Abstract
The Ras/mitogen-activated protein kinase (MAPK) pathway regulates a variety of cellular processes by activating specific transcriptional and translational programs. Ras/MAPK signaling promotes mRNA translation and protein synthesis, but the exact molecular mechanisms underlying this regulation remain poorly understood. Increasing evidence suggests that the mammalian target of rapamycin (mTOR) plays an essential role in this process. Here, we show that Raptor, an essential scaffolding protein of the mTOR complex 1 (mTORC1), becomes phosphorylated on proline-directed sites following activation of the Ras/MAPK pathway. We found that ERK1 and ERK2 interact with Raptor in cells and mediate its phosphorylation in vivo and in vitro. Using mass spectrometry and phosphospecific antibodies, we found three proline-directed residues within Raptor, Ser8, Ser696, and Ser863, which are directly phosphorylated by ERK1/2. Expression of phosphorylation-deficient alleles of Raptor revealed that phosphorylation of these sites by ERK1/2 normally promotes mTORC1 activity and signaling to downstream substrates, such as 4E-BP1. Our data provide a novel regulatory mechanism by which mitogenic and oncogenic activation of the Ras/MAPK pathway promotes mTOR signaling.
Keywords: MAP Kinases (MAPKs), mTOR, Protein Phosphorylation, Ras, Signal Transduction
Introduction
The Ras/mitogen-activated protein kinase (MAPK) pathway regulates a variety of cellular processes, including cell growth, proliferation, survival, and motility, through the activation of specific transcriptional and translational programs (1–3). The first evidence linking Ras/MAPK signaling to the regulation of mRNA translation stemmed from the finding that inhibition of MEK1/2 blocks growth factor-stimulated global protein synthesis (4). Subsequently, the MAPK-activated protein kinases MNK1/2 were found to phosphorylate the translation initiation factor eIF4E (5, 6), providing a potential mechanism by which MAPK signaling regulates mRNA translation (reviewed in 7, 8). However, more recent evidence indicates that the MNKs are dispensable for global protein synthesis (9). Thus, the functional impact of eIF4E phosphorylation remains unclear and a matter of debate (8, 10).
Increasing evidence suggests that Ras/MAPK signaling largely promotes protein synthesis through the regulation of the mammalian target of rapamycin (mTOR),6 an important regulator of cell growth and proliferation (reviewed in 11–13). mTOR, a member of the phosphoinositide 3-kinase-related protein kinase family, senses and integrates signals from diverse environmental cues to regulate crucial biosynthetic processes involved in cell growth and proliferation (reviewed in 14–16). mTOR associates with different protein partners to form two functionally distinct signaling complexes, the rapamycin-sensitive and rapamycin-insensitive mTOR complex (mTORC) 1 and 2, respectively (17–19). mTORC1 contains the catalytic subunit mTOR, mLST8/GβL, PRAS40, and Raptor (17, 20–24). Raptor directly binds mTOR and is thought to function as a scaffolding protein that recruits mTORC1 substrates through their TOR signaling motif (25, 26). When activated, mTORC1 phosphorylates two main regulators of mRNA translation and ribosome biogenesis, S6K1 (p70 ribosomal S6 kinase 1) and 4E-BP1 (eIF4E-binding protein 1), and thus stimulates cell growth and proliferation (13, 27, 28).
The GTP-bound form of the small GTPase Rheb (Ras homolog enriched in brain) promotes mTORC1 signaling via mechanisms that remain incompletely defined (reviewed in 29, 30). Rheb is reported to bind mTOR directly (31–33), leading to increased phosphotransferase activity of immunoprecipitated mTORC1 (34). Recently, Rheb was found to enhance binding of 4E-BP1 to mTORC1 (35), suggesting an additional mechanism by which Rheb may promote mTORC1 signaling (36). A number of studies have determined that Rheb is a substrate for the TSC1/TSC2 GTPase-activating protein complex (reviewed in 37). PI3K and Ras signaling promote Rheb-mediated activation of mTORC1 through direct phosphorylation and inactivation of TSC2 by Akt and ERK/RSK, respectively (38–43). Hypoxia and energy depletion were shown to activate the TSC1/2 complex through transcriptional up-regulation of REDD (regulated in development and DNA damage) (44–46) and activation of AMP-activated protein kinase and glycogen synthase kinase 3 (47, 48). mTORC1 signaling is also regulated by amino acids; removal of extracellular amino acids was found to inactivate mTORC1 signaling (49). Recent findings revealed that a Ras-related GTPase heterodimer binds to Raptor in an amino acid-dependent manner, thereby recruiting mTORC1 to Rheb-containing endomembrane compartments (50, 51). These data provide a molecular mechanism by which amino acids enable Rheb-mediated activation of mTORC1 signaling.
Although intense effort has focused on understanding the upstream regulation of mTOR, the molecular functions of most mTOR-associated proteins and how they participate in mTOR activation and signaling remain poorly characterized. Activated Akt was found to phosphorylate PRAS40, resulting in the dissociation of PRAS40 from mTORC1, preventing its ability to suppress mTORC1 signaling (23, 34). mTORC1 activity is also regulated by energy sufficiency and growth signaling pathways at the level of Raptor, through its direct phosphorylation by AMP-activated protein kinase and RSK, respectively (52, 53). More recent evidence has indicated that mTOR itself regulates the phosphorylation of Raptor in response to insulin stimulation (54, 55), providing potential feedforward mechanisms that prolong mTORC1 activation. Mitosis-specific alteration of mTORC1 activity has recently been shown to occur through cyclin-dependent kinase 1- and glycogen synthase kinase 3-mediated phosphorylation of Raptor (52, 56), highlighting the close link between mTORC1 activity and the status of Raptor phosphorylation.
