Abstract
Prior work indicates that IL-6 can stimulate c-Myc expression in multiple myeloma (MM) cells, which is independent of effects on transcription and due to enhanced translation mediated by an internal ribosome entry site in the 5′-UTR of the c-Myc RNA. The RNA-binding protein hnRNP A1 (A1) was also critical to IL-6-stimulated translation. Because A1 shuttles between nucleus and cytoplasm, we investigated whether the ability of IL-6 to enhance Myc translation was mediated by stimulation of A1 shuttling. In MM cell lines and primary specimens, IL-6 increased A1 cytoplasmic localization. In contrast, there was no effect on the total cellular levels of A1. Use of a dominant negative A1 construct, which prevents endogenous A1 from nucleus-to-cytoplasm transit, prevented the ability of IL-6 to enhance Myc internal ribosome entry site activity, Myc protein expression, and MM cell growth. IL-6-stimulated cytoplasmic localization was mediated by alterations in the C-terminal M9 peptide of A1, and this correlated with the ability of IL-6 to induce serine phosphorylation of this domain. A p38 kinase inhibitor prevented IL-6-induced A1 phosphorylation. Thus, IL-6 activates c-Myc translation in MM cells by inducing A1 phosphorylation and cytoplasmic localization in a p38-dependent fashion. These data suggest A1 as a potential therapeutic target in MM.
Keywords: Cytokine Action, Interleukin, Myc, Translation Control, Translation Regulation, hnRNP A1, Interleukin 6, Multiple Myeloma
Introduction
Our previous work (1) demonstrated that IL-6 could enhance c-Myc protein expression in multiple myeloma (MM)2 cells independent of any effect on Myc transcription. This was not due to altered c-Myc protein stability or the mammalian target of rapamycin-dependent stimulation of cap-dependent translation. In contrast, it resulted from an up-regulation of Myc IRES activity and cap-independent translation. IRESes are structures in the 5′-UTR of transcripts that can promote translation and are especially important when RNA leaders are highly structured or when cap-dependent translation is inhibited (2, 3). The three-dimensional IRES structure recruits the 40 S ribosomal subunit to the mRNA for translation initiation. The Myc IRES is well characterized (4, 5). In addition, there are trans-acting proteins that facilitate Myc IRES function via binding to the RNA at sites in the 5′-UTR and inducing conformational changes that facilitate recruitment of the IRES to the ribosome (4). Several publications (6, 7) document a myeloma-specific up-regulation of Myc IRES function.
The ability of IL-6 to stimulate Myc IRES function in MM cells, up-regulate rapamycin-resistant Myc translation and expression, and stimulate MM cell proliferation were all prevented by knockdown of hnRNP A1, an RNA-binding protein (1). Because other studies (8–10) demonstrate that hnRNP A1 (A1) can function as an IRES-trans-activating factor (ITAF), regulating Myc IRES activity, these results collectively indicate that IL-6-up-regulated c-Myc translation in MM cells was dependent on enhanced A1 ITAF function.
hnRNP A1 is a shuttling protein with movement from nucleus to cytoplasm (11), and a recent study (12) demonstrated that the degree of cytoplasmic localization of A1 can serve as a regulatory control over IRES activity. A previous study (13) also documented that the nucleocytoplasmic shuttling of A1 in myeloid cells was integral to promoting BCR-ABL leukemogenesis. We thus undertook the present study to investigate whether the ability of IL-6 to enhance A1-dependent Myc translation in MM cells was mediated by a stimulation of A1 shuttling.
EXPERIMENTAL PROCEDURES
Cell Lines, Constructs, and Reagents
The ANBL-6 wild type and K-RAS-transfected cell lines were gifts from Dr. Brian Van Ness (University of Minnesota, Minneapolis, MN). All other MM lines were obtained from ATCC (Manassas, VA). The pRF construct was a kind gift of Dr. A. Willis (University of Leicester, Leicester, UK). pRmF and pRp27F were generated as described previously (1) by cloning the Myc and p27 IRESes into the intercistronic region of pRF. The wild type A1 construct in PCGT-A1 plasmid was a kind gift of Dr. Adrian Krainer (Cold Spring Harbor Laboratory) (14). The NLS-A1 construct was a kind gift from Dr. Danilo Perrotti (Ohio State University, Columbus, OH). The mutant A1, where serines of the M domain are mutated into alanines, was generated by inserting full-length wild type A1 into P3xFLAG-CMV-10 mammalian expression plasmid between BamHI and XbaI restriction sites. With wild type A1 as a template, the mutant was generated by PCR with a forward primer (5′-gaatggatcccttcACCATGGACTACAAAGACCATGACGGTGATTATAAAG-3′) and reverse primer (5′-cccgctcgagTTAAAATCTTCTGCCAGCGCCATATGCAGCGGCAGCTGCGGCACCGCCATAGCCACCTTGGTTT-3′). The serine-to-aspartic acid phosphomimetic mutant was generated by PCR with the same forward primer and a different reverse primer (5′-cccgctcgagTTAAAATCTTCTGCCGTCGCCATAATCGTCATCGTCATCGTCACCGCCATAGCCACCTTGGTTT-3′). The mutants as well as the NLS A1 and wild type A1 were inserted into pLenti6/V5-D-TOPO plasmids.
Transfections
pLenti6/V5-D-TOPO was co-transfected with ViraPower DNA into 293 cells, and after 48 h, viral supernatant was collected. MM cells (2.5 × 105) were infected with 1 ml of viral supernatant overnight and then selected with 2.5 μg/ml blasticidin to generate stable cell lines.
Primary Myeloma Specimens
Primary MM cells were purified from bone marrow of patients as described (1). The purity by microscopy and CD138 flow cytometric analysis was >99% plasma cells. The project was approved by the institutional review board of the Veterans Affairs Healthcare Center, and all of the participants gave written informed consent.
Dicistronic Reporter Assay
The reporter constructs were transfected into cell lines using Lipofectamine Plus (Invitrogen) and normalized for transfection efficiency by co-transfection with pSVbetaGal (Promega). Transfection efficiency was 5–10%. A transfection efficiency of at least 5% was required for carrying out a reporter assay. After 12 h, the cells were treated with or without IL-6. The cells were harvested and Renilla luciferase, firefly luciferase, and β-galactosidase activities were determined (1). All luciferase activity is normalized to the luciferase values (both Renilla and firefly) obtained for pRF in the absence of IL-6 stimulation, which is designated as a value of “1.” The data are presented in the figures as fold change in luciferase activity induced by IL-6.
Separation of Cytoplasmic from Nuclear Protein
Nuclear protein was separated from cytoplasmic protein with reagents and kit from Thermo Fisher Scientific Inc. (Rockford, IL), using nuclear and cytoplasmic extraction reagents. Briefly, the cells were washed with cold PBS and resuspended in CER I buffer and incubated on ice for 10 min. CER II buffer was then added. The cells were centrifuged (16,000 × g for 5 min) to separate supernatant (cytoplasmic extract) from the nuclei. The pellet (nuclei) was suspended in NER buffer and vortexed for 15 s on the highest setting. The sample was placed on ice with continued vortexing for 15 s every 10 min for a total of 40 min. The tube was then centrifuged at 16,000 × g for 10 min. The supernatant (nuclear extract) was transferred to a prechilled tube.
Evaluation of Protein and RNA Expression
Western blot was performed as described (1). Real time PCR for Myc RNA and GAPDH RNA was performed as described (1). Briefly, gene amplifications for real time PCR were performed with an ABI PRISM 7700 sequence detection system. Each 20-μl reaction in a 96-well plate comprised 9 μl of cDNA template, 1 μl of 20× primer mixtures for c-Myc or GAPDH, and 10 μl of 2× TaqMan Universal PCR Master mixed with AmpErase® UNG. After an initial 2 min at 50 °C to activate ampErase® and a denaturation step of 10 min at 95 °C, 60 cycles of amplification were performed with denaturation for 15 s at 95 °C, and annealing/extension for 1 min at 60 °C. All of the samples were run in triplicate, and no template controls were included in all plates for both c-Myc and GAPDH.
