Abstract
In the yeast Saccharomyces cerevisiae, mitochondrial cytochrome c oxidase (COX) biogenesis is translationally regulated. Mss51, a specific COX1 mRNA translational activator and Cox1 chaperone, drives the regulatory mechanism. During translation and post-translationally, newly synthesized Cox1 physically interacts with a complex of proteins involving Ssc1, Mss51, and Cox14, which eventually hand over Cox1 to the assembly pathway. This step is probably catalyzed by assembly chaperones such as Shy1 in a process coupled to the release of Ssc1-Mss51 from the complex. Impaired COX assembly results in the trapping of Mss51 in the complex, thus limiting its availability for COX1 mRNA translation. An exception is a null mutation in COX14 that does not affect Cox1 synthesis because the Mss51 trapping complexes become unstable, and Mss51 is readily available for translation. Here we present evidence showing that Cox25 is a new essential COX assembly factor that plays some roles similar to Cox14. A null mutation in COX25 by itself or in combination with other COX mutations does not affect Cox1 synthesis. Cox25 is an inner mitochondrial membrane intrinsic protein with a hydrophilic C terminus protruding into the matrix. Cox25 is an essential component of the complexes containing newly synthesized Cox1, Ssc1, Mss51, and Cox14. In addition, Cox25 is also found to interact with Shy1 and Cox5 in a complex that does not contain Mss51. These results suggest that once Ssc1-Mss51 are released from the Cox1 stabilization complex, Cox25 continues to interact with Cox14 and Cox1 to facilitate the formation of multisubunit COX assembly intermediates.
Keywords: Cytochrome Oxidase, Mitochondria, Mitochondrial Metabolism, Protein Assembly, Translation Regulation, Yeast, COX14, COX25, MSS51, SSC1
Introduction
Eukaryotic cells rely on the mitochondrial respiratory chain and oxidative phosphorylation system for aerobic ATP production. Cytochrome c oxidase (COX)3 is a heme A-copper terminal oxidase. It is the last enzyme of the respiratory chain and plays fundamental roles both in electron transfer from reduced cytochrome c to molecular oxygen and in proton pumping through the inner mitochondrial membrane to contribute to the generation of a proton gradient in the intermembrane space that is subsequently used by the F1F0-ATP synthase to drive synthesis of ATP. COX biogenesis is complicated by its dual genetic origin, with subunits (11 in yeast and 13 in mammals) encoded both in the organelle and in the nucleus. In most cases, the three subunits forming the catalytic core of the enzyme (subunits 1–3) are encoded in the mitochondrial DNA. In the yeast Saccharomyces cerevisiae, COX assembly requires the assistance of at least 30 nuclear gene products acting at all stages of the assembly process (1, 2).
COX assembly requires the accumulation of its constitutive subunits in a defined stoichiometric ratio. Previous studies led to the notion of two mechanisms responsible for the concerted accumulation of COX subunits in yeast mitochondria. First, most unassembled COX subunit 1 and the other highly hydrophobic core subunits 2 and 3 are very efficiently post-translationally degraded (3). Second, Cox1 is subject to assembly-controlled translational auto-regulation (4–9). This kind of translational regulation was initially found to operate in the assembly of photosynthetic complexes in chloroplasts from the alga Chlamydomonas reinardthii (10, 11) and in higher plants (12) and termed control by epistasis of synthesis. A distinctive characteristic of these organellar translational auto-regulatory systems is the involvement of ternary factors, mRNA-specific translational activators, whose availability would be regulated by the specific gene products. In the case of yeast COX, the ternary factor is Mss51, a specific translational activator of COX1 mRNA (4–9).
Mss51 acts on the 5′-UTR of COX1 mRNA to promote translation initiation (4, 7) and additionally acts on a target in the protein coding sequence of COX1 mRNA, perhaps to promote elongation (4). During Cox1 synthesis on the mitoribosomes, Mss51 and newly synthesized Cox1 form a transient complex (4, 6) that is stabilized by Cox14 (6), the mitochondrial hsp70 chaperone Ssc1, and its co-chaperone Mdj1 (8). Following Cox1 synthesis, the Ssc1-Mss51-Cox1-Cox14 complex remains stable until Cox1 proceeds to downstream assembly steps. We have postulated that these interactions down-regulate Cox1 synthesis when COX assembly is impaired by trapping Mss51 and limiting its availability for COX1 mRNA translation (6, 8). The C-terminal residues of Cox1 have recently been shown to be essential for Mss51 sequestration and to stabilize the Mss51-Cox14 interaction (9). We have shown that when Mss51 is released from the complex, it is still in a very stable binary complex with Ssc1 (8). According to this model, the release of Mss51-Ssc1 from the post-translational complex and Mss51 availability for Cox1 synthesis (8) probably occur when Cox1 acquires its prosthetic groups or interacts with other COX subunits, a step possibly catalyzed by Shy1, a protein involved in maturation and/or assembly of Cox1 (6, 13, 14). Coa1 could also participate in Cox1 maturation. Coa1 has been proposed to stabilize the Cox1-Ssc1-Mss51-Cox14 complex prior to its interaction with Shy1 (13, 15); however, we and others did not find Coa1 as part of Mss51-containing complexes (8, 16). Independently, once Mss51 is released from the Cox1 preassembly complex, Cox14 still interacts with increasingly matured COX assembly intermediates (13, 15).
To gain insight into how Mss51 is recycled from its post-translational function to become available for COX1 mRNA translation and to fully clarify how this regulatory mechanism operates, we recently analyzed protein-interacting partners of Mss51 in wild-type and a collection of COX assembly mutants (8). These studies allowed us to identify Ssc1 as an important Mss51 partner, a partnership that could mediate the coordination of the translational and post-translational Mss51 functions (8). Mass spectrometric analyses of Mss51-containing complexes allowed us to identify additional proteins that could be potential candidates to participate in these processes. In the present study, we provide evidence that one of them, encoded in open reading frame YJL062W-A and here termed Cox25, is part of Mss51-containing Cox1 translational and preassembly complexes and is essential for their stability. Following Mss51 release from the Cox1 preassembly complex, Cox25 remains involved and becomes part of complexes containing Shy1 and the nuclear encoded subunit Cox5. We conclude that Cox25 and Cox14 work together toward coordinating the assembly line of Cox1 with the incorporation of additional subunits during COX biogenesis.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
The genotypes and sources of the S. cerevisiae strains carrying null alleles of COX-related genes are listed in Table 1. A Δcox25 mutant in the BY4741 background created by replacement of the full COX25 gene with a KANMX cassette was obtained from Open Biosystems/Thermo Scientific (Huntsville, AL). The KANMX cassette and flanking sequences of the COX25 gene were PCR-amplified using primers: 5′-GCGCCTTGTCCCTAGAGAG-3′ and 5′-GAAGTGTGCGTGGTAGATGAAC-3′. The amplicon was transformed into a W303-1A wild-type strain to create W303Δcox25. The Δmss51 and Δcox14 null mutants transformed with an integrative plasmid containing fusion genes expressing Mss51-GST and Cox14-GST, respectively, were previously reported (6). The Δcox25 null mutant was transformed with an integrative plasmid containing fusion gene expressing functional Cox25-GST (supplemental Fig. S1). Wild-type and mutant strains were transformed with an episomal plasmid (YEplac181) overexpressing COX25. Double mutants involving Δcox25 were constructed by crosses of the single mutants. The compositions of the growth media have been described elsewhere (17). The following media were used routinely to grow yeast: YPD (2% glucose, 1% yeast extract, 2% peptone), YPGal (2% galactose, 1% yeast extract, 2% peptone), and YPEG (2% ethanol, 2% glycerol, 1% yeast extract, 2% peptone).
TABLE 1.