Here, we demonstrate an additional mechanism by which the Ras/MAPK pathway leads to increased mTORC1 signaling. We demonstrate that Ras/MAPK signaling leads to Raptor phosphorylation on proline-directed sites in an ERK1/2-dependent manner. Reconstitution of mTORC1 with phosphorylation site-deficient alleles of Raptor impairs mTORC1 kinase activity and downstream signaling to 4E-BP1, suggesting that ERK1/2-mediated phosphorylation of Raptor positively regulates mTORC1. Our data reveal a novel mechanism by which oncogenic activation of the Ras/MAPK pathway promotes mTORC1 signaling and cell growth.
EXPERIMENTAL PROCEDURES
DNA Constructs
The plasmids encoding myc-tagged human Raptor were provided by Dr. David Sabatini (MIT) and described previously (22, 53). All human Raptor mutants were generated using the QuikChange methodology (Stratagene, La Jolla, CA). The vectors encoding FLAG-MEK1-DD, RasG12V, RasS17N, HA-S6K1 and AU1-mTOR (wild-type, kinase-inactive (D2338A), and rapamycin-resistant (S2035I)) were described previously (40, 57). The plasmids encoding FLAG-mTOR and GST-4E-BP1 were kindly provided by Dr. Nahum Sonenberg (McGill University, Montreal QC), and the plasmids encoding HA-tagged ERK1 and HA6-tagged ERK2 were a kind gift from Dr. John Blenis (Harvard Medical School) and Dr. Sylvain Meloche (IRIC, Université de Montréal), respectively.
Antibodies
Antibodies targeted against proline-directed sites (pS/T-P), Raptor, phospho-rpS6 (Ser240/244 and Ser235/236), phospho-Akt (Ser473), ERK1/2, phospho-4EBP1 (Thr37/Thr46), and mTOR were purchased from Cell Signaling Technologies (Beverly, MA). For endogenous ERK1/2 immunoprecipitation assays, ERK1/2 antibodies were purchased from Millipore (Billerica, MA). Anti-HA, anti-myc, anti-FLAG, and phospho-ERK1/2 monoclonal antibodies were purchased from Sigma-Aldrich. The phospho-Raptor Ser696, Thr706, Ser863, and Ser877 antibodies were kindly provided by Millipore. All secondary horseradish peroxidase (HRP)-conjugated antibodies used for immunoblotting were purchased from Chemicon (Temecula, CA).
Cell Culture, Treatment, and Extract Preparation
HEK293, U2OS, and HeLa cells were maintained at 37 °C in Dulbecco's modified Eagle's medium (DMEM) with 4.5 g/liter glucose supplemented with 10% fetal bovine serum (FBS) and antibiotics. HEK293 cells were transfected, serum-starved, and treated with different agonists as described previously (53). Starved cells were pretreated with 10 μm U0126, 10 μm PD184352 (also termed CI-1040), or 25 nm rapamycin) (Biomol, Plymouth Meeting, PA) and stimulated with either 10% FBS, 100 nm insulin, 25 ng/ml EGF (Invitrogen), or 50–100 ng/ml phorbol 13-myristate 12-acetate (PMA; Biomol) before harvesting.
Immunoprecipitations and Immunoblotting
Cell lysates were prepared as described (40, 53). Briefly, cells were lysed in an Nonidet P-40-containing buffer (10 mm KPO4, 1 mm EDTA, 5 mm EGTA, 10 mm MgCl2, 50 mm β-glycerophosphate, 0.5% Nonidet P-40, 0.1% Brij 35, 0.1% deoxycholic acid, 1 mm sodium orthovanadate (Na3VO4), 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 10 μg/ml pepstatin). For mTORC1 immunoprecipitations, cells were harvested in CHAPS lysis buffer (40 mm HEPES, pH 7.4, 2 mm EDTA, 10 mm sodium pyrophosphate, 10 mm β-glycerophosphate, 0.3% CHAPS) as described (53). If indicated, total cell lysates were incubated for 45 min at room temperature with the cleavable cross-linker DTSSP (Thermo Scientific) at a final concentration of 0.5 mg/ml. The cross-linking reactions were quenched by addition of Tris-HCl, pH 7.4 (20 mm final concentration), followed by an additional 30-min incubation at room temperature. Immunoprecipitations were carried out with the indicated antibody for 2 h followed by a 1-h incubation with protein A-Sepharose CL-4B beads (GE Healthcare). Immunoprecipitates were washed three times in lysis buffer and along with total cell lysates were subjected to SDS-PAGE and electroblotted onto nitrocellulose. The data presented are representative of at least three independent experiments.
RNA Interference (RNAi)
For the small interfering RNA (siRNA) studies, validated 21-nucleotide complementary RNA with symmetrical two-nucleotide overhangs were obtained from Qiagen (Valencia, CA). HEK293 cells were transfected using calcium-phosphate and 50 nm siRNA/35-mm dishes. Transfection efficiency was determined to be >90% using a fluorescently labeled mock siRNA. Twenty-four hours following transfection, cells were serum-starved for 16–18 h and stimulated with mitogens.
Protein Phosphotransferase Assays
For ERK assays, beads from immunoprecipitations were washed twice in lysis buffer and twice in kinase buffer (25 mm Tris-HCl, pH,7.4, 10 mm MgCl2, 5 mm β-glycerophosphate). If indicated, recombinant activated ERK1 purchased from Cell Signaling Technologies was used instead of immunopurified proteins. Kinase assays were performed with immunopurified full-length myc-tagged Raptor as substrates, under linear assay conditions. Assays were performed for 10 min at 30 °C in kinase buffer supplemented with 5 μCi of [γ-32P]ATP. All samples were subjected to SDS-PAGE, and incorporation of cold or radioactive 32P was determined by immunoblotting or autoradiography using a Fuji PhosphorImager with ImageQuant software, respectively. The data presented are representative of at least three independent experiments.
