Abstract
Characteristically for a regulatory protein, the IRF-1 tumor suppressor turns over rapidly with a half-life of between 20–40 min. This allows IRF-1 to reach new steady state protein levels swiftly in response to changing environmental conditions. Whereas CHIP (C terminus of Hsc70-interacting protein), appears to chaperone IRF-1 in unstressed cells, formation of a stable IRF-1·CHIP complex is seen under specific stress conditions. Complex formation, in heat- or heavy metal-treated cells, is accompanied by a decrease in IRF-1 steady state levels and an increase in IRF-1 ubiquitination. CHIP binds directly to an intrinsically disordered domain in the central region of IRF-1 (residues 106–140), and this site is sufficient to form a stable complex with CHIP in cells and to compete in trans with full-length IRF-1, leading to a reduction in its ubiquitination. The study reveals a complex relationship between CHIP and IRF-1 and highlights the role that direct binding or “docking” of CHIP to its substrate(s) can play in its mechanism of action as an E3 ligase.
Keywords: Protein Domains, Transcription Factors, Tumor Suppressor, Ubiquitin Ligase, Ubiquitination, CHIP, Chaperone, Docking, IRF-1
Introduction
IRF-1 (interferon regulatory factor-1) is a transcription factor initially identified as an activator of IFNβ (interferon-β gene) (1), which has subsequently been intimately linked to the antiviral response and the response to DNA damage (2, 3). Additionally, IRF-1 is a tumor suppressor protein, and deletions of IRF-1 are associated with the development of gastric and esophageal tumors as well as some leukemias (4–6). The IRF-1 protein is short lived and has a half-life in the region of 30 min in cultured cells (7–9). It is primarily degraded via the 26 S proteasome (8, 10), and the rate of degradation can be regulated in response to cellular conditions (7). For example, agents such as ionizing radiation increase steady state levels of the IRF-1 protein through a concerted mechanism that includes a decrease in its rate of degradation (2, 7).
Like other proteins degraded via the proteasome, IRF-1 is polyubiquitinated prior to degradation (8, 10). The ubiquitination process itself involves at least three distinct enzymes. A ubiquitin (Ub)4-activating enzyme, or E1, activates Ub and forms a Ub-thiol ester in an ATP-dependent process subsequently. A ubiquitin-conjugating enzyme (E2), which associates with a ubiquitin ligase (E3), is involved in ubiquitin transfer from the E1 to the substrate through the E3. The E3 and occasionally the E2·E3 complex give specificity to the system because they are involved in substrate recognition (11). Although IRF-1 has been characterized as a substrate of the ubiquitination system, the E3 ligase(s) involved in IRF-1 ubiquitination have not yet been identified.
In the current study, we describe an interaction between IRF-1 and CHIP (C terminus of Hsc70-interacting protein), leading to the identification of CHIP as an E3 ligase for IRF-1. CHIP is thought to provide a link between the protein folding pathway(s) and the pathways within a cell that lead to protein degradation. Structurally, CHIP comprises an N-terminal tetratricopeptide repeat (TPR) domain, through which it can bind to the Hsp70 and Hsp90 families of molecular chaperones; a central charged domain that is required for dimerization but otherwise has a largely unknown function; and a C-terminal U-box structure that binds E2 enzymes and mediates CHIP function as an E3 ubiquitin ligase (12–15). The U-box is structurally similar to the RING (really interesting new gene) domain present in the RING family of E3 Ub ligases, although it is stabilized by intramolecular hydrogen bonds and salt bridges rather than Zn2+ ions (16). It is commonly believed that CHIP binds to Hsp70 and targets misfolded client proteins for degradation, bypassing the need for a direct interaction with its substrate (17, 18). Recently, however, a number of studies have suggested an alternate CHIP ubiquitination pathway in which the substrate binding activity of CHIP may play a key role in determining its specific E3 ligase function (19, 20). Here we present evidence in support of diverse roles for CHIP in the regulation of IRF-1. Although CHIP has a positive effect on IRF-1 protein levels in unstressed cells, in response to specific stresses, such as heat and heavy metal stress, CHIP binds directly to a docking site in the central region of IRF-1 facilitating IRF-1 ubiquitination. Thus, (i) CHIP and IRF-1 can form a stable complex in vitro and in cells that does not require Hsp70, (ii) CHIP binding to an intrinsically disordered domain of IRF-1 is required for its ubiquitination because this domain can act in trans to interact with CHIP and inhibit ubiquitination of IRF-1, and (iii) CHIP·IRF-1 complex formation is regulated in cells exposed to selective stress conditions and correlates with an increase in IRF-1 ubiquitination and a decrease in its steady state levels. Therefore, direct binding of both Hsp70 (21) and CHIP can regulate IRF-1, highlighting the intimate link between the molecular chaperones and IRF-1 function.
EXPERIMENTAL PROCEDURES
Chemicals, Antibodies, and Peptides
Antibodies were used at the concentrations indicated by the supplier and were anti-IRF-1 mAb (catalog no. 612047, BD Biosciences), anti-GFP mAb and pAb (catalog nos. 632380 and 632459, Clontech), anti-GAPDH pAb (catalog no. ab9483, Abcam), anti-FLAG mAb, anti-Myc pAb, anti-GST mAb, anti-β-actin mAb, and anti-vimentin mAb (catalog nos. F3165, C3956, G1160, A5441, and V5255, Sigma), anti-caspase-3 pAb (catalog no. sc7148, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-HP1α mAb (catalog no. 05-689, Millipore), anti-ubiquitin FK-1 and FK-2 (Biomol BML-PW8805-0500 and BML-PW8810-0500), anti-histone H3 and anti-calreticulin mAbs (catalog nos. 5192 and 2891, Cell Signaling), and anti-His mAb (catalog no. 70796-3, Novagen). Anti-CHIP mAb (MBC3) was from Moravian Biotechnology, and anti-Myc mAb was obtained from CRUK. Secondary antibodies were purchased from DakoCytomation. MG-132 (Calbiochem) was dissolved in DMSO to 10 mm and used as indicated. Peptides were from Chiron Mimotopes and were synthesized with a biotin tag and an SGSG spacer at the N terminus.
Plasmids and Protein Purification
The human IRF-1 sequence was codon-optimized for Escherichia coli expression (Genscript) and inserted into pDEST-15 using Gateway technology (Invitrogen) to generate GST-IRF-1. FLAG-IRF-1 was generated by amplifying an EcoRI-IRF-1-BamHI fragment from pcDNA3-IRF-1 (8) and ligating it into p3xFLAG-Myc-CMV-24 (Sigma). For IRF-1 Δ106–140, the BlpI internal site on human IRF-1 was used. A BlpI-IRF-1(141–325)-BamHI fragment was amplified from FLAG-IRF-1 WT and ligated with FLAG-IRF-1 WT that was digested with BlpI and BamHI to give IRF-1 Δ106–140; this was inserted into the Gateway system by introducing attB1 and attB2 sites according to the manufacturer's instructions. pDEST53-IRF-1 (GFP-IRF-1) was as described previously (8). pET15bmod-CHIP was a kind gift from Prof. Alicja Zylicz and Renata Filipek, and pMBC1-IRF-1(115–140) (GFP-IRF-1(115–140)) was from the Hauser group (22). pcDNA3.1-His/Myc-CHIP WT and domains were kind gifts from Prof. Cam Patterson and Dr. Holly McDonough. GST-IRF-1 expressed in BL21-AI (Invitrogen) was purified using glutathione-Sepharose beads (GE Healthcare) according to the manufacturer's instructions. His-CHIP was expressed in BL21-DE3 and purified using Ni2+-NTA-agarose (Qiagen) according to the manufacturer's instructions. In both cases, Mg2+-ATP washes were incorporated prior to elution to remove any bound chaperones. Additionally, untagged human IRF-1 was expressed and purified in a cell-free environment using the PURExpress in vitro protein synthesis kit (New England Biolabs) according to the manufacturer's instructions.
