Abstract
Voltage-dependent potassium (Kv) channels are tetramers of six transmembrane domain (S1–S6) proteins. Crystallographic data demonstrate that the tetrameric pore (S5–S6) is surrounded by four voltage sensor domains (S1–S4). One key question remains: how do voltage sensors (S4) regulate pore gating? Previous mutagenesis data obtained on the Kv channel KCNQ1 highlighted the critical role of specific residues in both the S4-S5 linker (S4S5L) and S6 C terminus (S6T). From these data, we hypothesized that S4S5L behaves like a ligand specifically interacting with S6T and stabilizing the closed state. To test this hypothesis, we designed plasmid-encoded peptides corresponding to portions of S4S5L and S6T of the voltage-gated potassium channel KCNQ1 and evaluated their effects on the channel activity in the presence and absence of the ancillary subunit KCNE1. We showed that S4S5L peptides inhibit KCNQ1, in a reversible and state-dependent manner. S4S5L peptides also inhibited a voltage-independent KCNQ1 mutant. This inhibition was competitively prevented by a peptide mimicking S6T, consistent with S4S5L binding to S6T. Val254 in S4S5L is known to contact Leu353 in S6T when the channel is closed, and mutations of these residues alter the coupling between the two regions. The same mutations introduced in peptides altered their effects, further confirming S4S5L binding to S6T. Our results suggest a mechanistic model in which S4S5L acts as a voltage-dependent ligand bound to its receptor on S6 at rest. This interaction locks the channel in a closed state. Upon plasma membrane depolarization, S4 pulls S4S5L away from S6T, allowing channel opening.
Keywords: Heart, Ion Channels, Peptides, Potassium Channels, Protein-Protein Interactions
Introduction
Voltage-gated ion channels are ubiquitously expressed in human tissues where they play diverse physiological functions such as generation and modulation of the electrical activity in excitable tissues, myocyte contraction, modulation of neurotransmitter and hormone release, and electrolyte transport in epithelia. Crystallization and x-ray diffraction of both a prokaryote and a mammalian voltage-gated potassium (Kv) channel provided a lot of information on the structure of Kv channels (1–3). Even though these data initiated controversies on the dynamics of the voltage sensor, S4 (4), they provided a new template for investigations on Kv channel molecular characteristics. For example, the structure of the Kv pore domain (S5–S6) turned out to be similar to the pore domain of two-transmembrane domain potassium channels like KcsA (5) and KirBac1.1 (6). Besides structure, the crystallographic analyses of KvAP and Kv1.2 gave insights on the dynamics of channel voltage dependence. Notably, the crystal structure of Kv1.2 is believed to represent the channel in an open state (3), and in this conformation, the S4-S5 linker (S4S5L)7 is interacting with the S6 C terminus (S6T). The authors (3) used homology modeling to infer a closed state structure from the open state structure. In the closed state, the model shows that S4S5L and S6T are also in contact. Those results pointed to a mechanism by which S4S5L are permanently linked to S6, and this link is critical in translating the voltage sensor movement into gate opening or closure (3).
In a few other channels, the S4S5L/S6T interaction seems rather state-dependent. A second-site suppressor yeast screen in the hyperpolarization-activated channel KAT1 suggested that S4S5L and S6T are interacting only in the channel open state (7). In another hyperpolarization-activated channel, cross-linking of S4S5L and S6T prevented the channel from deactivating (8), suggesting that S4S5L and S6T are interacting only when the channel is open. In hERG too, a cross-linking study suggested a state-dependent S4S5L and S6T interaction but, this time, in the closed state (9). In the cited studies, the state-dependent interaction between S4S5L and S6T suggests that this interaction may stabilize the open state of hyperpolarization-activated channels (KAT1, HCN) and the closed state of depolarization-activated channels (hERG).
Scanning mutagenesis studies were performed in a cardiac voltage-gated potassium channel, KCNQ1 (10, 11) to study the role of S4S5L (see companion article (31)) and S6T (12) in the voltage modulation of this channel. These studies identified residues implicated in the S4S5L/S6T interaction. In particular, the V254A mutation in S4S5L and L353A mutation in S6T led to a similar phenotype, i.e. the appearance of a voltage-independent current component, suggesting that mutation of one of these residues decreases the interaction between S4S5L and S6T in the closed state. Thus, as in hERG, specific interaction between S4S5L and S6T residues may also stabilize KCNQ1 channel closed state. This would suggest that S4S5L acts as a ligand, stabilizing the channel closed state, such as ATP stabilizes the KATP channel closed state (13). To test whether S4S5L by itself is a ligand and S6T is its receptor, we have designed plasmids coding for the potential ligand (S4S5L, 16 amino acids) and receptor (S6T, 13 amino acids) and tested their effects on KCNQ1 function. Our results suggest that S4S5L stabilizes the channel in a closed conformation by interacting with S6T. Upon depolarization of the plasma membrane, S4 pulls S4S5L away from S6T, allowing channel opening.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Complementary DNAs encoding KCNQ1 peptides were amplified by polymerase chain reaction using specific primers corresponding to KCNQ1 S4S5L or S6T. The PCR products were cloned into pCRII-TOPO, sequenced, and then cloned into pIRES2-EGFP (Clontech). The 5′-end of the forward primers contained a Kozak consensus sequence followed by an initiation codon (ATG) and a glycine (GGA) to protect the ribosome binding site (14). To construct the pCB6-L353A hKCNQ1, a Not1-BamH1 fragment containing the mutation was cut out of the pIRES2-EGFP-L353A hKCNQ1 (12) and ligated into pCB6-hKCNQ1 to replace the WT sequence. Sequencing of the plasmid confirmed the presence of the mutation and the absence of unwanted mutations.
