conserved in the mouse promoter region. Surprisingly, they found that this element binds NR2F2 as well, which has a repressive effect on promoter activity. As another example, Nurr1, a nuclear orphan receptor involved in neuronal differentiation, binds a response element involved in regulating transcription of the tyrosine hydroxylase (TH) gene, a key regulator of synthesis of catecholamine neurotransmitters (49, 50). In the murine system, Nurr1 is required to transactivate mouse TH minimal promoters (51). However, Nurr1 has no effect on human TH gene regulation. In fact, there are no Nurr1 binding sites found in the human TH promoter (52).
Our data demonstrate that mutation at nucleotide −85 and −84 abolished NR2F2 protein binding, resulting in a ∼50% decrease in hNT/N promoter activity; potentially this is due to the release of a positive factor from this element. Furthermore, deletion from −122 to −95 of the hNT/N promoter resulted in a ∼60% decrease in hNT/N promoter activity, further indicating the release of a positive factor. These findings suggest that, in addition to NR2F2, other regulatory factors (or co-factors) play a role in transcriptional regulation of the hNT/N promoter. Jordan et al. (53) have demonstrated that mutations at different nucleotides of the NR2F2 binding site in the F1F0-ATP synthase α-subunit gene (ATPA) cis-acting regulatory element-1 disrupt NR2F2 protein binding, consequently leading to a decrease and/or increase in ATPA promoter activity due to the release of positive and/or negative factors, respectively. One such factor, upstream regulatory factor-2 (USF2), stimulates transcription of the ATPA gene through the same cis-acting regulatory element-1. Co-transfection assays demonstrated that NR2F2 inhibits the USF2-mediated activation of the wild type ATPA gene promoter, whereas overexpression of USF2 reversed the NR2F2-mediated repression of the ATPA promoter. EMSA analysis revealed that NR2F2 and USF2 compete for binding to the ATPA regulatory element-1. Identification of other such factors that interact with NR2F2 and elucidating their functional significance will allow a more complete understanding of the molecular mechanism underlying transcriptional regulation of the hNT/N gene.
In higher vertebrates, the NR2F family contains two highly conserved members, NR2F1 and NR2F2. NR2F2 was also cloned as the apolipoprotein regulatory protein-1 (ARP-1). NR2Fs are widely expressed and generally act as transcriptional repressors. Several mechanisms have been identified whereby NR2Fs inhibit gene transcription including active repression, transrepression, and competition for occupancy of binding sites (31–33). Active repression is dependent on the DNA binding site; NR2Fs have been shown to actively repress the basal transcriptional activity of a number of genes. Our data indicate that NR2F2 represses the basal activity of the hNT/N promoter by active repression under normal conditions. Evidence in support for this hypothesis includes the finding that the C terminus, which contains the active repression domain, is crucial for hNT/N repression. More importantly, mutation of the core area (−95 to −86) of the NR2F2 binding site prevented NR2F2 protein binding and attenuated repression (increased hNT/N promoter activity). Notably, mutation at nucleotides −98 and −96 abolished NR2F2 protein binding (resulting in a different protein binding pattern) but did not affect hNT/N promoter activity. We speculate that this effect was due to transrepression (which does not require direct DNA binding) as a result of heterodimerization with other co-factors. One of the mechanisms for transrepression by NR2Fs involves heterodimerization with RAR/RXR, thyroid hormone receptor, and other nuclear hormone receptors (31–33). In this regard, we found that co-expression of RARα or RXRα resulted in significant protein binding with labeled WTII probe, suggesting that NR2F2 is able to heterodimerize with RARα or RXRα to repress promoter activity. Future studies will be directed toward examining the role of these factors in regulating hNT/N promoter activity.
Previous studies have shown that fat ingestion induces a potent, dose-related increase in plasma concentrations of NT, whereas glucose and amino acids produce only minor effects (1, 2). The major physiological functions of NT include stimulation of pancreatic exocrine secretion, inhibition of gastroduodenal motility, and inhibition of gastric acid secretion (4–8). Moreover, NT has also been shown to play a role in the absorption of fatty acids and triglycerides from the intestinal lumen into the lymphatic system where they are transported to the liver and other organs for storage and energy production (9–12). Interestingly, NR2F2 has also been characterized molecularly as regulating the transcription of genes involved in fat metabolism including several apolipoproteins (apoA-I, apoA-II, apoA-IV, apoB, apoC-II, and apoC-III) that are involved in the transport of fatty acids in the bloodstream (31, 32, 35). In the present study, we have shown for the first time that the transcription of the gut hormone NT is regulated by the orphan receptor superfamily member NR2F2. It is interesting to speculate that NR2F2 may be a master regulator of lipid metabolism that acts to regulate several different steps in the process including the uptake of fatty acids (by regulating the level of hNT/N expression) and the transport of these fatty acids to their target organs via the bloodstream (by regulating the level of apolipoprotein expression).