Cell Culture and Immunoblotting
A375, MCF-7, MDA-MB-231, and HeLa cells were cultured in DMEM (Invitrogen), whereas H1299, OVCAR8, HCC-827, DU-145, A549, and ACHN were cultured in RPMI 1640 (Invitrogen). All media were supplemented with 10% (v/v) fetal bovine serum (Autogen Bioclear) and 1% (v/v) penicillin/streptomycin mix (Invitrogen). A375 and HeLa cells were maintained at 10% CO2; all remaining cells were maintained at 5% CO2. Cells were seeded 24 h before transfection. DNA was transfected into the cells using Attractene (Qiagen) and siRNA (OnTargetPlus siRNA pools from Dharmacon; catalogue no. D-001810-10-20 for control siRNA pool and L-007201-00-0020 for CHIP siRNA pool) using Dharmafect (Thermo Scientific) as described in the manufacturer's instructions. If required, cells were treated as follows. For serum starvation, serum was withdrawn from the media at the time of transfection 24 h prior to harvesting; heat shock was carried out at 43 °C for 30 min immediately prior to harvesting unless otherwise indicated; for heavy metal stress, cells were treated with 1 mm ZnCl2 for 90 min. MG-132 treatment (50 μm) was for 4 h prior to harvesting. Post harvesting, cells were lysed in Triton Lysis Buffer (50 mm HEPES, pH 7.5, 0.2% (v/v) Triton X-100, 150 mm NaCl, 10 mm NaF, 2 mm DTT, 0.1 mm EDTA, protease inhibitor mix (20 μg/ml leupeptin, 1 μg/ml aprotinin, 2 μg/ml pepstatin, 1 mm benzamidine, 10 μg/ml soybean trypsin inhibitor, 2 mm pefabloc)) unless otherwise indicated. Immunoblots were performed as described previously (21). To detect endogenous IRF-1, 75 μg of protein was loaded per lane, whereas 25 μg was sufficient to detect exogenous IRF-1.
Immunoprecipitation and Affinity Chromatography Using Recombinant GST-IRF-1
FLAG-IRF-1 or GFP-IRF-1 (or corresponding empty vector) was transfected into A375 cells as described above. Post-transfection (24 h), cells were harvested and lysed in Triton Lysis Buffer. Following this, the lysates were precleared using Sepharose 4B beads (Sigma). FLAG-tagged complexes from 3 mg of total cellular lysate were purified using anti-FLAG-M2-agarose (35 μl; Sigma) according to the manufacturer's instructions and analyzed by SDS-PAGE/immunoblot. GFP-tagged complexes were isolated by incubating precleared lysate (7.5 mg) with protein G-Sepharose beads (40 μl; GE Healthcare) and anti-GFP pAb (5 μl) overnight at 4 °C. The beads were washed extensively with PBS supplemented with 0.4% Triton X-100 and eluted by heating at 90 °C in SDS-PAGE sample buffer for 5 min. For affinity chromatography, GST-tagged IRF-1 and GST alone were purified as described above; however, bound GST-proteins were not eluted from the glutathione-Sepharose column. Instead, precleared A375 cell lysate (1 mg) was added and incubated for 1 h at 4 °C. The column was washed five times with PBS containing 0.2% (v/v) Triton X-100 and once with Buffer W (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mm benzamidine). Bound proteins were eluted by heating at 90 °C in sample buffer and analyzed by SDS-PAGE/immunoblot.
Protein Binding Assay
Purified recombinant His-CHIP (100 ng) was immobilized on a microtiter plate in 0.1 m NaHCO3 buffer (pH 8.6) at 4 °C. Non-reactive sites were blocked using PBS containing 3% BSA. A titration of the protein of interest was added in 1× Reaction Buffer (25 mm HEPES, pH 7.5, 50 mm KCl, 10 mm MgCl2, 5% (v/v) glycerol, 0.1% (v/v) Tween 20, 2 mg/ml BSA) for 1 h at room temperature. After washing in PBS containing 0.1% (v/v) Tween 20, binding was detected using anti-GST and HRP-tagged anti-mouse antibodies, and electrochemical luminescence was quantified using a luminometer (Fluoroskan Ascent FL, Labsystems). For peptide competition assays, His-CHIP (100 ng) was preincubated with a titration of peptide (or DMSO control) in 1× reaction buffer for 10 min at room temperature, after which the mix was incubated for 1 h at room temperature with GST-IRF-1 (100 ng) immobilized on a microtiter plate as above. Washing and detection were as described above, except using anti-His mAb. For peptide binding assays, microtiter plates were coated with streptavidin (1 μg/well), following which enough biotin-tagged peptide to saturate the streptavidin (∼60 pmol) was added. Non-reactive sites were blocked using PBS containing 3% BSA as above. His-CHIP (25 ng) in 1× reaction buffer was added for 1 h at room temperature. Washing and detection were as above using anti-His mAb.
Ubiquitination Assay
In vitro ubiquitination assays were carried out essentially as previously described (23) except using 25 nm GST-IRF-1 (WT or mutant) as substrate. Reactions were started with His-CHIP (60–360 nm as indicated; usually 60 nm), incubated for 20 min at 30 °C, and analyzed using 4–12% NuPAGE gels in a MOPS buffer system/immunoblot. In vivo ubiquitination assays were carried out as described (8).
In Vitro Pull-down
His/Myc-CHIP (WT or mutant) was expressed in a TNT coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. The CHIP proteins were isolated using Ni2+-NTA-agarose (Qiagen) and washed extensively according to the manufacturer's recommendations. Mg2+-ATP washes were incorporated to remove any bound chaperones. Post washing, 5 μl of untagged IRF-1 (see “Plasmids and Protein Purification” for details) was added to the column and incubated for 1 h at 4 °C. Columns were then washed extensively with 20 mm Tris, pH 8.0, and 150 mm NaCl containing 0.2% Triton X-100, and bound proteins were eluted in 20 mm Tris, pH 8.0, 150 mm NaCl, and 300 mm imidazole. Eluates were analyzed by SDS-PAGE/immunoblot.
In-Cell Western
Cells were seeded in a 96-well sterile black plate with a clear bottom (Costar) as required. In-Cell WesternTM assays were subsequently performed on a Licor Odyssey SA scanner according to the manufacturer's instructions. Anti-IRF-1 mAb (BD Biosciences) and anti-CHIP mAb (MBC3) were used at 1:500 and 1:50, respectively. The DNA stain (DRAQ5) was used as a control to normalize for cell number, as recommended by the manufacturer.
Subcellular Fractionation
Subcellular fractionation was carried out using the ProteoExtract kit (Calbiochem) or the subcellular protein fractionation kit (Thermo Scientific) as indicated, according to the manufacturer's instructions.
RESULTS
Identification of the Ubiquitin E3 Ligase CHIP as a Novel IRF-1-binding Protein
We have previously shown that IRF-1 is polyubiquitinated and degraded via the proteasome and that the C terminus of IRF-1, which is required for the efficient ubiquitination of the protein, binds directly to the molecular chaperone Hsp70 (8, 21). Given that the outcome of Hsp70-substrate interactions is governed by a host of co-chaperones, we were interested in extending our previous study (21) in order to determine whether the co-chaperone and E3 ubiquitin ligase CHIP (24) played a role in the regulation of IRF-1.
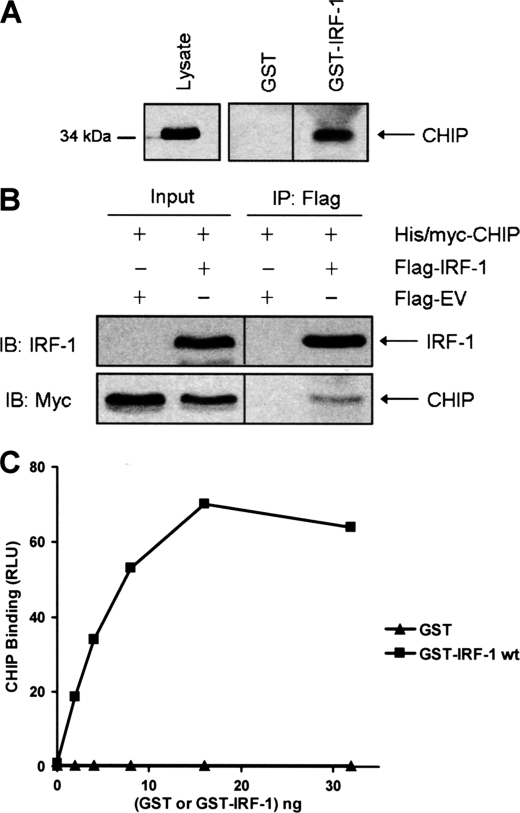
When A375 cell lysate was passed through a column prepared by immobilizing GST-IRF-1 on glutathione-Sepharose beads, endogenous CHIP bound specifically to GST-IRF-1 and not to a GST alone control column (Fig. 1A). Additionally, CHIP was co-immunoprecipitated with IRF-1 from A375 cells in which both proteins were overexpressed (Fig. 1B). The above experiments suggest that CHIP and IRF-1 can form a complex; however, they do not address whether the proteins interact directly or whether additional factors, such as Hsp70, are required. In order to examine whether CHIP could bind IRF-1 in the absence of other factors, recombinant proteins purified from E. coli were used. When CHIP was immobilized on a microtiter well and incubated with IRF-1 that was in the mobile phase, it bound specifically to GST-IRF-1 but not GST alone (Fig. 1C). This shows that CHIP has the potential to bind directly to IRF-1 and that Hsp70 or other cellular factors are not required to mediate the interaction.