Cell Culture and Transfection
The African green monkey kidney-derived cell line COS-7 was obtained from the American Type Culture Collection (CRL-1651) and cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum and antibiotics (100 international units/ml penicillin and 100 μg/ml streptomycin) at 5% CO2 and 95% air at 37 °C in a humidified incubator. Cells were transfected in 35-mm plates when the culture reached 60–80% confluence, with DNA (2–5 μg total DNA as described below) complexed with FuGENE 6 (Roche Molecular Biochemical) according to the standard protocol recommended by the manufacturer. In most of our experiments, the peptide effects were studied on a fusion protein of the human KCNE1 linked to the N terminus of the human KCNQ1 (a kind gift of Dr Robert S. Kass, Department of Pharmacology, College of Physicians and Surgeons, Columbia University, New York, NY). In the present study, this pCDNA3.1-KCNE1-KCNQ1 construct is named the KCNE1-KCNQ1 concatemer (E1-Q1). GFP allowed the detection of cells expressing the peptide. For experiments with WT E1-Q1 or S140G E1-Q1 (WT KCNE1 fused to S140G KCNQ1), pIRES-GFP plasmids encoding S4S5L, S6T, or pEGFP plasmids for control cells (1.8 μg per plate) were cotransfected with the human pCDNA3.1-KCNE1-KCNQ1 (0.2 μg per plate) concatemer. In pIRES-GFP plasmids, the second cassette (GFP) is less expressed than the first cassette, guaranteeing high levels of peptides expression in fluorescent cells. For experiments with WT or L353A KCNQ1, the pIRES-GFP plasmids (4 μg per plate) were cotransfected with the human pCB6-KCNQ1 (1 μg per plate). In experiments in which two peptides were simultaneously expressed (see Fig. 7), DNA composition was 0.2 μg of pCDNA3.1-S140G E1-Q1, 0.9 μg pIRES-GFP plasmids encoding S4S5L, and 0.9 μg of pIRES-GFP plasmids encoding S6T (or pCB6-CD4 (15) for control cells). In hERG experiments, COS-7 cells were co-transfected with 0.4 μg of pSI hERG (16) and 1.6 μg of pEGFP (control) or pIRES-GFP plasmids encoding a peptide (S4S5L or S6T).
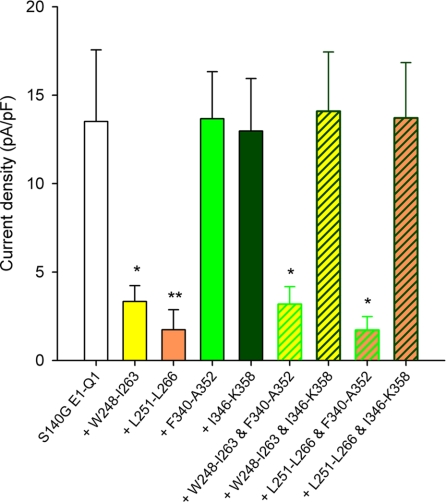
FIGURE 7.
Interaction between S4S5L and S6T peptides probed by S140G. S140G E1-Q1 tail current densities in the absence (S140G E1-Q1) and in the presence of various S4S5L or/and S6T peptides suggesting interactions between Leu251–Leu266 and Ile346–Lys358, Trp248–Ile263 and Ile346–Lys358 and the lack of interaction between Leu251–Leu266 and Phe340–Ala-352, and between Trp248–Ile263 and Phe340–Ala352 (n = 9–13). *, p < 0.05; **, p < 0.01 versus S140G E1-Q1.
