Abstract
We previously presented evidence that transmembrane domain (TM) IV and TM X-XI are important for inhibitor binding and ion transport by the human Na+/H+ exchanger, hNHE1 (Pedersen, S. F., King, S. A., Nygaard, E. B., Rigor, R. R., and Cala, P. M. (2007) J. Biol. Chem. 282, 19716–19727). Here, we present a structural model of the transmembrane part of hNHE1 that further supports this conclusion. The hNHE1 model was based on the crystal structure of the Escherichia coli Na+/H+ antiporter, NhaA, and previous cysteine scanning accessibility studies of hNHE1 and was validated by EPR spectroscopy of spin labels in TM IV and TM XI, as well as by functional analysis of hNHE1 mutants. Removal of all endogenous cysteines in hNHE1, introduction of the mutations A173C (TM IV) and/or I461C (TM XI), and expression of the constructs in mammalian cells resulted in functional hNHE1 proteins. The distance between these spin labels was ∼15 A, confirming that TM IV and TM XI are in close proximity. This distance was decreased both at pH 5.1 and in the presence of the NHE1 inhibitor cariporide. A similar TM IV·TM XI distance and a similar change upon a pH shift were found for the cariporide-insensitive Pleuronectes americanus (pa) NHE1; however, in paNHE1, cariporide had no effect on TM IV·TM XI distance. The central role of the TM IV·TM XI arrangement was confirmed by the partial loss of function upon mutation of Arg425, which the model predicts stabilizes this arrangement. The data are consistent with a role for TM IV and TM XI rearrangements coincident with ion translocation and inhibitor binding by hNHE1.
Keywords: Biophysics, Cell pH, Crystal Structure, Electron Paramagnetic Resonance (EPR), Trafficking, Exchanger, Ion Transport, Structure, pH Regulation
Introduction
The ubiquitous plasma membrane Na+/H+ exchanger isoform 1 (NHE1)4 plays central roles in cellular pH and volume homeostasis, cell migration, proliferation, and survival, and increased NHE1 activity contributes to ischemia-reperfusion injury as well as tumor growth and proliferation (1, 2). Hence, the ability to selectively block NHE1, although of high clinical relevance, is hampered by a lack of detailed understanding of NHE1 structure and mechanism of ion translocation.
Hydropathy analyses and accessibility studies indicate that NHE1 has 12 transmembrane (TM) segments and a large C-terminal cytoplasmic region (3). Cysteine accessibility studies suggest the presence of two small re-entrant loops between TM IV and TM V (intracellular loop (IL) II) and TM VIII and TM IX (IL IV), respectively, and a larger re-entrant loop between TM IX and TM X (extracellular loop V) (1, 3). Portions of IL II and IL IV are located within the membrane and accessible from either side of the membrane, suggesting that they may form structures lining an aqueous pore and could be involved in ion translocation by NHE1 (3). Extracellular loop V is also interesting in this regard, because it resembles the P-loops found in voltage-gated ion channels (1, 3). These putative re-entrant loops are highly conserved among several NHE1 homologs, consistent with the notion that they are critical for NHE1 function (3–5).
A number of regions within the NHE1 protein have been implicated in inhibitor binding, e.g. TM IV and TM IX (6–11); however, the mechanism(s) of interaction between NHE1 and its commonly used inhibitors, amiloride and benzoyl guanidine type compounds, remain to be fully elucidated.
Using a comparative approach based on chimeras generated using human NHE1 (hNHE1) and two NHE1 homologs (flounder paNHE1 and Amphiuma tridactylum NHE1) with high sequence homology to hNHE1 yet markedly different inhibitor profiles (4, 5), we previously obtained novel information on the regions of NHE1 important for inhibitor binding and ion transport (12). These studies confirmed that TM IV plays a central role in inhibitor binding (12) as suggested by earlier point mutation studies (6–11). Moreover, we demonstrated that regions in TM X-XI and/or IL V and extracellular loop VI are important determinants of inhibitor sensitivity (12).
The three-dimensional structure of NHE1 is unknown; however, the structure of the distantly related bacterial (Escherichia coli) Na+/H+ antiporter NhaA was recently solved at 3.45 Å resolution (13). Similar to NHE1, NhaA has 12 membrane-spanning domains and intracellular N and C termini (13). As for NHE1, NhaA is important for cellular pH regulation and electrolyte homeostasis (1). Despite the low sequence homology (<15% similarity as calculated by ClustalW analysis) and different Na+:H+ transport stoichiometry (NHE1 = 1:1, NhaA = 1:2), comparison of NHE1 and NhaA is relevant given their similar topology and the fact that structure tends to be far better conserved than sequence. A hNHE1 lacking N-glycosylation sites and expressed in Saccharomyces cerevisiae was recently used to create a 22-Å resolution structure (14). However, because glycosylation is important for NHE1 trafficking (15), it is uncertain whether this structure is representative of the mature NHE1.
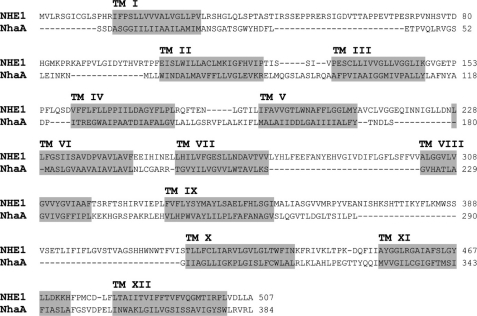
The low sequence homology between NhaA and NHE1 makes homology modeling highly challenging. A structural model of hNHE1 based on threading on NhaA has recently been published (16). This model was constructed from multiple sequence alignments, fold recognition, and evolutionary conservation analysis. However, the assignment of TM regions in this model is inconsistent with experimental evidence from earlier cysteine scanning accessibility studies of hNHE1 (3), and the model was not validated by experimental measurements of interhelix distances in hNHE1.
We have therefore created a three-dimensional structural model of the N-terminal region of hNHE1 based on threading (17) on the NhaA structure, in which we constrained our alignment of TM domains to regions of NHE1 that were experimentally determined to be in a membrane-like environment. In the NhaA structure, and thus in our model, TM IV and TM XI are in close proximity, in agreement with our experimental evidence for hNHE1 (12). The hypothesis that these helices are involved in ion translocation and inhibitor binding by NHE1 was tested (i) through functional analysis of NHE1 mutants and (ii) by experimentally determining the relative positions of TM IV and TM XI and their conformational changes during activation and inhibition. Accordingly, cysteine residues were introduced at the desired positions, followed by the addition of site-directed spin labels. The labeled protein was then used for EPR spectroscopy (18). The EPR spectra provide information on side chain dynamics (19), and thus on protein topography and conformational changes, as well as on secondary and tertiary structure (20, 21). Introduction of a second paramagnetic center allows distance measurements within the protein (18, 21).
We present here a three-dimensional model of hNHE1 threaded on the NhaA structure, in which TM IV and TM XI are in close proximity. EPR analyses of hNHE1 and the Pleuronectes americanus homolog, paNHE1, combined with point mutations and NHE1 function analyses confirmed the close proximity of TM IV and TM XI and were consistent with a major role for these regions in ion translocation and inhibitor binding by NHE1.
EXPERIMENTAL PROCEDURES
Materials
Unless otherwise stated, reagents were from Sigma-Aldrich or Fisher. CompleteTM protease inhibitor was from Roche Applied Science. Cariporide was a kind gift from Sanofi-Aventis. 5′-(N-Ethyl-N-isopropyl) amiloride (EIPA) was from Sigma-Aldrich. n-Dodecyl β-d-maltopyranoside (DDM) was from Anatrace. (1-Oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethio-sulfonate (MTS spin label) was from Toronto Research Chemicals. HiTrap chelating nickel column and CaM-agarose beads for affinity chromatography were from GE Healthcare and Sigma-Aldrich, respectively.
Structural Modeling of the N-terminal Domain of hNHE1
The structural homology model of the N-terminal domain of NHE1 protein was generated from its primary sequence (residues 1–507) using several local structure and fold recognition methods at the MetaServer (22). The established structure of the protein with the highest scores (NhaA; Protein Data Bank accession code 1ZCD), as evaluated by the 3D-Jury method at the same server, was used as template, and the comparison was done with the Swiss-Model program (23).
Threading of NHE1 on the NhaA template was limited to the N-terminal domain of NHE1 (residues 1–507), which includes the membrane-spanning domains. Structurally, the N-terminal domain of NHE1 is distinguished from that of NhaA by a much greater fraction of hydrophilic (extramembrane) sequence. Thus, nearly all of the assigned structure in the model for NHE1 concerns the transmembrane regions of the protein. We therefore constrained our alignment of NHE1 and NhaA to sequences (and their flanking 10 residues) that have been found experimentally (3) to reside in a membrane-like environment. Alignments between the known NhaA TM regions and the hNHE1 TM regions (including the flanking 10 residues) suggested by Wakabayashi et al. (3) based on cysteine accessibility analyses were then carried out independently using the ClustalW algorithm. The resultant TM alignments were then used to match the regions of low homology and ensure that gaps fell within the hydrophilic loops connecting the TM segments.