Protected region I, located between nucleotides −55 and −39 of the proximal hNT/N promoter, is remarkably well conserved in both rat and hNT/N genes (24). It contains a near consensus CRE/AP-1-like motif that binds a complex of proteins including c-Jun, JunD, CREB, ATF-1, and ATF-2. This multifunctional element serves a pivotal role in the regulation of NT/N gene expression and also the tissue-specific or developmental expression of several other genes including the c-jun proto-oncogene, T-cell receptors α and β, and CD3, β-interferon, and E-selectin (24). The Jun, ATF, and CREB proteins belong to the leucine zipper family of proteins (26). Jun proteins can heterodimerize with proteins of the Fos family (26, 27). Moreover, the different CREB/ATFs selectively heterodimerize with each other or with other members of the leucine zipper-rich family such as c-Jun to generate either stimulatory or inhibitory complexes (45, 54). This functional cross-talk between different signaling pathways may be a mechanism by which members of various transcription factor families can limit the expression of certain genes. Furthermore, there are also qualitative and quantitative tissue type specificities in the distribution of the various AP-1 and CREB/ATF-binding proteins, suggesting that these factors may play a role in tissue-specific gene expression (54).
In summary, we utilized the novel BON endocrine cell line to delineate the regulatory elements involved in cell-specific expression of the hNT/N gene. We have identified two crucial cis-regulatory elements, one that results in hNT/N activation and another that functions to repress hNT/N expression. The former region contains a CRE/AP-1-like element that binds both AP-1 and CREB/ATF proteins and is crucial for constitutive hNT/N expression. Meanwhile, the latter region binds to the orphan hormone receptor, NR2F2; the C terminus of NR2F2 strongly represses transcription of the hNT/N gene. Importantly, we have shown regulation of NT/N expression by the NR2F family of receptors, which may play an important role in regulating different steps in the cascade of lipid absorption and transport.
Acknowledgments
We thank Karen Martin for manuscript preparation and Nathan L. Vanderford for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R37 AG10855 and R01 DK48489.
- NT
- neurotensin
- N
- neuromedin N
- hNT/N
- human neurotensin/neuromedin N
- GI
- gastrointestinal
- CRE
- cAMP-responsive element
- CREB
- cAMP-responsive element-binding protein
- ATF
- activating transcription factor
- NR2F
- nuclear receptor subfamily 2, group F
- HNF
- hepatocyte nuclear factor
- RAR
- retinoic acid receptor
- NTC
- nontargeting control
- EIA
- enzyme immunoassay
- FSK
- forskolin
- TH
- tyrosine hydroxylase
- RXR
- retinoid X receptors.