Electrophysiology
24 to 72 h after transfection, COS-7 cells were mounted on the stage of an inverted microscope and constantly perfused at a rate of 2 ml/min. The bath temperature was maintained at 24.0 ± 1.0 °C. Stimulation, data recording and analysis were performed by Acquis1 (Bio-Logic Science Instruments) through an analog-to-digital converter (Tecmar TM100 Labmaster; Scientific Solution). Electrodes were connected to a patch-clamp amplifier (RK-400; Bio-Logic Science Instruments). Currents were recorded in the whole-cell configuration. KCNQ1 and hERG activation curves were obtained from the tail currents and fitted by Boltzmann equations. KCNE1-KCNQ1 activation and deactivation kinetics were obtained by a monoexponential fit. If channels are spending more time in deeper closed states, they need to go through more states before reaching the open state. This is reflected by a prolonged delay in channel activation (17). This delay in channel activation was calculated from the shift in the monoexponential fit of the current measured at +80 mV.
Solutions
In ruptured patch experiments, pipettes (Kimble; Vineland, NJ) were filled with the following solution: 150 mm KCl, 10 mm HEPES, 5 mm EGTA and 0.5 mm MgCl2, pH 7.2, with KOH. In permeabilized patch experiments, the pipette solution contained the following: 120 mm K-gluconate, 25 mm KCl, 10 mm HEPES, 1 mm EGTA, 0.8 μg/ml amphotericin B, pH 7.2, with KOH. The standard Tyrode perfusion solution contained the following: 145 mm NaCl, 4 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 5 mm HEPES and 5 mm glucose, pH 7.4, with NaOH.
Kinetic Model
The KCNE1-KCNQ1 kinetic model was derived from the model described by Silva and Rudy (18). This model has been optimized using Model Maker (version 4.0, AP Benson) to fit our experimental data, with an additional constraint: Po is <0.65 at potentials where the channel is fully activated. This constraint comes from the observation that S6 peptide Ile346–Lys358 can increase the maximal current by 50% (see Fig. 2). Consistent with the hypothetical mechanism, endogenous S4S5L linkers are pulled away from the binding site in the closed state preceding channel opening (Fig. 8, A, panel a). Two other models can be then designed (Fig. 8, A, panels b and c), with either S4S5L or S6T peptide binding to this closed state. The three models (control, S4S5L, S6T) were simultaneously optimized for current density, half-activation potential, activation, and deactivation kinetics using Mathematica (Wolfram Research). Optimized transition rates are presented in Table 1.
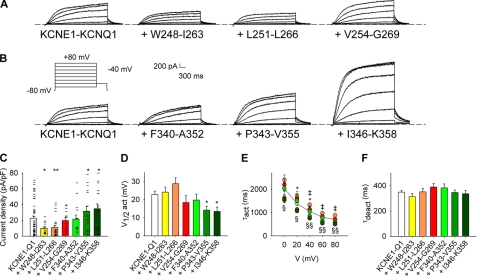
FIGURE 2.
Effects of the “S4S5L linker” or the “S6 C terminus” peptides on KCNE1-KCNQ1 currents. A and B, representative whole-cell recordings of the WT KCNE1-KCNQ1 current in the absence (KCNE1-KCNQ1) and in the presence of S4S5L (A) or S6T peptides (B). Inset, voltage protocol used in A and B, one epoch every 7 s. C–F, average KCNE1-KCNQ1 tail current density at −40 mV after a prepulse at +80 mV (C), half-activation potential (V½act, D), activation time constant at potential from 0 to +80 mV (τact, E), and deactivation time constant at −40 mV (τdeact, F) in the absence and in the presence of various S4S5L or S6T peptides (n = 15–58). In C, current amplitude scatter data is also presented (−). *, Trp248–IIe263; ‡, Leu251–Leu266; §, Pro343–Val355; ¶, Ile346–Lys358. One symbol, p < 0.05; two symbols, p < 0.01. Error bars show the S.E.
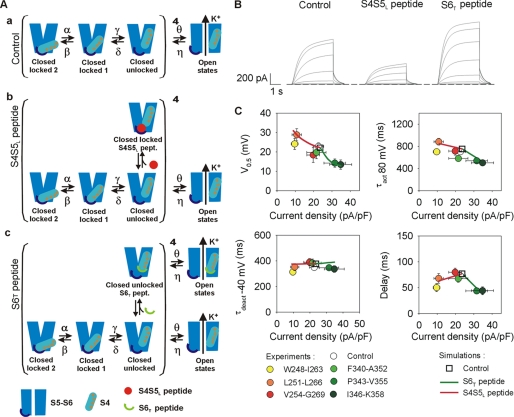
FIGURE 8.
Kinetic model of KCNE1-KCNQ1 and its interactions with S4S5L or S6T peptide. A, kinetic model scheme. This model is based on Rudy's model for the KCNE1-KCNQ1 tetramer (18). We hypothesize that the unbinding of the S4S5L and S6T domains unlocks the channel and corresponds to the last transition of the closed channel (closed locked 1 state to closed unlocked state). Peptides are likely to interact with each monomer in the unlocked states. The model covers three experimental conditions: absence of peptide (panel a), presence of S4S5L peptides (panel b) and presence of S6T peptides (panel c). Optimized transition rates are presented in Table 1. B, simulated currents during step protocols in the absence (Control) or the presence of S4S5L or S6T peptide. C, comparison between simulated and experimental half-activation potentials, activation kinetics, deactivation kinetics, and delay of activation versus current density. The changes of these parameters driven by the peptides are well described by the model. The continuum of simulated results in presence of peptides (solid lines) is obtained by varying the binding rates of the peptides.