Genotypes and sources of yeast strains carrying null alleles of COX-related genes
The categories indicate the general function of the deleted genes in COX biogenesis.
| Strain | Genotype | Source |
|---|---|---|
| Wild-type | ||
| BY4741 | MATa his3D1 leu2D0 met15D0, ura3D0 | a |
| W303-1A | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | b |
| COX structural subunits | ||
| W303Δcox1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1, cox1 | Ref. 7 |
| W303Δcox5a | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox5a::HIS3 | Ref. 38 |
| COX1 expression | ||
| W303Dpet309 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dpet309::HIS3 | Ref. 38 |
| W303Dmss51 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dmss51::HIS3 | Ref. 5 |
| COX2 expression | ||
| W303Dpet111 | ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dpet111::HIS3 | Ref. 39 |
| Heme A biosynthesis | ||
| W303Dcox10 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox10::HIS3 | Ref. 40 |
| W303Dcox15 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox15::HIS3 | Ref. 41 |
| Maturation of CuA or CuB centers | ||
| W303Dcox11 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox11::HIS3 | Ref. 42 |
| W303Dcox17 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox17::TRP1 | Ref. 43 |
| W303Dsco1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dsco1::URA3 | Ref. 44 |
| COX assembly/unknown | ||
| W303Dshy1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dshy1::URA3 | Ref. 5 |
| W303Doxa1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Doxa1::HIS3 | Ref. 45 |
| W303Dcoa1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcoa1::KanMX4 | Ref. 15 |
| W303Dcox16 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox16::URA3 | Ref. 46 |
| W303Δpet100 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dpet100::HIS3 | Ref. 6 |
| W303Dpet191 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dpet191::HIS3 | Ref. 6 |
| W303Δcox14 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Dcox14::TRP1 | Ref. 6 |
| BYDcox25 | MATa his3Δ1 leu2Δ0 met15Δ0, ura3Δ0 Dcox25::KanMX | a |
| W303Dcox25 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1Dcox25::KanMX | This work |
| Other strains | ||
| W303Dcox25Δshy1 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox25::KanMX Dshy1::URA3 | This work |
| W303Δcox11Δmss51 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 Δcox11::KanMX Dmss51::HIS3 | This work |
| W303Dcox25Δcox14 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1Δcox25::KanMX Dcox14::TRP1 | This work |
| W303Dcox25Δcox11 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1Δcox25::KanMX Δcox11::HIS3 | This work |
| W303-1A/rho0 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1/rho0 | Ref. 7 |
a Strains purchased from Open Biosystems.
b Dr. R. Rothstein (Department of Human Genetics, Columbia University, New York, NY).
Characterization of Yeast Mitochondrial Respiratory Chain
Endogenous cell respiration was assayed in whole cells in the presence of galactose using a Clark type polarographic oxygen electrode from Hansatech Instruments (Norfolk, UK) at 24 °C as described (5). Mitochondria were prepared from strains grown in media containing 2% galactose plus 0.5% glucose according to the method of Faye et al. (18) except that zymolyase 20T (ICN Biochemicals Inc., Aurora, OH) instead of glusulase was used for the conversion of cells to spheroplasts. Mitochondria were used to measure oxygen consumption in the presence of either NADH or succinate as the substrates as reported (14). The specific activities reported were corrected for KCN-insensitive respiration.
Mitochondria prepared from the different strains were used for spectrophotometric assays carried out at 24 °C to measure KCN-sensitive COX activity, antimycin A-sensitive NADH cytochrome c reductase, and succinate cytochrome c reductase activities and oligomycin-sensitive ATP synthase activity, measured as described (5, 19). Total mitochondrial cytochrome spectra were obtained as reported (20).
In Vivo and in Organello Mitochondrial Protein Synthesis
Mitochondrial gene products were labeled with [35S]methionine in whole cells at 30 °C in the presence of cycloheximide (5). For in organello translation, mitochondria were prepared by the method of Herrmann et al. (21) and labeled with [35S]methionine as described (14). Equivalent amounts of total cellular or mitochondrial proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to x-ray film.
Preparation of Antibodies to Cox25
A rabbit polyclonal antibody was obtained against a C-terminal Cox25 peptide (DPQELEALKKEYEAKKKA) using the services of Open Biosystems/Thermo Scientific (Huntsville, AL).
Sucrose Gradients
The sedimentation properties of Cox25 and Mss51 in sucrose gradients were analyzed essentially as described (6). Mitochondria were prepared by the method of Herrmann et al. (21). Four mg of protein were solubilized in 400 μl of extraction buffer (20 mm HEPES, pH 7.4, 0.5 mm PMSF, 1% digitonin, 1.2 mm MgCl2, and 150 mm KCl) on ice for 30 min. The clarified extract obtained by centrifugation at 50,000 × gav for 15 min was mixed with hemoglobin and lactate dehydrogenase and applied to 5 ml of linear 7–20% sucrose gradient containing 20 mm HEPES, 0.5 mm PMSF, 0.1% digitonin, 1.2 mm MgCl2, and 150 mm KCl. Following centrifugation for 12 h at 28,000 r.p.m. in a Beckman 55Ti rotor, the gradients were collected in 14 equal fractions. Each fraction was subsequently assayed for hemoglobin by absorption at 409 nm, and for lactate dehydrogenase activity by measuring NADH-dependent conversion of pyruvate to lactate. The distribution of Cox25, Cox14, and Mss51 was assayed by Western blot analysis. The mass of Cox25 was determined from the positions of the respective peaks relative to those of the markers (22). All of the gradients were performed at least in triplicate using independent mitochondrial preparations. The gradients reported are representative of each strain because the patterns observed were reproducible.
Construction of COX25-GST Chimera
The construction of a plasmid containing chimeric COX25-GST was performed in two steps. First, the GST gene was amplified by PCR and second, cloned in-frame to the COX25 gene into a plasmid already containing it. The GST gene plus a thrombin cleavage site at its N terminus was amplified from pGEX-3X (Amersham Biosciences) with primers 5′-GGCTGCAGCTGGTTCCGCGTGGATCCGGAGGAATGTCCCCTATACTAGGT and 5′-CCGGGAGCTCGGATCCACGCGGAACCAGATCC. The ∼600-bp product was digested with PstI/SacI and kept until used for further cloning. The COX25 gene was amplified with the primers 5′-GGCCGAATTCGACTTGACTAACAGCCACATC-3′ and 5′-GGCCCTGCAGTGCCTTCTTCTTGGCTTCGTACTCC-3′ using total DNA isolated from wild-type W303 cells as the template and cloned as an EcoRI/PstI fragment into YIp352 (23). The previously amplified GST gene was subsequently cloned into this plasmid as PstI-SacI, in-frame with the 3′-end of COX25 to yield YIp352/COX25-Thr-GST.
GST Pulldown
A functional Mss51 fused with GST with an intercalated thrombin site was expressed from an integrative plasmid in a strain carrying a null mutant allele of mss51 (Δmss51/MSS51-GST) and in the double mutant Δmss51Δcox11 (Δmss51Δcox11/MSS51-GST) as previously reported (6). Mitochondria were prepared from the Δmss51/MSS51-GST and the Δmss51Δcox11/MSS51-GST strains by the method of Herrmann et al. (21). Mitochondrial proteins (24 mg) were solubilized in 20 mm HEPES, pH 7.4, 0.5 mm PMSF, 1% digitonin, 1.2 mm MgCl2, and 150 mm KCl, and the extracts were loaded in six sucrose gradients. After centrifugation, 14 equal fractions were collected from each gradient; the equivalent fractions were pooled and tested for Mss51 distribution by Western blot analyses. Three fractions around each peak were pooled and used for GST pulldown experiments as reported (8). Each set of pooled fractions were incubated in a rotator with glutathione-Sepharose beads for 4 h at 4 °C. After centrifugation at 500 × gav for 5 min, the beads were washed three times with cold PBS. The Mss51-GST fusion protein was eluted with 10 mm reduced glutathione, 50 mm Tris-base, pH 8.0. Samples from the different pulled down materials were used for Western blot analyses using antibodies against Cox25 and Mss51.
Cox25 fused with GST with an intercalated thrombin site was expressed from YIp352/COX25-Thr-GST in a W303Δcox25 strain. This strain was respiratory competent and grew on nonfermentable carbon sources with a doubling time similar to the parental wild-type strain (supplemental Fig. S1). A functional Cox14 fused with GST with an intercalated thrombin site was expressed from an integrative plasmid in a strain carrying a null mutant allele of cox14 (Δcox14/COX14-GST) as previously reported (6). Mitochondria were prepared from W303Δcox14/COX14-Thr-GST, W303Δcox25/COX25-Thr-GST, and W303Δmss51/MSS51-Thr-GST strains by the method of Herrmann et al. (21), solubilized as mentioned above and directly used for GST pulldown followed by Western blot analyses of the pulled down material.