FIGURE 1.
Identification of CHIP as a novel IRF-1-binding protein. A, recombinant GST and GST-IRF-1 were immobilized on glutathione-Sepharose beads and incubated with A375 cell lysate (1 mg). Bound proteins and the input (Lysate; 2.5% loaded on gel) were analyzed by SDS-PAGE/immunoblot using anti-CHIP mAb. The data are representative of two individual experiments. B, A375 cells were co-transfected with FLAG-empty vector (EV) or FLAG-IRF-1 (2 μg) and His/Myc-CHIP (2 μg), and FLAG conjugates were immunoprecipitated using anti-FLAG-M2-agarose. Eluates (IP), and 1.5% of the lysate used for the immunoprecipitation (Input) were analyzed by SDS-PAGE/immunoblot (IB) using anti-IRF-1 and anti-Myc mAbs. Results are representative of at least three separate experiments. C, His-CHIP (100 ng) was immobilized and incubated with a titration (0–32 ng) of GST-IRF-1 or GST alone. Binding was detected using anti-GST mAb. Protein amount against binding, expressed as relative light units (RLU) is shown. The results are representative of two separate experiments.
CHIP Binds to an Arg-Lys-Ser-rich Motif in the Mf2 Domain of IRF-1
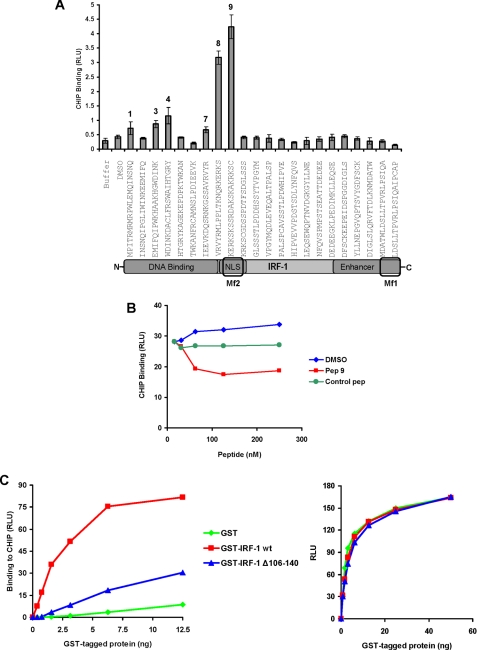
Having established that CHIP can interact with IRF-1 in cells and in a cell-free environment, we sought to map the CHIP binding interface on IRF-1 using a library of biotin-tagged overlapping peptides spanning the entire length of the IRF-1 protein. When the peptides were immobilized on streptavidin-coated microtiter wells and incubated with purified CHIP, the CHIP bound stably to an Arg-Lys-Ser-rich region in the Mf2 (multifunctional 2) domain of IRF-1 (aa 106–140; Fig. 2A, peptides 8 and 9). In addition, CHIP bound to a lesser extent to a number of peptides from the DNA-binding domain of IRF-1 (Fig. 2A, peptides 1, 3, 4, 7). When the Mf2-derived peptide 9 was used in a competition assay to determine if it could reduce the binding of CHIP to full-length IRF-1, peptide 9, but not a control peptide, significantly inhibited CHIP·IRF-1 complex formation (Fig. 2B). The above data suggest that CHIP binds to a complex interface on IRF-1 and that a region from the IRF-1 Mf2 domain, aa 106–140, is sufficient to form a stable interaction and to partially compete with the full-length protein for binding to CHIP.
FIGURE 2.
CHIP binds to an Arg-Lys-Ser-rich motif in the Mf2 domain of IRF-1. A, biotin-tagged IRF-1 peptides (60 pmol) spanning the entire length of the protein were immobilized on streptavidin-coated microtiter wells and incubated with His-CHIP (25 ng); binding was detected using anti-His mAb. CHIP binding to the IRF-1 peptides, expressed as relative light units (RLU) is shown. The results are representative of four separate experiments. B, His-CHIP (100 ng) was preincubated with a titration of the indicated IRF-1 peptides and added to immobilized GST-IRF-1 (100 ng). CHIP binding was detected and expressed as above. A C-terminal IRF-1 peptide (sequence MDATWLDSLLTPVRLPSIQA) that did not bind CHIP (see Fig. 2A) was used as a control. C, His-CHIP (100 ng) was immobilized on microtiter wells and incubated with a titration (0–12.5 ng) of GST-IRF-1 WT, GST-IRF-1 Δ106–140, or GST alone. Binding was detected using an anti-GST monoclonal antibody. The results are representative of two independent experiments. Right, normalization of protein levels using anti-GST antibody.
To further establish a requirement for the Mf2 domain of IRF-1 in CHIP binding, a deletion mutant (IRF-1 Δ106–140) was generated and purified from E. coli. A comparison of CHIP binding to GST-tagged full-length and mutant IRF-1 showed a marked loss of binding to the mutant protein lacking the Arg-Lys-Ser motif (Fig. 2C). Fig. 2C (right) shows that the mutant and wild-type protein were normalized on the well. The fact that Mf2-derived peptides do not completely block IRF-1·CHIP complex formation (Fig. 2B) and that IRF-1 Δ106–140 retains partial CHIP binding activity supports the idea that the IRF-1·CHIP interface is complex, involving points of contact in the DNA-binding domain as well as the Mf2 region (Fig. 2A). Together, the results suggest a high affinity interaction between CHIP and aa 106–140 of IRF-1, with additional weaker contact sites in the IRF-1 DNA-binding domain.
IRF-1 Binding to CHIP Is Independent of Hsp70
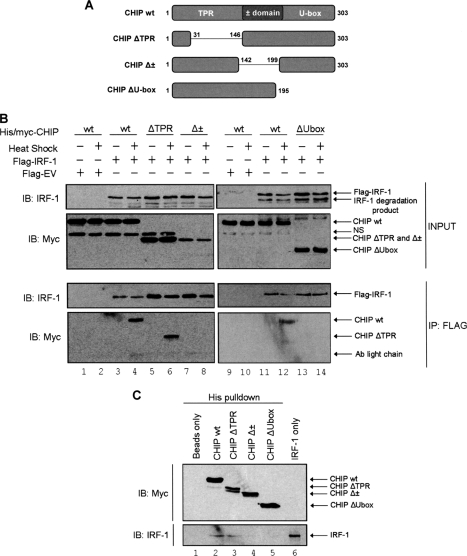
In order to identify the IRF-1 binding region of CHIP, a series of CHIP mutants lacking its functional domains were used (Fig. 3A) (25). When the mutants were transfected into A375 cells together with IRF-1 and subjected to heat stress as described previously (25), both full-length CHIP (CHIP WT) and a CHIP mutant lacking the Hsp70 binding domain (CHIP ΔTPR) co-immunoprecipitated with IRF-1 (Fig. 3B, see lanes 4 and 6). In contrast, when either the charged domain (CHIP Δ±) or the U-Box (ΔUbox) was deleted, binding to IRF-1 was below the level of detection (Fig. 3B, compare lanes 8 and 14 with lanes 4, 6, and 12). However, the interpretation of the cell-based assays was complicated by the fact that, (i) different amounts of IRF-1 were pulled down in the presence of the various CHIP constructs, (ii) A375 cells express endogenous CHIP, and (iii) the ΔUbox CHIP mutant runs at the same size as the α-FLAG antibody light chain on SDS-polyacrylamide gels. CHIP has been shown to form dimers in vitro (12) with the interface extending over the U-box and the charged domain (13), whereas dimerization occurs independently of the TPR domain (12, 13). Thus, the data presented in Fig. 3B may reflect the ability of the various mutants to form complexes with endogenous CHIP and its associated proteins. To address our concerns, a cell-free assay was developed using CHIP constructs purified from a TNT coupled reticulocyte lysate expression system under conditions designed to remove Hsp70, and IRF-1 was generated using the Hsp70-free PURExpress system. All purified protein samples were shown to have Hsp70 levels that were below the levels of detection using immunoblot analysis (data not shown). Consistent with the data obtained using the cell-based assay, when IRF-1 was added to the various CHIP proteins immobilized on Ni2+-NTA agarose, it bound to both the WT and ΔTPR CHIP proteins but not Δ± and ΔUbox deletion mutants (Fig. 3C compare lanes 2 and 3 with lanes 4 and 5). Together the data suggest that (i) IRF-1 binding does not require the TPR domain, (ii) IRF-1 and CHIP can interact independently of Hsp70 function (because Hsp70 cannot bind to the TPR domain mutant), and (iii) IRF-1 binding possibly requires both the charged and U-box domains of CHIP.