For mTORC1 kinase assays, Raptor immunoprecipitates were washed twice in CHAPS buffer followed by two more washes in CHAPS buffer containing 150 mm NaCl, as described (34). Assays were performed for 30 min at 30 °C with bacterially purified recombinant GST-4E-BP1 (3 μg/assay) and 10 μCi of [γ-32P]ATP in mTOR kinase buffer (25 mm HEPES, pH 7.4, 50 mm NaCl, 50 mm β-glycerophosphate, 10 mm MnCl2, 100 μm cold ATP). All samples were analyzed as for ERK assays, and the data presented are representative of at least three independent experiments.
Mass Spectrometry
To identify phosphorylation sites in Raptor, myc-tagged Raptor was immunoprecipitated from ∼5 × 107 HEK293 cells stimulated with PMA. Following SDS-PAGE, Coomassie Blue-stained gel bands corresponding to immunoprecipitated Raptor were excised and subjected to in-gel trypsin or V8 protease digestion, as described previously (53). The resulting peptides were extracted and subjected to capillary LC-tandem MS using a high resolution hybrid mass spectrometer LTQ-orbitrap XL (Thermo Fisher Scientific). All candidate tandem MS spectra were manually inspected.
RESULTS
Activation of the Ras/MAPK Pathway Induces ERK-dependent Phosphorylation of Raptor on Proline-directed Sites
To determine whether Raptor was phosphorylated on proline-directed sites, we used a phospho motif antibody that recognizes phosphorylated Ser or Thr residues when followed by a Pro at the p + 1 position (pS/T-P). Most protein kinases disfavor a Pro residue at this position, but activated ERK1/2 and cyclin-dependent kinase 1 can accommodate such consensus sequences (58). HEK293 cells transfected with myc-tagged human Raptor were stimulated with different agonists of the Ras/MAPK pathway, and immunoprecipitated Raptor was analyzed for phosphorylation by immunoblotting. Using this method, we found that strong agonists of the Ras/MAPK pathway, such as the phorbol ester PMA and EGF, robustly induced Raptor phosphorylation on pS/T-P consensus sites (Fig. 1A). Conversely, we found that cell treatment with serum, which is a stronger PI3K/Akt pathway stimulus, led to a weak increase in Raptor phosphorylation. Treatment of immunoprecipitated Raptor samples with λ-phosphatase resulted in a complete loss of immunoreactivity (Fig. 1B), confirming that the phospho motif antibody recognized a phosphorylated form of Raptor.
FIGURE 1.
Activation of the Ras/MAPK pathway induces ERK1/2-dependent phosphorylation of Raptor on proline-directed sites. A, HEK293 cells were transfected with empty vector or myc-tagged Raptor, serum-starved, and stimulated for 10 or 20 min with agonists of the Ras/MAPK pathway. Immunoprecipitated (IP) Raptor was then assayed for phosphorylation using a phospho motif antibody that recognizes the pS/T-P consensus motif. B, HEK293 cells were transfected with myc-tagged Raptor, serum-starved, and stimulated with 100 ng/ml PMA. Immunoprecipitated Raptor was then treated with 400 units of λ-phosphatase for 60 min at 30 °C and immunoblotted with the pS/T-P phospho motif antibody. C, model depicting the Ras/MAPK pathway and inhibitors and activators used in this study is shown. D, as in A, but cells were pretreated with 10 μm U0126, 10 μm PD184352, or 10 μm BI-D1870 for 30 min prior to 50 ng/ml PMA stimulation. E, HEK293 cells were cotransfected with control vector or myc-tagged Raptor, and siRNA duplexes were targeted against a scrambled sequence (Scr), ERK1 or ERK2, serum-starved, and stimulated with 50 ng/ml PMA. Immunoprecipitated Raptor was then assayed as in A. F, cells were cotransfected with myc-tagged Raptor and constitutively activated (G12V) or dominant negative (S17N) Ras, serum-starved overnight. Immunoprecipitated Raptor was then assayed as in A.
To identify the proline-directed Ser/Thr kinase(s) involved in Raptor phosphorylation, cells were pretreated with different pharmacological inhibitors (Fig. 1C). Both U0126 and PD184352 are small molecule inhibitors of MEK1/2 that prevent activation of the proline-directed kinases ERK1/2, and treatment of cells with these inhibitors prevented Raptor phosphorylation at pS/T-P consensus, suggesting that ERK1/2 are the likely Raptor kinases activated by Ras/MAPK signaling (Fig. 1D). To confirm a requirement for ERK1/2 in the phosphorylation of Raptor, we used RNA interference to deplete ERK1 or ERK2 specifically. Knockdown of ERK1 or ERK2 was found to partially reduce Raptor phosphorylation at pS/T-P consensus sites (Fig. 1E). When both ERK isoforms were knocked down simultaneously, PMA-stimulated Raptor phosphorylation was completely abolished. Thus, both ERK1 and ERK2 are required for Raptor phosphorylation on proline-directed sites.
The Ras GTPases are often mutated in human cancers, leading to constitutive activation of growth signaling cascades, including the MAPK and mTOR pathways (reviewed in 59). To determine whether oncogenic forms of Ras lead to constitutive Raptor phosphorylation, cells were transfected with active (G12V) and inactive (S17N) mutants of H-Ras (Fig. 1F). Interestingly, expression of RasG12V, but not RasS17N, in serum-starved cells strongly stimulated Raptor phosphorylation on pS/T-P consensus sites. These results indicated that oncogenic activation of the Ras/MAPK cascade is sufficient to promote Raptor phosphorylation in the absence of serum and growth factors.