Pulldown of Newly Synthesized Mitochondrial Products with Cox25-GST
Mitochondria were prepared from a cox25 null mutant with a chromosomally integrated plasmid expressing the Cox25-GST fusion protein. Mitochondria were labeled with [35S]methionine for 30 min as described (6) and extracted with 1% digitonin, 20 mm HEPES, pH 7.4, 1.2 mm MgCl2, 150 mm KCl, and 0.5 mm PMSF. The extract was clarified by centrifugation at 50,000 × gav for 30 min and incubated with glutathione-Sepharose beads for 4 h at 4 °C. Following centrifugation at 500 × gav for 5 min, the supernatant was collected, the beads were washed three times with PBS and eluted with Laemmli buffer (24). Mitochondria corresponding to 20 μg of protein, equivalent volumes of the extract, the supernatant from the glutathione-Sepharose beads, and the washed beads were separated on a 17.5% polyacrylamide gel by SDS-PAGE.
Cox25 Localization and Topology in Isolated Mitochondria
A sample of mitochondria at 4 mg/ml was gently sonically irradiated and centrifuged at 50,000 × gav for 30 min at 4 °C to separate the soluble and membrane proteins. The membrane pellet was resuspended in 0.6 m sorbitol, 20 mm HEPES buffer containing 0.1 m Na2CO3, pH 11, and 50 mm EDTA. After 30 min on ice, the sample was centrifuged at 100,000 × gav for 15 min at 4 °C to separate the soluble membrane extrinsic from the insoluble intrinsic membrane proteins. Equivalent volumes of each fraction were analyzed by Western blotting.
Miscellaneous Procedures
Standard procedures were used for the preparation and ligation of DNA fragments and for transformation and recovery of plasmid DNA from Escherichia coli (25). Yeast were transformed by the method of Schiestl and Gietz (26). The proteins were separated by SDS-PAGE in the buffer system of Laemmli (24), and Western blots were treated with antibodies against the appropriate proteins followed by a second reaction with anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Sigma). The SuperSignal chemiluminescent substrate kit (Pierce) was used for the final detection. In vivo mitochondrial protein synthesis was performed as previously described (5, 8). The one-step gene insertion method (27) was used to integrate linear plasmids at the URA3 or LEU2 locus of yeast chromosomal DNA.
Statistical Analysis
All of the experiments were done at least in triplicate. The data are presented as the means ± S.D. of absolute values or percentages of control. The values obtained for wild-type and cox25 mutant strains for the different parameters studied were compared by Student's t test. p < 0.05 was considered significant.
RESULTS
Identification of Cox25
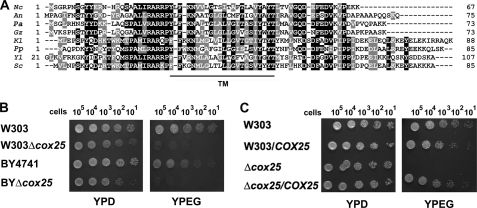
Mass spectrophotometry analyses of a 450-kDa protein complex containing Mss51, Cox14, and newly synthesized Cox1 allowed us to identify the presence of the mitochondrial Hsp70 chaperone Ssc1 in this complex (8). Peptides from the protein encoded in the gene corresponding to ORF YJL062W-A were also detected in some of our replica samples. Recently, Merz and Westermann (28) reported a genome-wide deletion mutant analysis that revealed a number of new genes required for respiratory growth in S. cerevisiae. One of these genes is encoded in ORF YJL062W-A, and it was termed RRG10 (28). Rrg10 was proposed to be involved in the expression of mitochondrial proteins based on the attenuation of [35S]methionine incorporation into Cox1 during in vivo mitochondrial protein synthesis using a deletion mutant of rrg10 (28). The precise function of Rrg10 is not known at present. As explained below, we have shown that Rrg10 is a COX assembly factor that acts early in Cox1 metabolism and, together with Mss51, Cox14, and Ssc1, operates in translational regulation of COX1 mRNA. For these reasons, we have renamed the gene COX25. A Blast search identified homologues of the COX25 product only among fungi (Fig. 1A).
FIGURE 1.
S. cerevisiae COX25 codes for a protein required for respiratory growth. A, sequence alignment of COX25 from S. cerevisiae (Sc) and other fungi: Neurospora crassa (Nc), Aspergillus nidulans (An), Podospora anserina (Pa), Gibberella zeae (Gz), Kluiveromyces lactis (Kl), Pichia pastoris (Pp), and Yarrowia lipolytica (Yl). TM indicates a predicted transmembrane domain. B, the respiratory competent wild-type strains BY4741 and W303 and strains carrying a null allele of cox25 in both genetic backgrounds were grown overnight in liquid YPD media. 10-fold serial dilutions of the four strains were plated on solid YPD or YPEG media and incubated at 30 °C. The pictures were taken after 3 days of incubation. C, wild-type W303 and mutant Δcox25 cells overexpressing or not COX25 were grown overnight in liquid YPD media. Serial dilution growth test was performed as for B.
COX25 Is Essential for Respiration, Cytochrome c Oxidase Assembly, and Function
We obtained a BYΔcox25 strain from Open Biosystems in which the COX25 gene was replaced by a KANMX cassette in the BY4741 background. The cox25 deletion cassette was transferred to a W303-1A strain to create a W303Δcox25 strain, which was used for most of the experiments. The cox25 mutants in either background are able to use glucose (Fig. 1B) and less efficiently other fermentable nonrepressive carbon sources (supplemental Fig. S2) but are unable to utilize respiratory substrates for growth (Fig. 1B and supplemental Fig. S2). Overexpression of Cox25 did not affect the growth of wild-type cells and fully restored the respiratory growth of Δcox25 cells (Fig. 1C).
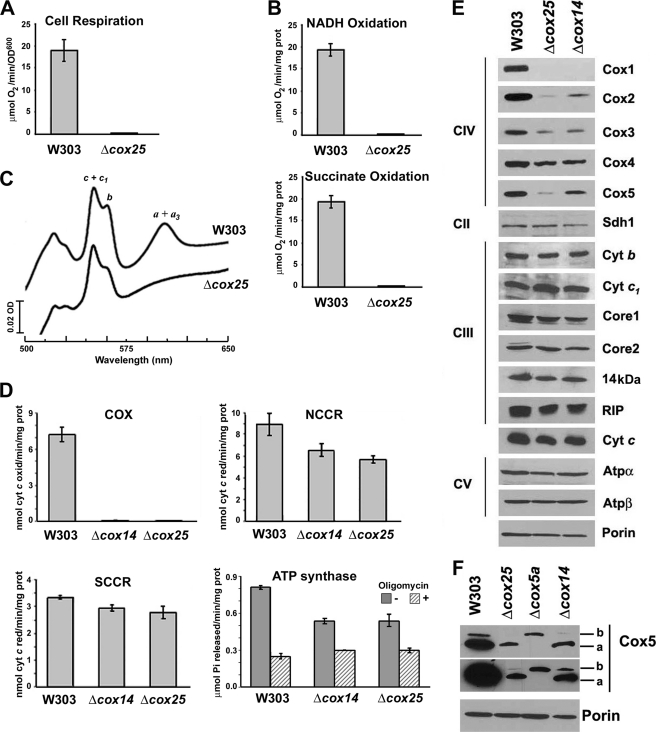
The respiratory growth defect of Δcox25 cells is due to a virtually complete abolishment of the capacity of the cells to respire as shown by a polarographic measurement of KCN-sensitive endogenous cell respiration (Fig. 2A). The ability of the mutant cells to respire was restored by reinsertion of recombinant COX25 (data not shown). The respiratory defect was also evident by the complete inability of mitochondria isolated from W303Δcox25 cells to oxidize either NADH or succinate (Fig. 2B).
FIGURE 2.