FIGURE 3.
CHIP binding to IRF-1 is Hsp70-independent. A, schematic depicting the domain organization of CHIP WT and the mutants used in this study. B, A375 cells were co-transfected with FLAG-IRF-1 (2 μg) and the indicated His/Myc-CHIP constructs (2 μg) and heat-stressed at 43 °C for 30 min as required. FLAG conjugates were immunoprecipitated (IP) using anti-FLAG-M2-agarose and analyzed, along with the input (1.5% of the total lysate), by SDS-PAGE/immunoblot (IB) using anti-IRF-1 mAb and anti-Myc pAb. Results are representative of at least three separate experiments. NS, nonspecific band detected with anti-Myc pAb. C, the indicated His/Myc-CHIP proteins were expressed using a TNT coupled reticulocyte lysate system and purified by metal affinity chromatography. Purified untagged IRF-1 was added to the CHIP proteins immobilized on Ni2+-NTA-agarose, and bound proteins were eluted using 300 mm imidazole. Eluates were analyzed by SDS-PAGE/immunoblot using anti-Myc and anti-IRF-1 antibodies as above.
Cellular CHIP Ubiquitinates IRF-1
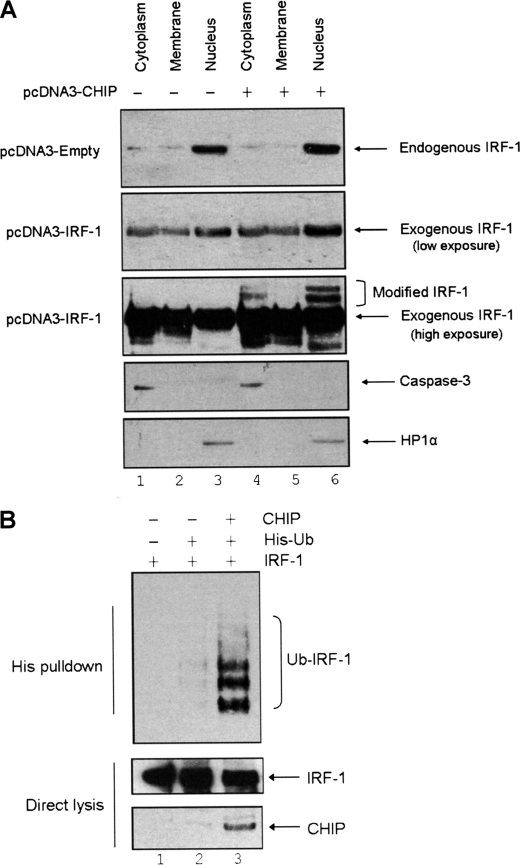
In order to examine the effects of CHIP on IRF-1 in a cellular environment, A375 cells were transfected with pcDNA3-CHIP alone or pcDNA3-CHIP and pcDNA3-IRF-1 and then fractionated. Interestingly, when exogenous CHIP was present, high molecular weight IRF-1 proteins were detected in the nuclear and cytosolic fractions, indicative of post-translational modification (Fig. 4A, panel 3, compare lanes 4 and 6 with lanes 1 and 3). To determine whether these high molecular weight bands represented ubiquitinated forms of IRF-1, a cell-based ubiquitination assay was utilized (8). H1299 cells were transfected with pcDNA3-IRF-1, His-Ub, and pcDNA3-CHIP as indicated in Fig. 4B. In this assay, although endogenous E3 ligase activity is sufficient for IRF-1 modification, overexpression of CHIP results in a significant increase in the amount of ubiquitinated IRF-1 detected (Fig. 4B, His pulldown, compare lanes 2 and 3). These results demonstrate that CHIP-dependent ubiquitination of IRF-1 can occur in a complex cellular background.
FIGURE 4.
CHIP ubiquitinates IRF-1 in cells. A, A375 cells were transiently transfected with pcDNA3-empty vector (for endogenous IRF-1 blots) or pcDNA3-IRF-1 and pcDNA3-CHIP (0.5 and 2 μg, respectively) as indicated. Post-transfection, the cells were fractionated using the ProteoExtract kit. The fractions were analyzed by SDS-PAGE/immunoblot developed using anti-IRF-1 mAb. Endogenous IRF-1 was detected using 75 μg of total protein/lane, whereas 25 μg was used to detect exogenous IRF-1. Caspase-3 and HP1α (heterochromatin protein 1α) were used as markers for the cytoplasmic and nuclear fractions, respectively. B, H1299 cells were transfected with pcDNA3-IRF-1 (0.5 μg), His-ubiquitin (0.5 μg), and pcDNA3-CHIP (2 μg) as detailed. Post-transfection (20 h), cells were treated with MG132 (50 μm) for 4 h, and His-ubiquitinated protein was isolated by metal affinity chromatography and analyzed by SDS-PAGE/immunoblot. Immunoblots show total IRF-1 and CHIP (lower panels) and His-ubiquitinated IRF-1 (upper panel). The data are representative of at least four separate experiments.
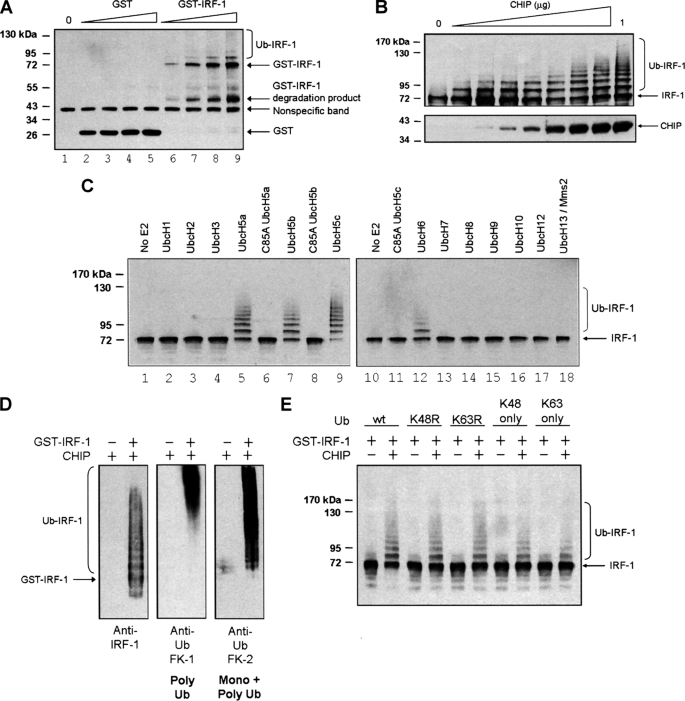
CHIP Cooperates with E2 Enzymes of the UbcH5 Family and UbcH6 to Ubiquitinate IRF-1
In order to determine if direct binding of CHIP to IRF-1 was sufficient to signal ubiquitination or whether additional cellular components, such as Hsp70, were required, a stopped enzyme assay using purified components was developed in which CHIP was rate-limiting (Fig. 5, A and B). Under these conditions, CHIP specifically ubiquitinates GST-IRF-1 but not GST alone (Fig. 5A, compare lanes 6–9 with lanes 2–5). To further dissect the IRF-1 ubiquitination pathway, we used a library of purified E2 ubiquitin-conjugating enzymes to determine which E2s could catalyze CHIP ubiquitination of IRF-1. Our data suggest that E2 enzymes belonging to the UbcH5 family and UbcH6 can cooperate with CHIP to efficiently ubiquitinate IRF-1 in vitro (Fig. 5C). As expected, mutation of the active site Cys (C85A) of the UbcH5 family members completely blocked IRF-1 ubiquitination by CHIP (Fig. 5C, compare lanes 6, 8, and 11 with lanes 5, 7, and 9). Additionally, when GST-IRF-1 (total protein, including ubiquitinated and unmodified IRF-1) was isolated from the ubiquitination reaction mix in which UbcH5a was used as the E2, immunoblot analysis showed that CHIP·UbcH5a can both monoubiquitinate and form polyubiquitin chains on IRF-1 (Fig. 5D). Moreover, CHIP·UbcH5a forms both Lys48-linked and Lys63-linked ubiquitin chains on IRF-1 because Ub mutants with either Lys48 or Lys63 mutated to Arg or with all lysine residues except Lys48 or Lys63 mutated to Arg cause no gross change in the pattern of IRF-1 modification by CHIP (Fig. 5E).