Use of Inhibitors
MM cells were exposed to an ERK inhibitor, U0126 (Promega); a p38 inhibitor, SB203580 (Calbiochem); and a MNK kinase inhibitor, MNK1 inhibitor (Calbiochem) for 30 min prior to stimulation with IL-6. Control groups without inhibitors had the same concentration of the Me2SO solvent as the inhibitor groups.
Assays to Measure Effect of NLS on A1 Splice Function
The RT-PCR-based assay for pyruvate kinase RNA splicing was performed according to the method published by Clower et al. (15) with some modifications. To improve the correlation of band intensity with the quantity of DNA, we applied the FAM labeling to replace ethidium bromide staining in the quantification process. The sequence of primers were the same as described (15) except the 5′ primer was labeled with FAM. 2 μg of total RNA was extracted from respective cell lines with RNeasy mini kit (Qiagen). The cDNAs were produced with a high capacity cDNA reverse transcription kit (Applied Biosystem). Semi-quantitative PCRs using PuReTag Ready-To-Go PCR beads (GE Healthcare) were carried out with annealing temperature set at 62 °C. After 25 amplification cycles, the products were equally separated into two tubes: one left undigested and another one digested with PST1 (Invitrogen). After initial electrophoresis on a 2% agarose gel to check for the successful amplification of PKMs, the products were further analyzed on a 5% polyacrylamide gel. The FAM-labeled PCR products were visualized with Fuji imager system (LAS-4000). Densitometry was done with the QuantityOne program from Bio-Rad. The ratio of intensity of 185 bp from PKM2 digested to that of the 318-bp undigested M1 band were calculated. RT-PCR analysis of CD44 alternative splicing was also performed. Total RNA were isolated using RNeasy kit from Qiagen. cDNA were prepared from 1 μg of RNA with a high capacity cDNA archive kit from Applied Biosystems. Semi-quantitative PCR of CD44 cDNA were performed using the forward primer in exon 5 and the reverse primer in exon 17, so that the amplified PCR product covered all of the potential variable exons. Totally, 28 cycles of amplification were finished before the products were separated in a 1.5% agarose gel.
Immunofluorescence
Glass coverslips were coated with 0.01% poly-l-lysine (Sigma) overnight at 4 °C and rinsed with PBS before use. The cells were plated onto coverslips in complete medium. After 2 h in 37 °C, adhered cells were fixed with 3.7% formaldehyde for 10 min and rinsed with PBS. The cell membranes were permeabilized by treating with PBS, 0.25% Triton X-100 for 5 min. The cells were blocked with PBS, 10% BSA and incubated with monoclonal anti-FLAG M2 (1:1000) in 3% BSA, PBS for 2 h at 37 °C. After two washes, anti-mouse Alexa Fluor 488 secondary antibody (Molecular Probes) was added. 45 min later, the cells were washed, and the coverslips were mounted on slides in Prolong Gold anti-fade reagent (Invitrogen). The cells were visualized with a Leica Leitz DMRBE microscope using a 40× objective lens (Leica PL Fluotor), and fluorescent and corresponding bright field images were captured with a Hamamatsu C4742-95 camera and OpenLab program 3.1.5.
Cell Growth Assay
Parental or transfected ANBL-6 cells were depleted of IL-6 for 24 h and then seeded at 3.3 × 105/ml in complete medium. IL-6 was added, and 48 h later, viable cell counts were enumerated by trypan blue staining. The data are presented as IL-6-induced fold increase in viable cell count relative to no IL-6. The data are the means ± S.D. (n = 3).
Statistics
The t test was used to determine significance of differences between groups.
RESULTS
IL-6 Enhances hnRNP A1 Localization to Cytoplasm
Our previous work demonstrated that IL-6 significantly increased c-Myc translation in MM cells, and this effect was mediated by hnRNP A1 (A1), which is an ITAF for the Myc IRES (1). A recent study (12) provided evidence that alterations in cytoplasmic distribution of A1 could regulate rhinovirus or apaf-1 IRES function. We thus treated MM cell lines and primary specimens with or without IL-6, separated nucleoprotein from cytoplasmic protein, and performed immunoblot experiments to test whether IL-6 enhanced cytoplasmic localization of A1 (Fig. 1). Because A1 is primarily a nuclear protein with much less cytoplasmic localization, in all Western blot experiments shown in Fig. 1, 5-fold greater amounts of cytoplasmic protein extract were electrophoresed relative to nuclear protein extract.
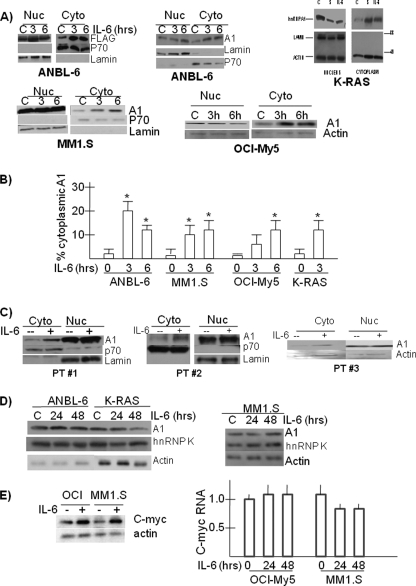
FIGURE 1.
IL-6 enhances cytoplasmic localization of hnRNP A1 in MM cells. A, MM cell lines were treated with or without IL-6 (100 units/ml) for 3 or 6 h, and nuclear (Nuc) versus cytoplasmic (Cyto) extract was immunoblotted for identified proteins. In one experiment with K-RAS cells, sorbitol (S) was also used at 600 mm. B, percentage of A1 localized to cytoplasm is calculated in cell lines ± IL-6. The results are the means ± S.D. (n = 3). The asterisk denotes statistically different (p < 0.05). C, three 1° MM bone marrow samples were treated with or without IL-6 for 3 h. D, cell lines were exposed with or without IL-6 (100 units/ml) for 24 or 48 h, and Western blot was performed on whole cell lysates for total expression of hnRNP A1, hnRNP K, or actin. Because ANBL-6 and K-RAS cells were continuously maintained in IL-6, all of the experiments with those cell lines were performed following prior depletion of IL-6 for 24 h. E, OCI-My5 or MM1.S cell lines were stimulated with or without IL-6 (100 units/ml) for 24 or 48 h. Immunoblot assay at 48 h for expression of c-Myc or actin is shown. An increase in c-Myc/actin ratios (by densitometry) stimulated by IL-6 was significant (p < 0.05) (2.1 ± 0.3 for OCI-My5 and 1.9 ± 0.2 for MM1.S, fold mean increase ± S.D., n = 3). Compared with no IL-6 after 24 or 48 h, IL-6 had no effect on c-Myc RNA expression in those cell lines (relative RNA expression by real time PCR where control (no IL-6) is kept at “1” (mean ± S.D., n = 3)).
Initial experiments utilized the IL-6-dependent ANBL-6 MM cell line, which demonstrates a significant IL-6-induced increase in Myc translation and Myc IRES function (1). This increased translation was dependent on A1 function, although total cellular A1 levels were not increased by IL-6 (1). Thus, IL-6-starved ANBL-6 cells were transiently transfected with FLAG-tagged hnRNP A1 and then re-exposed to IL-6. As shown in Fig. 1A (first panel), IL-6 significantly increased cytoplasmic levels of transfected A1 with little effect on nuclear levels. p70 and lamin immunoblots demonstrated equal protein loading of fractions and ruled out contamination of cytoplasmic extracts with nuclear material. When nontransfected ANBL-6 cells and other MM cell lines were studied to investigate effects on endogenous A1, a similar enhancement of A1 localized to cytoplasm was induced by IL-6 (Fig. 1A). In addition to wild type ANBL-6, we also studied ANBL-6 cells stably expressing a mutant K-RAS allele (K-RAS cells in Fig. 1A), MM1.S cells, and OCI-My5 cells. In each experiment there was a significant increase in A1 localization to cytoplasm induced by IL-6 exposure. In K-RAS cells, the effect of IL-6 was compared with sorbitol, which had been previously shown (16) to enhance nuclear-to-cytoplasmic movement of A1, presumably secondary to osmotic stress. IL-6 was comparable with sorbitol in its ability to enhance cytoplasmic localization of A1. For these cell lines, three identical experiments were performed, and the percentage of cytoplasmic A1 was calculated by densitometry. The results in Fig. 1B demonstrate a significant IL-6-induced increase in cytoplasmic localization to levels between 12 and 20% of total A1.