Biochemical properties of Δcox25 cells. Respiratory assays A, KCN-sensitive endogenous cell respiration measured polarographically in the presence of galactose. B, rate of NADH and succinate oxidation in isolated mitochondria. C, total mitochondrial cytochrome spectra. Mitochondria from the wild-type strain W303 and the null mutant Δcox25 cells were extracted at a protein concentration of 5 mg/ml with potassium deoxycholate under conditions that quantitatively solubilize all of the cytochromes (36). Difference spectra of the reduced (sodium dithionite) versus oxidized (potassium ferricyanide) extracts were recorded at room temperature. The absorption bands corresponding to cytochromes a and a3 have maxima at 603 nm (a and a3); the maxima for cytochrome b (b) and for cytochrome c and c1 (c and c1) are 560 and 550 nm, respectively. D, mitochondrial respiratory chain enzyme spectrophotometric measurements in isolated mitochondria. COX, NADH cytochrome c reductase (NCCR), succinate cytochrome c reductase (SCCR), and ATP synthase activities were measured as described under “Experimental Procedures.” E, steady state concentrations of mitochondrial respiratory chain complexes IV (COX), II (Sdh), and III (bc1 complex) and ATP synthase subunits estimated by Western blot analyses of proteins separated in a 12% Tris-glycine SDS-PAGE. F, steady state concentrations of COX subunit 5 isoforms estimated by Western blot analyses of proteins separated in a 16.5% Tris-Tricine SDS-PAGE. Two expositions of the film are shown. In E and F, an antibody against porin was used to normalize the signals for protein loading.
To ascertain whether the origin of the respiratory defect stemmed from a defect in the respiratory chain enzymes, we analyzed the oxidized minus reduced total mitochondrial cytochrome spectra. Comparison of the wild-type and mutant spectra revealed slight differences in the levels of cytochromes b, c, or c1. More importantly, cytochromes a and a3 were absent in the Δcox25 mutant compared with the wild-type strain (Fig. 2C), indicating a complete COX impairment. The enzymatic defect was confirmed by measuring COX activity in isolated mitochondria, which indicated a complete absence of functional COX in the mutant as compared with the isogenic wild type (Fig. 2D). The Δcox25 mutation also induced a pleiotropic decrease in other segments of the OXPHOS system, particularly in NADH cytochrome c reductase and ATP synthase activities (Fig. 2D). Although the precise mechanisms for these pleiotropic effects remain to be explored, they were similar to the alterations measured in a Δcox14 strain (Fig. 2D) and several other COX assembly mutants (29).
Steady state levels of Cox1 and Cox2 were barely detected in the Δcox25 mutant, and those of Cox3 were significantly reduced (Fig. 2E). This is most likely due to the rapid turnover of the COX catalytic core subunits when the assembly process is hindered. Like in most COX mutants, including Δcox14, a less significant difference was detected in the more stable nuclear encoded subunits such as Cox4 (Fig. 2E). However, the steady state levels of Cox5 were particularly low in the absence of Cox25 (Fig. 2E), and the two subunit isoforms, Cox5a and Cox5b, were similarly affected (Fig. 2F).
A Null Mutation in COX25 Does Not Repress Cox1 Synthesis
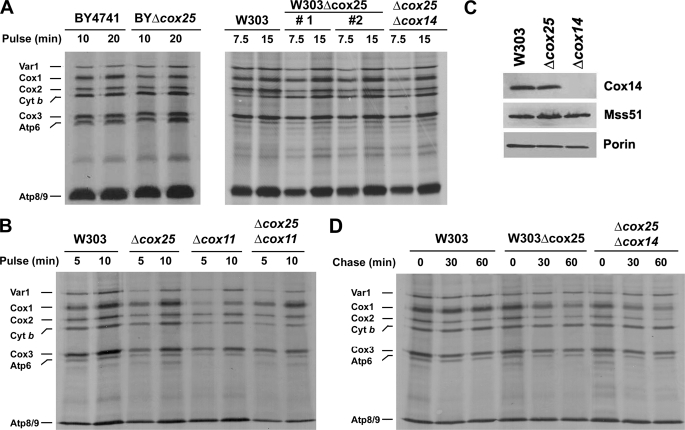
The Δcox25 mutation in the BY4741 background was previously reported to induce a Cox1 synthesis defect (28). This phenotype likely results from a down-regulation of Cox1 synthesis when normal COX assembly is compromised as has been reported for most COX assembly mutants (6). However, in vivo pulse labeling with [35S]methionine of either BYΔcox25 or W303Δcox25 cells in the presence of cycloheximide showed that the amount of detectable newly synthesized Cox1 is similar in Δcox25 and wild-type cells (Fig. 3A). To date, the two reported exceptions of genes that when mutated bypass Cox1 synthesis down-regulation in the absence of COX assembly are Cox14 (6) and Coa1 (13, 15).
FIGURE 3.
In vivo synthesis and turnover of mitochondrial gene products in Δcox25 cells. A, wild type (either BY4741 or W303), null mutants of cox25 in both backgrounds and a double mutant Δcox25Δcox14 were labeled with [35S]methionine at 30 °C for the indicated times in the presence of cycloheximide. Equivalent amounts of protein were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and analyzed by autoradiography. #1 and #2 indicate two independent Δcox25 clones. B, in vivo labeling of mitochondrial products from the indicated strains with [35S]methionine at 30 °C for 5 and 10 min in the presence of cycloheximide. C, steady state levels of Cox14 and Mss51 in mitochondria isolated from wild-type, Δcox25, and Δcox14 cells analyzed by Western blotting. D, the indicated strains were labeled for 15 min at 30 °C with [35S]methionine in the presence of cycloheximide. Labeling was terminated by the addition of 80 μmol of cold methionine and 12 μg/ml puromycin (0 time). The samples were collected after the indicated times of incubation at 30 °C and processed as in A.
The lack of effect of the cox25 mutation on Cox1 synthesis (Fig. 3, A and B) suggested that similar to Cox14, Cox25 might contribute to negatively regulate translation of this COX subunit. This was tested by measuring Cox1 synthesis in strains carrying mutations in cox25 and other COX-specific genes including cox11 (Fig. 3B), mss51, pet309, and shy1 (data not shown). The cox25 mutation restored normal Cox1 synthesis in all the COX mutants except in strains carrying null alleles of mss51 and pet309, genes that encode the two translational activators essential for Cox1 synthesis. Despite being epistatic with respect to Cox1 expression, the cox25 mutation did not rescue either respiration or the ability of the mutants to assemble COX (data not shown). The observed phenotype is not the result of a decrease in Cox14 steady state levels, which were not modified by the absence of Cox25 (Fig. 3C).
Stability of Newly Synthesized Cox1p in Wild-type and COX Mutants
We have reported that in the absence of Cox14, newly synthesized Cox1 is rapidly degraded because the presence of Cox14 is required to stabilize a high molecular mass protein complex additionally containing newly synthesized Cox1, the COX1 mRNA translational activator Mss51, and the mitochondrial Hsp70 chaperone Ssc1 (6, 8). To assess the stability of unassembled Cox1 in the absence of Cox25, wild-type W303, Δcox25, and double mutant Δcox14Δcox25 cells were pulsed with [35S]methionine for 15 min in the presence of cycloheximide and chased for different times following addition of puromycin and excess of cold methionine. Most of the translation products in the wild-type strain, including the three COX subunits, were stable during 1 h of chase (Fig. 3D). In the mutants, most of the Cox1 was degraded during the chase, and a significant fraction of Cox2 and Cox3 was also degraded during this period (Fig. 3D). This is in contrast with the pattern of most COX assembly mutants in which Cox1 synthesis is down-regulated, but the small amount of labeled Cox1 detected in these mutants remains unchanged during the chase, whereas Cox2 and Cox3 are labile (6). The greater instability of Cox1 in the cox25 mutant mimics that in the cox14 mutant and is also supported by the results of Western analysis showing barely detectable Cox1 steady state concentrations in these two mutants (Fig. 2E) even lower than in other COX assembly mutants (6).
A double Δcox25Δcox14 mutation produced a bypass of Cox1 translational autoregulation similar to that produced by the single mutations (Fig. 3, A and D) and a similar instability of newly synthesized Cox1 (Fig. 3D), suggesting that both Cox25 and Cox14 contribute to establish Cox1 translational control jointly or through the same mechanism.
Overexpression of Cox25 Does Not Alter Synthesis of Cox1
Normal labeling of Cox1 in cox25 single and double mutants could indicate that Cox25 acts to increase degradation of unassembled or incompletely assembled Cox1. If this were the case, overexpression of Cox25 in a wild-type or mutant background might be expected to affect the amount of newly synthesized Cox1. This was examined by transforming wild-type and Δcox25 cells with an episomal plasmid containing a wild-type COX25 gene. Overexpression of Cox25 in these strains was more than 10-fold (supplemental Fig. S3A). It did not affect the growth of wild-type cells, fully restored the respiratory growth of Δcox25 cells (Fig. 1C), and did not affect in vivo labeling of Cox1, similar to Cox14 overexpression in the wild-type strain (supplemental Fig. S3B).