Analysis of the NHE1 N-terminal domain structural model was performed by use of the DeepView/Swiss-PdbViewer and by use of Insight II software (version 2005) on the Octane work station by Silicon Graphics. The figures were produced using University of California, San Francisco Chimera software. Calculation of the distance between Ala173 and Ile461 based on our homology model and that of Landau et al. (16) was made using the same Chimera software. We are grateful to M. Landau (Tel Aviv University, Tel Aviv, Israel) for providing us with the Protein Data Bank file that allowed us to calculate this value for their model.
Cell Culture
AP-1 cells (a Chinese hamster ovary-derived cell line with no endogenous NHE activity (24) were a kind gift from Dr. S. Grinstein (Hospital for Sick Children, Toronto, Canada). These cells have previously been shown to exhibit no recovery from an acid load in the nominal absence of HCO3− (12, 24). AP-1 cells were grown at 37 °C, 5% CO2, 95% humidity in α-minimum essential medium with Eagle's salts (Mediatech, Inc., Manassas, VA), supplemented with 10% fetal bovine serum, 1% l-glutamine, 1% penicillin/streptomycin, and 600 μg/ml geneticin (G418) sulfate (Invitrogen). Every 3 or 4 days, the cells were passaged by gentle trypsinization, and only passages 5–30 were used for experiments.
Constructs and Stable Transfection of Mutant hNHE1 and paNHE1
The full-length Cys-less hNHE1 was constructed by replacing all of the native cysteine residues with alanines. Using restriction digest, hNHE1 was cloned into the mammalian expression vector pcDNA3.1(+) (Invitrogen). The desired residues were altered using site-directed mutagenesis (QuikChange XL; Stratagene). To facilitate affinity chromatography purification of the constructs, a polyhistidine tag was added to the C-terminal end by three-way ligation. This Cys-less construct was found to be fully functional, in agreement with earlier reports (3). From the Cys-less construct, three different hNHE1 constructs containing Cys replacements at Ala173 and/or Ile461 were prepared. All of the constructs were verified by DNA sequencing prior to transfection (DBS Sequencing Facility, University of California Davis). Positive transfectants were selected for resistance to 600 μg/ml G418. hNHE1 expression was verified by immunoblotting as previously described (25). Briefly, protein homogenates were separated on 7.5% SDS-PAGE gels and electrotransferred to nitrocellulose or PVDF membranes. Mouse monoclonal NHE1 antibody (MAB3140 Millipore/Chemicon) or rabbit polyclonal NHE1 antibody (a kind gift from Mark Musch, University of Chicago) and horseradish peroxidase-conjugated goat secondary antibodies were used to label NHE1, followed by visualization by enhanced chemiluminescence (GE Healthcare). Clonal selection was carried out by limiting dilution, and stably transfected clones were used in all experiments. The corresponding set of constructs for paNHE1, i.e. Cys-less paNHE1 with reintroduction of cysteines in Ala164 (TM4), Ile452 (TM11), and Ala164/Ile452, were prepared in a similar manner.
Functional Analysis of NHE1 Mutants Expressed in AP-1 Cells
All of the constructs were tested for NHE1 function using a Zeiss Axiovert S100 microscope or a PTI fluorescence spectrophotometer, employing the pH-sensitive fluorescent probe 2′,7′-bis-(2-carboxyethyl)-5,6-carboxyfluorescein, tetra-acetoxymethylester to monitor pHi, and the ammonium prepulse technique to acid load the cells, as previously described in detail (12). Isotonic Ringer solution for pHi measurements contained 130 mm NaCl, 3 mm KCl, 20 mm HEPES, 1 mm MgCl2, 0.5 mm CaCl2, 10 mm NaOH, 10 mm glucose, pH 7.4, osmolarity 300 mOsm.
Immunofluorescence Analysis of Wild Type and Mutant NHE1 Expressed in AP1 Cells
Immunofluorescence labeling of NHE1 in AP-1 cells was carried out as described previously (26). Briefly, cells grown on glass coverslips were washed in isotonic Ringer solution (130 mm NaCl, 3 mm KCl, 20 mm HEPES, 1 mm MgCl2, 0.5 mm CaCl2, 10 mm NaOH, 10 mm glucose, pH 7.4), fixed in 2% paraformaldehyde, washed in TBS (150 mm NaCl, 10 mm Tris-HCl, 1 mm MgCl2, 1 mm EGTA), permeabilized (0.2% Triton X-100 in TBS), blocked in 5% BSA in TBST, incubated with primary antibody against NHE1 (Xb-17, 1:100 in TBST), washed in TBST, incubated with Alexa488-conjugated anti-mouse secondary antibody (1:600 in TBST, 1 h), washed in TBST, and mounted. Fluorescence was visualized using a 100×/1.4 NA plan apochromat objective, pinhole size 1 airy disc, and the 488-nm laser line of a Leica DM IRB/E microscope and Leica TSC NT confocal laser scanning unit (Leica Lasertechnik GmbH, Heidelberg, Germany). The images shown are frame-averaged and presented in RGB pseudocolor. No or negligible labeling was detectable in the absence of primary antibody. The relative plasma membrane levels of WT and R425A hNHE1 were estimated from the NHE1 immunofluorescence images by placing regions of interest arbitrarily in the plasma membrane and cytosolic regions (four of each per cell), and calculating the mean plasma membrane/cytosolic pixel intensity ratio per cell. The ratios obtained were 1.72 ± 0.110 (WT) and 0.67 ± 0.048 (R425A), resulting in an estimated relative plasma membrane expression level of 0.67/1.72 = 0.39 (n = 12 cells in three independent experiments for each cell type).
Expression and Functional Analysis of R425A NHE1 in Xenopus Oocytes
Expression and functional analysis of R425A hNHE1 was carried out as previously described (27). Briefly, the human R425A NHE1 was cloned into a vector optimized for oocyte expression (pDEST SML). The cDNA was linearized and in vitro transcribed with T7 RNA polymerase using the T7 mMessage mMachine kit (catalog number AM1344; Ambion). 50 ng of cRNA was injected into defoliculated Xenopus laevis oocytes that were incubated in Kulori medium (90 mm NaCl, 1 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH 7.4) at 19 °C for 3–7 days before experiments were performed. The two-electrode voltage clamp method was used to control the membrane potential and monitor the current in oocytes expressing R425A hNHE1. The recordings were performed at room temperature with a Dagan clampator interfaced to an IBM compatible PC using a DigiData 1320 A/D converter and pCLAMP 9.0 (Axon Instruments). Currents were low pass-filtered at 500 Hz and sampled at 2 kHz. The electrodes were pulled from borosilicate glass capillaries to a resistance of 0.5–2 megaOhm and were filled with 1 m KCl. Generally, the membrane potential (Vm) was held at −50 mV, and the experimental chamber was continuously perfused by a NaCl buffer (100 mm NaCl, 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, pH 7.4). The I/V relations were obtained by changing the membrane potential from +50 to −150 mV in 20-mV increments and measuring the resulting steady state current at each membrane potential. The I/V relationships were obtained in the NaCl buffer at normal pH (pH 7.4) and at low pH (pH 5.8) and in Na+ free buffers (as above but with 100 mm Na+ replaced with 100 mm NMDG-Cl) at the two pH values. The actual I/V curves were obtained by subtracting currents in NMDG-Cl buffer from currents obtained in NaCl buffer. To stimulate NHE1, the oocytes were incubated for 60 min in the low pH buffer before the I/V relationships were determined.
Preparation of Membrane Fractions
Preparation of membrane fractions was based on Ref. 14, with several modifications. Unless otherwise indicated, subsequent procedures were carried out at 4 °C. One day post-confluence, the cells were washed in PBS (60 mm K2HPO4, 30 mm KH2PO4, 145 mm NaCl, pH 7.5) and harvested in lysis buffer (25 mm HEPES, 1 mm EGTA, 1 mm EDTA, 1 mm PMSF, pH 7.5) containing a protease inhibitor mixture (CompleteTM; Roche Applied Science). The cells were pelleted and resuspended in 4 ml of lysis buffer, and complete lysis was obtained by sonication. The lysates were centrifuged at 6000 × g (15 min), and the supernatant (S1) was saved. The pellet was resuspended in lysis buffer and centrifuged at 7500 × g (10 min), and the supernatant (S2) was combined with S1. S1 + S2 were centrifuged at 150,000 × g (60 min), and the pelleted membrane fractions were resuspended in lysis buffer (25 mm HEPES, 1 mm EGTA, 1 mm EDTA, 1 mm PMSF, pH 7.5) containing protease inhibitor, snap frozen in liquid nitrogen, and stored at −80 °C. The membrane fraction was diluted with stripping buffer (25 mm HEPES, 1750 mm KCl, 1 mm EGTA, 1 mm EDTA, 1 mm PMSF, pH 7.5) containing protease inhibitor to a final KCl concentration of 0.7 m and incubated at 4 °C for 30 min, and the stripped membrane fraction was pelleted by centrifugation at 175,000 × g (30 min). To solubilize NHE1, the pellets were resuspended in solubilization buffer (25 mm HEPES, 100 mm NaCl, 1 mm PMSF, 2% Fos-Cholin 14, 1% Triton X-100, 0.05% DDM, pH 7.5) with EDTA-free protease inhibitor (CompleteTM protease inhibitor EDTA-free; Roche Applied Science) and incubated with mild stirring at room temperature (2 × 15 min). The suspension was centrifuged at 150,000 × g to pellet debris (30 min), and the supernatant was used for further purification.