ERK Interacts with and Phosphorylates Raptor in Vitro
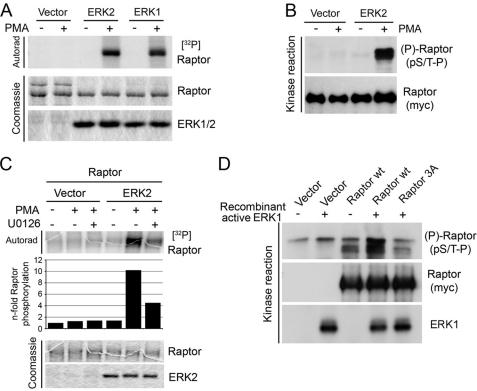
To determine whether ERK can phosphorylate Raptor, we performed in vitro kinase assays using purified proteins. HEK293 cells were transiently transfected with single HA-tagged ERK1 or HA6-tagged ERK2, and the anti-HA immunoprecipitates from unstimulated or PMA-treated cells were incubated with myc-tagged full-length Raptor immunopurified from serum-starved HEK293 cells. Although no incorporation of 32P label was seen in purified Raptor incubated with immunoprecipitates from unstimulated cells, PMA-activated ERK1 and ERK2 were found to increase 32P incorporation robustly in Raptor (Fig. 2A). In a similar experiment without radioactivity where Raptor samples were analyzed using the pS/T-P phospho motif antibody, ERK2 potently stimulated the incorporation of phosphate in purified Raptor at proline-directed sites (Fig. 2B). This effect was abrogated by inhibition of MEK1/2 using U0126 (Fig. 2C), suggesting that ERK activity was required for Raptor phosphorylation. Consistent with this, we found that a recombinant activated form of ERK1 was able to phosphorylate myc-tagged full-length Raptor immunopurified from HEK293 cells (Fig. 2D), confirming the involvement of ERK in Raptor phosphorylation.
FIGURE 2.
ERK1/2 phosphorylate Raptor in vitro. A, immunoprecipitated HA-tagged WT ERK1 or HA6-tagged ERK2 from serum-starved or PMA-stimulated cells lysed in a Nonidet P-40 containing buffer was incubated with immunopurified Raptor in a kinase reaction with [γ-32P]ATP. The resulting samples were subjected to SDS-PAGE, and the dried Coomassie Blue-stained gel was autoradiographed. B, samples were treated as in A, but kinase reactions were performed without radioactivity. The resulting samples were immunoblotted for Raptor phosphorylation on pS/T-P consensus sites. C, as in A, but ERK2 was immunoprecipitated from cells treated with 50 ng/ml PMA in the presence or absence of 10 μm U0126. The levels of 32P incorporation into Raptor were quantified and are shown below the autoradiogram. D, recombinant activated ERK1 was incubated with immunopurified WT or S3A Raptor in a kinase reaction, and the resulting samples were immunoblotted for Raptor phosphorylation on pS/T-P consensus sites.
To determine whether ERK physically interacts with Raptor, HEK293 cells were cotransfected with Raptor and ERK2, and Raptor immunoprecipitates were assayed for the presence of ERK2 (Fig. 3A). Cells were lysed either in CHAPS-containing buffer (Fig. 3A, left), previously shown to maintain integrity of mTORC1 (22), or in Nonidet P40-containing buffer (Fig. 3A, right), which disrupts binding between mTOR and Raptor. Under both conditions, we found that ERK2 readily associated with Raptor, suggesting that ERK2 and Raptor interact in an mTOR-independent manner. This interaction was also detected when ERK2 was directly immunoprecipitated (data not shown) and was not modulated by growth factor stimulation (Fig. 3A). To confirm ERK binding to Raptor, similar experiments were performed using antibodies directed against endogenous forms of ERK1 and ERK2. Although no interaction could be detected in the absence of cross-linker, we found that endogenous ERK1/2 readily interacted with transfected Raptor in the presence of DTSSP (Fig. 3B). These results indicate that ERK binding to Raptor is transient and do not suggest the presence of docking motifs that would stabilize their interaction.
FIGURE 3.
ERK2 interacts with Raptor in a mTOR-independent manner. A, HEK293 cells were transfected with myc-tagged Raptor and HA-tagged ERK2, stimulated with PMA, and lysed in either CHAPS or Nonidet P-40 buffer (NP-40). The presence of ERK2 was assayed within Raptor immunoprecipitates. B, cells were transfected with myc-tagged Raptor and lysed in Nonidet P-40 buffer. Cell extracts were incubated or not with 0.5 mg/ml DTSSP cross-linker for 45 min at room temperature. Cross-linking reactions were quenched by adding Tris, pH 7.4, and the presence of Raptor was assayed within endogenous ERK1/2 immunoprecipitates.
Identification of Ras/MAPK-dependent Proline-directed Phosphorylation Sites in Raptor
We next wanted to identify the specific proline-directed residues phosphorylated in Raptor in response to Ras/MAPK pathway activation. We performed MS experiments on immunoprecipitated Raptor from PMA-stimulated cells and identified eight phosphorylation sites (Ser696, Thr706, Ser719, Ser721, Ser722, Ser859, Ser863, and Ser877), all located between the HEAT repeats and the WD40 domain of Raptor (Fig. 4A). Among these in vivo phosphorylation sites, four were found to have a Pro at the p + 1 position (Ser696, Thr706, Ser863, and Ser877), suggesting that they could be targeted for phosphorylation by ERK1/2. In addition to these residues, we decided to determine the potential role of seven additional sites that were not detected by MS but that lay within a proline-directed consensus motif (Ser8, Ser188, Thr269, Thr363, Ser367, Thr714, and Thr1035). We generated the corresponding unphosphorylatable alanine mutants and analyzed their immunoreactivity with the pS/T-P antibody in response to PMA treatment (data not shown). As demonstrated in Fig. 4B, the only three mutants to display reduced immunoreactivity with the anti-pS/T-P antibody were Raptor S8A, S696A, and S863A.