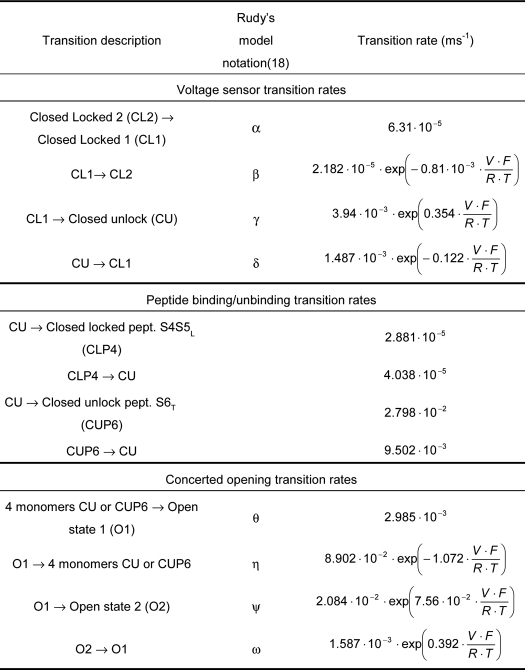
TABLE 1.
Optimized transition rates used in the model presented in Fig. 8
Values are as follows: F = 96,485 C · mol−1 (Faraday constant); R = 8.314 J · mol−1 · K−1 (Gas constant); T = 297 K; and V (membrane potential).
Statistics
All data are expressed as means ± S.E. Statistical differences between samples were determined using Student's t tests, rank sum tests (when data were not normally distributed), and two-way analysis of variance associated with a Tukey test when needed (SigmaStat, version 2.03, SPSS, Inc.). A value of p < 0.05 was considered significant.
RESULTS
S4S5L Linker Peptides Inhibit KCNE1-KCNQ1 Channels
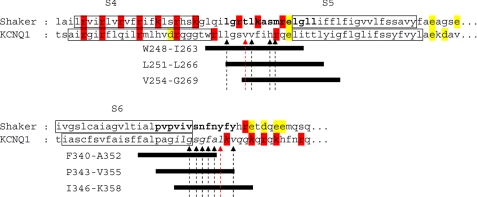
S4S5L and S6T plasmids were designed based on sequence alignment with Shaker (Fig. 1), in which interacting areas in the S4S5L and the S6T were suggested (19, 20). For each region, three different plasmids were designed, with the same length but different starting positions, to compensate for the inaccuracy of the alignment due to the poor similarity between KCNQ1 and Shaker sequences. Black arrows in Fig. 1 show the high impact residues studied in KCNQ1, and red arrows show the interacting residues Val-254 and Leu-353 (12 and see accompanying article (31)). At least one of these residues is lacking in Val254–Gly269 and Phe340–Ala352. For that reason, one can suppose that these peptides will be the least efficient. To study the effect of peptides on channel activity, cDNA coding for a peptide was cotransfected with the KCNE1-KCNQ1 fusion protein cDNA (21). This fusion protein limits variability in the current characteristics caused by variable KCNQ1/KCNE1 expression ratio (22).
FIGURE 1.
Determination of the hypothetical voltage-dependent ligand and its binding site. Shown is an alignment with the Shaker sequence. Boldface letters, interacting area in the S4S5L and the S6 C-terminal parts as suggested by Lu et al. (19, 20). Black bars represent the peptides selected to probe the ligand/binding site hypothesis. Positively and negatively charged residues are presented in red and yellow, respectively. Arrows show the high impact mutations (Ref. 12 and see companion article (31)).
If the endogenous S4S5L peptide is acting like a ligand modulating channel gating, then the exogenous S4S5L peptide should also modulate the channel activity. We examined the peptide effects using the whole-cell configuration of the patch clamp technique, right after patch rupture. We show that two of the three S4S5L peptides (Trp248–Ile263 and Leu251–Leu266) significantly reduced the WT KCNE1-KCNQ1 current (Fig. 2, A and C). Moreover, activation was slowed by both peptides (Fig. 2E), but deactivation was unaffected (Fig. 2F). The absence of effect of peptide Val254–Gly269 is consistent with the fact that this peptide is missing Leu251. This residue seems to play a major role in the channel voltage modulation, as its mutation to an alanine has a high impact on the channel voltage dependence (cf. Fig. 1 and see companion article (31)).