Protein Purification and Spin Labeling
To obtain highly purified NHE1, two sequential purification steps were employed. First, a HiTrap Chelating Nickel column was charged with Ni2+ according to the manufacturer's protocol and equilibrated with binding buffer (BB; 25 mm HEPES, 100 mm NaCl, 5 mm imidazole, 0.05% DDM, pH 7.5). The membrane protein extract was applied to the column at 1.0 ml/3 min. The column was subsequently washed with 3 volumes of BB containing 0.05% Triton X-100 and 6 volumes of BB without Triton X-100 at a flow rate of 1 ml/min. Spin labeling was carried out by applying 3 volumes BB containing 0.2 mm MTS spin label to the column containing bound NHE1 protein (for the use of MTS-SL labels for EPR measurements; see “Discussion”). The MTS-SL chemically modifies the engineered cysteines by forming covalent interactions with the thiol group on the cysteine side chains. The on-column labeling procedure with bound and enriched target protein allows for efficient labeling and facilitates the subsequent removal of unreacted MTS-SL label. After 30 min of MTS labeling, the column was extensively washed with BB and washing buffer (as BB but with 50 mm imidazole) to remove unreacted MTS-SL and then eluted with elution buffer (as BB but with 300 mm imidazole) at 1 ml/3 min. The eluates were collected for further purification according to OD 280 measurements. In the second purification step, 1.5 ml of CaM-agarose affinity resin was pre-equilibrated by rocking for 10 min in 6 ml of CaM binding buffer (CaM-BB; 25 mm HEPES, 100 mm NaCl, 2.5 mm CaCl2, 2.5 mm MgCl2, 0.05% DDM, pH 7.5) two or three times. To allow NHE1 protein to bind to the CaM resin, the eluted sample from the nickel column was incubated with the resin in the presence of 2.5 mm CaCl2 and 2.5 mm MgCl2 for 1 h at room temperature. The column was washed four times with 6 ml CaM-BB and once or twice with 6 ml of HEPES buffer (25 mm HEPES, 100 mm NaCl, 0.05% DDM). To elute NHE1 from the CaM resin, the washed resin was rocked in 2.5 ml of CaM elution buffer (25 mm HEPES, 100 mm NaCl, 2.5 mm EGTA, 2.5 mm EDTA, 0.05% DDM, pH 7.5) containing 0.1 mm of the CaM-binding domain of endothelial NOS (AnaSpec, Freemont, CA) for 1 h at room temperature and overnight at 4 °C. The eluates were collected, and the column was further eluted with 1.6 ml of CaM elution buffer containing 0.05 mm endothelial NOS peptide and 0.05% of FC-14 by rocking for 1 h at room temperature and 1.5 h at 4 °C. The two eluates were pooled and concentrated to 40–60 μl (from ∼4 ml), using a 30-kDa Vivaspin 2 concentrator (Vivascience).
Coomassie Fluor Orange Staining
The presence of hNHE1 in each purified protein sample was confirmed by immunoblotting as described above, and the purity was assessed by Coomassie Fluor Orange protein gel staining according to the manufacturer's (Invitrogen/Molecular Probes) protocol. Briefly, a minigel was incubated with 100 ml of the staining solution by rocking for 45 min. The stained gel was washed first with 7.5% acetic acid solution briefly and then with water for 5 min once or twice. The gel was imaged using a Fujifilm Imager LAS-4000.
Functional Analysis of Reconstituted hNHE1
Reconstitution of NHE1 Protein
Purified polyhistidine-tagged NHE1 protein was reconstituted into liposomes formed of E. coli polar lipids (Avanti Polar Lipids, Inc., Alabaster, AL) essentially as in Refs. 28 and 29. Briefly, liposomes to be assayed for either 22Na+ uptake or H+ flux were mixed in a protein:lipid ratio of 1 μg/mg or 5 μg/mg, respectively. The lipids were prepared by nitrogen streaming to dryness, washed in pentane, and reconstituted in appropriate intravesicular buffers (for 22Na+ uptake: 400 mm NaCl, 10 mm MES at pH 6.0; for H+ flux: 300 mm KCl, 100 mm citrate, 40 mm KH2PO4, pH 4.0). 34 mm CHAPS was added to uniform opacity under sonication, followed by a 1-h preincubation before adding NHE1 protein. Vesicles were gel-filtered with Sephadex beads saturated with intravesicular buffer and eluted with the same buffer. Vesicle-containing fractions were snap frozen in liquid nitrogen and stored at −80 °C in 110-μl aliquots.
22Na+ Uptake and H+ Flux in Reconstituted Liposomes
For 22Na+ uptake assays, the liposomes were centrifuged at low speed through extravesicular medium (EVM1: 10 mm MES, 800 mm sucrose/sorbitol, pH 6.0)-saturated Sephadex beads as in Ref. 28, suspended in 100 μl of EVM1, and immediately combined with 700 μl of EVM1 containing 22Na+ (0.5 μCi/ml). 100-μl aliquots were sampled by washing through DOWEX cation exchange resin with 15 volumes of cold EVM1. Sample γ radioactivity was quantified by Cerenkov counting. Valinomycin permeabilization was used to obtain isotope saturation of the liposomes, and empty liposomes were used to obtain nonspecific 22Na+ uptake (subtracted as background). For H+ flux assays, liposomes were washed in a similar fashion with Sephadex (EVM2: 0.3 mm HEPES, 400 mm KCl, pH 7.4), and 100 μl was removed to a small glass chamber with a micro-stir bar. A precalibrated pH microelectrode (Microelectrodes, Inc., Bedford, NH) was placed into the solution, under continuous sampling to a PowerLab data acquisition system (ADInstruments, Inc., Colorado Springs, CO), and the solution was adjusted to 7.4. 10 μl of poorly buffered NaCl solution (4 m NaCl, 0.3 mm HEPES) was added to initiate H+ flux. H+ flux was determined by conversion and correction for the measured buffer capacity of EVM2. In both assays, liposomes treated with inhibitor were suspended in EVM in the presence of 50 μm EIPA.
EPR Spectroscopy
EPR measurements were performed using a JEOL X-band spectrometer fitted with a loop-gap resonator (30, 31). A 6-μl aliquot of the purified, spin-labeled protein, at a final concentration of ∼10 μm (1.0 μg/μl) in CaM elution buffer (pH 7.5) containing 0.01% DDM, was placed in a sealed quartz capillary contained in the resonator. Spectra (averages of three 2-min scans) were acquired at room temperature at a microwave power of 4 milliwatts and with the amplitude optimized to the natural line width of the individual spectrum. The spectra were normalized according to their double integrated intensity. To narrow the broad spectra and improve integration quality, normalization was performed on each sample in the presence of 92% SDS. After the double integration, the total area was equalized for the two single-labeled constructs, TM IV (A173C) and TM XI (I461C), respectively, which thus normalized them to the same number of spins. The spectra of the two single-labeled constructs were summed and compared with the spectrum for the double-labeled construct, using the same process as described above. The identically treated Cys-less protein that was used as a negative control did not show a signal above background (see Fig. 6, pink trace).
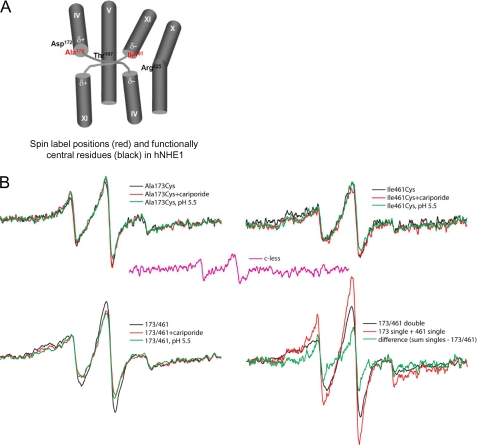
FIGURE 6.
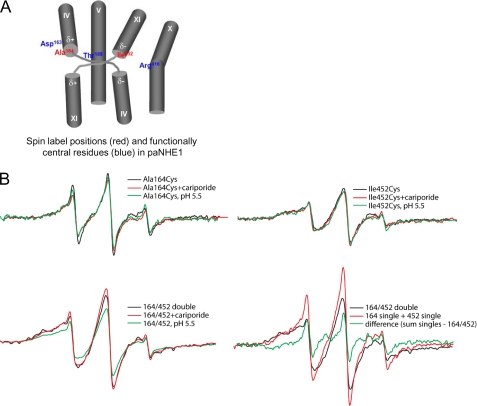
EPR analyses of hNHE1 A173C, I461C, and A173C/I461C. The spectra were collected from samples of purified hNHE1 in CaM elution buffer (pH 7.5) containing 0.01% DDM, and the protein concentration (normalized to 20 μm spin for singles, 40 μm for double and sum of singles) is normalized to the same value for all samples. A, spin label positions (red) and functionally central residues (black) in hNHE1. See text for details. B, top panels, effects of cariporide (0.1 mm) and low pH (5.1) on the EPR spectra of samples containing a spin label at position A173C or I461C, respectively. Bottom panels, effects of cariporide (0.1 mm) and low pH (5.1) on the EPR spectra of double labeled A173C/I461C samples, and the signal from the double labeled sample compared with the sum of singles. Center in pink, signal from the cysteine-less hNHE1, which was incubated with spin label and examined at a concentration similar to the preparations containing a single- or double-Cys substitution (∼20 μm NHE1). This background signal was subtracted from all spectra in panels A–D. Each spectrum shows a scan over a field of 100 G.