FIGURE 5.
CHIP-dependent ubiquitination of IRF-1 does not require Hsp70 and uses E2 enzymes of the UbcH5 and H6 families. A, an in vitro ubiquitination assay was assembled with purified E1, E2, His-CHIP, ubiquitin, and a titration (0, 25, 50, 75, and 100 ng) of GST-IRF-1 (or GST alone) in the presence of ATP. Ubiquitinated protein was analyzed by SDS-PAGE/immunoblot using anti-GST mAb. B, ubiquitination assays were assembled as above using a fixed amount of GST-IRF-1 (40 ng) and a titration of His-CHIP (0, 12.5, 25, 50, 100, 250, 500, 750, and 1000 ng). Immunoblots show total CHIP (bottom) and ubiquitinated IRF-1 (top) detected using anti-CHIP mAb and anti-IRF-1 mAb. C, ubiquitination assays were assembled as in A, using a constant amount of GST-IRF-1 and various E2 enzymes as indicated. Ubiquitinated protein was analyzed by SDS-PAGE/immunoblot, using anti-IRF-1 mAb. D, a ubiquitination assay was assembled as above with UbcH5a as E2, and total GST-IRF-1 was purified from the reaction mix using glutathione-Sepharose. Purified GST-IRF-1 was analyzed by SDS-PAGE/immunoblot using anti-IRF-1 mAb and anti-ubiquitin FK1 and FK2 antibodies. E, ubiquitination assays were assembled as above using WT ubiquitin and various ubiquitin mutants. Immunoblots were probed with anti-IRF-1 mAb.
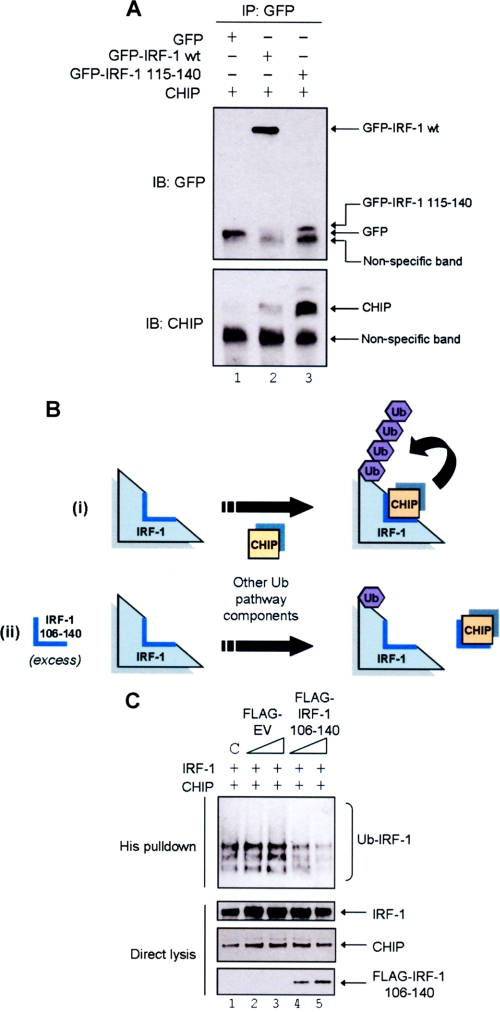
CHIP Binding to IRF-1 aa 106–140 Is Essential for IRF-1 Ubiquitination
The data presented above show that CHIP binds stably to IRF-1 aa 106–140 (Fig. 2A) and that CHIP can efficiently ubiquitinate IRF-1 both in cells and in vitro (Figs. 4B and 5B). However, the experiments do not demonstrate whether the direct binding of CHIP to the Mf2 domain is sufficient to signal ubiquitination of IRF-1. To address this question, we performed a series of cell-based experiments using a GFP fusion protein containing IRF-1(115–140) that has been previously described (22). When the Arg-Lys-Ser motif on IRF-1 was fused with GFP (GFP-IRF-1(115–140)) and transiently transfected into A375 cells, the fusion protein was sufficient to capture exogenous CHIP (Fig. 6A). In fact, IRF-1(115–140) had a much higher affinity for CHIP when expressed directly as a GFP fusion protein than in the context of the full-length protein (GFP-IRF-1 WT; Fig. 6A, compare lane 3 with lane 2). Because GFP-IRF-1(115–140) was sufficient for CHIP binding in a cellular environment, we hypothesized that if this interaction was important in the mechanism of CHIP-mediated IRF-1 ubiquitination, the fragment should compete with full-length IRF-1 for CHIP binding, leading to a decrease in the level of IRF-1 modification by ubiquitin (Fig. 6B (ii)). To overcome nonspecific effects of GFP alone on IRF-1 ubiquitination (data not shown) residues 106–140 of IRF-1, comprising the CHIP binding site, were expressed as a FLAG fusion (FLAG-IRF-1(106–140)). When a titration of FLAG-IRF-1(106–140) was transfected into H1299 cells together with fixed amounts of IRF-1, CHIP, and His-Ub, a dose-dependent decrease in the ubiquitination of full-length IRF-1 was observed in accordance with our hypothesis (Fig. 6C, compare lanes 4 and 5 with lanes 2 and 3). Additional support comes from in vitro data (supplemental Fig. S1) showing that Δ106–140 IRF-1 is less efficiently ubiquitinated by CHIP than the wild-type protein. It should be noted that we cannot rule out the possibility that the loss of lysine residues in the Mf2 domain may contribute to the inefficient ubiquitination of the Δ106–140 IRF-1 mutant; however, analysis to date suggests that IRF-1 can be ubiquitinated out with the Mf2 domain (data not shown). These data suggest that although CHIP can ubiquitinate IRF-1 in the absence of its docking site, the reaction is inefficient when compared with ubiquitination of wild-type IRF-1. Together, the above experiments demonstrate that the Mf2 domain of IRF-1 is required for efficient CHIP-dependent ubiquitination of the protein in cells and in vitro.
FIGURE 6.
IRF-1 aa 106–140 can act in trans to inhibit CHIP-dependent ubiquitination of full-length IRF-1. A, A375 cells were co-transfected with GFP alone, GFP-IRF-1 WT or GFP-IRF-1(115–140), and pcDNA3-CHIP. GFP conjugates were immunoprecipitated using anti-GFP pAb (IP) and analyzed by SDS-PAGE/immunoblot (IB) using anti-GFP mAb and anti-CHIP mAb. Results are representative of two separate experiments. B, model showing the requirement of aa 106–140 for IRF-1 ubiquitination. i, CHIP directly binds to IRF-1 aa 106–140 and ubiquitinates IRF-1. ii, the addition of IRF-1 106–140 in trans would compete with full-length IRF-1 for CHIP binding, resulting in a decreased ubiquitination of the full-length protein. C, H1299 cells were transfected with pcDNA3-IRF-1 WT (0.5 μg), His-ubiquitin (0.5 μg), pcDNA3-CHIP (2 μg), and a titration of FLAG-empty vector (EV) or FLAG-IRF-1(106–140) (1 or 2 μg); DNA was normalized using pcDNA3 empty vector, and C represents a control sample that was transfected with pcDNA3-EV but no FLAG constructs. Post-transfection (20 h) cells were treated with MG132 (50 μm) for 4 h. His-ubiquitinated protein was isolated and analyzed by SDS-PAGE/immunoblot developed using anti-IRF-1, anti-CHIP, and anti-FLAG mAbs.