As we had previously shown (1), exposure to IL-6 did not significantly increase total cellular A1 expression in these experiments (Fig. 1D). Additional experiments demonstrated no significant effect of IL-6 on expression levels of other proteins previously identified as possible Myc IRES ITAFs (PTB, hnRNP E1/E2, hnRNP C1/C2, and unr; data not shown). In particular, there were no changes in expression of hnRNPK (Fig. 1D), an RNA-binding protein known to induce Myc IRES function in BCR-ABL-expressing hematopoietic cells (17).
Primary MM cells from three consecutively studied patients (Fig. 1C) were also tested. After isolation of MM cells by positive selection of CD138 (>99% pure), MM cells were treated with or without IL-6, and the cell fractions were studied for A1 presence. As in the cell line studies, 5-fold greater amounts of cytoplasmic extract were electrophoresed relative to nuclear extracts. As shown in Fig. 1C, IL-6 increased the cytoplasmic levels of hnRNP A1 in these primary cell specimens.
In our previous report (1), we only studied ANBL-6 and K-RAS MM cell lines for IL-6-induced effects on Myc translation. To extend those findings, we additionally examined the MM cell lines described above. As shown in Fig. 1E, there was a concurrent increase in c-Myc protein expression following IL-6 exposure in MM1.S and OCI-My5 MM cell lines without associated increase in c-Myc RNA levels. Thus, the enhanced translation of c-Myc stimulated by IL-6 is not singular to the IL-6-dependent ANBL-6 cell line and is found in other lines that are responsive to IL-6, although not completely dependent on the cytokine for their survival.
Effects of a Nucleus-localized, Shuttling-deficient hnRNP A1 Mutant
To test whether the enhanced cytoplasmic localization of A1 induced by IL-6 participated in the stimulation of Myc IRES function and Myc expression, we utilized an A1 shuttling-deficient mutant, NLS-A1 (generous gift of D. Perrotti) (13). The NLS-A1 construct contains the bipartite basic type nuclear localization signal (NLS) of hnRNPK fused in-frame with the N terminus of a FLAG-tagged hnRNPA1 mutant (A1-G274A), which lacks both nuclear export and import activities. The NLS localizes newly made protein to the nucleus from which it cannot escape because of the Gly-274 mutation. The mutant inhibits A1-dependent mRNA export when microinjected into nuclei of Xenopus oocytes (18). It has also been used in mammalian hematopoietic cells as a dominant negative to prevent endogenous hnRNPA1 nuclear-to-cytoplasmic shuttling and to inhibit A1-mediated nuclear export of mRNAs (13). The nucleus-localized, shuttling-deficient mutant presumably out-competes endogenous A1 for nuclear factors required for A1 mRNA nuclear export.
Initial experiments confirmed that the NLS-A1-FLAG mutant was nucleus-localized in MM cells. ANBL-6 MM cells were transiently transfected with either FLAG-tagged wild type A1 (FLAG-WT) or the FLAG-NLS-A1 mutant. Nucleoprotein was separated from cytoplasmic protein, and extracts were immunoblotted for FLAG, total A1, and lamin (nuclear marker). As shown in Fig. 2A, transfected wild type A1 (FLAG-WT) is found expressed in cytoplasm (mean cytoplasmic FLAG-A1 = 9.1 ± 0.4%, n = 3) as well as nucleus (FLAG immunoblot, second gel). However, the NLS mutant is almost completely localized to the nucleus (mean cytoplasmic FLAG-NLS = 2.1 ± 0.2%, n = 3). Immunoblot assay for lamin demonstrates no contamination of cytoplasmic extracts with nuclear protein. Immunofluorescent analysis of these transfected ANBL-6 cells using anti-FLAG antibody (Fig. 2B) confirmed the ability of wild type A1 to move to cytoplasm during exposure to actinomycin-D (as described previously (13)), whereas the NLS mutant remained confined to the nucleus.
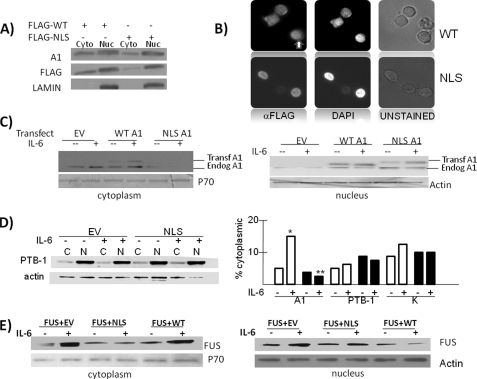
FIGURE 2.
The NLS-A1 mutant functions as a dominant negative construct in MM cells. A, ANBL-6 cells were transiently transfected with FLAG-WT A1 or FLAG-NLS A1, and nuclear (Nuc) versus cytoplasmic (Cyto) extracted protein was immunoblotted for total A1, FLAG, or lamin expression. B, immunofluorescence of WT A1- versus NLS A1-transfected cells treated with actinomycin-D (2.5 μg/ml). Immunodetection of transfected FLAG-tagged WT A1 shown to identify cytoplasmic (white arrow) as well as nuclear signal. In contrast, NLS A1 is only found in nuclei. C, EV, WT A1, or NLS A1 stably expressed in ANBL-6 cells that are treated with or without IL-6 (100 units/ml) for 3 h, after which cytoplasmic versus nuclear extracts are immunoblotted for presence of A1, p70, or actin. D, effect of IL-6 or NLS expression on nuclear-to-cytoplasmic shuttling of PTB-1 or hnRNPK. The left panel shows immunoblot of cytoplasmic (C) or nuclear (N) PTB-1 in stably EV- or NLS-expressing MM cells treated with or without IL-6. To the right of the gel is the percentage of cytoplasmic localization of A1, PTB-1, or hnRNPK in EV-transfected (white bars) or NLS-transfected (black bars) cells. *, significantly increased value versus no IL-6, p < 0.05; **, significant decreased value versus IL-6 treatment of EV-transfected cells (p < 0.05). E, ANBL-6 stably co-transfected with FUS+EV, FUS+NLS A1, or FUS+WT A1 constructs and treated with or without IL-6, after which nuclear versus cytoplasmic extracts are immunoblotted for the presence of FUS, p70, and actin.
To confirm that the NLS-A1 mutant could function as a dominant negative in MM cells, we tested its effect on IL-6-induced nuclear export of endogenous hnRNP A1 and the RNA-binding protein FUS. The larger size of the expressed WT or NLS-A1 transgenes allowed us to differentiate the transfected proteins from endogenous A1 when a 12% gel was used. Thus, ANBL-6 cells were stably transfected with empty vector (EV), wild type hnRNP A1 (WT A1), or the shuttling-deficient mutant (NLS A1). The three cell lines were treated with or without IL-6 for 6 h, nuclear protein was separated from cytoplasmic protein, and extracts were immunoblotted for hnRNP A1. As shown in Fig. 2C, IL-6 was successful in inducing cytoplasmic localization of endogenous A1 in EV- and WT-transfected cells without significant effect on nuclear levels. In contrast, little endogenous A1 was localized to cytoplasm in NLS-A1-transfected cells, and there was no significant increase when these latter cells were exposed to IL-6. The immunoblot also demonstrates that IL-6 can also induce nuclear export of transfected wild type A1, similar to what we had shown after transient transfections (Fig. 1A). To show the specificity of the NLS dominant negative effect, we also tested its ability to alter cytoplasmic localization of other shuttling proteins. As shown in Fig. 2D, there was little effect of either IL-6 or the NLS on cytoplasmic localization of PTB-1. This experiment was repeated, as were similar ones investigating the localization of hnRNPK. The mean cytoplasmic localization calculated from three independent experiments is shown on the graph in the right panel of Fig. 2D (white bars are EV-transfected cells, and black squares are NLS-transfected cells). As can be seen, IL-6 significantly (p < 0.05) enhanced A1 cytoplasmic localization as described above but had no significant effect on shuttling of PTB-1 or hnRNPK, indicating relative specificity for the effect of the cytokine. Furthermore, the NLS dominant negative prevented the enhanced A1 cytoplasmic shuttling but had no effect on the cytoplasmic levels of the other two shuttling proteins. Thus, the NLS-A1 can operate as an effective and specific dominant negative to prevent nuclear export of endogenous A1.