RESULTS
Structural Model of hNHE1
To create the tertiary structural model of the transmembrane N-terminal domain of hNHE1, we used in silico homology structure building, using the known structure of NhaA as a template. The primary structure of the N-terminal domain (residues 1–507) of hNHE1 was analyzed for structural homologs at the MetaServer (22). Despite low primary sequence identity, substantial structural similarities between hNHE1 and NhaA (Protein Data Bank accession code 1ZCD) were identified.
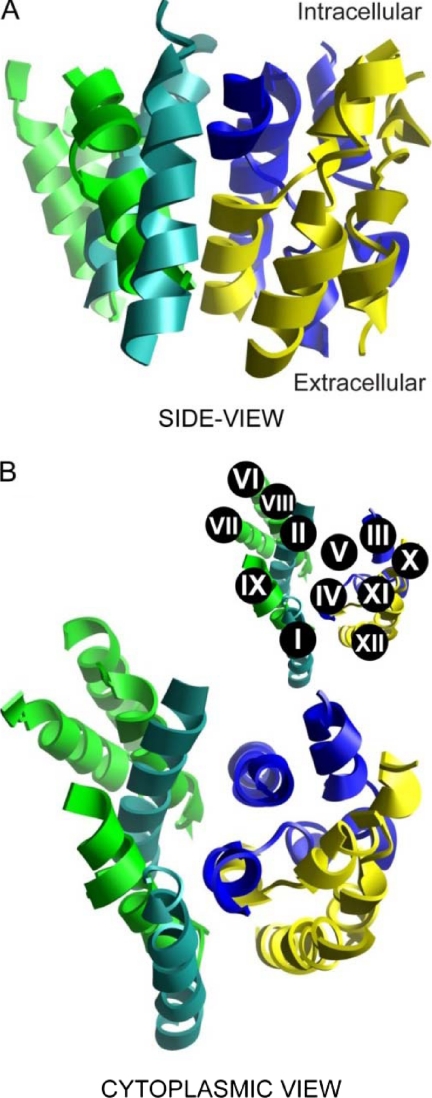
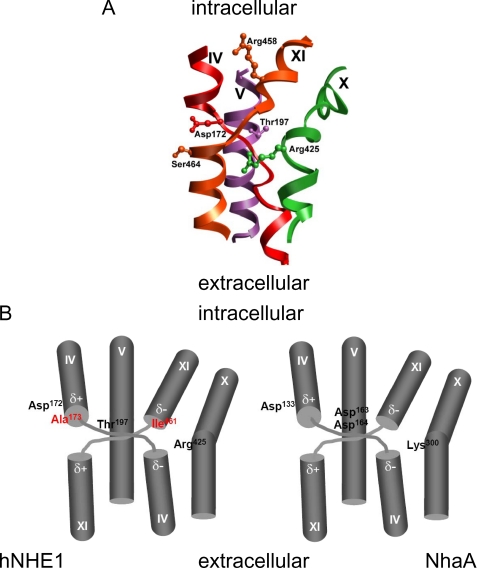
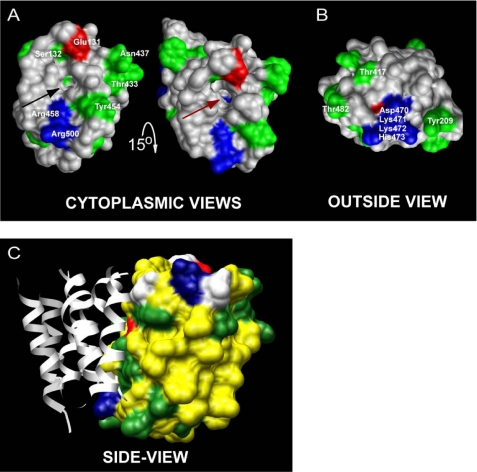
Based on previously published experimental findings (3) and sequence analysis, the primary sequence alignment was manually optimized prior to threading the hNHE1 sequence onto the NhaA tertiary structure (Fig. 1). The resulting structural homology model of hNHE1 (Fig. 2) is limited to the N-terminal (largely membrane-spanning) domain and encompasses amino acid residues Pro12–Ala507. In accordance with the general finding that prokaryotic membrane proteins possess shorter extramembrane loops and terminal extensions than eukaryotic members of the same superfamily (32), NhaA is seen to have smaller extramembrane loops than NHE1 (Fig. 1). Consequently, several of the intra- and extracellular loops connecting the transmembrane regions were excluded from our model of hNHE1 (see also “Experimental Procedures”). In the NhaA structure, and thus in our model, TM IV and TM XI are in close proximity (Fig. 2; see close-up view in Fig. 4). It is noteworthy that the architecture and antiparallel arrangement of TMs IV and XI are unusual in that both helices are extended and positioned such that the partial charges of the N- and C-terminal dipoles are in close apposition in the membrane interior. Closer examination of the threaded model allowed us to identify amino acid residues that are important elements of the putative catalytic core of hNHE1 (Figs. 3 and 4). Briefly, from the solid surface representation of the hNHE1 model shown in Fig. 3 (A and B), it is clearly seen that several charged and polar residues are located near this cavity. Importantly, Arg425 (corresponding to Lys300 in NhaA, which is assigned a central role in the catalytic core of NhaA; see Ref. 13) and presumably then in the catalytic core of NHE1, is accessible from the cytoplasmic side, despite being positioned approximately in the central plane of the lipid bilayer. Fig. 3C shows a side view of the model, illustrating that primarily hydrophobic side chains are pointed into the interior of the lipid bilayer.
FIGURE 1.
Alignment of hNHE1 and NhaA. hNHE1 was predicted to be similar to the structure of NhaA by a number of well established secondary structure and fold recognition methods at the MetaServer (25). The alignment of the sequences (14% identical residues and 23% conserved substitutions in the aligned regions) is from the joint evaluation of the outcome by the 3D-Jury system at the same server and by manual adjustments as detailed under “Experimental Procedures.” The predicted TM helices of NHE1 are shaded in gray (note the lack of fit for some loop regions).
FIGURE 2.
Full structural model of the N-terminal region of hNHE1. The structure model is based on the known structure of the smaller bacterial Na+/H+ exchanger NhaA. The shown representations include the transmembrane domains only (residues 15–31 (TM I), 104–123 (TM II), 130–147 (TM III), 160–179 (TM IV), 191–209 (TM V), 228–246 (TM VI), 255–272 (TM VII), 300–317 (TM VIII), 333–353 (TM IX), 417–437 (TM X), 453–473 (TM XI), and 481–502 (TM XII)) and exclude connecting loops and terminal extensions. The color code used is Turquoise for TMs I and II; dark blue for TMs III, IV and V; green for TMs VI, VII, VIII, and IX; and yellow for TMs X, XI and XII. A, side view of the hNHE1 structure in the plane of the lipid bilayer. B, cytoplasmic view. The inset shows the numbering of the individual helices.
FIGURE 4.
Structure of the Na+/H+ exchanger catalytical core. A, the central parts of TM domains IV, V, X, and XI, the presumed catalytical core for hNHE1-catalyzed Na+/H+ exchange, are represented as a ribbon diagram. B, schematic depiction of the central core of NHE1 (left panel) and NhaA (right panel). The amino acid side chains suggested to directly participate in ion translocation are shown. The positions of the main spin labels are shown in red in hNHE1. Note that the characteristic crossover by the extended structures of helices IV and XI results in energetically unfavorable dipole-dipole pairings (dipoles shown as δ+ and δ−) at the ends of the disrupted α-helices.
FIGURE 3.
Solid surface structure representations of TMs III, IV, V, X, XI, and XII (referred to as the catalytical core in the text) in the hNHE1 model. Positively charged residues are shown in blue, negatively charged residues are in red, and polar residues are in green. A, the cytoplasmic view (the structure on the right in A is tilted 15° downwards compared with the structure on the left) reveals a cavity (arrows) in the structure that reaches down to Arg425 (brown arrow). Several charged and polar residues that may be involved in the ion translocation are located near this cavity. These include Arg458 and Arg500 (positively charged); Glu131 (negatively charged); and Ser132, Thr433, Asn437, and Tyr454 (polar). B, the face of the protein exposed to the outside of the cell does not exhibit any obvious cavity in the structure. However, a cluster of charged residues (Asp470, Lys471, Lys472, and His473) at the end of TM XI and also some scattered polar residues (e.g. Tyr209, Thr417, and Thr482) are accessible on the outside surface of the transmembrane domain. C, the side view representation shows that primarily hydrophobic side chains (yellow) are pointed into the interior of the lipid bilayer. TMs I, II, VI, VII, VIII, and IX are shown as gray ribbons for orientation (compare with Fig. 2).
Functional Analysis of Cys-replaced Mutants and Reconstituted NHE1 Protein
The residues Ala173 and Ile461 were chosen for cysteine replacements for site-directed spin labeling, because of their locations in TM IV and TM XI, respectively, which have been assigned important roles in ion translocation (Fig. 4). Importantly, the conservation of these residues between NHEs is low, and replacement was therefore not likely to interfere with protein function.