S6 C Terminus Peptides Activate KCNE1-KCNQ1 Channels
If the S6 C terminus is the receptor for S4S5L, then the S6T peptides should decoy the endogenous S4S5L linker, prevent its binding to S6T, and increase channel activity. Indeed, two of them (Pro343–Val355 and Ile346–Lys358) up-regulated the WT channel and shifted the half-activation potential toward negative values (Fig. 2, B–D). Activation was accelerated (Fig. 2E), but deactivation was unaffected (Fig. 2F). The absence of effect of peptide Phe340–Ala352 is consistent with the fact that this peptide is missing Leu353 and Val355. These residues seem to play a major role in the channel voltage modulation because their substitutions to an alanine have a high impact on channel voltage dependence (cf. Fig. 1 and Ref. 12).
Use- and State-dependent Reversibility of Peptide Effects
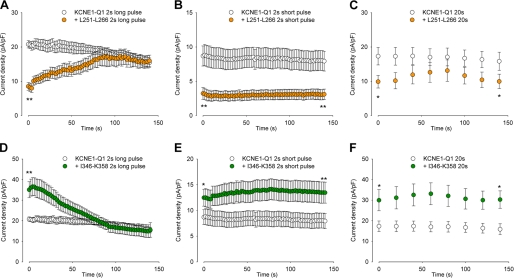
In the following experiments, we studied the most potent inhibiting peptide Leu251–Leu266 and the most potent activating peptide Ile346–Lys358 in greater details. We first analyzed the reversibility of the effects, as peptides were diffusing out of the cytosol and into the pipette after patch rupture (Fig. 3, A and D). In cells expressing KCNE1-KCNQ1 alone, the current was quite stable with a slight rundown (23). In cells coexpressing the channel and the Leu251–Leu266 peptide, the current density increased to reach control values measured in absence of peptide (Fig. 3A). Conversely, in cells coexpressing the channel and the Ile346–Lys358 peptide, the current density decreased to reach control values (Fig. 3D). This suggests that both peptides do not act on channel trafficking but strictly on gating.
FIGURE 3.
Reversibility of the peptides effects. A–C, average KCNE1-KCNQ1 tail-current density measured at −40 mV after a 250-ms to 1-s prepulse at +80 mV, as a function of time after patch rupture, in cells expressing no peptide (KCNE1-KCNQ1) or the Leu251–Leu266 peptide. Both voltage pulse duration (A, 1 s; B, 250 ms) and depolarization frequencies were varied (A, a depolarization every 2 s; C, a depolarization every 20 s). Resting potential was −80 mV (n = 8–38). D–F, same experiments as in A–C, with the Ile346–Lys358 peptide (n = 8–28). *, p < 0.05; **, p < 0.01 versus KCNE1-KCNQ1 alone.
Because any bound peptide (S4S5L or S6T) is most probably intercalated between the endogenous S4S5L and S6T (cf. scheme in Fig. 8A), membrane depolarization, which pulls on the endogenous S4S5L, may free the peptide that is otherwise trapped in the core of the protein. In other words, long and frequent depolarization should favor reversion; shorter or less frequent depolarization should have less effect. To test this, a shorter depolarizing pulse was applied with maintained frequency. In this condition, no reversion was observed for both the inhibiting peptide Leu251–Leu266 (Fig. 3C) and the activating peptide Ile346–Lys358 (Fig. 3E). We also tested a depolarizing pulse with maintained duration but lower frequency. Again, no reversion was observed (Fig. 3, D and F). Altogether, these results suggest that peptide unbinding is favored by membrane depolarization, consistent with the state-dependent channel/peptide interaction.
Effect of Peptides on KCNQ1 without KCNE1
Several studies suggest that KCNE1 interacts with S4S5L (24) and S6T (24, 25). To evaluate the role of KCNE1 in the ligand/receptor mechanism, we coexpressed the most potent peptides (Leu251–Leu266 for S4S5L and Ile346–Lys358 for S6T) with KCNQ1 alone. Like in the presence of KCNE1, we observed a current inhibition with Leu251–Leu266 and an activating effect with Ile346–Lys358 (Fig. 4, A and C). These results suggest that the S4S5L/S6T state-dependent interaction is independent of the presence/absence of KCNE1.
FIGURE 4.
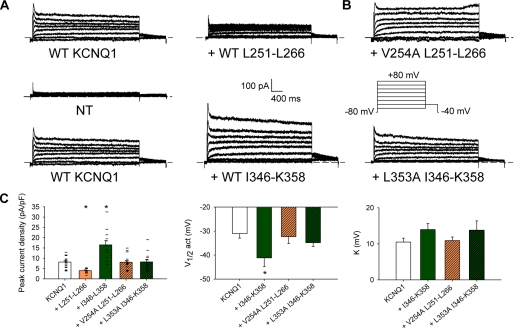
Effects of WT and mutant S4S5L or S6T peptides on KCNQ1 currents. A, representative whole-cell recordings of nontransfected cells (NT) and WT KCNQ1 current in the absence (KCNQ1) and in the presence of the S4S5L peptide (+ Leu251–Leu266) or the S6T peptide (+ Ile346–Lys358). Inset, voltage protocol, one epoch every 7 s. B, representative whole-cell recordings of the WT KCNQ1 current in the presence of a mutant S4S5L peptide (+ V254A Leu251–Leu266) or a mutant S6 C terminus peptide (+ L353A Ile346–Lys358). C, average KCNQ1 peak current density at +80 mV, half-activation potential (V½) and slope (K) in the conditions shown in A and B (n = 5 for nontransfected cells, 10–18 otherwise). In C, current amplitude scatter data are also presented (−). *, p < 0.05 versus KCNQ1 alone.