CHIP-dependent Effects on IRF-1 Steady State Levels
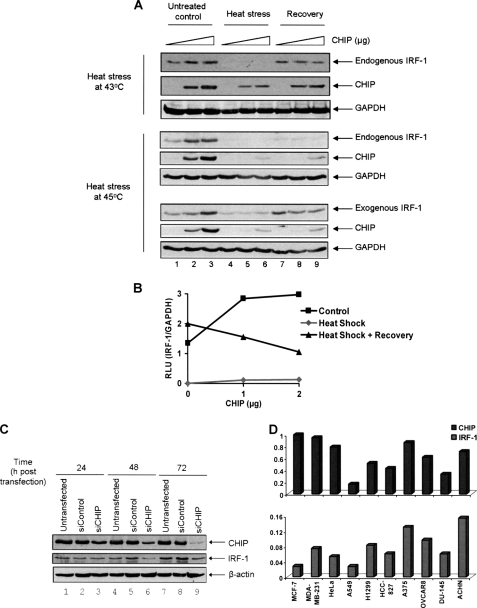
Having established that IRF-1 is a substrate for CHIP and that CHIP-mediated ubiquitination of IRF-1 proceeds through a stable interaction between CHIP and the Mf2 domain, we sought to determine the cellular conditions under which CHIP-dependent regulation of IRF-1 may be relevant. We began by examining the effect of expressing exogenous CHIP on IRF-1 steady state levels in unstressed cells and in cells exposed to heat stress. Of interest was the fact that steady state levels of both endogenous (Fig. 7, A (lanes 1–3, top and middle) and B) and exogenous (Fig. 7A, lanes 1–3, bottom) IRF-1 increased in a titrative manner when CHIP was transfected into A375 cells. Although we could not determine the effect of CHIP on IRF-1 during heat stress because the protein was below detectable levels (Fig. 7A, lanes 4–6), we were able to demonstrate that expression of CHIP during recovery from a heat stress of either 43 °C (Fig. 7, A (top) and B) or 45 °C (Fig. 7A, bottom) led to a decrease in both endogenous and exogenous IRF-1 steady state levels (Fig. 7A, lanes 7–9). The data are consistent with the hypothesis that CHIP is an E3 ligase for IRF-1 under certain physiological conditions (i.e. in cells that have undergone heat stress). In support of this, we found that depletion of CHIP using siRNA, like treatment with MG132 (Fig. 8C), resulted in a partial rescue from the dramatic IRF-1 loss detected during the response to a 43 °C heat stress (supplemental Fig. S2, compare lane 4 with lane 3). This demonstrates a role for CHIP in the degradation of IRF-1 in heat-treated cells but suggests that other IRF-1 degradation pathways may also operate under these conditions.
FIGURE 7.
Dual role of CHIP as a chaperone and Ub-E3 ligase for IRF-1. A, A375 cells were transiently transfected with a titration of His/Myc-CHIP (top; 0, 1, and 2 μg) or pcDNA3-CHIP (middle; 0, 2.5, and 5 μg; DNA levels were normalized using pcDNA3 empty vector in all cases). Bottom, the cells were co-transfected with pcDNA3-CHIP as above and pcDNA3-IRF-1 (0.5 μg). 24 h post-transfection, the cells were heat-shocked at 43 °C for 30 min (top) or 45 °C for 60 min (middle and bottom) and harvested immediately (Heat stress) or allowed to recover at 37 °C for 1 h (Recovery) prior to harvesting. Control cells were maintained at 37 °C until harvested (Untreated control). Cell lysate was analyzed by SDS-PAGE/immunoblot using anti-IRF-1 and anti-Myc mAbs (top) or anti-IRF-1 and anti-CHIP antibodies (middle and bottom). GAPDH was used as a loading control. To detect endogenous IRF-1, 75 μg of total protein was loaded per lane (top and middle), whereas 25 μg was loaded to detect exogenous IRF-1 (bottom). B, IRF-1 and GAPDH signals from the experiment in A (top) were quantified using the Scion Imaging software, and a graph of the intensity of the IRF-1/GAPDH signal expressed in relative light units (RLU) under the various conditions is shown. C, H1299 cells were left untransfected or were transiently transfected with 50 nmol of nonspecific control siRNA (siControl) or CHIP siRNA (siCHIP) as indicated. Cells were harvested at the indicated time points, and the lysate was analyzed by SDS-PAGE/immunoblot as in B. β-Actin was used as a loading control. D, the indicated cells were seeded in a 96-well plate, and In-Cell WesternTM assays were performed with anti-IRF-1 and anti-CHIP mAbs to measure the relative CHIP (top) and IRF-1 (bottom) levels in situ using a Licor Odyssey SA scanner. Antibody signals across samples were normalized for cell number, which was reflected in the DRAQ5 (DNA stain) signal (see supplemental Fig. S3). Normalized signals, expressed as relative fluorescence units, are shown.
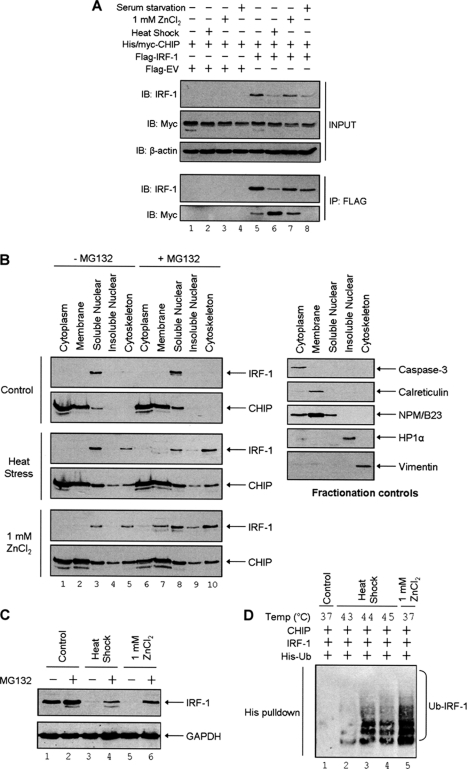
FIGURE 8.
CHIP-dependent IRF-1 ubiquitination is enhanced by heat and heavy metal stress. A, A375 cells were transfected with FLAG-empty vector (EV) or FLAG-IRF-1 (2 μg) plus His/Myc-CHIP (2 μg) and subjected to the indicated stresses (heat shock 43 °C for 30 min, 1 mm ZnCl2 for 90 min, or serum withdrawal for 24 h). FLAG conjugates were immunoprecipitated (IP) using anti-FLAG-M2-agarose and analyzed by SDS-PAGE/immunoblot (IB) using anti-IRF-1 and anti-Myc mAbs. Also shown is the input (2% of total lysate) used for the IP. β-Actin was used as a loading control. Results are representative of at least three independent experiments. B, A375 cells were treated with MG132 (50 μm) or DMSO carrier for 4 h and then stressed as in A. Cells were fractionated using the subcellular protein fractionation kit. The fractions were analyzed by SDS-PAGE/immunoblot developed using IRF-1 mAb and anti-CHIP mAb. Caspase-3, calreticulin, NPM/B23, HP1α, and vimentin were used as markers for the various fractions. C, A375 cells were treated with MG132 (50 μm) or DMSO carrier and heat-stressed or treated with ZnCl2 as in A. Cell lysates were analyzed by SDS-PAGE/immunoblot developed using IRF-1 mAb and GAPDH pAb as a loading control. D, H1299 cells were transfected with pcDNA3-IRF-1 WT (0.5 μg), His-ubiquitin (0.5 μg), and pcDNA3-CHIP (2 μg) as indicated. Post-transfection (20 h), cells were treated with MG132 (50 μm) for 4 h and heat-stressed at the indicated temperatures for 30 min or treated with ZnCl2 as above. His-ubiquitinated protein was isolated using Ni2+-NTA-agarose, and immunoblots show His-ubiquitinated IRF-1 detected using anti-IRF-1 mAb.