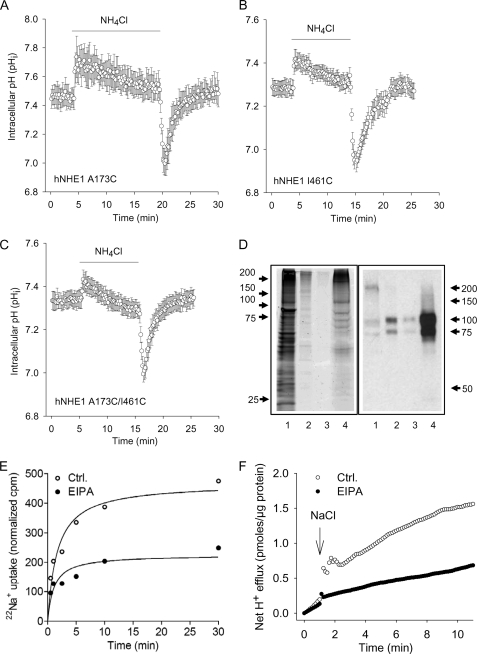
To ascertain that the introduction of cysteines at these positions had not compromised NHE1 function, which would render interhelix distance measurements unreliable, the function of each construct was tested after expression in AP-1 cells, by monitoring pHi recovery after acidification induced by a NH4Cl prepulse. As seen in Fig. 5 (A–C), the three Cys-replaced constructs (A173C, I461C, and A173C/I461C) were all functional, and their regulation by pHi appeared normal. It is noteworthy that this contrasts with a previous study in which an A173C mutation was found to reduce NHE1 function (33). The reason for this discrepancy is not clear; however, it may be significant that virtually all Cys mutants studied by Slepkov et al. (33) exhibited strongly reduced function, including Leu163 and Gly174, whereas comparable mutations have been reported by others to be fully functional (34). Similarly, all of the corresponding paNHE1 constructs were fully functional (not shown, n = 2–3 independent experiments/condition).
FIGURE 5.
Functional evaluation of the NHE1 constructs in AP1 cells and liposomes. A–C, regulation of pHi after an acid load in AP1 cells expressing the A173C (A), I461C (B), or A173C/I461C hNHE1 (C). AP1 cells were loaded with 2′,7′-bis-(2-carboxyethyl)-5,6-carboxyfluorescein and mounted on a Zeiss Axiovert S100 microscope. The cells were perfused with nominally HCO3−-free HEPES-buffered isotonic Ringer solution that, where indicated by the bar, additionally contained 10 mm NH4Cl. Calibration to pHi was carried out as previously described (12). The data shown are representative of six or seven independent experiments/condition. The rates of pHi recovery, obtained at similar starting pHi values but not normalized to expression levels, were 0.13 ± 0.010 (A173C, n = 7), 0.10 ± 0.0039 (I461C, n = 6), and 0.17 ± 0.0037 (A173C/I461C, n = 6). D, Coomassie Fluor Orange staining and Western blot of the A173C/I461C hNHE1 mutant after purification. Purification, staining, and immunoblotting were carried out as described under “Experimental Procedures.” Coomassie Fluor Orange staining is shown in the left panel, and Western blotting for NHE1 is shown in the right panel. Lane 1, the solubilized membrane fraction before transfer to the Ni2+ column; lane 2, eluate from Ni+ column with Ni+ elution buffer (containing 300 mm imidazole); lane 3, eluate from the CaM column with CaM elution buffer; lane 4, purified A173C/I461C hNHE1 sample obtained from the concentration of the sample shown in lane 3. E and F, 22Na+ uptake and H+ flux by hNHE1 reconstituted in liposomes. E, 22Na+ uptake was measured over time in hNHE1 liposome suspensions. The assays were performed in high osmolarity buffered sucrose solution and initiated by the addition of tracer 22Na+ containing solution. F, hNHE1 liposomes were suspended in a poorly buffered KCl medium, monitored with a pH microelectrode in the external bath solution, and expressed as H+ flux. The H+ flux reaction was initiated by adding 10 μl of 4 m NaCl solution at the time indicated. Where indicated, the liposomes were suspended in EVM in the presence of 50 μm EIPA. The difference caused by EIPA represents the NHE1-specific flux. Ctrl., control.
Fig. 5D shows a representative Coomassie Fluor Orange stain and corresponding Western blot of an A173C/I461C hNHE1 sample purified for EPR. As seen, the final eluted sample is highly enriched in NHE1 protein. Although the final eluate is greatly purified compared with the starting material, it contains several bands as revealed by Coomassie Fluor Orange staining. However, comparison of the Western blot and Coomassie data strongly indicates that the majority of the protein in the sample is NHE1: the ∼100- and ∼85-kDa bands correspond to glycosylated and immature NHE1, respectively, and the ≥200-kDa smear is likely to be NHE1 dimers or oligomers.
Another concern was whether the purification and reconstitution of NHE1 might affect function. We therefore monitored NHE1 function following reconstitution in liposomes, i.e. a treatment and environment similar to those of the EPR experiments. As illustrated in Fig. 5 (E and F), acid-activated H+ and Na+ transport sensitive to the NHE1 inhibitor EIPA was clearly evident following NHE1 reconstitution. That NHE1 was functional after purification and reconstitution is a strong indicator that, in our hands, structure was preserved. A third possible caveat would be if spin labeling significantly altered protein folding, stability, or behavior. However, this is highly unlikely to be the case to any detectable extent (see “Discussion”).
EPR Analysis Supports the Structural Model of hNHE1
EPR Line Shape of Single-labeled hNHE1
As noted above, it is necessary to obtain EPR analyses of the noninteracting individual spin-labeled proteins to measure the distance between two spin labels. Hence, hNHE1 constructs with single-labeled TM IV (A173C) and single-labeled TM XI (I461C), respectively, were purified, labeled, and analyzed by EPR. Unless otherwise stated, these analyses were performed at pH 7.5, i.e. under conditions where NHE1 is expected to be in its inactive conformation. Comparison of the EPR spectra of the two single-labeled constructs reveals different spectral shapes (Fig. 6B, top panels). The spectrum for the single-labeled TM IV (A173C) construct exhibits less broadening compared with the single-labeled TM XI construct (I461C), which displays a strongly immobilized component in the low field peak. This reflects that position 173 experiences greater motional freedom, possibly arising from a more disordered backbone. Because the presence of trace amounts of proteins other than NHE1 in the purified sample cannot be ruled out (Fig. 5D), an important control is the EPR signal from the Cys-less NHE1, which was purified and labeled in exactly the same manner as the remaining samples. This is the background signal, a minor contribution that was subtracted from the signal for each of the Cys-containing mutants (Fig. 6, pink trace).
Distance Measurements between the Two Spin Labels on TM IV and TM XI, Respectively
The EPR spectrum for the double-labeled TM IV·TM XI construct containing spin labels at residues 173 and 461 is shown in Fig. 6B (bottom panels, black traces). In the absence of spin-spin interaction in the double-labeled construct, the spectrum would approximate the spectral sum of the two corresponding single-labeled constructs. However, when comparing the trace for the double-labeled versus the sum of the single-labeled constructs, the latter shows less spectral broadening (Fig. 6B), indicating a dipolar component in the double-labeled sample. Because dipolar broadening of spin-labeled proteins is only appreciable for distances within 20 Å (35), the moderate level of broadening seen here is consistent with nitroxide moieties separated by a distance on the order of 13–17 Å, assuming a narrow distribution of distances separating the spin pairs (18, 36, 37). Given the low expression levels of the proteins in cell culture, and the low purification yield, the limited signal:noise ratio of the EPR spectra makes the quantitative calculation of the interspin distance unreliable. However, a qualitative determination within ±2 Å is sufficient for the identification of neighboring TM domains.
Effect of the Functional State of hNHE1 on the Distance between TM IV and TM XI
The double-labeled construct was next studied to observe whether manipulations shown to activate or inhibit the protein result in positional changes in TM IV and TM XI as reflected in altered interaction between the spin labels. Reducing buffer pH to 5.1, which is expected to activate hNHE1 (yet which may also exert inhibitory effects caused by competition with Na+; see “Discussion”), induces a significant additional broadening of the spectrum for the double-labeled construct resulting in the apparent reduction of its signal height (Fig. 6B, bottom left panel, green trace). This indicates that the distance between residues 173 and 461 of hNHE1 is reduced in the acidic buffer. Similarly, the addition of 10 μm of the NHE1 inhibitor cariporide to the purified protein also increases the spectral broadening (Fig. 6B, bottom left panel, red trace). Thus, similar to the effect of reducing pH, the inhibitor induced a conformation where the two positions experienced stronger interaction, although still within the intermediate regime for dipolar coupling (13–17 Å). The inhibitory effect of cariporide on NHE1 function in AP-1 cells is shown in supplemental Fig. S1.