The Function of Cox25 Does Not Overlap the Functions of Other COX Assembly Factors
Because Cox25 seems to act early in the biogenesis of Cox1, we asked whether overexpression of COX assembly factors could at least partially restore respiratory growth in Δcox25 cells. Overexpression of Mss51, Cox14, Cox10, Shy1, Cox11, Cox17, and Sco1 as well as overexpression of cytochrome c, the presence of which is required for COX assembly, and overexpression of Hap4, which partially rescues the COX deficiency of shy1 mutants by inducing overexpression of nuclear encoded subunits Cox5 and Cox6 (14), failed to suppress the Δcox25 respiratory defect (data not shown). Reciprocally, overexpression of COX25 in strains carrying null alleles of cox14, mss51, cox10, cox15, oxa1, pet309, pet111, pet100, pet191, shy1, sco1, cox11, cox16, cox17, and cox5a did not rescue the respiratory defect of these strains (data not shown).
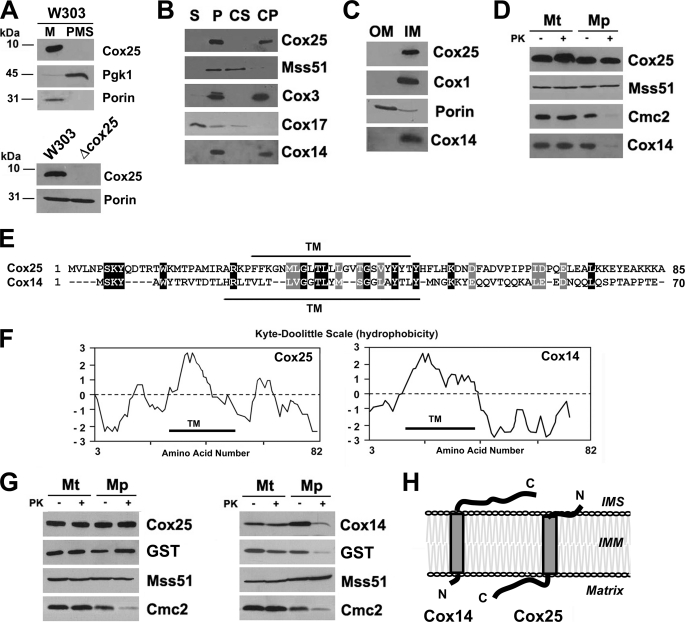
Cox25 Is an Integral Mitochondrial Inner Membrane Protein
The phenotype of Δcox25 cells strongly suggested that Cox25 must be a mitochondrial protein. Cox25 is a 10-kDa protein predicted to have a single transmembrane domain. To determine the subcellular localization of Cox25, we have generated an affinity-purified peptide antibody that recognizes this protein. Cell fractionation and Western blot analyses enabled us to find Cox25 in the mitochondrial fraction but not in the postmitochondrial supernatant containing cytoplasmic proteins such as Pgk1 (3-phosphoglycerate kinase) (Fig. 4A, upper panel). The protein was absent in the Δcox25 strain (Fig. 4A, lower panel). We have determined the submitochondrial localization of Cox25, its topology, and its solubility properties. Sonic irradiation of wild-type mitochondria solubilized Cox17, a soluble protein of the intermembrane space, but not Cox25, Cox3, Cox14, or Mss51 (Fig. 4B). Mss51, however, was solubilized by treatment of mitochondria with alkaline carbonate because it is a peripheral protein, whereas Cox3 was recovered in the membrane fraction, because it is an intrinsic protein of the inner membrane. Cox14 was previously shown to be associated with the inner membrane of mitochondria (30) and proposed to be a peripheral protein (6). However, similar to Cox25, Cox14 has a predicted transmembrane domain (Fig. 4E), and when reassessing its solubility properties we consistently found that both Cox14 and Cox25 were not extracted with alkaline carbonate (Fig. 4B). To discern to which membrane Cox25 is integrated, we sonicated mitochondria and separated inner and outer membranes by sucrose gradient fractionation. Porin and Cox1 were used as markers of the outer and inner mitochondrial membranes, respectively, to test the purity of the fractions. Similar to Cox1, Cox14 and Cox25 are found exclusively in the inner membrane fraction (Fig. 4C).
FIGURE 4.
Mitochondrial localization of Cox25. A, Cox25 is a mitochondrial protein. Mitochondria (M) and the post-mitochondrial supernatant (PMS) fraction were isolated from the wild-type W303 strain. Samples of the two fractions corresponding to 40 μg of protein were analyzed by Western blotting using antibodies against Cox25, the cytosolic marker 3-phosphoglycerate kinase subunit 1 (Pgk1), and the mitochondrial marker porin. The specificity of the signal detected with the anti-Cox25 antibody was tested by including a sample of Δcox25 mitochondria. B, Cox25 is a membrane protein. As described under “Experimental Procedures,” soluble (S) and membrane-bound (P) mitochondrial proteins were separated from 40 μg of total wild-type mitochondria. The pellet was submitted to alkaline extraction to allow the separation of the extrinsic proteins present in the supernatant (CS) from the intrinsic proteins in the pellet (CP). The samples were analyzed by Western blotting using antibodies against Cox25, Cox14, the inner membrane extrinsic protein Mss51, the soluble intermembrane space protein Cox17, and the inner membrane intrinsic protein Cox3. C, Cox25 is an inner mitochondrial membrane protein. Isolated mitochondria were fractionated into inner and outer membranes by sonication plus sucrose gradient sedimentation as described (37). Inner membranes (IM) and outer membranes (OM) were analyzed by Western blotting using antibodies against porin (outer membrane marker), Cox1 (inner membrane marker), Cox14, and Cox25. D, Cox25 is a membrane protein facing the matrix. Four aliquots of 40 μg of mitochondrial protein were pelleted and resuspended in buffer containing either 20 mm HEPES or 0.6 m sorbitol with 20 mm HEPES. One aliquot in each buffer was supplemented with final 100 μg/ml proteinase K (PK) and incubated on ice for 60 min. The reaction was stopped with 2 mm PMSF. Mitochondria (Mt) and mitoplasts (Mp) were recovered by centrifugation at 50,000 × gav for 15 min at 4 °C. The samples were analyzed by Western blotting using antibodies against Cox25, Cox14, Cmc2 (protein facing the inner membrane space), and Mss51 (protein facing the matrix). E, sequence alignment of S. cerevisiae Cox25 and Cox14. F, Kyte-Doolittle hydrophobicity plots for these proteins. TM indicates a predicted transmembrane domain. G, topology of Cox25 and Cox14 in the inner membrane. Mitochondria were prepared from Δcox25 or Δcox14 strains, respectively, expressing Cox25 or Cox14 fused with GST at their C terminus and used for proteinase protection assays as in D. H, cartoon depicting the topology of Cox25 and Cox14 in the mitochondrial inner membrane.
Similar to Mss51, a peripheral inner membrane protein facing the matrix, Cox25 has no significant hydrophilic portions located on the intermembrane space side of the inner membrane as evidenced by its resistance to proteinase K in mitochondria and in mitoplasts prepared by hypotonic swelling of mitochondria (Fig. 4D). In contrast, similar to Cmc2, a peripheral inner membrane protein facing the intermembrane space, Cox14 is protected against proteinase K degradation in mitochondria but not in mitoplasts, indicating that it must contain some hydrophilic domain exposed to the intermembrane space.
Cox25 and Cox14 are both small proteins with similar predicted structural features. An alignment of both proteins does not show a major homology beyond the predicted transmembrane domains in each one (Fig. 4E). The Kyte-Doolittle hydrophobicity plot (31) shows that both proteins contain a relatively hydrophilic C terminus (Fig. 4F). To elucidate the topology of these proteins, we prepared mitochondria from Δcox25 cells expressing functional Cox25-GST (supplemental Fig. S2) or Δcox14 cells expressing functional Cox14-GST (6) and used them in proteinase protection assays. In both cases, the GST tag was fused to the C terminus of the proteins. Cox25 and its GST tag were proteinase K protected in both mitochondria and mitoplasts, whereas Cox14 and its GST tag were protected from proteinase K digestion in mitochondria but not in mitoplasts (Fig. 4G). Taken together and as depicted in the cartoon in Fig. 4H, our results demonstrate that both Cox14 and Cox25 have a transmembrane domain and a relatively hydrophilic C-terminal domain that protrudes either into the intermembrane space (Cox14) or into the matrix (Cox25).