Additional support for the effectiveness of the NLS-A1 as a dominant negative comes from experiments where we stably co-expressed (by lentiviral infection) A1 constructs with FLAG-tagged FUS. FUS is an RNA-associated protein that binds to A1 in the nucleus. Although FUS is also a shuttling protein, it lacks nuclear export or import signals (19), suggesting that its nuclear export is regulated by its association with hnRNP A1. Prior studies demonstrated a dominant negative effect of NLS-A1 on cytoplasmic export of FUS (13). Thus, MM cells were stably co-transfected with tagged FUS + WT A1, FUS + NLS-A1, or FUS + EV. The cells were then stimulated with or without IL-6, nuclear versus cytoplasmic extracts were obtained, and immunoblot assays were performed for the presence of transfected FUS (FLAG immunoblots; Fig. 2E). As shown, IL-6 exposure significantly increased cytoplasmic levels of transfected FUS in cells co-transfected with EV or with wild type A1. However, co-expression of the NLS-A1 mutant prevented nuclear export of FUS to cytoplasm. Thus, expression of the NLS mutant in MM cells is an efficient dominant negative that prevents cytoplasm localization of endogenous A1 as well as its FUS cargo.
Effects of the NLS-A1 Mutant on IL-6 Stimulation of IRES Function, c-Myc Protein Levels, and Enhanced Proliferation
To test the effects of the NLS mutant on the response of Myc IRES activity to IL-6, ANBL-6 MM cells were stably transfected with the wild type A1, the dominant negative NLS, or EV. The cells were subsequently transiently transfected with the dicistronic reporter constructs pRF or pRmF (1) (shown in Fig. 3A). These reporter constructs contain an upstream Renilla luciferase gene (RLuc) and a downstream firefly luciferase gene (FLuc). The pRF construct contains no IRES, but the pRmF contains the Myc IRES cloned into the intracistronic space. In the latter vector, firefly luciferase translation is driven by the Myc IRES and is a measure of cap-independent translation. In contrast, expression of the upstream Renilla luciferase gene is an estimate of cap-dependent translation. The cells were then treated with or without IL-6, and luciferase expression was evaluated. The results were normalized to transfection efficiency by co-transfection with a β-galactosidase construct. The presence of the Myc 5′-UTR IRES in pRmF increased firefly luciferase expression (versus pRF) 32-fold (mean, n = 3) while having no effect on Renilla luciferase expression. When exposed to 100 units/ml of IL-6, a further increase in firefly luciferase expression was demonstrated in EV-transfected cells (3.7-fold, mean, n = 3; Fig. 3A) and in wild type A1-transfected cells (3.1-fold). As a negative control, Fig. 3A also shows that similar exposure to IL-6 did not increase firefly luciferase driven by the p27 IRES (pRp27F in Fig. 3A). There was a very modest increase in Renilla luciferase expression also seen after IL-6 exposure (1.3–1.5-fold increase; Fig. 3A), which is consistent with a mild enhancing effect of IL-6 on cap-dependent translation secondary to activation of mammalian target of rapamycin (20). In contrast to these effects on Myc IRES activity in WT and EV cells, IRES activity is significantly depressed in NLS-expressing MM cells (only 8.5-fold increased versus pRF), and IL-6 had no enhancing effect (Fig. 3A). As a corollary to the ability of the NLS shuttling-deficient mutant to inhibit IL-6-induced up-regulation of Myc IRES activity, we compared EV- and NLS-transfected ANBL-6 cells for IL-6-induced effects on Myc protein expression. As shown in Fig. 3B, the dominant negative mutant prevented the ability of IL-6 to stimulate Myc protein expression. Finally, the shuttling-defective NLS also prevented the ability of IL-6 to stimulate cell growth (Fig. 3C).
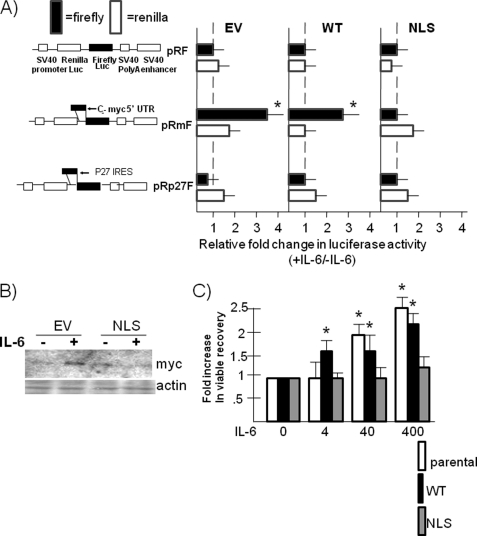
FIGURE 3.
The NLS A1 dominant negative mutant prevents IL-6-induced increases in Myc IRES activity, Myc protein expression, and cell growth. A, ANBL-6 cells, stably expressing either empty vector, WT A1, or NLS A1 genes, were transfected with pRF, pRmF, or pRp27F reporter plasmids, treated with or without IL-6 for 6 h, and firefly versus Renilla luciferase expression was evaluated. The results shown are mean increases in luciferase expression induced by IL-6 (means ± S.D., n = 3). *, statistically significant increase induced by IL-6, p < 0.05. B, EV or NLS A1-transfected ANBL-6 cells were depleted of IL-6 for 24 h followed by readdition of IL-6 (100 units/ml) or none for 24 h, and Western blot was performed for c-Myc and actin expression. C, ANBL-6 parental, WT A1-transfected, or NLS A1-transfected cells were depleted of IL-6 for 24 h and then restimulated with 0, 4, 40, or 400 units/ml IL-6 for 48 h, after which the viable cell count was enumerated. The data are the fold increases in viable cell numbers induced by IL-6 versus no IL-6 (means ± S.D., n = 3). *, significant increase, p < 0.05.
Because ANBL-6 MM cells also respond to IL-6 with enhanced Myc RNA expression (1), it was possible that the NLS mutant curtailment of Myc protein expression could also be mediated by prevention of transcription or decrease in RNA stability. The latter is a distinct possibility because hnRNP A1 is also implicated in regulation of RNA stability (21). However, real time PCR analysis demonstrated a very minimal effect of the NLS mutant on IL-6 stimulation of Myc RNA. The cytokine increased the amount of Myc RNA by 2.05-fold (mean, n = 3) in EV-transfected cells and increased Myc RNA amounts by 1.75-fold in NLS-transfected cells. To provide further support that the shuttling-deficient NLS inhibitory effect on IL-6 responses was specifically due to prevention of IRES-dependent translation, we also studied K-RAS cells (Fig. 4). Previous work (1) documented that IL-6 enhanced c-Myc protein expression in K-RAS cells but had no effect on RNA expression. K-RAS-mutated ANBL-6 cells may express heightened constitutive Myc RNA expression resulting from the RAS mutation without room for additional stimulation by IL-6. Thus, the K-RAS cells are good models for studying effects of IL-6 purely on translation. In addition, IL-6 effectively enhances A1 cytoplasmic localization in K-RAS cells (Fig. 1A). Thus, K-RAS cells were also stably transfected with EV or the NLS mutant and treated with or without IL-6. Fig. 4A demonstrates a significant increase in Myc protein expression induced by IL-6 in EV control cells, which was prevented in NLS-transfected cells. Similar to wild type RAS ANBL-6 cells, the EV-transfected K-RAS cells respond to IL-6 with a significant increase in firefly luciferase expression when the pRmF reporter construct is used, whereas the NLS-transfected cells demonstrate a marked blunted effect (Fig. 4B, mean, n = 3). Real time PCR analysis demonstrated that the NLS mutant had no significant inhibitory effect on Myc RNA expression in IL-6-treated cells (Fig. 4C). These results collectively demonstrate that the NLS shuttling-deficient mutant prevents IL-6 increases in Myc IRES activity as well as enhanced responses in protein expression and cell growth. The latter inhibitory effects are independent of any inhibition of RNA expression.