Analysis of TM IV·TM XI Distance and Dynamics in paNHE1
If the TM IV·TM IX complex conserved from NhaA to NHE1 is central to ion translocation by NHE1, then these helices should also be in similarly close proximity in NHE1 from other vertebrate species. Moreover, if the effect of cariporide seen in Fig. 6B indeed reflects interaction of the inhibitor with NHE1, it should not be seen in the flounder protein (paNHE1), which we have previously shown to be insensitive to cariporide (12). We therefore carried out the corresponding experiment in paNHE1, which shares high sequence homology to hNHE1 in both regions. Thus, all endogenous cysteines were removed, and Cys was substituted at the corresponding TM IV or TM XI location. In paNHE1 these positions are Ala164 (TM IV) and Ile452 (TM XI). Likewise, the double Cys mutant (Ala164/Ile452) was also generated to determine the proximity of TMs IV and XI. The EPR analysis of the spin-labeled paNHE1 protein is shown in Fig. 7. As seen in Fig. 7, the two single-labeled constructs behaved similar to the corresponding hNHE1 constructs, i.e. the spin label in TM11 exhibited greater mobility compared with that in TM4. Similarly, the spectrum for the double-labeled construct was clearly broadened compared with the sum of the two single-labeled spectra (Fig. 7B, bottom panels), confirming the finding from the hNHE1 experiments that the two helices are close to one another (on the order of 15 Å). The effects of acidic pH and cariporide on the EPR spectrum of the double-labeled paNHE1 are shown in Fig. 7B (bottom left panel). As illustrated, acidic pH (green trace) resulted in broadening of the spectrum, i.e. the same effect seen for hNHE1. In marked contrast, cariporide (10 μm, red trace) had no detectable effect on the paNHE1, consistent with the lack of functional effect of cariporide on this NHE1 homolog (12). Collectively, the EPR data thus indicate that spin labels placed approximately in the center of TM IV and TM IX of NHE1 are within 20 Å of each other and that the distance between these spin labels is affected by acidification and by cariporide binding, the latter in the cariporide-sensitive NHE1 homolog only.
FIGURE 7.
EPR analyses of paNHE1 A164C, I452C, and A164C/I452C. The spectra were collected from samples of purified paNHE1 in PBS buffer (pH 7.5) containing 0.01% DDM, and the protein concentration (normalized to 20 μm spin for singles, 40 μm for double and sum of singles) is normalized to the same value for all samples. A, spin label positions (red) and functionally central residues (blue) in paNHE1. See text for details. B, top panels, effects of cariporide (1 mm) and low pH (5.1) on the EPR spectra of samples containing a spin label at position A164C or I452C, respectively. Bottom panels, effects of cariporide (1 mm) and low pH (5.1) on the EPR spectra of double labeled A164C/I452C samples and the signal from the double labeled sample compared with the sum of singles. The residual signal was subtracted from all spectra. Each spectrum shows a scan over a field of 100 G.
Effect of a Neutralizing Mutation in the Putative Screen Residue Arg425
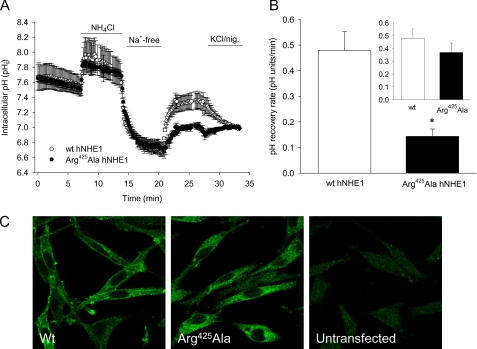
As alluded to above, the NhaA model suggests that Lys300 plays a central role as a “screen residue” that permits the energetically unfavorable dipole-dipole arrangement of the catalytic core of NhaA (13). The corresponding residue in hNHE1 is Arg425, which in the homology model is positioned approximately in the central plane of the lipid bilayer yet accessible from the cytoplasmic side (Figs. 3 and 4). We therefore hypothesized that replacement of Arg425 with a neutral amino acid residue would destabilize the catalytic core and strongly affect NHE1 function. Introduction of an R425A mutation indeed reduced NHE1-dependent pHi recovery after an acid load (Fig. 8, A and B). Confocal imaging verified that this at least in part reflected a strongly reduced targeting of the mutant NHE1 to the plasma membrane (Fig. 8C).
FIGURE 8.
Effect of R425A mutation on hNHE1 expression, localization, and function. A, recovery of pHi after an NH4Cl prepulse-induced acid load in AP-1 cells stably transfected with WT hNHE1 (open circles) or R425A hNHE1 (filled circles). The experiments were carried out essentially as described in the legend to Fig. 5, except that the measurements were carried out in a PTI fluorescence spectrophotometer. Where indicated, HEPES-buffered isotonic Ringer solution was replaced with NH4Cl-containing and Na+-free Ringer solution, respectively, as indicated by the bars. The graphs shown are representative of at least three independent experiments per condition. B, summary of the experiments shown in A, showing the initial rate of pHi recovery for WT hNHE1 and R425A hNHE1, respectively. The inset shows the pHi recovery rates corrected for the relative levels of WT and R425A hNHE1 in the plasma membrane, calculated as described under “Experimental Procedures.” C, confocal images of AP-1 cells transfected with WT hNHE1, R425A hNHE1, or untransfected as indicated. The cells were paraformaldehyde-fixed, labeled for NHE1 followed by secondary, Alexa488-coupled antibody, and imaged using a Leica confocal microscope as detailed under “Experimental Procedures.” The images shown are representative of at least three independent experiments per condition.
One possible effect of the R425A mutation that would not have been picked up in the pHi recovery measurements is a switch of the ion transport mode of NHE1 from one of Na+/H+ exchange to conductive Na+ transport. To test this possibility, we expressed the R425A mutant in Xenopus oocytes, allowing us to monitor mutant-related Na+ currents. In the series of experiments performed, we were unable to distinguish currents in the NHE1 R425A mutant-injected oocytes from those of the noninjected oocytes (supplemental Fig. S2). It should be noted that the voltage-dependent currents exhibited substantial variation, most likely caused by batch-dependent endogenous Cl− and K+ channel activity (38, 39). Thus, Arg425 appears to serve a crucial function in stabilizing NHE1 structure, but its mutation to alanine does not seem to induce a Na+ current via NHE1.
DISCUSSION
Based on the crystal structure of NhaA (13) and experimental evidence from cysteine accessibility studies (3), we created a threaded structural model of hNHE1. The TM IV·TM XI complex of NhaA, by virtue of its unlikely architecture (charges and partial charges in the membrane interior) and its apparent conservation in NHE1, could be the catalytic core of the ion translocation pathway (13). If this inference is correct, it is likely that NHE1 and NhaA mediate Na+ and H+ translocation through a very similar mechanism. The inferred structural model of hNHE1 resembles that recently proposed by Landau et al. (16) in many respects. The two models agree on the location of several of the helical transmembrane domains. In addition, the NhaA crystal structure indicates a critical role for a basic side chain on TM X, and both predictions identify Arg425 as the residue occupying this position. To further compare the two models, we calculated the distance between the ends of the amino acid side chains of Ala173 and Ile461 obtained for each model. This value was very similar for the two models, with a distance of 8.46 Å obtained for our model compared with 7.91 Å for the Landau model. However, there are significant differences between the TM assignments in the two models. Because of the very low homology of NHE1 to NhaA, we constrained our alignment of TM segments to regions of NHE1 that were experimentally determined (3) to be in a membrane-like environment, whereas Landau et al. (16) achieved their alignment solely from homology-based predictions. For example, the first TM segment in our model starts at residue 15. In contrast, the N-terminal extension in the Landau model is more than 100 residues longer, and the first TM segment instead starts at residue 128 (TM I of the Landau model co-localizes with TM III of the structure we propose). This arrangement, however, does not appear to take into account that residues 126 and 127 have previously been shown to be inaccessible for MTSET labeling even in permeabilized cells, strongly indicating that they are embedded in the bilayer (3). Consequently, the negatively charged, or polar, residues facing the ion-binding pocket are different in the two models, with Asp172 and Thr197 (Fig. 4) being postulated to be part of the critical residues for translocation in our model, whereas Landau et al. (16) depict Asp238 and Asp267 (on TM IV and TM V, respectively, in the Landau model) to provide the negative charges in the core of the translocation pathway. Furthermore, the PX3D motif is highly conserved in all NHE proteins, and multiple hydropathy and experimental analyses has placed this motif in TM segment IV (e.g. (3, 7, 25, 40). The NhaA structure highlights the significance of this motif, because it facilitates a distinctive extension and crossover of TM domains IV and XI. In our alignment, this motif is assigned to TM IV in agreement with previous experimental evidence (3, 40), whereas in Landau et al. (16) this motif falls within TM II. Moreover, in the Landau model, residue 173 (located in TM II in their model) is much farther apart from residue 461 than the distance that we observed experimentally using EPR (∼15 Å). Thus, although further validation of both models is obviously needed, the model that we present here is supported by multiple lines of experimental evidence.
Validation of the Structural Model and Ion Translocation Hypothesis by Site-directed Spin Labeling and EPR
Our structural model of hNHE1 thus supported our recent evidence that TM IV and TM XI play important roles in inhibitor binding and ion translocation by NHE1 (12). We further validated this notion, first by determining the distance between TM IV and TM XI and the conformational changes in these domains in response to manipulations known to activate or inhibit hNHE1 and paNHE1 by site-directed spin labeling and EPR spectroscopy. Demonstrating the feasibility of this approach for assessing NHE1 structure, a recently published study employed EPR spectroscopic distance measurements between spin-labeled side chains on two NhaA monomers to confirm NhaA dimerization (41). To estimate TM IV·TM XI distances in hNHE1, we created two single-labeled mutants, A173C and I461C, and the double-labeled mutant A173C/I461C. It may be noted that although a strict kinetic analysis was not performed, the function of these constructs was comparable with that of normal hNHE1. Comparison of the spectra for the two single-labeled constructs indicates that residue 173 is in a less constrained environment than residue 461 (Figs. 6 and 7). This is recapitulated in the model structure, where both residues face the inside of the protein, yet Ile461 seems to be more enclosed in the protein than Ala173. The distance between Ala173 in TM IV and Ile461 in TM XI obtained from the double- and single-labeled constructs was determined to ∼15 Å, confirming that TM IV and TM XI are in close proximity.