FIGURE 4.
Identification of Ras/MAPK-dependent proline-directed phosphorylation sites in Raptor. A, schematic representation of Raptor and phosphorylation sites identified by mass spectrometric analysis (proline-directed sites are indicated in boldface characters) is shown. B, cells transfected with wild-type (wt) Raptor, S8A, S696A, or S863A mutant, serum-starved, and stimulated with 50 ng/ml PMA for 20 min. Immunoprecipitated (IP) Raptor was assayed for phosphorylation using the pS/T-P phospho motif antibody. C, tandem MS spectrum of Glu-C peptide precursor ion m/z 727.32+. Labeled ions correspond to b-type fragment ions. The location of Ser8 phosphorylation is confirmed by the observation of a y8 ion at m/z 865.4 and a b5 ion at m/z 589.1 D, cells transfected with wild-type or S3A mutant of Raptor (S8A/S696A/S863A) and treated as in B. E, primary sequence alignment showing conservation of the three proline-directed sites within different Raptor orthologs.
Whereas phosphorylation of Ser696 and Ser863 has been demonstrated in several large scale MS studies (60–64), Ser8 has never been identified as a site of phosphorylation. This may be due to the high reliance on trypsin for polypeptide cleavage and the fact that Ser8 lies within a region poor in Arg and Lys residues. To obtain a more suitable digestion pattern, we used the V8 protease (also called Glu-C) which cleaves peptide bonds on the carboxyl side of Asp and Glu residues. Using this strategy, we detected a peptide phosphorylated at a position corresponding to Ser8 in PMA-treated cells (SEMLQ[pSP]LLGLGE; see spectrum in Fig. 4C). Interestingly, when all three mutations were combined into a single Raptor mutant (Ser8A/S696A/S863A; termed S3A), Raptor phosphorylation following PMA stimulation was abolished (Fig. 4D). Accordingly, we observed the same loss of phosphorylation with Raptor S3A compared with wild-type Raptor when using a recombinant activated form of ERK1 (Fig. 2D). Together, these results confirmed that ERK1/2 mediate the phosphorylation of Raptor on three Ser residues, which are conserved in vertebrate species (Fig. 4E), upon stimulation of the Ras/MAPK pathway.
Characterization of ERK1/2-dependent Phosphorylation of Raptor
To analyze further the regulation of Raptor phosphorylation by ERK1/2, we used rabbit polyclonal antibodies directed against phosphorylated Ser696 and Ser863, as well as two Ras/MAPK-independent phosphorylated residues (Thr706 and Ser877). As we identified Ser8 phosphorylation subsequent to the identification of the other sites, we have not yet generated phosphospecific antibodies against Ser(P)8. Using our panel of phosphospecific antibodies, we found that PMA treatment significantly increased ectopic Raptor phosphorylation at Ser696 and Ser863 compared with serum-starved cells, whereas phosphorylation of Thr706 and Ser877 was unchanged (Fig. 5A). The increase in Raptor phosphorylation was abrogated by the MEK inhibitor U0126, suggesting that Ser696 and Ser863 are phosphorylated by ERK1/2 in cells following activation of the Ras/MAPK pathway.
FIGURE 5.
ERK1/2 phosphorylate Raptor on Ser696 and Ser863in vivo. A, cells were transfected with myc-tagged Raptor and stimulated with 50 ng/ml PMA following pretreatment with 10 μm U0126 or vehicle for 30 min. Immunoprecipitated Raptor was then assayed for phosphorylation using phosphospecific antibodies. B, cells were transfected with myc-tagged Raptor and aui-tagged mTOR, serum-starved, and stimulated with 50 ng/ml PMA or 100 nm insulin following pretreatment with 10 μm PD184352 or 25 nm rapamycin and treated as in A. C, endogenous Raptor phosphorylation at Ser696 and Ser863 was determined in U2OS and HeLa cells. Both cell types were serum-starved overnight, and total cell lysates were assayed for Raptor phosphorylation using phosphospecific antibodies against Ser696 and Ser863. D, phosphorylation of endogenous Raptor in HEK293 cells stimulated with 50 ng/ml PMA or 25 μg/ml EGF for 20 and 10 min, respectively, is shown. Cells were either transfected with an siRNA with a scrambled sequence (Scr), or targeting both ERK1 and ERK2. Phosphorylation of endogenous Raptor in total cell lysates was performed as in A.
Because Ser863 was previously found to be targeted by activated mTOR (54, 55), we next determined the role of mTOR in regulating this site. Although treatment of cells with insulin induced rapamycin-sensitive Raptor phosphorylation at Ser863 (Fig. 5B), we found that both Ser863 and Ser696 were not significantly affected by mTOR inhibition (using rapamycin or PI-103) in response to Ras/MAPK pathway activation. Conversely, treatment of cells with the MEK1/2 inhibitor PD184352 completely abrogated Ser696 and Ser863 phosphorylation, indicating that these sites are predominantly targeted by ERK1/2 in cells. Next, we wanted to validate these findings by looking at endogenous Raptor in other cell types. We found that PMA treatment of serum-starved U2OS and HeLa cells induced endogenous Raptor phosphorylation at both Ser696 and Ser863 (Fig. 5C). Moreover, Raptor phosphorylation in these cells was completely inhibited by the MEK1/2 inhibitor U0126, indicating again the likely involvement of ERK1/2. To demonstrate further the requirement for these kinases, HEK293 cells subjected to RNAi-mediated knockdown of ERK1/2 were stimulated with EGF or PMA (Fig. 5D). Depletion of ERK1/2 strongly decreased PMA- and EGF-induced phosphorylation of endogenous Raptor on Ser696 and Ser863, further confirming that Raptor is directly targeted by these two proline-directed kinases.