REFERENCES
- 1. Evers B. M. (2006) Peptides 27, 2424–2433 [DOI] [PubMed] [Google Scholar]
- 2. Ferris C. F., Carraway R. E., Hammer R. A., Leeman S. E. (1985) Regul. Pept. 12, 101–111 [DOI] [PubMed] [Google Scholar]
- 3. Polak J. M., Sullivan S. N., Bloom S. R., Buchan A. M., Facer P., Brown M. R., Pearse A. G. (1977) Nature 270, 183–184 [DOI] [PubMed] [Google Scholar]
- 4. Baca I., Feurle G. E., Schwab A., Mittmann U., Knauf W., Lehnert T. (1982) Digestion 23, 174–183 [DOI] [PubMed] [Google Scholar]
- 5. Sakamoto T., Newman J., Fujimura M., Greeley G. H., Jr., Townsend C. M., Jr., Thompson J. C. (1984) Surgery 96, 146–153 [PubMed] [Google Scholar]
- 6. Wood J. G., Hoang H. D., Bussjaeger L. J., Solomon T. E. (1988) Pancreas 3, 332–339 [DOI] [PubMed] [Google Scholar]
- 7. Andersson S., Rosell S., Hjelmquist U., Chang D., Folkers K. (1977) Acta Physiol. Scand. 100, 231–235 [DOI] [PubMed] [Google Scholar]
- 8. Thor K., Rosell S. (1986) Gastroenterology 90, 27–31 [DOI] [PubMed] [Google Scholar]
- 9. Armstrong M. J., Parker M. C., Ferris C. F., Leeman S. E. (1986) Am. J. Physiol. 251, G823–G829 [DOI] [PubMed] [Google Scholar]
- 10. Kitabgi P. (2002) Curr. Opin. Drug Discov. Devel. 5, 764–776 [PubMed] [Google Scholar]
- 11. Thomas R. P., Hellmich M. R., Townsend C. M., Jr., Evers B. M. (2003) Endocr. Rev. 24, 571–599 [DOI] [PubMed] [Google Scholar]
- 12. Slogoff M., Evers B. M. (2003) in Encyclopedia of hormones (Henry H. L., Norman A. W. eds) pp. 19–23, Academic Press, San Diego [Google Scholar]
- 13. Feurle G. E., Müller B., Rix E. (1987) Gut 28, (suppl.) 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evers B. M., Izukura M., Chung D. H., Parekh D., Yoshinaga K., Greeley G. H., Jr., Uchida T., Townsend C. M., Jr., Thompson J. C. (1992) Gastroenterology 103, 86–91 [DOI] [PubMed] [Google Scholar]
- 15. Chung D. H., Evers B. M., Shimoda I., Townsend C. M., Jr., Rajaraman S., Thompson J. C. (1992) Gastroenterology 103, 1254–1259 [DOI] [PubMed] [Google Scholar]
- 16. Evers B. M., Izukura M., Rajaraman S., Parekh D., Thakore K., Yoshinaga K., Uchida T., Townsend C. M., Jr., Thompson J. C. (1994) Am. J. Physiol. 267, G180–G186 [DOI] [PubMed] [Google Scholar]
- 17. Evers B. M., Izukura M., Townsend C. M., Jr., Uchida T., Thompson J. C. (1992) Dig. Dis. Sci. 37, 426–431 [DOI] [PubMed] [Google Scholar]
- 18. Wood J. G., Hoang H. D., Bussjaeger L. J., Solomon T. E. (1988) Am. J. Physiol. 255, G813–G817 [DOI] [PubMed] [Google Scholar]
- 19. Bean A. J., Dagerlind A., Hökfelt T., Dobner P. R. (1992) Neuroscience 50, 259–268 [DOI] [PubMed] [Google Scholar]
- 20. Kislauskis E., Bullock B., McNeil S., Dobner P. R. (1988) J. Biol. Chem. 263, 4963–4968 [PubMed] [Google Scholar]
- 21. Evers B. M., Ehrenfried J. A., Wang X., Townsend C. M., Jr., Thompson J. C. (1994) Am. J. Physiol. 267, G875–G882 [DOI] [PubMed] [Google Scholar]
- 22. Evers B. M., Rajaraman S., Chung D. H., Townsend C. M., Jr., Wang X., Graves K., Thompson J. C. (1993) Am. J. Physiol. 265, G482–G490 [DOI] [PubMed] [Google Scholar]
- 23. Muraki K., Mitra S. P., Dobner P. R., Carraway R. E. (1993) Peptides 14, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 24. Evers B. M., Wang X., Zhou Z., Townsend C. M., Jr., McNeil G. P., Dobner P. R. (1995) Mol. Cell. Biol. 15, 3870–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kislauskis E., Dobner P. R. (1990) Neuron 4, 783–795 [DOI] [PubMed] [Google Scholar]
- 26. Busch S. J., Sassone-Corsi P. (1990) Trends Genet. 6, 36–40 [DOI] [PubMed] [Google Scholar]
- 27. Curran T., Franza B. R., Jr. (1988) Cell 55, 395–397 [DOI] [PubMed] [Google Scholar]