Although CHIP has a negative effect on IRF-1 protein levels in cells undergoing or recovering from heat stress, we were intrigued by the observation that CHIP expression increased, rather than decreased, IRF-1 levels in unstressed cells. To investigate this further, we looked at the effect of depleting CHIP from cells using siRNA. Fig. 7C shows the decrease in the level of CHIP protein after 48 and 72 h with CHIP siRNA (lanes 6 and 9) compared with the siRNA control lanes (lanes 5 and 8). When IRF-1 steady state levels were determined, they correlated with those of CHIP (i.e. IRF-1 levels were lower in the siRNA CHIP lanes (lanes 6 and 9) than in untransfected cells (lanes 4 and 7) or siRNA control-treated cells (lanes 5 and 8)). Thus, depletion of CHIP in unstressed cells led to a concomitant decrease in IRF-1 steady state levels. Next, we used In-Cell WesternTM assays to obtain an accurate determination of the relative levels of IRF-1 and CHIP across a range of cell lines (Fig. 7D; secondary antibody and total DNA controls are given in supplemental Fig. S3). This showed that there is a positive correlation between the protein levels of CHIP and IRF-1 across 7 of the 10 cell lines tested. In fact, only in MCF7 cells was there a strong negative correlation. We conclude from these experiments that CHIP-dependent down-regulation of IRF-1 operates predominantly under conditions of cellular stress, whereas CHIP may play a positive role in the regulation of IRF-1 levels in a range of unstressed cells.
Heat Stress and Heavy Metal Stress Induce CHIP Binding and Ubiquitination of IRF-1
Because CHIP has been shown to associate with substrates like luciferase and Daxx under conditions of heat stress (25, 26), we asked whether the CHIP-dependent down-regulation of IRF-1 levels in heat stress conditions (Fig. 7, A and B, and supplemental Fig. S2) was accompanied by changes in CHIP·IRF-1 complex formation. We also looked at the formation of CHIP·IRF-1 complexes in heavy metal and serum-starved cells, both of which, like heat stress, lead to down-regulation of IRF-1 protein levels (Fig. 8A and supplemental Fig. S4). Interestingly, in the presence of both heat and heavy metal stress, although IRF-1 protein levels were significantly reduced, its association with CHIP was enhanced (Fig. 8A, compare lanes 6 and 7 with lane 5). On the other hand, although a reduction in the steady state levels of IRF-1 was also observed in cells subject to serum withdrawal, an association of IRF-1 and CHIP was not promoted, suggesting that complex formation is signal-specific (Fig. 8A, compare lane 8 with lane 5). To gain further insight into the stress-dependent association of IRF-1 with CHIP, subcellular fractionation experiments were performed in the presence and absence of the proteasome inhibitor MG132. In the absence of any cellular stress, IRF-1 was predominantly nuclear, whereas CHIP was largely cytosolic and membrane-associated, although measurable levels were consistently detected in the nucleus (Fig. 8B, Control). Interestingly, under conditions of heat and heavy metal stress, IRF-1 accumulated in the cytoskeletal fraction, although it was also detected in the membrane and nuclear compartments (Fig. 8B, compare lanes 7–10 in control, heat stress, and ZnCl2). Under these stress conditions, CHIP also started to accumulate in the cytoskeleton (Fig. 8B). Thus, IRF-1 and CHIP have a similar distribution in heat-stressed cells. In order to determine whether the reduced steady state levels of IRF-1 under these stress conditions was a consequence of the observed increase in CHIP binding (Fig. 8A), we first examined whether the levels of IRF-1 were affected by proteasome inhibition in whole cell lysate (Fig. 8C). In the presence of the inhibitor MG-132, the decrease in IRF-1 protein seen under heat and metal stress was significantly less (Fig. 8C, compare lanes 4 and 6 with lanes 3 and 5). This suggests that IRF-1 protein levels decrease in a proteasome-dependent manner under conditions of heat stress or in response to heavy metal ions. In support of this conclusion, we have seen that depletion of CHIP using siRNA can partially rescue IRF-1 down-regulation in heat-stressed cells (supplemental Fig. S2), whereas in the presence of exogenous CHIP, IRF-1 ubiquitination is markedly enhanced during both heat and metal stress (Fig. 8D, compare lanes 2–5 with lane 1). Thus, the interaction of CHIP with IRF-1 is favored under conditions such as heat stress and heavy metal stress, which results in increased IRF-1 ubiquitination.
DISCUSSION
Steady state expression of the IRF-1 tumor suppressor protein is maintained at low levels allowing a rapid response to environmental conditions through changes in its rate of synthesis and/or rate of degradation (7, 27, 28). IRF-1 is turned over quickly in unstressed cells, with a half-life of between 20 and 40 min, dependent on the cell line (8–10), and changes in the rate of IRF-1 degradation are required for maximal activation under some stress conditions (7). Given this, surprisingly little is known about the nature of the proteins and enzymes responsible for maintaining homeostatic levels of IRF-1 and how these are modulated under conditions of intracellular or environmental stress. In the current study, we show that the steady state levels of IRF-1 decrease under conditions of heat stress or during the response to heavy metal ions and that these changes are associated with the formation of complexes between IRF-1 and the E3 ubiquitin ligase CHIP.
CHIP appears to form a link between the processes that lead to protein folding and those that mediate the degradation of incorrectly or partially folded proteins. In the canonical pathway, CHIP is targeted to Hsp90-bound client proteins by Hsp70, and the clients are then ubiquitinated for delivery to and degradation by the proteasome (17, 18). However, in recent years, the picture has become more complex, with evidence beginning to emerge that (i) there is a role for direct binding of CHIP to some of its substrates (19, 20), (ii) CHIP can mediate ubiquitination of target substrates by Ubc13, an E2-enzyme that generates Lys63-linked ubiquitin chains (29), and (iii) CHIP can function as a molecular chaperone to promote or maintain folded protein conformation(s) (30, 31). The latter activity is of interest in the current study because we found that the levels of IRF-1 were increased by the expression of CHIP in unstressed cells (Fig. 7A) and that conversely, IRF-1 levels decreased when CHIP was depleted using siRNA (Fig. 7C). These data suggest that CHIP may act as a chaperone for IRF-1 in unstressed cells. This idea is supported by the recent observation that CHIP can bind directly to wild-type p53, folding it into a “native” conformation and promoting p53 DNA binding activity (31). Alternatively, because we have recently shown that manipulation of Hsp70/Hsp90 affects IRF-1 levels in a similar manner to CHIP (21), CHIP may form part of a multiprotein IRF-1 chaperone complex in unstressed cells. Interestingly, CHIP is not unique in acting as both a chaperone and an E3 ligase. The MDM2 E3 ligase is also known to possess an intrinsic chaperone activity that can be utilized to protect p53 from thermal denaturation in vitro and to promote folding of p53 to its native conformation in cells (32).
A number of recent studies have identified substrates that bind directly to CHIP in the absence of Hsp70, suggesting that under some conditions CHIP might bypass the requirement for a chaperone partner. For example, the death domain-associated protein Daxx binds to the charged domain of CHIP in heat-stressed cells, and this is accompanied by Daxx ubiquitination (25). This interaction appears to occur independently of Hsp70 because the TPR domain of CHIP is not needed for Daxx binding (25). Similarly, Runx1 binds to CHIP independently of Hsp70/Hsp90 (19). Unlike the CHIP-Daxx interaction, however, CHIP is reported to interact with Runx1 under normal cellular conditions (19). Although studies on Daxx and Runx1 suggest that CHIP can ubiquitinate substrates in an Hsp70-independent manner, they do not demonstrate a direct relationship between the ability of CHIP to bind to a protein and the protein's ability to act as a substrate for CHIP-dependent ubiquitination. In the current study, we have shown that IRF-1 binds to CHIP through a complex interface involving a high affinity interaction with the Mf2 domain of IRF-1 and additional lower affinity sites in the IRF-1 DNA binding domain (Fig. 2A). More specifically, an Arg-Lys-Ser-rich region in the Mf2 domain is required for maximal binding of IRF-1 to CHIP and is sufficient to form a stable complex with CHIP both in vitro and in cells (Figs. 2, A and C, and 6A). In common with Daxx and Runx1, IRF-1 binding involves the central charged domain of CHIP (Fig. 3, B and C); however, the U-box domain of CHIP may also be required to form a stable complex with IRF-1. This suggests that (i) IRF-1 may bind directly to both the charged and U-box domains by, for example, contacting the conformationally flexible hinge region of CHIP (33), (ii) binding may require CHIP to be present as a dimer (13, 29), or (iii) the U-box may not be involved in direct contact with IRF-1 but may be required to maintain the charged domain in a favorable conformation for substrate binding.