ERK1/2-mediated Phosphorylation of Raptor Positively Regulates mTORC1 Activity
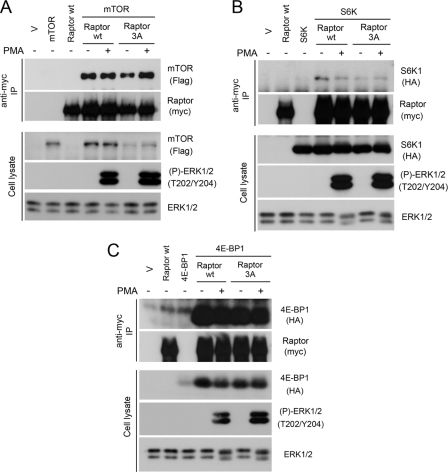
We next wanted to determine the consequences of ERK1/2-dependent phosphorylation of Raptor on mTORC1 function. To assess whether Raptor phosphorylation regulates its association with mTOR, we transfected cells with wild-type Raptor or the S3A mutant and tested their interaction with mTOR by coimmunoprecipitation experiments. The Raptor S3A mutant bound as efficiently as wild-type Raptor to mTOR (Fig. 6A), suggesting that ERK1/2-dependent phosphorylation of Raptor does not regulate mTORC1 assembly and/or stability of the complex. To determine whether Raptor phosphorylation regulates recruitment of mTORC1 substrates, we performed coimmunoprecipitation experiments from cells expressing Raptor and S6K1 or 4E-BP1. As for mTOR interaction, we found that the S3A mutant bound as efficiently as wild-type Raptor to S6K1 (Fig. 6B) and 4E-BP1 (Fig. 6C), indicating that Raptor phosphorylation does not regulate TOR signaling motif-mediated interaction.
FIGURE 6.
Raptor phosphorylation by ERK1/2 does not regulate its ability to interact with mTOR or mTORC1 substrates. A, cells were cotransfected with FLAG-tagged mTOR and either wild-type (wt) Raptor or the S3A mutant, serum-starved, and stimulated with PMA. Associated mTOR was assayed within Raptor immunoprecipitates by immunoblotting. B, cells were cotransfected with HA-tagged S6K1 and either wild-type Raptor or the S3A mutant, serum-starved, and stimulated with PMA. Associated S6K1 was assayed within Raptor immunoprecipitates. C, cells were cotransfected with HA-tagged 4E-BP1 and either wild-type Raptor or the S3A mutant, serum-starved, and stimulated with PMA. Associated 4E-BP1 was assayed within Raptor immunoprecipitates.
Next, we determined whether Raptor phosphorylation regulates mTORC1 activity using an in vitro mTOR phosphotransferase activity assay and recombinant GST-4E-BP1 as substrate (53). Under these conditions, activation of the Ras/MAPK pathway with PMA stimulated mTORC1 activity. Importantly, we found that mTOR kinase activity associated with the Raptor S3A mutant was lower than that associated with wild-type Raptor (Fig. 7A). To confirm this finding, we measured the in vivo kinase activity of the expressed mTORC1 using a rapamycin-resistant mutant of mTOR (S2035I)(Fig. 7B). This mutant was expressed together with either wild-type Raptor or the S3A mutant in HEK293 cells. After serum starvation, cells were treated with rapamycin to block endogenous mTORC1 kinase activity and then stimulated with PMA. Consistent with in vitro activity assays, mutation of the ERK1/2-dependent sites in Raptor decreased PMA-stimulated mTORC1 activity in vivo as shown by reduced phosphorylation of 4E-BP1 at the mTORC1-dependent Thr37 and Thr46 residues (Fig. 7B). Consistent with these observations, we found that treatment of serum-growing cells with MEK1/2 inhibitors completely inhibited mTORC1 kinase activity (Fig. 7C). Altogether, these results suggest that Raptor phosphorylation by ERK1/2 positively stimulates mTORC1 activity. These data support a direct role for the Ras/MAPK pathway in mTORC1 activation, whereby ERK1/2-mediated phosphorylation of Raptor promotes mTORC1 activity independently of the PI3K/Akt pathway (Fig. 8).
FIGURE 7.
ERK1/2-mediated phosphorylation of Raptor is important for mTORC1 kinase activity. A, cells were cotransfected with wild-type (wt) mTOR and wild-type Raptor or the S3A mutant and serum-starved, and mTORC1 kinase activity was assayed within Raptor immunoprecipitates using GST-4E-BP1 as a substrate. The kinase reaction was performed in the presence of [γ-32P]ATP, and the resulting samples were subjected to SDS-PAGE, and the dried Coomassie Blue-stained gel was autoradiographed. B, wild-type Raptor or the S3A mutant was transfected together with wild-type mTOR or a rapamycin-resistant mutant (S2035I) into HEK293 cells. After serum starvation, cells were treated with 20 nm rapamycin for 20 min to block endogenous mTORC1 activity and then stimulated with PMA for 20 min. Phosphorylation of 4E-BP1 on the mTORC1-dependent residues Thr37/Thr46 was detected by immunoblotting. The histogram shows quantifications of phosphorylated 4E-BP1 from three independent experiments. C, as in A, but cells were cotransfected with wild-type mTOR and Raptor and treated with 10 μm U0126, 10 μm PD184352, or vehicle for 30 min.
FIGURE 8.
Schematic representation of the molecular mechanisms employed by the Ras/MAPK pathway to regulate mTORC1 signaling. Activation of the Ras/MAPK pathway stimulates mTORC1 activity through the coordinated action of the kinases ERK1/2 and RSK on the TSC protein complex upstream mTORC1, and on Raptor, an important partner of mTOR within mTORC1.