When pH was reduced to 5.1, the A173C/I461C spectrum revealed stronger interaction of the spin labels, indicating that in this presumably active conformation, the spin-labeled side chains undergo stronger dipolar coupling, although still within the intermediate regime of 13–17 Å (18, 36, 37). Similarly, in the presence of the NHE1 inhibitor cariporide, the A173C/I461C spectrum exhibited increased spin label interaction. This indicates that interaction of cariporide with hNHE1 causes a conformational change resulting in decreased distance between Ala173 and Ile461. Again, this would be in congruence with our recent findings that TMs IV and XI play a key role in inhibitor binding and therefore are strong candidate domains for participation in the ion translocation process (12). EPR analysis of the corresponding set of residues in the cariporide-insensitive paNHE1 homolog showed that the spin label distance as well as the effect of acidification were similar to those observed for hNHE1, whereas introduction of cariporide had no effect on the EPR spectrum. This difference in the effect of cariporide on the distance between TMs IV and XI in hNHE1 and paNHE1 shows that the TM IV·TM XI complex is conserved among vertebrate NHE1s and provides a strong indication that the effect of cariporide on the EPR spectrum in fact reflects an inhibitory interaction of this compound with the transporter.
These findings correlate very well with the NhaA translocation mechanism proposed by Hunte et al. (13). In the NhaA structure (and thus in our hNHE1 model), the TM IV·TM XI helices are extended, cross over each other, and exhibit similar conformational changes in response to activating stimuli. Given the homology between the presumed catalytic core in NhaA and NHE1, an equivalent mechanism for ion translocation in hNHE1 seems highly probable. Specifically, Asp133 of NhaA aligns with Asp172 of hNHE1, Asp163 of NhaA aligns with Thr197 of hNHE1, and Lys300 of NhaA aligns with that of Arg425 of hNHE1 in our model (Fig. 4). Asp163 of NhaA is suggested to act as a molecular switch, such that its protonation state determines whether the Na+-binding site (Asp164) is accessible to the periplasm or the cytoplasm (42). In hNHE1, Thr197 is likely to carry out the same function as an accessibility control site. In the NhaA structure, the energetically unfavorable negative/negative and positive/positive dipole-dipole pairings caused by the antiparallel extended arrangement of the TM IV·TM XI helices are stabilized by electrostatic screening provided by the negative Asp133 and the positive Lys300 (13). These residues are conserved among bacterial NhaA homologs and have been shown to be essential to NhaA activity (43, 44). We hypothesized that the corresponding residues in hNHE1, Asp172 (negative charge) and Arg425 (positive charge), fulfill similar dipole “screening” functions (Fig. 9). If this is correct, a mutation in one of the screening residues should severely disrupt the catalytic core of hNHE1. Arg425 is located in TM X, which we in our previous studies implicated in ion transport and inhibitor binding (12) and which has been assigned as central in ion translocation by NhaA (44). Moreover, in our structural model, Arg425 is located at the bottom of the open cavity and is thus directly accessible from the cytoplasmic side of the membrane (Fig. 3). Consistent with the hypothesis, an R425A mutation in hNHE1 resulted in marked reduction in NHE1 plasma membrane targeting and pH regulatory ion transport. Our data indicate that membrane targeting of NHE1 is more strongly affected by the R425A mutation than its transport function. However, regardless of which of these defects is more prominent, they support the hypothesis that Arg425 is of central importance in the functionally organized NHE1 protein.
FIGURE 9.
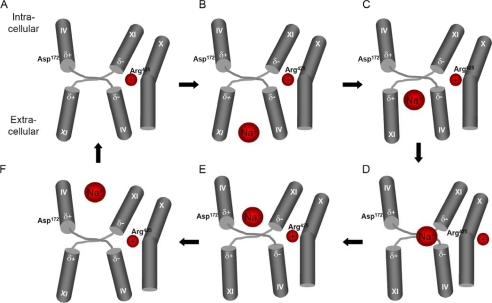
Tentative working model of ion translocation in the catalytic core of NHE1. The figure depicts possible conformational changes in the TM IV·TM XI crossover arrangement during NHE1 ion translocation induced by an acidic pH change. A, charge compensation from Arg425 located in TM X stabilizes the energetically unfavorable negative/negative and positive/positive dipole-dipole pairings in the arrangement of the TM IV·TM XI helices. B, an acidic pH change results in alteration of the protonation state of the region of TM IX (not shown) located at the entrance of the proposed funnel, eliciting a conformational change in TM IX, which causes a direct contact between TM IV and TM IX (not shown). This rearrangement of TM IV results in a reorientation of TM IV and TM XI such that a Na+-binding site is exposed to the extracellular space. C–F, binding of Na+ causes a charge imbalance, triggering a movement of the TM IV and TM XI helices, exposing Na+ to the cytoplasm. The release of Na+ results in protonation of the Na+-binding site, causing a conformational change leading back to the original arrangement of TM IV and TM XI.
Furthermore, we hypothesize that Arg425 not only stabilizes the helices by screening the partial charges from the dipoles on the extended helix C-terminal ends but functions as a “check valve” permitting Na+ coordination and or gating (Fig. 9). Although Arg425 on TM X is clearly well positioned for screening the negative dipoles at the C-terminal breaks in TMs IV and XI, intuitively, it seems that this charge-screening role could also be fulfilled through a basic residue in the extended region of TM XI (similar to Asp172). However, the crossover structure of TMs IV and XI constrains the degree to which this positive charge can be repositioned during the transport cycle. On the other hand, the localization of Arg425 along the straight TM X permits axial rotations that can alter the proximity of this charge relative to the catalytic core by several angstroms. In this regard, we postulate that Arg425 may function as a gate-controlling Na+ transport.
Critical Assessment of the Validity of the EPR Data
Functional analysis of both Cys-replaced NHE1 mutants in AP-1 cells and of purified and reconstituted NHE1 protein strongly indicated that NHE1 function, and hence structure, was retained during protein purification and modification for EPR analyses. Another obvious concern in site-directed spin labeling EPR experiments is that engineering cysteine substitutions and subsequent modification by the spin label may significantly alter protein folding, stability, or behavior. However, substantial work has revealed that the nitroxide ring is well tolerated in proteins and assumes a limited number of rotamers, facilitating the modeling of the spin label within the three-dimensional structure (19, 45, 46). The method has been applied to a wide assortment of protein types, with very few examples showing a major functional or structural consequence resulting from this modification. Direct evidence for how the incorporated nitroxide is accommodated in protein structures has been obtained in high resolution crystal structures of T4 lysozyme containing spin-labeled side chains. Even at buried sites, no significant perturbation of the backbone is evident (19, 45). Finally, others have reported modest or negligible effects of spin labeling on protein folding (47) or backbone structure of peptides as measured by NMR (48). This can be attributed to the relatively compact size of the modified Cys residue (a molecular volume on the order of Tyr) and its ambivalent chemical nature, which does not favor highly polar or nonpolar environments. Distortions of a few angstroms are possible; however, because confidence within ±5 Å is more than enough to map the correct TM arrangement, this should not cause concern.
Possible Mechanism of H+ Sensing by NHE1
In NhaA, TM IX is located at the entrance of the cytoplasmic funnel, where it has been proposed to function as a “pH sensor” (13). This arrangement is recapitulated in our hNHE1 model (Fig. 9) based upon NhaA crystal structure, our chimera, and EPR studies. Thus, the mechanism of pH-regulated ion translocation proposed for NhaA could also be expected for hNHE1 (Fig. 9): an acidic pH change could result in alteration of the protonation state of the region of TM IX (not shown) located at the entrance of the proposed funnel, which could elicit a conformational change in TM IX, causing a direct contact between TM IV and TM IX (not shown). This rearrangement of TM IV could result in a reorientation of the TM IV·TM XI arrangement such that a Na+-binding site is exposed to the extracellular space. Binding of Na+ would cause a charge imbalance that would trigger a movement of the TM IV and TM XI helices, exposing Na+ to the cytoplasm. The release of Na+ would result in protonation of the Na+-binding site, causing a conformational change leading back to the original arrangement of TM IV and TM XI (Fig. 9). At least in NhaA, this mechanism only requires small conformational changes of the helices (13); thus, the proposed TM IV·TM XI arrangement would be suited for a relatively high turnover rate of Na+/H+ exchange.
In conclusion, we present here a structural model of hNHE1 that places TM IV and TM XI in close proximity. This architecture was confirmed by EPR analyses, from which the distance between TM IV and TM XI was determined at ∼15 Å. This distance was decreased both at acidic pH and in the presence of cariporide, consistent with a role for TM IV and TM XI rearrangements in ion translocation and inhibitor binding by hNHE1.