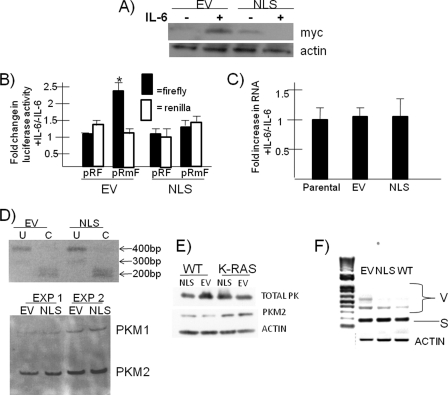
FIGURE 4.
The NLS A1 dominant negative prevents IL-6 responses in K-RAS cells. A, K-RAS cells transfected with EV or NLS A1, depleted of IL-6 for 24 h, and then treated with or without IL-6 (100 units/ml) for 24 h, after which immunoblot was performed for c-Myc and actin expression. B, cells described above for A were transiently transfected with pRF or pRmF reporter plasmids and treated with or without IL-6 for 6 h, and then firefly and luciferase reporter expression was assessed. The data are the mean increases in luciferase expression induced by IL-6 (means ± S.D., n = 3). *, statistically significant increases, p < 0.05. C, real time PCR analysis of c-Myc RNA levels in parental K-RAS cells or those stably expressing EV or NLS A1. Cell lines are stimulated with IL-6 (100 units/ml) for 24 h. The data are the mean increases in RNA levels induced by IL-6 (means ± S.D., n = 3). D, the upper panel shows agarose gel resolving PKM PCR products uncut (U) or cut (C) by PST1. The lower panel is a polyacrylamide gel used to semi-quantify the FAM-labeled PCR products after PST1 digestion. E, WT ANBL-6 or K-RAS-mutated ANBL-6 (K-RAS) cells stably transfected with either NLS dominant negative A1 or EV and immunoblot performed for expression of total pyruvate kinase (TOTAL PK), pyruvate kinase isoform 2 or actin. F, PCR amplification of all CD44 splice isoforms resolved by agarose gel electrophoresis. S, standard isoform; V, variant isoforms.
Recent work (22) has also implicated hnRNP A1 as a splice factor that promotes the conversion of the pyruvate kinase adult isoform, PKM1, to the embryonic pyruvate kinase isoform, PKM2 (23), which facilitates aerobic glycolysis. Exons 9 and 10 are alternatively spliced in a mutually exclusive fashion, resulting in PKM1 (exon 9 inclusion) or PKM2 (exon 10 inclusion) isoforms. hnRNP A1 is a splicing repressor that represses exon 9 inclusion, giving rise to relatively increased amounts of PKM2. Thus, one alternative explanation for the ability of the NLS dominant negative to inhibit IL-6-stimulated MM cell growth is that, by inhibiting endogenous A1 activity, it relieves the repression on exon 9 inclusion and reverts the PKM2 isoform back to PKM1. This could theoretically prevent the switch in metabolism that favors an increase in tumor cell biomass, which may be needed for tumor cell expansion. To examine this possibility, we assessed splicing in the pyruvate kinase transcript comparing the EV- and NLS-transfected isogenic ANBL-6 cell lines using a semi-quantitative assay described previously (15). RNA was harvested, reverse transcribed, and PCR-amplified using primer sets that anneal to exons 8 and 11. After amplification, the reactions were separated into aliquots for digestion with PST1 or no digestion. The products were first analyzed on a 2% agarose gel as shown in Fig. 4D (upper panel). PST1 specifically cleaves exon 10, resulting in 213- and 185-bp fragments. This is shown in the lanes where PST1 was used to cut the products versus uncut lanes. As shown in Fig. 4D, the predominant isoform in both cell lines is the PKM2 isoform. To semi-quantify the relative amounts of PKM2 to PKM1 RNA, we used the same primers, but the 5′ primer was labeled with FAM. The FAM-labeled PCR products were cut with PST1 and then analyzed on a 5% polyacrylamide gel and assessed by densitometry. As shown in Fig. 4D, the PKM2 isoform is again shown to predominate in both cell lines in the two experiments depicted. Calculation by densitometry showed that the EV cell line contained 91 ± 3% PKM2, and the NLS cell line contained 88 ± 4% PKM2 (n = 3). This small difference is not significant. We also immunoblotted extracts from EV versus NLS-transfected wild type ANBL-6 and K-RAS-mutated ANBL-6 MM cells with an antibody specific for the PKM2 isoform. Immunoblot was also performed with an antibody detecting both isoforms. As shown in Fig. 4E, there was no decrease in PKM2 expression in NLS-expressing myeloma cells, ruling out the notion that curtailed growth in NLS-expressing cells is due to alteration of the PKM1/PKM2 ratio. This is consistent with the notion that endogenous A1 nuclear splicing activity is unaffected by the NLS dominant negative and further supports that the inhibitory action of NLS is specifically due to its prevention of A1 nuclear-to-cytoplasm export. Interestingly, as further detailed under “Discussion,” the K-RAS mutated ANBL-6 cells demonstrated an increased expression of the PKM2 isoform compared with wild type ANBL-6 cells (Fig. 4E).
To further assess whether the NLS dominant negative might prevent the role of endogenous A1 in alternative splicing, we also analyzed expression of CD44 splice variants in EV-, NLS-, and WT A1-transfected MM cells. Alternative pre-mRNA splicing generates variant CD44 isoforms by the inclusion of up to 10 variant exons (V1–V10), and previous work (24) implicates hnRNP A1 function in repressing inclusion of variant exons. Thus, cDNAs were amplified using the forward primer located at exon 5 and reverse primer in exon 17 such that all variant isoforms would be amplified (as described in Ref. 25). Fig. 4F shows that ectopic expression of both WT and NLS A1 repressed inclusion of variant exons. Thus, the NLS can function as a CD44 splice regulator and is not likely to inhibit endogenous A1 splice activity.
Effects of M Peptide Phosphorylation of A1
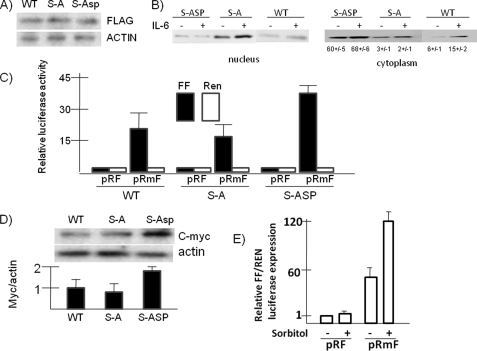
Cytoplasmic shuttling of hnRNP A1 is regulated by phosphorylation of A1 on 4 of 7 serine residues in the M9 domain, a 38-amino acid segment in the C terminus of the protein (10, 14, 26). To ascertain whether these serines play a role in the ability of IL-6 to enhance nuclear export of A1, MM cells were stably transfected with tagged wild type A1 versus an A1 mutant where all 7 serines in the M9 peptide are converted to alanine to prevent phosphorylation versus a phosphomimetic mutant where all serines are converted to aspartic acids. As shown (Fig. 5A), significant and comparable expression of all three transgenes was achieved. Although IL-6 increased WT A1 cytoplasmic localization, there was little effect of IL-6 on nuclear-to-cytoplasm transit of the Ser-to-Ala mutant (Fig. 5B). The abrogation of the ability of IL-6 to induce cytoplasmic localization in the Ser-to-Ala mutant underscores the importance of the M9 domain in the effect of cytokine. In contrast to the WT and Ser-to-Ala mutant protein, the S to Asp phosphomimetic mutant was predominantly localized in cytoplasm. However, there was also no further increase in cytoplasmic localization of this latter mutant upon exposure to IL-6, presumably because a potential IL-6-stimulatory effect was already achieved by the phosphomimetic mutations. This experiment was repeated three times, and the mean ± S.D. percentage of transfected A1 localized to cytoplasm (relative to nuclear A1) in the three cell lines is shown below the cytoplasmic immunoblot in Fig. 5B.