DISCUSSION
The data presented here demonstrate a novel mechanism employed by the Ras/MAPK pathway to promote mTORC1 signaling. Previous studies by our group as well as others indicated that both ERK and RSK stimulate Rheb GTP loading via the phosphorylation of TSC2 and subsequent inactivation of the TSC complex (38, 40). Our more recent findings demonstrated that Ras/MAPK signaling also directly activates mTORC1 via RSK-mediated phosphorylation of Raptor (53). Here, we show that the MAP kinases ERK1 and ERK2 also directly phosphorylate Raptor and thereby promote mTORC1 signaling upon Ras/MAPK pathway activation. Together with previous findings, our study indicates that the Ras/MAPK pathway activates mTORC1 signaling via a 2-fold mechanism, the phosphorylation of both TSC2 and Raptor via both ERK1/2 and RSK.
Using complementary approaches, we found that ERK1/2 phosphorylate Raptor on three proline-directed residues (Ser8, Ser696, and Ser863) following Ras/MAPK pathway activation. Interestingly, Ser863 has previously been found to be phosphorylated by mTOR following insulin stimulation or under conditions of Rheb overexpression (54, 55). Here, we demonstrate that the kinase responsible for Ser863 phosphorylation depends on the particular cell stimulus. We confirmed that insulin stimulation, which strongly activates the PI3K pathway, leads to rapamycin-sensitive Ser863 phosphorylation (Fig. 5B), thereby implicating mTOR in phosphorylation of this site. Our data also indicate that when the stimulus primarily engages the Ras/MAPK pathway, ERK1/2 become the predominant kinases involved in Raptor phosphorylation (Figs. 4 and 5). This conclusion was demonstrated using rapamycin (Fig. 5) and PI-103 (data not shown), which did not significantly block Raptor phosphorylation induced by PMA and EGF. Consistently, inhibition of MEK1/2 using PD184352 or U0126 strongly prevented Raptor phosphorylation upon PMA and EGF treatments, thus showing that Raptor phosphorylation can be achieved by different protein kinases. In agreement, ERK1/2 interacted with Raptor and RNAi against these kinases efficiently blocked Raptor phosphorylation (Figs. 1E and 5D). These findings suggest that although mTOR-mediated phosphorylation of Raptor plays an important role in mTORC1 activation by insulin, this can be mediated in part by ERK1/2 when cells are treated with Ras/MAPK pathway agonists.
A recent study indicated that Raptor Ser863 and Ser696 are phosphorylated during mitosis, possibly by Cdk1 and glycogen synthase kinase 3 (56). Although ERK1/2 are thought to be inactive during mitosis, the fact that Ser863 and Ser696 are phosphorylated during this phase highlights the potential importance of regulating the phosphorylation status of these sites. In addition, our results are consistent with the idea that several protein kinases other than mTOR regulate Raptor phosphorylation. In addition to known phosphorylation sites, we also identified Ser8 as a predominant ERK1/2-dependent phosphorylation site (Fig. 4, B and C). Whereas Ser696 and Ser863 lie within a central region of Raptor with no homology to known functional domains, Ser8 localizes near the Raptor N-terminal conserved domain (22), which mediates binding to TOR signaling motif-containing substrates (65). Although this structural insight initially suggested that Ser8 phosphorylation could be involved in regulating substrate binding to Raptor, mutation of this site alone or in combination did not affect the ability of Raptor to interact with 4E-BP1 and S6K1 (Fig. 6, B and C, and data not shown). Although our data indicate that Ras/MAPK pathway-dependent phosphorylation of Raptor does not affect its ability to interact with mTOR, we found that mutation of these phosphorylation sites impairs mTORC1 kinase activity (Fig. 7). These findings are in accordance with studies implicating phosphorylation of Ser863 and Ser696 in mTORC1 activity (54–56), but our study specifically implicates ERK1/2 as the proline-directed kinases involved following Ras/MAPK pathway activation by growth factors and oncogenes.
Based on our data and those of others, we propose that Raptor functions as a molecular sensor that responds to the cellular environment via complex phosphorylation governed by distinct kinases, thus enabling tight control of mTORC1 activity (Fig. 8). Although the molecular mechanisms underlying such regulation remain poorly defined, Raptor phosphorylation may impose conformational changes in mTORC1 components that would modulate the way mTOR and its partners function. Our findings also highlight the crucial role of the Ras/MAPK pathway in regulating mTORC1 activity and suggest that rapamycin or second-generation mTOR inhibitors may be therapeutically effective for treating neoplasms with high mutation rates in components of the Ras/MAPK pathway, such as colorectal cancer and melanoma.
Acknowledgments
We thank Dr. Arnold Kristof for helpful comments on the manuscript and Drs. John Blenis, Sylvain Meloche, and David Sabatini for providing DNA constructs.
This work was supported by Terry Fox Foundation Grant 018311from the Canadian Cancer Society Research Institute. IRIC core facilities are supported by the Fondation pour la recherche en santé du Québec.
- mTOR
- mammalian target of rapamycin
- DTSSP
- 3,3′-dithiobis[sulfosuccinimidylpropionate]
- 4E-BP1
- eIF4E-binding protein 1
- mTORC
- mTOR complex
- PMA
- phorbol 13-myristate 12-acetate
- Rheb
- Ras homolog enriched in brain
- S6K1
- p70 ribosomal S6 kinase 1
- TSC
- tuberous sclerosis complex
- RSK
- p90 ribosomal S6 kinase.