Supplementary Material
Acknowledgments
We thank S. Grinstein (Hospital for Sick Children, Ontario, Canada) for the kind gift of AP-1 cells, C. Altenbach (Department of Ophthalmology, the Jules Stein Eye Institute, UCLA, Los Angeles, California) for providing us with the Basephase program, and M. Landau (Tel Aviv University, Tel Aviv, Israel) for providing us with the Protein Data Bank file for their model. Cariporide was a kind gift from Sanofi-Aventis. Anni Bech Nielsen (University of Copenhagen) and Steven E. Anderson (University of California Davis) are gratefully acknowledged for skilled experimental assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-21179 (to P. M. C.). This work was also supported by funds from the Danish National Research Council (to S. F. P.) and the Swedish Research Council (to J. O. L.) and a Biocampus Scholarship from University of Copenhagen (to E. B. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- NHE1
- Na+/H+ exchanger isoform 1
- PaNHE1
- P. americanus NHE1
- hNHE1
- human NHE1
- TM
- transmembrane
- IL
- intracellular loop
- EIPA
- 5′-(N-ethyl-N-isopropyl)amiloride
- DDM
- n-dodecyl β-d-maltoside
- CaM
- calmodulin
- BB
- binding buffer
- MES
- 4-morpholineethanesulfonic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- EVM
- extravesicular medium.
REFERENCES
- 1. Orlowski J., Grinstein S. (2004) Pflugers Arch. 447, 549–565 [DOI] [PubMed] [Google Scholar]
- 2. Pedersen S. F., O'Donnell M. E., Anderson S. E., Cala P. M. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1–R25 [DOI] [PubMed] [Google Scholar]
- 3. Wakabayashi S., Pang T., Su X., Shigekawa M. (2000) J. Biol. Chem. 275, 7942–7949 [DOI] [PubMed] [Google Scholar]
- 4. McLean L. A., Zia S., Gorin F. A., Cala P. M. (1999) Am. J. Physiol. 276, C1025–C1037 [DOI] [PubMed] [Google Scholar]
- 5. Pedersen S. F., King S. A., Rigor R. R., Zhuang Z., Cala P. M. (2003) Bull. Mt. Desert Island 42, 38–39 [Google Scholar]
- 6. Counillon L., Franchi A., Pouysségur J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khadilkar A., Iannuzzi P., Orlowski J. (2001) J. Biol. Chem. 276, 43792–43800 [DOI] [PubMed] [Google Scholar]
- 8. Touret N., Poujeol P., Counillon L. (2001) Biochemistry 40, 5095–5101 [DOI] [PubMed] [Google Scholar]
- 9. Noël J., Germain D., Vadnais J. (2003) Biochemistry 42, 15361–15368 [DOI] [PubMed] [Google Scholar]
- 10. Orlowski J., Kandasamy R. A. (1996) J. Biol. Chem. 271, 19922–19927 [DOI] [PubMed] [Google Scholar]
- 11. Wang D., Balkovetz D. F., Warnock D. G. (1995) Am. J. Physiol. 269, C392–C402 [DOI] [PubMed] [Google Scholar]
- 12. Pedersen S. F., King S. A., Nygaard E. B., Rigor R. R., Cala P. M. (2007) J. Biol. Chem. 282, 19716–19727 [DOI] [PubMed] [Google Scholar]
- 13. Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. (2005) Nature 435, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 14. Moncoq K., Kemp G., Li X., Fliegel L., Young H. S. (2008) J. Biol. Chem. 283, 4145–4154 [DOI] [PubMed] [Google Scholar]
- 15. Coupaye-Gerard B., Bookstein C., Duncan P., Chen X. Y., Smith P. R., Musch M., Ernst S. A., Chang E. B., Kleyman T. R. (1996) Am. J. Physiol. 271, C1639–C1645 [DOI] [PubMed] [Google Scholar]
- 16. Landau M., Herz K., Padan E., Ben-Tal N. (2007) J. Biol. Chem. 282, 37854–37863 [DOI] [PubMed] [Google Scholar]
- 17. Lagerstedt J. O., Voss J. C., Wieslander A., Persson B. L. (2004) FEBS Lett. 578, 262–268 [DOI] [PubMed] [Google Scholar]
- 18. Hubbell W. L., Cafiso D. S., Altenbach C. (2000) Nat. Struct. Biol 7, 735–739 [DOI] [PubMed] [Google Scholar]
- 19. Langen R., Oh K. J., Cascio D., Hubbell W. L. (2000) Biochemistry 39, 8396–8405 [DOI] [PubMed] [Google Scholar]
- 20. Voss J., He M. M., Hubbell W. L., Kaback H. R. (1996) Biochemistry 35, 12915–12918 [DOI] [PubMed] [Google Scholar]
- 21. Zhao M., Zen K. C., Hubbell W. L., Kaback H. R. (1999) Biochemistry 38, 7407–7412 [DOI] [PubMed] [Google Scholar]
- 22. Ginalski K., Elofsson A., Fischer D., Rychlewski L. (2003) Bioinformatics 19, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 23. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rotin D., Grinstein S. (1989) Am. J. Physiol. 257, C1158–C1165 [DOI] [PubMed] [Google Scholar]
- 25. Pedersen S. F., King S. A., Rigor R. R., Zhuang Z. P., Cala P. M. (2003) Biophysical J. 84, 151A [Google Scholar]
- 26. Rasmussen M., Alexander R. T., Darborg B. V., Møbjerg N., Hoffmann E. K., Kapus A., Pedersen S. F. (2008) Am. J. Physiol. Cell Physiol. 294, C197–C212 [DOI] [PubMed] [Google Scholar]
- 27. Meinild A. K., Loo D. D., Skovstrup S., Gether U., MacAulay N. (2009) J. Biol. Chem. 284, 16226–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nimigean C. M. (2006) Nat. Protoc. 1, 1207–1212 [DOI] [PubMed] [Google Scholar]
- 29. Accardi A., Kolmakova-Partensky L., Williams C., Miller C. (2004) J. Gen. Physiol. 123, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Froncisz W., Hyde J. S. (1982) J. Magn. Reson. 47, 515–521 [Google Scholar]
- 31. Hubbell W. L., Froncisz W., Hyde J. S. (1987) Rev. Sci. Instrum. 58, 1879 [Google Scholar]
- 32. Pao S. S., Paulsen I. T., Saier M. H., Jr. (1998) Microbiol. Mol. Biol. Rev. 62, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slepkov E. R., Rainey J. K., Li X., Liu Y., Cheng F. J., Lindhout D. A., Sykes B. D., Fliegel L. (2005) J. Biol. Chem. 280, 17863–17872 [DOI] [PubMed] [Google Scholar]
- 34. Counillon L., Noël J., Reithmeier R. A., Pouysségur J. (1997) Biochemistry 36, 2951–2959 [DOI] [PubMed] [Google Scholar]
- 35. Altenbach C., Oh K. J., Trabanino R. J., Hideg K., Hubbell W. L. (2001) Biochemistry 40, 15471–15482 [DOI] [PubMed] [Google Scholar]
- 36. Columbus L., Hubbell W. L. (2004) Biochemistry 43, 7273–7287 [DOI] [PubMed] [Google Scholar]
- 37. Mchaourab H. S., Oh K. J., Fang C. J., Hubbell W. L. (1997) Biochemistry 36, 307–316 [DOI] [PubMed] [Google Scholar]
- 38. Kowdley G. C., Ackerman S. J., John J. E., 3rd, Jones L. R., Moorman J. R. (1994) J. Gen. Physiol. 103, 217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu L., Montrose-Rafizadeh C., Hwang T. C., Guggino W. B. (1990) Biophys. J. 57, 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shrode L. D., Gan B. S., D'Souza S. J., Orlowski J., Grinstein S. (1998) Am. J. Physiol. 275, C431–C439 [DOI] [PubMed] [Google Scholar]
- 41. Hilger D., Polyhach Y., Padan E., Jung H., Jeschke G. (2007) Biophys. J. 93, 3675–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arkin I. T., Xu H., Jensen M. Ø., Arbely E., Bennett E. R., Bowers K. J., Chow E., Dror R. O., Eastwood M. P., Flitman-Tene R., Gregersen B. A., Klepeis J. L., Kolossváry I., Shan Y., Shaw D. E. (2007) Science 317, 799–803 [DOI] [PubMed] [Google Scholar]
- 43. Galili L., Herz K., Dym O., Padan E. (2004) J. Biol. Chem. 279, 23104–23113 [DOI] [PubMed] [Google Scholar]
- 44. Kozachkov L., Herz K., Padan E. (2007) Biochemistry 46, 2419–2430 [DOI] [PubMed] [Google Scholar]
- 45. Guo Z., Cascio D., Hideg K., Kálái T., Hubbell W. L. (2007) Protein Sci. 16, 1069–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fajer M. I., Li H., Yang W., Fajer P. G. (2007) J. Am. Chem. Soc. 129, 13840–13846 [DOI] [PubMed] [Google Scholar]
- 47. DeWeerd K., Grigoryants V., Sun Y., Fetrow J. S., Scholes C. P. (2001) Biochemistry 40, 15846–15855 [DOI] [PubMed] [Google Scholar]
- 48. Bolin K. A., Hanson P., Wright S. J., Millhauser G. L. (1998) J. Magn. Reson. 131, 248–253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.