Cox25 Co-sediments with Mss51 in Sucrose Gradient
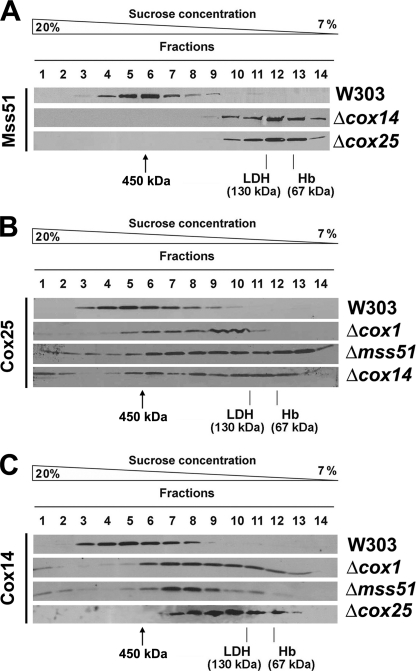
The similar phenotypes of Cox25 and Cox14 suggest that both proteins could cooperate with Mss51 and Ssc1 to coordinate Cox1 synthesis with its assembly into the COX holoenzyme. We have previously shown that in wild-type cells, most Mss51 is part of a 450-kDa complex (Ref. 8 and Fig. 5A) that contains Cox1, Cox14, Mss51, and Ssc1 (8). In the absence of Cox14, the 450-kDa complex is destabilized, and Mss51 accumulates in a 120-kDa heterodimer (Ref. 8 and Fig. 5A) in partnership with Ssc1 (8). Similarly, the absence of Cox25 results in the accumulation of Mss51 in the 120-kDa complex (Fig. 5A), supporting the fact that Cox25 is also part of the 450-kDa complex. This result is in agreement with the detection of Cox25 in the 450-kDa complex by mass spectrometry mentioned earlier. As expected, analyses of the sedimentation properties of Cox25 extracted from wild-type mitochondria in a sucrose gradient showed that Cox25, Cox14, and Mss51 co-sediment in the 450-kDa complex (Fig. 5). Although the absence of Cox1, Mss51, and Cox14 destabilizes this complex (Ref. 8 and Fig. 5A), both Cox25 and Cox14 remain part of the high molecular mass material (Fig. 5, B and C), presumably interacting with other COX subunits or assembly factors.
FIGURE 5.
Native molecular mass and steady state levels of Cox25. A, sedimentation properties of Mss51 in a linear 7–20% sucrose gradient using mitochondrial extracts from the indicated strains. The gradient was calibrated with hemoglobin (Hb, 67 kDa) and lactate dehydrogenase (LDH, 130 kDa). The distribution of Mss51 was assayed by Western blot analysis. B and C, sedimentation properties of Cox25 (B) and of Cox14 (C) in a linear 7–20% sucrose gradient using mitochondrial extracts from the indicated strains, analyzed as in A.
It is particularly noticeable that in the absence of Cox1, Mss51, or Cox14, a small portion of Cox25 accumulates in heavy fractions (Fig. 5B). Also Cox14 is detected in these fractions in the absence of Cox1 and Mss51 but not in the absence of Cox25 (Fig. 5C). In these fractions we have previously detected ribosomal molecules (8). The absence of Cox1, Mss51, Cox14, Cox2, and Cox5a or the large number of COX assembly factors tested does not alter the steady state levels of Cox25 (supplemental Fig. S4). Unexpectedly, Cox25 levels are enhanced by ∼2.5-fold in mitochondria isolated from cells devoid of mitochondrial DNA (ρ0) and thus of mitochondrial ribosomes (supplemental Fig. S4). Future studies will be devoted to explore a possible association of Cox25 with mitochondrial ribosomes.
Cox25 Is Part of High Molecular Mass Complexes Containing Mss51, Cox14, Ssc1, and Newly Synthesized Cox1
Following our recently reported model of translational regulation of Cox1 synthesis, Mss51 is a component of a high molecular mass Cox1 translational complex (HMW-T), a 450-kDa Cox1 stabilization complex, and finally a 120-kDa complex containing translationally competent Mss51 (8). We have now asked whether Cox25 is physically part of these complexes.
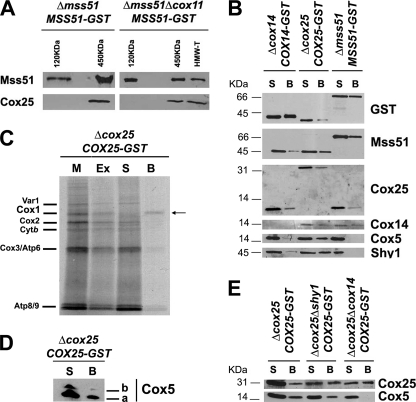
Mss51-containing 450- and 120-kDa complexes were purified from mitochondrial extracts from a Δmss51 strain expressing MSS51-GST using a combination of sucrose gradients and GST pulldown as reported (8). In this strain, the steady state level of Mss51-GST is 2-fold that of Mss51 in a wild-type strain, and the excess of Mss51 accumulates in the 120-kDa complex (8). Mss51-containing HMW-T and 450- and 120-kDa complexes were similarly purified from mitochondrial extracts from a Δmss51Δcox11 strain expressing MSS51-GST as reported (8). In the later strain, in the absence of COX assembly, a significant portion of Mss51 remains trapped in the HMW-T and 450-kDa complexes, thus limiting its availability for translation (8). Using Western blot analyses, we have detected Cox25 in both the HMW-T and 450-kDa complexes but not in the 120-kDa complex as expected (Fig. 6A). The interaction among Cox25, Mss51, and Cox14 was further tested by GST pulldown assays of extracts prepared from mitochondria isolated from Δcox25 cells expressing Cox25-GST, Δcox14 cells expressing Cox14-GST, and Δmss51 cells expressing Mss51-GST, respectively. In each case, the tagged protein and the other two untagged proteins were detected by Western blot in the material adsorbed to the GST-Sepharose beads, indicating physical interactions among them (Fig. 6B).
FIGURE 6.
Cox25 physically interacts with Cox1 and Cox1 biogenesis and assembly factors. A, Cox25 interacts with Mss51 in the 450-kDa complex and in the high molecular mass Cox1 translational complex (HMW-T) but not in the 120-kDa complex previously described (8). Complexes containing Mss51 were purified by sucrose gradient followed by GST pulldown of mitochondrial extracts from either a Δmss51 or a Δmss51Δcox11 both expressing a GST-tagged version of Mss51 (8). Samples from the different complexes were analyzed by Western blotting using antibodies against Mss51 and Cox25. B, mitochondria were prepared from Δmss51 cells expressing Mss51-GST, Δcox14 cells expressing Cox14-GST and Δcox25 cells expressing Cox25-GST, extracted with 1% digitonin, 150 mm KCl, and 1.2 mm MgCl2 in buffer containing 20 mm HEPES and 0.5 mm PMSF and used for GST pulldown assays. Samples of material bound to GST-Sepharose beads (lanes B) or remaining in the supernatant (lanes S) were separated in a 12% Tris-glycine SDS-PAGE and analyzed by Western blotting using specific antibodies against the indicated proteins. C, interaction of Cox25 with newly synthesized Cox1. Mitochondria isolated from a Δcox25 strain with a chromosomally integrated plasmid expressing Cox25-GST fusion protein were labeled with [35S]methionine for 30 min, extracted, and submitted to GST pulldown as described under “Experimental Procedures.” Mitochondria (M) corresponding to 20 μg of protein, equivalent volumes of the extract (Ex), the supernatant from the glutathione-Sepharose beads (S), and the washed beads (B) were separated on a 17.5% polyacrylamide gel by SDS-PAGE. D, Cox25-GST pulldown samples were also separated in a 16.5% Tris-Tricine SDS-PAGE and analyzed by Western blotting using and anti-Cox5 antibody. E, GST pulldown analyses performed as in C, using mitochondrial extracts from Δcox25, Δcox25Δcox14, and Δcox25Δshy1 cells all expressing COX25-GST. Material unbound (S) or bound to the GST-Sepharose beads was analyzed by Western blotting using antibodies against Cox25 and Cox5.