We exploited the interaction between CHIP and amino acids 106–140 of IRF-1 to demonstrate a direct link between the ability of CHIP to bind to IRF-1 and the ability to utilize it as a substrate. Thus, we were able to demonstrate that in addition to being essential for maximal IRF-1·CHIP binding in vitro (Fig. 2C), amino acids 106–140 could bind stably to CHIP in a cellular environment and in doing so could inhibit CHIP-dependent ubiquitination of full-length IRF-1 in trans (Fig. 6, A and C).
Although the data presented in the current study show that direct binding of CHIP to IRF-1 is sufficient to signal its ubiquitination, it is likely that cellular CHIP can also interact with IRF-1 through Hsp70. We have previously shown that Hsp70 binds to the C-terminal Mf1 domain of IRF-1 and mediates the effects of Hsp90 inhibition using 17AAG on IRF-1 steady state levels (21). Thus, we can envisage situations where CHIP could interact with IRF-1 both directly (i.e. in an Hsp70-independent manner), and through Hsp70 binding to the C terminus of IRF-1, dependent on the prevailing cellular conditions or the required outcome. Alternately, Hsp70 could act as a loading factor, delivering CHIP to IRF-1 and then dissociating from the complex because it is not required for modification of IRF-1 with ubiquitin. In this way, Hsp70 would be a regulator of the CHIP-IRF-1 interaction and therefore of IRF-1 ubiquitination. In this respect, it is interesting to look at the relationship between p53 and CHIP. Several studies have shown that CHIP together with Hsp70 and Hsp90 can ubiquitinate both wild-type and mutant forms of the protein, leading to degradation (34, 35). However, binding of CHIP to the C terminus of p53 can maintain it in an active DNA-binding conformation (31). Thus, CHIP appears to play a more complex role in protein folding and degradation pathways than previously assumed. In addition, the observation that CHIP can mediate the transfer of Lys63-linked ubiquitin chains suggests that we are likely to uncover further complexities because Lys63-linked chains do not usually target proteins for degradation but rather are involved primarily in cell signaling (36–38). It is therefore likely that CHIP can function in a variety of modes, allowing it to impact on the structure, function, and activity of its substrates to provide diverse outcomes.
It will therefore be of interest to determine the precise relationship between CHIP, Hsp70, and IRF-1 as well as the mechanism by which CHIP regulates IRF-1 steady state levels in non-stress conditions.
Supplementary Material
Acknowledgments
We thank Ted Hupp and Borivoj Vojtesek for advice, Cam Patterson for generously providing CHIP plasmids for mammalian cell expression, Alicja Zylicz and Renata Filipek from the International Institute of Molecular and Cell Biology in Warsaw for the kind gift of a CHIP plasmid for expression in E. coli, and H. Hauser for the GFP-IRF-1(115–140) construct.
This work is funded by Cancer Research UK Programme Grant C377/A6355 (to K. L. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- Ub
- ubiquitin
- TPR
- tetratricopeptide repeat
- pAb
- polyclonal antibody
- aa
- amino acids.
REFERENCES
- 1. Fujita T., Sakakibara J., Sudo Y., Miyamoto M., Kimura Y., Taniguchi T. (1988) EMBO J. 7, 3397–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanaka N., Ishihara M., Lamphier M. S., Nozawa H., Matsuyama T., Mak T. W., Aizawa S., Tokino T., Oren M., Taniguchi T. (1996) Nature 382, 816–818 [DOI] [PubMed] [Google Scholar]
- 3. Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001) Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 4. Nozawa H., Oda E., Ueda S., Tamura G., Maesawa C., Muto T., Taniguchi T., Tanaka N. (1998) Int. J. Cancer 77, 522–527 [DOI] [PubMed] [Google Scholar]
- 5. Tamura G., Sakata K., Nishizuka S., Maesawa C., Suzuki Y., Terashima M., Eda Y., Satodate R. (1996) J. Pathol. 180, 371–377 [DOI] [PubMed] [Google Scholar]
- 6. Willman C. L., Sever C. E., Pallavicini M. G., Harada H., Tanaka N., Slovak M. L., Yamamoto H., Harada K., Meeker T. C., List A. F. (1993) Science 259, 968–971 [DOI] [PubMed] [Google Scholar]
- 7. Pamment J., Ramsay E., Kelleher M., Dornan D., Ball K. L. (2002) Oncogene 21, 7776–7785 [DOI] [PubMed] [Google Scholar]
- 8. Pion E., Narayan V., Eckert M., Ball K. L. (2009) Cell. Signal. 21, 1479–1487 [DOI] [PubMed] [Google Scholar]
- 9. Watanabe N., Sakakibara J., Hovanessian A. G., Taniguchi T., Fujita T. (1991) Nucleic Acids Res. 19, 4421–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakagawa K., Yokosawa H. (2000) Eur. J. Biochem. 267, 1680–1686 [DOI] [PubMed] [Google Scholar]
- 11. Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 12. Nikolay R., Wiederkehr T., Rist W., Kramer G., Mayer M. P., Bukau B. (2004) J. Biol. Chem. 279, 2673–2678 [DOI] [PubMed] [Google Scholar]
- 13. Xu Z., Devlin K. I., Ford M. G., Nix J. C., Qin J., Misra S. (2006) Biochemistry 45, 4749–4759 [DOI] [PubMed] [Google Scholar]
- 14. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J., Patterson C. (2001) J. Biol. Chem. 276, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 16. Aravind L., Koonin E. V. (2000) Curr. Biol. 10, R132–134 [DOI] [PubMed] [Google Scholar]
- 17. Murata S., Minami Y., Minami M., Chiba T., Tanaka K. (2001) EMBO Rep. 2, 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 19. Shang Y., Zhao X., Xu X., Xin H., Li X., Zhai Y., He D., Jia B., Chen W., Chang Z. (2009) Biochem. Biophys. Res. Commun. 386, 242–246 [DOI] [PubMed] [Google Scholar]
- 20. Parsons J. L., Tait P. S., Finch D., Dianova I. I., Allinson S. L., Dianov G. L. (2008) Mol. Cell 29, 477–487 [DOI] [PubMed] [Google Scholar]
- 21. Narayan V., Eckert M., Zylicz A., Zylicz M., Ball K. L. (2009) J. Biol. Chem. 284, 25889–25899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schaper F., Kirchhoff S., Posern G., Köster M., Oumard A., Sharf R., Levi B. Z., Hauser H. (1998) Biochem. J. 335, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
- 24. Tsai Y. C., Fishman P. S., Thakor N. V., Oyler G. A. (2003) J. Biol. Chem. 278, 22044–22055 [DOI] [PubMed] [Google Scholar]
- 25. McDonough H., Charles P. C., Hilliard E. G., Qian S. B., Min J. N., Portbury A., Cyr D. M., Patterson C. (2009) J. Biol. Chem. 284, 20649–20659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marques C., Guo W., Pereira P., Taylor A., Patterson C., Evans P. C., Shang F. (2006) FASEB J. 20, 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Flodström M., Eizirik D. L. (1997) Endocrinology 138, 2747–2753 [DOI] [PubMed] [Google Scholar]
- 28. Fujita T., Reis L. F., Watanabe N., Kimura Y., Taniguchi T., Vilcek J. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9936–9940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 30. Rosser M. F., Washburn E., Muchowski P. J., Patterson C., Cyr D. M. (2007) J. Biol. Chem. 282, 22267–22277 [DOI] [PubMed] [Google Scholar]
- 31. Tripathi V., Ali A., Bhat R., Pati U. (2007) J. Biol. Chem. 282, 28441–28454 [DOI] [PubMed] [Google Scholar]
- 32. Wawrzynow B., Pettersson S., Zylicz A., Bramham J., Worrall E., Hupp T. R., Ball K. L. (2009) J. Biol. Chem. 284, 11517–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graf C., Stankiewicz M., Nikolay R., Mayer M. P. (2010) Biochemistry 49, 2121–2129 [DOI] [PubMed] [Google Scholar]
- 34. Esser C., Scheffner M., Höhfeld J. (2005) J. Biol. Chem. 280, 27443–27448 [DOI] [PubMed] [Google Scholar]
- 35. Muller P., Hrstka R., Coomber D., Lane D. P., Vojtesek B. (2008) Oncogene 27, 3371–3383 [DOI] [PubMed] [Google Scholar]
- 36. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 37. Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 38. Spence J., Sadis S., Haas A. L., Finley D. (1995) Mol. Cell Biol. 15, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.