- 28. Haus-Seuffert P., Meisterernst M. (2000) Mol. Cell. Biochem. 212, 5–9 [PubMed] [Google Scholar]
- 29. Shibata H., Spencer T. E., Oñate S. A., Jenster G., Tsai S. Y., Tsai M. J., O'Malley B. W. (1997) Recent Prog. Horm. Res. 52, 141–165 [PubMed] [Google Scholar]
- 30. Tsai M. J., O'Malley B. W. (1994) Annu. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 31. Pereira F. A., Tsai M. J., Tsai S. Y. (2000) Cell. Mol. Life Sci. 57, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park J. I., Tsai S. Y., Tsai M. J. (2003) Keio J. Med. 52, 174–181 [DOI] [PubMed] [Google Scholar]
- 33. Achatz G., Hölzl B., Speckmayer R., Hauser C., Sandhofer F., Paulweber B. (1997) Mol. Cell. Biol. 17, 4914–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rottman J. N., Gordon J. I. (1993) J. Biol. Chem. 268, 11994–12002 [PubMed] [Google Scholar]
- 35. Mietus-Snyder M., Sladek F. M., Ginsburg G. S., Kuo C. F., Ladias J. A., Darnell J. E., Jr., Karathanasis S. K. (1992) Mol. Cell. Biol. 12, 1708–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carraway R. E., Mitra S. P., Evers B. M., Townsend C. M., Jr. (1994) Regul. Pept. 53, 17–29 [DOI] [PubMed] [Google Scholar]
- 37. Evers B. M., Ishizuka J., Townsend C. M., Jr., Rajaraman S., Thompson J. C. (1991) Ann. Surg. 214, 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evers B. M., Townsend C. M., Jr., Upp J. R., Allen E., Hurlbut S. C., Kim S. W., Rajaraman S., Singh P., Reubi J. C., Thompson J. C. (1991) Gastroenterology 101, 303–311 [DOI] [PubMed] [Google Scholar]
- 39. Kunkel T. A., Roberts J. D., Zakour R. A. (1987) Methods Enzymol. 154, 367–382 [DOI] [PubMed] [Google Scholar]
- 40. Hall C. V., Jacob P. E., Ringold G. M., Lee F. (1983) J. Mol. Appl. Genet. 2, 101–109 [PubMed] [Google Scholar]
- 41. Evers B. M., Zhou Z., Celano P., Li J. (1995) J. Clin. Invest. 95, 2822–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shapiro D. J., Sharp P. A., Wahli W. W., Keller M. J. (1988) DNA 7, 47–55 [DOI] [PubMed] [Google Scholar]
- 43. Li J., Chen L. A., Townsend C. M., Jr., Evers B. M. (2008) J. Biol. Chem. 283, 2614–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J., O'Connor K. L., Cheng X., Mei F. C., Uchida T., Townsend C. M., Jr., Evers B. M. (2007) Mol. Endocrinol. 21, 159–171 [DOI] [PubMed] [Google Scholar]
- 45. Lee K. A. (1992) J. Cell Sci. 103, 9–14 [DOI] [PubMed] [Google Scholar]
- 46. Everett L. M., Crabb D. W. (1999) J. Steroid Biochem. Mol. Biol. 70, 197–201 [DOI] [PubMed] [Google Scholar]
- 47. Kimura A., Nishiyori A., Murakami T., Tsukamoto T., Hata S., Osumi T., Okamura R., Mori M., Takiguchi M. (1993) J. Biol. Chem. 268, 11125–11133 [PubMed] [Google Scholar]
- 48. Hatzis P., Talianidis I. (2001) Mol. Cell. Biol. 21, 7320–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim J. H., Auerbach J. M., Rodríguez-Gómez J. A., Velasco I., Gavin D., Lumelsky N., Lee S. H., Nguyen J., Sánchez-Pernaute R., Bankiewicz K., McKay R. (2002) Nature 418, 50–56 [DOI] [PubMed] [Google Scholar]
- 50. Zetterström R. H., Solomin L., Jansson L., Hoffer B. J., Olson L., Perlmann T. (1997) Science 276, 248–250 [DOI] [PubMed] [Google Scholar]
- 51. Kim K. S., Kim C. H., Hwang D. Y., Seo H., Chung S., Hong S. J., Lim J. K., Anderson T., Isacson O. (2003) J. Neurochem. 85, 622–634 [DOI] [PubMed] [Google Scholar]
- 52. Jin H., Romano G., Marshall C., Donaldson A. E., Suon S., Iacovitti L. (2006) J. Cell. Physiol. 207, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jordan E. M., Worley T., Breen G. A. (2003) Biochemistry 42, 2656–2663 [DOI] [PubMed] [Google Scholar]
- 54. Habener J. F. (1990) Mol. Endocrinol. 4, 1087–1094 [DOI] [PubMed] [Google Scholar]