The ability of Cox25 to interact with Cox14 and Mss51 suggested that Cox25 might be directly or indirectly interacting with newly synthesized Cox1. To test for a physical interaction with Cox1, mitochondria from a Δcox25 strain expressing Cox25-GST from a chromosomally integrated plasmid were labeled in organello with [35S]methionine, extracted with 1% digitonin in the presence of 150 mm KCl and 1.2 mm MgCl2 and adsorbed onto glutathione-Sepharose beads. SDS-PAGE analysis of the proteins recovered from the beads indicated quite a selective pulldown of labeled Cox1 (Fig. 6C).
Cox25 Interacts with Cox5 and Shy
It has been recently reported that Shy1, Cox14, and Coa1 are part of large complexes and interact with Cox1-containing subassemblies downstream from the roles of Mss51 in COX biogenesis (6, 13). We have asked whether Cox25 also participates in COX assembly once Mss51 has been released from the 450-kDa Cox1 stabilization complex. Using samples from the same pulldown assays explained earlier, we have tested whether Cox25 interacts with Shy1 and the nuclear encoded subunit Cox5, probably the first subunit to form an assembly intermediate with Cox1 (14). Although Mss51-GST does not pull down Shy1 nor Cox5, these proteins are poorly co-precipitated with Cox14-GST and very efficiently with Cox25-GST (Fig. 6B). Noticeably, both Cox5 isoforms were co-precipitated with Cox25-GST (Fig. 6D). Furthermore, we tested whether the interaction of Cox25 with Cox5 depends on Shy1 and Cox14. For that purpose, we prepared Δcox25 strains expressing COX25-GST from a chromosomally integrated plasmid and carrying an additional mutation either in shy1 or in cox14. Mitochondrial extracts from these strains were used for pulldown assays. As shown in Fig. 6E, Cox5 was efficiently pulled down in the absence of Shy1, but only traces were detected in the absence of Cox14 when the film was overexposed (data not shown). In all the GST pulldown assays, none of the analyzed proteins were detected bound to plain (non-GST conjugated) Sepharose beads (data not shown). Our results indicate that in addition to interacting with Mss51, Cox14 and Cox25 also exist as part of large COX assembly intermediates not necessarily involving Shy1.
DISCUSSION
Our findings identify Cox25 as a new essential COX biogenesis factor. It is required to promote the stability of newly synthesized Cox1 and, in this capacity, to facilitate regulation of COX1 mRNA translation and couple this process to Cox1 assembly into multi-subunit intermediates.
In most COX assembly mutants, the rate of Cox1 synthesis is significantly lower than in wild-type cells. Experimentally, COX1 mRNA translational auto-regulation can be bypassed in the absence of COX assembly by increasing the effective concentration of Mss51 as a COX1 mRNA translational activator, thus preventing the trap of Mss51 with newly synthesized Cox1 in high molecular mass complexes. This can be accomplished in a COX mutant strain by introducing an additional copy of MSS51 or by substituting the endogenous MSS51 by certain mutant mss51F199I and mss51T167R alleles that encode Mss51 proteins with an increased ability to be released from the 450-kDa Mss51-trapping complex (6, 8). An alternative approach consists of destabilizing the Mss51-trapping complex by introducing mutations in either coa1 (13, 15) or cox14 (6). This later approach allowed us to identify Cox14 as a protein involved in Cox1 translational auto-regulation. Now we have identified Cox25 as part of the Cox1 translational complex and the 450-kDa Cox1 preassembly complex, and in this capacity, the absence of Cox25 destabilizes these complexes, increasing the availability of Mss51 for COX1 mRNA translation in the absence of COX assembly.
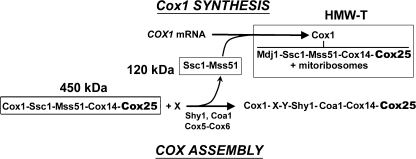
Following our recently reported model of translational regulation of Cox1 synthesis (8), depicted in Fig. 7, in wild-type cells, a small portion of total Mss51 is present in a binary 120-kDa complex with Ssc1. When required for COX assembly, Mss51 molecules contained in this pool would interact with the 5′-UTR of the COX1 mRNA to promote its translation (4, 7). During Cox1 synthesis, Mss51 would interact directly or indirectly with the mitochondrial ribosomes and with the nascent Cox1 polypeptide together with the chaperones Ssc1 and Mdj1, which would ensure the proper folding of the nascent polypeptide. Cox14 and Cox25 are imagined to be added to this complex at a later step, forming a HMW-T. Once Cox1 synthesis is completed, Cox14 and Cox25 serve to stabilize a 450-kDa Ssc1-Mss51-Cox1-Cox14-Cox25 complex. The release of Mss51 from this complex is possibly catalyzed by the incorporation of the COX assembly factors Shy1 and Coa1 as well as nuclear encoded subunits Cox5a-Cox6, thus allowing Cox1 to proceed to downstream events in the COX assembly process (4–6, 13, 15). In this way, Mss51 becomes available for new rounds of translation (Ref. 8 and Fig. 7).
FIGURE 7.
Scheme of a model depicting the role of Ssc1 and Cox14 on translational regulation of cytochrome c oxidase biogenesis by interacting with Mss51 in several high molecular mass complexes. Cox25 is a partner in some of these complexes. See the explanation in the text.
Noticeably, Cox25 and Cox14 are relatively similar proteins that seem to perform quite similar roles. However, their functions are essential for COX assembly and nonoverlapping because overexpression of COX25 in a Δcox14 mutant or of COX14 in a Δcox25 mutant does not suppress the COX assembly and respiratory growth defects of these strains. Both Cox25 and Cox14 have a similar predicted architecture, although their orientation in the inner membrane is different. Both have a transmembrane domain and a hydrophilic C terminus, but the latter protrudes into a different mitochondrial compartment for each protein. Although the C terminus of Cox14 resides in the intermembrane space and is glutamine-rich, the C terminus of Cox25 resides in the matrix milieu and contains a positively charged lysine-rich terminal domain. Because Cox25 is found in Cox1 translational complexes, it is tempting to speculate that Cox25 could mediate the interaction of Mss51 with the mitoribosome. Incidentally, we have noticed that ORF YJL062W-A containing COX25 is located in chromosome X “head to head” to ORF YJL063C containing MRPL8, which encodes a mitoribosomal protein of the large subunit. The proximity of these genes (less than 250 bp apart) suggests that their expression could be co-regulated. The topologies of Cox14 and Cox25 appear to be suitable to promote the stability of the Cox1-Mss51-Ssc1 complex by interacting with Cox1 through their transmembrane domains and holding the complex from both the intermembrane space and the matrix sides of the inner membrane.
A phenotypic trait that distinguishes Δcox25 and Δcox14 mitochondria is the different steady state level of Cox5. In the absence of Cox25, Cox5 accumulation is significantly lowered, suggesting a role for Cox25 in the stability of this subunit. COX assembly is thought to be a sequential process (32, 33) initiated by the synthesis of Cox1, which subsequently forms a subassembly with Cox5 and Cox6 (33, 34). These subunits are in direct contact with Cox1 in the mature enzyme (35). Cox5 interacts with Cox1 through its single transmembrane domain, whereas Cox6 caps the matrix side of Cox1 (35). It was previously reported that during the COX assembly process, upon release of Mss51 from the Cox1 preassembly complex, Cox14 still interacts with Cox1 and becomes part of higher order subassemblies involving Coa1, Shy1, and Cox5 (13, 15). Here we have shown that Cox25 also participates in these later assembly steps by interacting with Shy1 and Cox5. We were able to pull down Cox5 together with Cox25-GST with efficiency significantly higher than with Cox14-GST. It is tempting to speculate that Cox25 could directly interact with Cox5, thus providing stability to this protein in the absence of COX assembly. Additionally, the Cox25-Cox5 interaction requires Cox14. This could suggest that Cox25 does not exist in a complex with Cox5 prior to its role in Cox1 biogenesis. Contrarily, the interaction of Cox25 with Cox5 does not depend on Shy1, which suggests that Shy1 could interact with the Cox1 subassemblies concurrently or immediately after incorporation of subunit Cox5. Further studies are required to elucidate essential aspects of the COX biogenesis process involving the incorporation of Cox2 into the assembly intermediates and the precise timing for the maturation of both Cox1 and Cox2 by incorporation of their metal prosthetic groups.