FIGURE 5.
Effects of M peptide phosphorylation of A1. A, ANBL-6 cells, stably transfected with FLAG-tagged WT A1, serine-to-alanine mutant (S-A), or serine to aspartic acid (S-ASP) phosphomimetic mutant were assayed by immunoblot for expression of FLAG or actin. B, these transfected cell lines were treated with or without IL-6, and nuclear versus cytoplasmic extracts were immunoblotted for expression of FLAG. The percentage of cytoplasmic amount of transfected A1 is shown below cytoplasm immunoblot (means ± S.D., n = 3). C, the cell lines were assayed for Myc IRES activity by the dicistronic reporter assay as described for Fig. 3. The data represent firefly (FF, black bars) or Renilla (Ren, white bars) luciferase expression (means ± S.D., n = 3). The Ser-to-Asp mutant cell line had a significant (p < 0.05) increase in firefly expression compared with the other cell lines. D, the three cell lines were assayed for c-Myc protein expression by immunoblot assay for Myc versus actin expression. Nyc/actin ratios (by densitometry) are shown for the three cell lines where the WT line is arbitrarily set at “1.” The data are the means ± S.D. (n = 3). The Ser-to-Asp mutant had a significant increase in Myc protein expression (p < 0.05). E, 3T3 cells treated with or without sorbitol (0.6 m) for 2.5 h and Myc IRES function (firefly (FF)/Renilla (REN) luciferase) was assayed. The results are the means ± S.D. (n = 3).
We next asked whether there were any effects on c-Myc IRES activity and protein expression in these MM clones stably expressing the three different A1 constructs. As shown in Fig. 5C, Myc IRES activity in wild type A1-transfected cells was robust, demonstrated by a 25-fold increase in firefly luciferase expression when the Myc IRES is inserted in the intercistronic space (in pRmF). The firefly expression was comparable in the Ser-to-Ala mutant, suggesting that the mutant does not work effectively as a dominant negative. In contrast, a significantly higher degree of Myc IRES activity was observed in the Ser-to-Asp mutant-containing cell line. Because this latter mutant is predominantly localized to the cytoplasm (Fig. 5B), these data further support the contention that A1 subcellular locale has a regulatory effect on Myc IRES activity. As a corollary to these reporter expression data, c-Myc protein expression was modestly but significantly (p < 0.05) enhanced in the Ser-to-Asp mutant-containing MM cell line (Fig. 5D).
Additional support that the degree of A1 localization can regulate Myc IRES activity comes from experiments in osmotically stressed cells. As shown above in MM cells (Fig. 1) as well as in other cell models (14), osmotic shock induced by sorbitol results in accumulation of A1 in the cytoplasm. We attempted to test whether osmotic stress increased IRES activity in MM cells, but MM cell lines treated with 600 mm of sorbitol for 3 h began to show significant death. We thus tested IRES activity in sorbitol-treated NIH 3T3 cells after 2.5 h of exposure, a time when maximal A1 cytoplasmic localization has occurred (14). As shown in Fig. 5E, sorbitol treatment significantly enhanced Myc IRES activity.
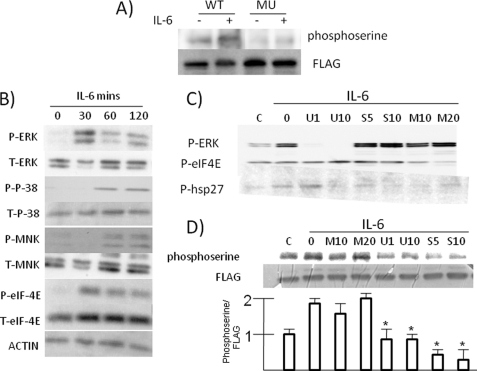
To test for serine phosphorylation of the M9 domain, the above MM cells expressing FLAG-tagged wild type A1 or the Ser-to-Ala mutant were treated with or without IL-6 followed by immunoprecipitation with anti-FLAG antibody and immunoblot with a phosphoserine antibody. As shown in Fig. 6A, there is some constitutive serine phosphorylation of transfected A1 in MM cells that is significantly increased by exposure to IL-6. This increased phosphorylation is abrogated by mutation of the seven M9 domain serines. We also tested whether individual serines are critical targets of IL-6 in the M9 peptide by creating serine-to-alanine point mutations at Ser-310, Ser-311, and Ser-312, as well as at Ser-192, a residue outside the M9 peptide previously implicated as a phosphorylation target of MNK kinase (27). However, none of these individual mutations could prevent serine phosphorylation of A1 (not shown) as was seen by serine-to-alanine mutations of the entire M9 peptide. It is likely that several or all of the serines in the M peptide are phosphorylated in tandem by IL-6 exposure. Nevertheless, these data, along with those above demonstrating that the M9 peptide is critical for cytoplasmic localization of A1, support the notion that IL-6-induced serine phosphorylation in the M peptide of A1 mediates the up-regulated translation of Myc.
FIGURE 6.
IL-6-induced phosphorylation of A1 is mediated by p38. A, ANBL-6 cells, transfected with either FLAG-tagged WT A1 or the serine-to-alanine mutant (MU), were treated with or without IL-6. Subsequently, transfected WT or mutant A1 was immunoprecipitated with anti-FLAG antibody and immunoblotted with an anti-phosphoserine or anti-FLAG antibody. B, ANBL-6 cells were depleted of IL-6 overnight and then re-exposed to IL-6 (100 units/ml) for 0, 30, 60, or 120 min with immunoblot assay performed for expression of phosphorylated ERK (P-ERK), phosphorylated p38 (P-P-38), phosphorylated MNK (P-MNK), phosphorylated eIF-4E (P-eIF-4E), or total (T) amounts of all these kinases. C, IL-6-depleted ANBL-6 MM cells were exposed to an ERK inhibitor at 1 or 10 μm (U1 or U10), a p38 inhibitor at 5 or 10 μm (S5 or S10), or a MNK inhibitor at 10 or 20 μm (M10 or M20) for 30 min followed by the addition of IL-6 for 3 h and immunoblot assay performed for phosphorylated ERK, eIF-4E, and Hsp27. D, IL-6-depleted ANBL-6 cells, stably expressing FLAG tagged A1, were exposed to MNK (M), ERK (U), or P38 (S) inhibitors at the given micromolar concentrations for 30 min followed by readdition of IL-6 (100 units/ml). After 3 h, protein was immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-phosphoserine or anti-FLAG antibody. The bar graph below the gel shows the ratio of phosphoserine:FLAG signal (means ± S.D., n = 3), where control (lane C) cells (no IL-6) are arbitrarily kept at “1.” *, significantly lower signal ratio versus no inhibitor group (second lane), p < 0.05.
Previous studies (16, 18, 28) have indicted a p38 MAPK/MNK kinase-dependent pathway for M9 domain phosphorylation. However, MNK kinases can also be activated by ERK (27). To investigate which pathway was involved in IL-6-stimulated MM cells, we first stimulated ANBL-6 cells with IL-6 and tested for activation of MAPK pathways by immunoblot assays. As shown in Fig. 6B, both p42 and p44 ERK MAPKs became phosphorylated by 30 min of exposure to IL-6. The p38 MAPK and MNK kinases became phosphorylated with similar kinetics, first identified by 60 min of exposure. Because serine 209 on eIF-4E is the specific substrate for MNK kinases, we also tested its phosphorylation, and as shown, phosphorylation was also increased by IL-6.