Mutations That Decrease S4S5L and S6T Interaction also Alter Peptide Effect
Altogether, the data presented above support the hypothesis that S4S5L peptides target the channel S6T and vice versa. To further address the specificity of the interaction between the peptides and the channel, we evaluated the effect of peptides bearing mutations known to decrease S4S5L and S6T interaction. In an accompanying paper, Snyders and co-workers (31) show that the V254A mutation in S4S5L and the L353A mutation in S6T generate an instantaneous current component, suggesting that S4S5L/S6T interaction is weakened by the mutations (see companion article (31)). Indeed, introduction of the V254A mutation into the S4S5L peptide Leu251–Leu266 abolished the peptide inhibition of the channel. Similarly, introduction of the L353A mutation into the S6T peptide Ile346–Lys358 abolished its activating effect. The suppression of both peptide effects (Fig. 4, B and C) supports that the S4S5L peptide is specifically targeting S6T, and vice versa. Looking now at the channel, introduction of the L353A mutation should also render the channel less sensitive to the WT S4S5L peptide. Indeed, the peak current measured in the mutant channel L353A was not decreased when the WT S4S5L peptide was coexpressed (Fig. 5, A and B), also suggesting that the S4S5L peptide is specifically targeting S6T.
FIGURE 5.
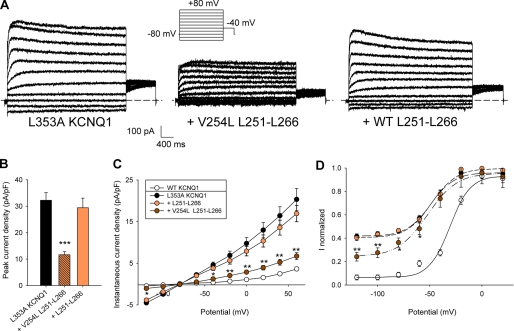
Effects of WT and mutant S4S5L peptides on L353A KCNQ1 currents. A, representative whole-cell recordings of L353A KCNQ1 current in the absence (KCNQ1 L353A) and in the presence of the mutant (+ Leu251–Leu266 V254L) or the WT S4S5L peptide (+ Leu251–Leu266). Inset, voltage protocol, one epoch every 7 s. B, mean peak current density, in the three conditions described in A (n = 10–12). C, instantaneous current density, measured at the beginning of the prepulse, as a function of prepulse potential (n = 10–12). D, activation curve built from the tail currents at −40 mV (n = 10–12). In C and D, besides the three conditions described in A, KCNQ1 data are shown for comparison. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus L353A KCNQ1 alone.
Rescue of L353A Channel Closure by V254L Peptide Further Supports Peptide Specificity
In the companion article (31), the partial open phenotype of the L353A mutant channel could be rescued by the V254L mutation in the channel S4S5L. In parallel, in the experiments with peptides, introduction of the V254L mutation in the S4S5L peptide restored its effect on the L353A mutant channel (Fig. 5, A and B). Moreover, the instantaneous current observed in L353A (cf. companion article (31)) was also reduced by the V254L S4S5L peptide but not by the WT S4S5L peptide (Fig. 5C). This reduction of the instantaneous current could also be observed in the activation curve (Fig. 5D). These experiments strictly mimicked the rescue of the S4S5L/S6T interaction observed within the channel by Labro et al. (see companion article (31)) and is a strong support for a specific interaction between the channel and the peptide.