REFERENCES
- 1. Karnoub A. E., Weinberg R. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajalingam K., Schreck R., Rapp U. R., Albert S. (2007) Biochim. Biophys. Acta 1773, 1177–1195 [DOI] [PubMed] [Google Scholar]
- 3. Mitin N., Rossman K. L., Der C. J. (2005) Curr. Biol. 15, R563–R574 [DOI] [PubMed] [Google Scholar]
- 4. Servant M. J., Giasson E., Meloche S. (1996) J. Biol. Chem. 271, 16047–16052 [DOI] [PubMed] [Google Scholar]
- 5. Waskiewicz A. J., Flynn A., Proud C. G., Cooper J. A. (1997) EMBO J. 16, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waskiewicz A. J., Johnson J. C., Penn B., Mahalingam M., Kimball S. R., Cooper J. A. (1999) Mol. Cell. Biol. 19, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roux P. P., Blenis J. (2004) Microbiol. Mol. Biol. Rev. 68, 320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buxade M., Parra-Palau J. L., Proud C. G. (2008) Front. Biosci. 13, 5359–5373 [DOI] [PubMed] [Google Scholar]
- 9. Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scheper G. C., Proud C. G. (2002) Eur. J. Biochem. 269, 5350–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anjum R., Blenis J. (2008) Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 12. Carriere A., Ray H., Blenis J., Roux P. P. (2008) Front. Biosci. 13, 4258–4275 [DOI] [PubMed] [Google Scholar]
- 13. Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 14. Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 15. Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 16. Hay N., Sonenberg N. (2004) Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 17. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 18. Jacinto E. (2008) IUBMB Life 60, 483–496 [DOI] [PubMed] [Google Scholar]
- 19. Bhaskar P. T., Hay N. (2007) Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 20. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 21. Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 22. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 23. Vander Haar E., Lee S. I., Bandhakavi S., Griffin T. J., Kim D. H. (2007) Nat. Cell Biol. 9, 316–323 [DOI] [PubMed] [Google Scholar]
- 24. Wang L., Harris T. E., Roth R. A., Lawrence J. C., Jr. (2007) J. Biol. Chem. 282, 20036–20044 [DOI] [PubMed] [Google Scholar]
- 25. Schalm S. S., Blenis J. (2002) Curr. Biol. 12, 632–639 [DOI] [PubMed] [Google Scholar]
- 26. Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 27. Proud C. G. (2009) Biochem. Soc. Trans. 37, 227–231 [DOI] [PubMed] [Google Scholar]
- 28. Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N. (2006) Oncogene 25, 6416–6422 [DOI] [PubMed] [Google Scholar]
- 29. Aspuria P. J., Tamanoi F. (2004) Cell. Signal. 16, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 30. Manning B. D., Cantley L. C. (2003) Trends Biochem. Sci. 28, 573–576 [DOI] [PubMed] [Google Scholar]
- 31. Urano J., Comiso M. J., Guo L., Aspuria P. J., Deniskin R., Tabancay A. P., Jr., Kato-Stankiewicz J., Tamanoi F. (2005) Mol. Microbiol. 58, 1074–1086 [DOI] [PubMed] [Google Scholar]
- 32. Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005) Curr. Biol. 15, 702–713 [DOI] [PubMed] [Google Scholar]
- 33. Smith E. M., Finn S. G., Tee A. R., Browne G. J., Proud C. G. (2005) J. Biol. Chem. 280, 18717–18727 [DOI] [PubMed] [Google Scholar]
- 34. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 35. Sato T., Nakashima A., Guo L., Tamanoi F. (2009) J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Avruch J., Long X., Lin Y., Ortiz-Vega S., Rapley J., Papageorgiou A., Oshiro N., Kikkawa U. (2009) Biochem. Soc. Trans. 37, 223–226 [DOI] [PubMed] [Google Scholar]
- 37. Huang J., Manning B. D. (2008) Biochem. J. 412, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 39. Ballif B. A., Roux P. P., Gerber S. A., MacKeigan J. P., Blenis J., Gygi S. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13489–13494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 42. Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002) Mol. Cell 10, 151–162 [DOI] [PubMed] [Google Scholar]
- 43. Rolfe M., McLeod L. E., Pratt P. F., Proud C. G. (2005) Biochem. J. 388, 973–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brugarolas J., Lei K., Hurley R. L., Manning B. D., Reiling J. H., Hafen E., Witters L. A., Ellisen L. W., Kaelin W. G., Jr. (2004) Genes Dev. 18, 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corradetti M. N., Inoki K., Guan K. L. (2005) J. Biol. Chem. 280, 9769–9772 [DOI] [PubMed] [Google Scholar]
- 46. Sofer A., Lei K., Johannessen C. M., Ellisen L. W. (2005) Mol. Cell. Biol. 25, 5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 48. Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L. (2006) Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 49. Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., Avruch J. (1998) J. Biol. Chem. 273, 14484–14494 [DOI] [PubMed] [Google Scholar]
- 50. Sancak Y., Sabatini D. M. (2009) Biochem. Soc. Trans. 37, 289–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008) Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 54. Wang L., Lawrence J. C., Jr., Sturgill T. W., Harris T. E. (2009) J. Biol. Chem. 284, 14693–14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Foster K. G., Acosta-Jaquez H. A., Romeo Y., Ekim B., Soliman G. A., Carriere A., Roux P. P., Ballif B. A., Fingar D. C. (2010) J. Biol. Chem. 285, 80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramirez-Valle F., Badura M. L., Braunstein S., Narasimhan M., Schneider R. J. (2010) Mol. Cell. Biol. 30, 3151–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. (2002) Genes Dev. 16, 1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ubersax J. A., Ferrell J. E., Jr. (2007) Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 59. Malumbres M., Barbacid M. (2003) Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 60. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 61. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 62. Munton R. P., Tweedie-Cullen R., Livingstone-Zatchej M., Weinandy F., Waidelich M., Longo D., Gehrig P., Potthast F., Rutishauser D., Gerrits B., Panse C., Schlapbach R., Mansuy I. M. (2007) Mol. Cell Proteomics 6, 283–293 [DOI] [PubMed] [Google Scholar]
- 63. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 64. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]