We next used inhibitors relatively specific for these kinases. Fig. 6C demonstrates their efficacy and specificity. The activity of MNK and p38 kinases was evaluated by assessment of phosphorylation of their respective substrates, eIF-4E and Hsp27. As shown, the U0126 ERK inhibitor prevented phosphorylation of ERK, had no effect on MNK activity (phosphorylation of eIF-4E), and had no effect on p38 activity (phosphorylation of Hsp27) when used at 1 μm. The p38 inhibitor SB203580 specifically inhibited p38 activity without any nonspecific effects on ERK or MNKs, and the MNK inhibitor inhibited MNK kinase activity when used at 20 μm but also nonspecifically inhibited p38 activity. When similar concentrations of these inhibitors were used to test for effects on phosphorylation of transfected A1 (Fig. 6D), it is apparent that the p38 inhibitor had the greatest prohibitive effect. The ERK inhibitor moderately curtailed A1 phosphorylation as well, although it is not clear whether this was due to its nonspecific inhibitory effect on p38 activity. In contrast, the MNK inhibitor had no effect.
DISCUSSION
Dysregulated expression of c-Myc occurs frequently in MM. Recent work (29) supports a role for c-Myc in the progression from the premalignant MGUS syndrome to overt myeloma. Myc expression would theoretically increase proliferation of tumor cells but has also been indicted in myeloma neo-angiogenesis and chemoresistance (30, 31). Dysregulated c-Myc expression in MM can occur as a result of gene translocation with juxtaposition to the Ig enhancer (32). However, such gene translocations occur in only ∼15% of newly diagnosed patients (33). Another potential mechanism of dysregulated Myc transcription is via stimulation of MM cells with the tumor growth factor IL-6. However, the ability of IL-6 to increase Myc RNA is cell type-specific, and IL-6 treatment of some cells results in decreased Myc transcription (34–36). Our previous work (1) documents that IL-6 can also enhance Myc translation in myeloma cells. This is consistent with an earlier report (6) demonstrating up-regulated Myc protein expression in the absence of up-regulated RNA expression in MM. Our earlier work focused on the critical role of hnRNP A1 in mediating IL-6-induced Myc translation in MM cells, which was dependent on the c-Myc IRES in the 5′-UTR of the Myc transcript.
Our results indicate that IL-6-induced activation of c-Myc IRES activity in MM cells is due to post-translational modification of hnRNP A1, resulting in enhanced cytoplasmic localization of the ITAF. Such an effect would increase transit of Myc RNA-bound A1 to a translationally competent subcellular locale and/or increase the accessibility of cytoplasmic hnRNP A1 for binding to the Myc IRES in the cytosol. In either case the result would be a significant increase in Myc translation. The ability of the cytoplasmically confined phosphomimetic Ser-to-Asp mutant to enhance Myc IRES activity and Myc expression is additional support for this contention. Increased IRES function was also present when osmotic stress enhanced cytoplasmic A1 localization. Previous work (8) has shown that hnRNP A1 significantly facilitates Myc IRES activity in vitro, indicating that the role of A1 is not only to participate in nuclear egress of Myc RNA but that the RNA-binding protein also has bona fide ITAF activity, presumably by altering conformation of the Myc IRES for enhanced ribosomal loading. The IL-6 MM growth factor enhances A1 nuclear egress by a p38 MAPK pathway-dependent phosphorylation of the M9 domain, a domain previously shown (14, 26) to regulate A1 transit.
Although our results employing the shuttling-deficient NLS A1 mutant support the notion that the cytoplasmic localizing effect of IL-6 participates in the up-regulation of Myc IRES activity, IL-6 may have additional positive effects on A1. For example, IL-6 treatment consistently increases A1-Myc IRES binding within myeloma cells (1). This is probably due to additional modifications of A1, possibly in its RNA-binding domain. In our preliminary experiments (not shown), the expression of the NLS dominant negative did not prevent this IL-6-induced increase in the ability of endogenous A1 to bind the Myc IRES.
The activation of hnRNP A1 by IL-6 and the requirement of nuclear-to-cytoplasmic shuttling of A1 for MM cell growth responses is reminiscent of the role of hnRNP A1 in BCR-ABL leukemogenesis (13). However, in the latter instance, BCR-ABL predominantly enhances A1 protein expression, and the pro-leukemic effect of A1 required shuttling. In our MM cell models, IL-6 does not up-regulate A1 expression but specifically enhances shuttling with subsequent enhanced translation.
The ability of the NLS dominant negative to prevent Myc protein up-regulation and cell growth was not due to any effects on Myc RNA expression. In addition, the NLS did not affect CD44 alternative splicing or pyruvate kinase isoform expression in parental or K-RAS-mutated ANBL-6 MM cells (potentially affected by pyruvate kinase RNA differential splicing). These results further support the notion that the NLS curtails IL-6 responses in MM cells because of its prevention of A1 transit to cytoplasm rather than any inhibitory effect on endogenous A1 function within the nucleus. Of interest, enhanced PKM2 expression was observed in K-RAS-mutated ANBL-6 cells versus parental ANBL-6 cells transfected with control empty vector (Fig. 4E). These cell lines were studied after depletion of IL-6, and the parental cells containing wild type RAS die off in culture within 14 days after depletion, whereas the K-RAS mutated cells continue to proliferate in the absence of IL-6. These provocative results suggest that pyruvate kinase isoform conversion occurs as a result of RAS mutation and participates in IL-6-independent growth. This possibility is being actively pursued.
Previous studies (18, 28) implicated a p38 pathway in A1 M9 peptide phosphorylation and cytoplasmic localization of hnRNP A1. Our results utilizing inhibitors are consistent with this notion because a p38 inhibitor prevented IL-6-induced phosphorylation. Inhibition of A1 phosphorylation was also demonstrated when MM cells were exposed to an ERK inhibitor. This could be due to the nonspecific effects of the ERK inhibitor on p38 activity, but inhibition with 1 μm, a concentration that did not affect p38 activity, suggests that the ERK MAPK cascade may also participate in the IL-6 response with M9 peptide phosphorylation. The inability of the MNK kinase inhibitor to prevent A1 phosphorylation was not anticipated, because the previous literature (27) suggests that the p38-dependent phosphorylation of A1 is mediated via MNK activation. One possible explanation for these results is that other p38-dependent kinases are regulating the IL-6-induced phosphorylation of A1 in the M9 domain. Genetic silencing of either MNK1 and/or MNK2 will be required to definitively rule out a role for the MNK kinases.
It was curious that our A1 mutant, where serine residues in the M9 peptide were converted to alanines, did not function as a dominant negative construct, at least for effects on endogenous A1-dependent Myc IRES activity (Fig. 5C). In contrast, the NLS mutant, which is retained in the nucleus to a comparable degree, is an effective dominant negative. We addressed the possibility that this was due to differences in binding to the Myc IRES, i.e. the NLS efficiently bound the Myc RNA, sequestering it in the nucleus, whereas the Ser-to-Ala mutant had diminished binding so that endogenous A1 was not restrained in its ability to transport the Myc IRES to the cytoplasm. However, the ability of these mutant A1s to bind the Myc IRES was comparable (supplemental Fig. S1). Although the NLS and Ser-to-Ala mutants are equally nuclear-localized, this nuclear localization probably occurs by different mechanisms. Wild type A1 accumulates in the cytoplasm in sorbitol-stimulated cells because of an osmotic stress-induced decreased interaction with transportin, the nuclear import receptor (14). This induced decrease of transportin binding in osmotic stress-stimulated cells does not occur in the Ser-to-Ala mutant (14), and thus, there is no modulation of nuclear import and an absence of cytoplasmic accumulation. A similar mechanism might explain the inability of the Ser-to-Ala mutant to cytoplasmically localize in IL-6-stimulated MM cells. In contrast, the NLS mutant retains normal nuclear import because of its NLS domain but is unable to exit the nucleus because of its G274A mutation (13). The effects of the nuclear-retained NLS may inhibit the ability of endogenous A1 to transport to the cytoplasm, resulting in the dominant negative effect. In contrast, if the deficiency of the Ser-to-Ala mutant is primarily in its inability to decrease transportin binding within the cytoplasm in IL-6-stimulated cells, this may have little effect on the normal modulation of endogenous A1 binding to transportin, thus explaining the absence of the dominant negative effect.