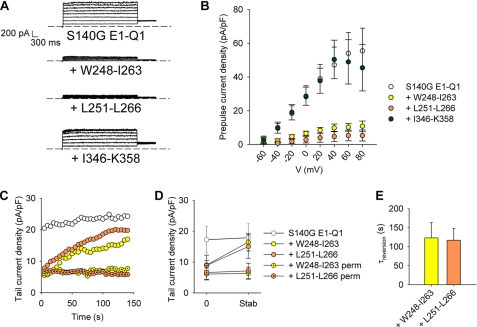
S4S5L Peptide Effect on a Voltage-independent Mutant
The S140G E1-Q1 mutant (WT KCNE1 fused to S140G KCNQ1) does not show any voltage dependence in the −80 to +80 mV range (26). If S4S5L peptides bind to S6T to stabilize the closed state, they should inhibit the mutant channel as well. Fig. 6 shows that coexpression of the S140G E1-Q1 mutant channel with the S4S5L peptides (Trp248–Ile263 or Leu251–Leu266) led to an almost complete inhibition of the current (Fig. 6, A and B), whereas the activating peptides had no effect on the already fully activated S140G channel. In cells expressing the inhibitory S4S5L peptides, patch rupture led to a gradual increase in current density (Fig. 6, C–E), consistent with the peptide dilution into the patch pipette, as observed for the WT channel above. Current recovery did not occur when peptide dilution was prevented using the permeabilized patch configuration (Fig. 6, C and D). It is worth noting that the current increased toward values similar to those observed in the absence of peptide indicating that the peptides affect channel gating but not trafficking. Importantly, the reversion of the peptide effects indicates a loose peptide/channel association consistent with a loose S4S5L/S6T interaction in the channel.
FIGURE 6.
Effects of the S4S5L or the S6T peptides on S140G E1-Q1 currents. A, representative ruptured patch recordings of the S140G E1-Q1 current in the absence (S140G E1-Q1) and in the presence of S4S5L peptides (+ Trp248–Ile263 or + Leu251–Leu266) or a S6T peptide (+ Ile346–Lys358). The voltage protocol was the same as described in the legend to Fig. 2. B, S140G E1-Q1 current density measured on the late current (n = 9–21). C, S140G E1-Q1 tail current density measured at −40 mV after a prepulse at +40 mV during dilution of S4S5L peptides and in permeabilized configuration (perm) (n = 8) in representative cells. Voltage protocol was from a holding potential of −80 mV, one voltage step to +40 mV (1000 ms) and then back to −40 mV (500 ms), every 5 s. D, mean tail-current density measured as in C, immediately after patch rupture/permeabilization (0) and after current stabilization (stab). E, reversion time constant (τreversion) determined from a monoexponential fit of the run-up in the conditions presented in C (n = 6–8).
S4S5L and S6T Peptides Interact with Each Other
Our observations suggest an interaction between S4S5L and S6T in the channel protein. Accordingly, when the two peptides are coexpressed, they should bind to each other and annihilate their respective effect. To confirm this, we used the S140G E1-Q1 mutant, which is sensitive to the S4S5L but not the S6T peptides. When both a S4S5L (Trp248–Ile263 or Leu251–Leu266) and a S6T peptide (Ile346–Lys358) were coexpressed with the channel, the inhibition of S140G E1-Q1 by a S4S5L peptide was not observed (Fig. 7). This loss of inhibition is consistent with the idea that the S6T peptide Ile346–Lys358 associates with the S4S5L peptide and competitively prevents S4S5L peptide association with the channel S6T. Hence, S4S5L and S6T peptides that bind the WT channel can also bind to each other. Interestingly, the effects of S4S5L peptides are not prevented by the S6T peptide Phe340–Ala352 that does not up-regulate the WT KCNE1-KCNQ1 channel either. This suggests that the S6T peptide that does not bind the WT KCNE1-KCNQ1 channel cannot bind the S4S5L peptide either, further confirming the specificity of the S4S5L/S6T interaction.
S4S5L/S6T Interaction Is Channel-specific
To establish the channel specificity of the S4S5L peptide Leu251–Leu266 and the S6T peptide Ile346–Lys358 for KCNE1-KCNQ1, we tested their potential effects on hERG, another voltage-gated potassium channel. As shown in the supplemental Fig. 1, KCNQ1 S4S5L and S6T peptides had no effect on hERG current amplitude and half-activation potential, confirming that peptides are channel-specific.
A Kinetic Model Describes Observed Data for All Peptides
To test the consistency between the hypothesis and the altered biophysical parameters in the presence of the peptide, we developed a kinetic model to describe the peptide-induced alterations in KCNE1-KCNQ1 channel properties (18). We deliberately made the peptides bind only the closed state immediately preceding the concerted opening (Fig. 8, A, panels b and c) because, in the other closed states, the proximity of the endogenous S4S5L and S6T is supposed to prevent peptides access. In the model, the S4S5L peptide mimics the endogenous S4S5L linker and locks the channel in a closed state. Conversely, the S6T peptide decoys the endogenous S4S5L peptide and limits this locking. This theoretical model fits the observed data; the peptides affect not only the current amplitude but also the half activation potential and the activation kinetics, and they do not affect the deactivation kinetics (Fig. 8, B and C). The kinetic model also illustrates that peptides binding prevents the channel from going to deep closed state (especially closed locked 2 in Fig. 8A). This should provoke a reduction in the delay of channel activation (17). Indeed, the delay measured in the current recordings was similar to the delay calculated in the model, further validating the model and the specificity of the peptides effects (Fig. 8C).