In summary, in this work we have described Cox25, a protein partner of Cox14, Mss51, and Ssc1, essential for COX assembly-dependent translational autoregulation of Cox1 synthesis. Our observations fit in a model in which Cox25 additionally participates in assembly steps beyond Cox1 biogenesis serving to promote the incorporation or stability of Cox5 within COX assembly intermediates. We conclude that Cox25 and Cox14 in cooperation coordinate the regulation of Cox1 synthesis and its assembly with partner subunits, thus facilitating the assembly of the COX holoenzyme.
Supplementary Material
Acknowledgments
We thank Dr. M. Bourens, Dr. C. Trivigno, and I. C. Soto for critically reading the manuscript. We are in debt to Dr. Alexander Tzagoloff (Columbia University, New York) for providing an antibody against Cox17.
Note Added in Proof
During the review process of our manuscript, it has come to our attention that a paper has been published in J. Cell. Biol. (47) presenting the identification of the gene encoded in open reading frame YJL062W-A, which we have called COX25, and which was called COA3 in that paper. COA3 is the standard name assigned in the SGD site. Both papers conclude that the protein encoded in this gene is involved in translational regulation of COX1 in mitochondria from S. cerevisiae.
This work was supported, in whole or in part, by National Institutes of Health Grant GM071775A (to A. B.). This work was also supported by a research grant from the Muscular Dystrophy Association (to A. B.) and a development grant from the Muscular Dystrophy Association (to F. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- COX
- cytochrome c oxidase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Fontanesi F., Soto I. C., Horn D., Barrientos A. (2006) Am. J. Physiol. Cell Physiol. 291, C1129–C1147 [DOI] [PubMed] [Google Scholar]
- 2. Fontanesi F., Soto I. C., Barrientos A. (2008) IUBMB Life 60, 557–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Langer T., Käser M., Klanner C., Leonhard K. (2001) Biochem. Soc. Trans. 29, 431–436 [DOI] [PubMed] [Google Scholar]
- 4. Perez-Martinez X., Broadley S. A., Fox T. D. (2003) EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrientos A., Korr D., Tzagoloff A. (2002) EMBO J. 21, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrientos A., Zambrano A., Tzagoloff A. (2004) EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zambrano A., Fontanesi F., Solans A., de Oliveira R. L., Fox T. D., Tzagoloff A., Barrientos A. (2007) Mol. Biol. Cell 18, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fontanesi F., Soto I. C., Horn D., Barrientos A. (2010) Mol. Cell. Biol. 30, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shingú-Vázquez M., Camacho-Villasana Y., Sandoval-Romero L., Butler C. A., Fox T. D., Pérez-Martínez X. (2010) J. Biol. Chem. 285, 34382–34389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wollman F. A., Minai L., Nechushtai R. (1999) Biochim. Biophys. Acta 1411, 21–85 [DOI] [PubMed] [Google Scholar]
- 11. Choquet Y., Wostrikoff K., Rimbault B., Zito F., Girard-Bascou J., Drapier D., Wollman F. A. (2001) Biochem. Soc. Trans. 29, 421–426 [DOI] [PubMed] [Google Scholar]
- 12. Wostrikoff K., Stern D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mick D. U., Wagner K., van der Laan M., Frazier A. E., Perschil I., Pawlas M., Meyer H. E., Warscheid B., Rehling P. (2007) EMBO J. 26, 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontanesi F., Jin C., Tzagoloff A., Barrientos A. (2008) Hum. Mol. Genet. 17, 775–788 [DOI] [PubMed] [Google Scholar]
- 15. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., Winge D. R. (2007) EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khalimonchuk O., Bestwick M., Meunier B., Watts T. C., Winge D. R. (2010) Mol. Cell. Biol. 30, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Myers A. M., Pape L. K., Tzagoloff A. (1985) EMBO J. 4, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faye G., Kujawa C., Fukuhara H. (1974) J. Mol. Biol. 88, 185–203 [DOI] [PubMed] [Google Scholar]
- 19. Soto I. C., Fontanesi F., Valledor M., Horn D., Singh R., Barrientos A. (2009) Biochim. Biophys. Acta 1793, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tzagoloff A., Akai A., Needleman R. B. (1975) J. Biol. Chem. 250, 8228–8235 [PubMed] [Google Scholar]
- 21. Herrmann J. M., Stuart R. A., Craig E. A., Neupert W. (1994) J. Cell Biol. 127, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin R. G., Ames B. N. (1961) J. Biol. Chem. 236, 1372–1379 [PubMed] [Google Scholar]
- 23. Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. (1986) Yeast 2, 163–167 [DOI] [PubMed] [Google Scholar]
- 24. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Maniatis T., Fritsch E. F., Sambrook J. (1982) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Schiestl R. H., Gietz R. D. (1989) Curr. Genet. 16, 339–346 [DOI] [PubMed] [Google Scholar]
- 27. Rothstein R. J. (1983) Methods Enzymol. 101, 202–211 [DOI] [PubMed] [Google Scholar]
- 28. Merz S., Westermann B. (2009) Genome Biol. 10, R95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horn D., Zhou W., Trevisson E., Al-Ali H., Harris T. K., Salviati L., Barrientos A. (2010) J. Biol. Chem. 285, 15088–15099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Glerum D. M., Koerner T. J., Tzagoloff A. (1995) J. Biol. Chem. 270, 15585–15590 [DOI] [PubMed] [Google Scholar]
- 31. Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 32. Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., Van den Bogert C. (1998) Eur. J. Biochem. 254, 389–394 [DOI] [PubMed] [Google Scholar]
- 33. Williams S. L., Valnot I., Rustin P., Taanman J. W. (2004) J. Biol. Chem. 279, 7462–7469 [DOI] [PubMed] [Google Scholar]
- 34. Church C., Goehring B., Forsha D., Wazny P., Poyton R. O. (2005) J. Biol. Chem. 280, 1854–1863 [DOI] [PubMed] [Google Scholar]
- 35. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) Science 272, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 36. Tzagoloff A., Akai A., Needleman R. B., Zulch G. (1975) J. Biol. Chem. 250, 8236–8242 [PubMed] [Google Scholar]
- 37. Manthey G. M., Przybyla-Zawislak B. D., McEwen J. E. (1998) Eur. J. Biochem. 255, 156–161 [DOI] [PubMed] [Google Scholar]
- 38. Glerum D. M., Tzagoloff A. (1997) FEBS Lett. 412, 410–414 [DOI] [PubMed] [Google Scholar]
- 39. Barros M. H., Tzagoloff A. (2002) FEBS Lett. 516, 119–123 [DOI] [PubMed] [Google Scholar]
- 40. Nobrega M. P., Nobrega F. G., Tzagoloff A. (1990) J. Biol. Chem. 265, 14220–14226 [PubMed] [Google Scholar]
- 41. Glerum D. M., Muroff I., Jin C., Tzagoloff A. (1997) J. Biol. Chem. 272, 19088–19094 [DOI] [PubMed] [Google Scholar]
- 42. Tzagoloff A., Capitanio N., Nobrega M. P., Gatti D. (1990) EMBO J. 9, 2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Glerum D. M., Shtanko A., Tzagoloff A. (1996) J. Biol. Chem. 271, 14504–14509 [DOI] [PubMed] [Google Scholar]
- 44. Glerum D. M., Shtanko A., Tzagoloff A. (1996) J. Biol. Chem. 271, 20531–20535 [DOI] [PubMed] [Google Scholar]
- 45. Hell K., Herrmann J., Pratje E., Neupert W., Stuart R. A. (1997) FEBS Lett. 418, 367–370 [DOI] [PubMed] [Google Scholar]
- 46. Carlson C. G., Barrientos A., Tzagoloff A., Glerum D. M. (2003) J. Biol. Chem. 278, 3770–3775 [DOI] [PubMed] [Google Scholar]
- 47. Mick D. U., Vukotic M., Piechura H., Meyer H. E., Warscheid B., Deckers M., Rehling P. (2010) J. Cell. Biol. 191, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.