In summary, our results indicate a critical role for A1 cytoplasmic localization in MM cells during the IL-6 induction of c-Myc translation. Thus, hnRNP A1 should be considered a potential target for therapy. It is overexpressed in malignant cells (37), and A1 silencing promotes cell death specifically in transformed cells with little toxicity to nontransformed cells (38, 39).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA109312 and RO1 CA111448. This work was also supported by research funds from the Department of Defense and the Department of Veterans Affairs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- MM
- multiple myeloma
- IRES
- internal ribosome entry site
- ITAF
- IRES-trans-activating factor
- NLS
- nuclear localization signal
- EV
- empty vector
- WT A1
- wild type hnRNP A1
- BCR-ABL
- breakpoint cluster region-Ableson oncogene
- MNK
- MAP kinase signal-integrating kinase
- FAM
- 5-carboxyfluorescein
- PKM
- pyruvate kinase M gene.
REFERENCES
- 1. Shi Y., Frost P. J., Hoang B. Q., Benavides A., Sharma S., Gera J. F., Lichtenstein A. K. (2008) Cancer Res. 68, 10215–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baird S. D., Turcotte M., Korneluk R. G., Holcik M. (2006) RNA 10, 1755–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hellen C. U., Sarnow P. (2001) Genes Dev. 15, 1593–1612 [DOI] [PubMed] [Google Scholar]
- 4. Stoneley M., Willis A. E. (2004) Oncogene 23, 3200–3207 [DOI] [PubMed] [Google Scholar]
- 5. Subkhankulova T., Mitchell S. A., Willis A. E. (2001) Biochem. J. 359, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paulin F. E., West M. J., Sullivan N. F., Whitney R. L., Lyne L., Willis A. E. (1996) Oncogene 13, 505–513 [PubMed] [Google Scholar]
- 7. Chappell S. A., LeQuesne J. P., Paulin F. E., deSchoolmeester M. L., Stoneley M., Soutar R. L., Ralston S. H., Helfrich M. H., Willis A. E. (2000) Oncogene. 19, 4437–4440 [DOI] [PubMed] [Google Scholar]
- 8. Jo O. D., Martin J., Bernath A., Masri J., Lichtenstein A., Gera J. (2008) J. Biol. Chem. 283, 23274–23287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonnal S., Pileur F., Orsini C., Parker F., Pujol F., Prats A. C., Vagner S. (2005) J. Biol. Chem. 280, 4144–4153 [DOI] [PubMed] [Google Scholar]
- 10. Lewis S. M., Veyrier A., HosszuUngureanu N., Bonnal S., Vagner S., Holcik M. (2007) Mol. Biol. Cell 18, 1302–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piñol-Roma S., Dreyfuss G. (1992) Nature 355, 730–732 [DOI] [PubMed] [Google Scholar]
- 12. Cammas A., Pileur F., Bonnal S., Lewis S. M., Lévêque N., Holcik M., Vagner S. (2007) Mol. Biol. Cell. 18, 5048–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iervolino A., Santilli G., Trotta R., Guerzoni C., Cesi V., Bergamaschi A., Gambacorti-Passerini C., Calabretta B., Perrotti D. (2002) Mol. Cell. Biol. 22, 2255–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allemand E., Guil S., Myers M., Moscat J., Cáceres J. F., Krainer A. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3605–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clower C. V., Chatterjee D., Wang Z., Cantley L. C., Vander Heiden M. G., Krainer A. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1894–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Houvenvan Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Cáceres J. F. (2000) J. Cell. Biol. 149, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Notari M., Neviani P., Santhanam R., Blaser B. W., Chang J. S., Galietta A., Willis A. E., Roy D. C., Caligiuri M. A., Marcucci G., Perrotti D. (2006) Blood 107, 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Michael W. M., Choi M., Dreyfuss G. (1995) Cell 83, 415–422 [DOI] [PubMed] [Google Scholar]
- 19. Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. (1997) J. Cell Sci. 110, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 20. Shi Y., Hsu J. H., Hu L., Gera J., Lichtenstein A. (2002) J. Biol. Chem. 277, 15712–15720 [DOI] [PubMed] [Google Scholar]
- 21. Hamilton B. J., Nagy E., Malter J. S., Arrick B. A., Rigby W. F. (1993) J. Biol. Chem. 268, 8881–8887 [PubMed] [Google Scholar]
- 22. David C. J., Chen M., Assanah M., Canoll P., Manley J. L. (2010) Nature 463, 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M., Cantley L. C. (2008) Nature 452, 181–1886 [DOI] [PubMed] [Google Scholar]
- 24. Matter N., Marx M., Weg-Remers S., Ponta H., Herrlich P., König H. (2000) J. Biol. Chem. 275, 35353–35360 [DOI] [PubMed] [Google Scholar]
- 25. Vincent T., Mechti N. (2004) Leukemia 18, 967–975 [DOI] [PubMed] [Google Scholar]
- 26. Siomi M. C., Eder P. S., Kataoka N., Wan L., Liu Q., Dreyfuss G. (1997) J. Cell. Biol. 138, 1181–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buxadé M., Parra J. L., Rousseau S., Shpiro N., Marquez R., Morrice N., Bain J., Espel E., Proud C. G. (2005) Immunity 23, 177–189 [DOI] [PubMed] [Google Scholar]
- 28. Guil S., Long J. C., Cáceres J. F. (2006) Mol. Cell. Biol. 26, 5744–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chesi M., Robbiani D. F., Sebag M., Affer M., Tiedemann R., Valdez R., Palmer S. E., Haas S. S., Stewart A. K., Fonseca R., Cattoretti G., Bergsagel P. L. (2008) Cancer Cell 13, 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J., Sattler M., Tonon G., Grabher C., Lababidi S., Zimmerhackl A., Raab M. S., Vallet S., Zhou Y., Cartron M. A., Hideshima T., Tai Y. T., Chauhan D., Anderson K. C., Podar K. (2009) Cancer Res. 69, 5082–5090 [DOI] [PubMed] [Google Scholar]
- 31. Greco D., D'Agnano I., Vitelli G. (2006) Int. J. Immunopathol. Pharmacol. 19, 67–79 [PubMed] [Google Scholar]
- 32. Bergsagel P. L., Kuehl W. M. (2001) Oncogene 20, 5611–5622 [DOI] [PubMed] [Google Scholar]
- 33. Avet-Loiseau H., Gerson F., Magrangeas F., Minvielle S., Harousseau J. L., Bataille R. (2001) Blood 98, 3082–3086 [DOI] [PubMed] [Google Scholar]
- 34. Kiuchi N., Nakajima K., Ichiba M., Fukada T., Narimatsu M., Mizuno K., Hibi M., Hirano T. (1999) J. Exp. Med. 189, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Melamed D., Tiefenbrun N., Yarden A., Kimchi A. (1993) Mol. Cell. Biol. 13, 5255–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheung W. C., Van Ness B. (2002) Leukemia 16, 1182–1188 [DOI] [PubMed] [Google Scholar]
- 37. Biamonti G., Bassi M. T., Cartegni L., Mechta F., Buvoli M., Cobianchi F., Riva S. (1993) J. Mol. Biol. 230, 77–89 [DOI] [PubMed] [Google Scholar]
- 38. Patry C., Bouchard L., Labrecque P., Gendron D., Lemieux B., Toutant J., Lapointe E., Wellinger R., Chabot B. (2003) Cancer Res. 63, 7679–7688 [PubMed] [Google Scholar]
- 39. Patry C., Lemieux B., Wellinger R. J., Chabot B. (2004) Mol. Cancer Ther. 3, 1193–1199 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.