DISCUSSION
Altogether, our results show that S4S5L acts as an inhibiting ligand that binds with low affinity to S6T only at negative potentials, locking the S5-S6 pore domain in a closed state. The state-dependent interaction between S4S5L and S6T has already been proposed by Ferrer et al. (9) for hERG, another delayed rectifier potassium channel. This idea was suggested by the fact that a covalent link between S4S5L and S6T stabilizes the hERG closed state (9). If S4S5L and S6T interact in both the closed and open states, a covalent interaction should not lead to channel closure. If, conversely, S4S5L interacts with S6 only in the closed state, one expects that forcing S4S5L interaction with S6 closes the channel.
In the present study, we propose a similar state-dependent interaction between S4S5L and S6T in KCNQ1 channels and provide a mechanistic scheme in which a ligand attached to S4 (S4S5L) interacts with the pore and locks it closed in a voltage-dependent fashion. This mechanism supports the functional model in which the six-transmembrane domain KCNQ1 channels may be regarded as the fusion of a voltage sensor domain (S1-S4) and a ligand-gated channel (S5-S6) very similar to a two-transmembrane domain channel.
In this model, the vicinity between S4S5L and S6T in the closed state suggests that the interaction between S4S5L and S6T is loose enough to allow S4S5L/S6T unbinding upon depolarization. We tried to confirm this by biochemical methods using synthetic MG Leu251–Leu266 and MG Ile346–Lys358 peptides, but we could not solubilize the peptides at concentrations >100 μm and did not observe any interaction using differential ultracentrifugation and native gels. One possibility is that the cytosol provides an environment to solubilize the peptide in the millimolar range and that this range must be reached to uncover the peptide-peptide or peptide-channel interaction. Importantly, proteins expressed under the control of a CMV promoter can reach expression levels in the millimolar range (27, 28). Yeast two hybrid technique was also used to probe the S4S5L and S6T peptides interaction in a “cytosolic” environment but no interaction between Leu251–Leu266 and Ile346–Lys358 could be detected. The later result is probably because of the weakness of the interaction and the poor sensitivity of the technique. Nevertheless, although chemical and biochemical approaches failed to demonstrate direct S4S5L/S6T interaction, our experiments analyzing the effects of Val254 and Leu353 point mutations (Figs. 4 and 5), the S4S5L/S6T peptide interaction (Fig. 7), and channel specificity (supplemental Fig. 1) strongly support this interaction. Importantly, all of these results are consistent with a loose interaction between S4S5L and S6T.
Interestingly, the ligand/receptor mechanism we propose in this study may explain the surprising observation that hyperpolarization-activated channels have a similar voltage sensor domain as depolarization-activated channels but an opposite response to depolarization. In the hyperpolarization-activated channel KAT1 (7) and HCN (29), S4S5L has also been suggested to couple voltage sensing and pore opening by a labile binding (29). Similar to our model where S4S5L binds S6T in the closed state, the S4S5L/S6 interaction seems present only in the open state of KAT1 and HCN (7, 8). This would explain why the latter channels are closed by membrane depolarization. The state-dependent interaction between S4S5L and S6T observed in hERG, KAT1, HCN, and now in KCNQ1 channels suggest that the mechanism described here may be applied to other voltage-gated potassium channels and probably sodium and calcium channels, which share the same overall structure with Kv tetramers.
In summary, the activation mechanism suggested here describes the pore domain as being locked closed at rest by the binding of a ligand (S4S5L) to the gate (S6T). Upon plasma membrane depolarization, S4 pulls S4S5L away from S6T, allowing channel opening. Other molecular mechanisms will have to be taken into account to fully understand the complex voltage dependence, like the molecular events accompanying the concerted channel opening triggered by S4 movement (30).
Generally, application of the ligand/receptor mechanism to voltage-gated ion channels may lead to the design of peptides specifically down- or up-regulating one specific type of ion channel. This may open new therapeutic avenues to treat channelopathies.
Supplementary Material
Acknowledgments
We thank Béatrice Leray, Marie-Joseph Louérat, and Agnès Carcouët for expert technical assistance.
This work was supported in part by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), from the Agence Nationale de la Recherche (ANR-05-JCJC-0160-01), the Hubert Curien Program (Tournesol program), and the Association Française contre les Myopathies (Grant 14120 to G. L.), and from Vaincre La Mucoviscidose (to J. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- S4S5L
- S4-S5 linker
- S6T
- S6 C terminus
- hERG
- human ether-a-go-go-related gene
- HCN
- hyperpolarization-activated cyclic nucleotide-gated channel.
REFERENCES
- 1. Jiang Y., Ruta V., Chen J., Lee A., MacKinnon R. (2003) Nature 423, 42–48 [DOI] [PubMed] [Google Scholar]
- 2. Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. (2003) Nature 423, 33–41 [DOI] [PubMed] [Google Scholar]
- 3. Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 903–908 [DOI] [PubMed] [Google Scholar]
- 4. Ahern C. A., Horn R. (2004) Trends Neurosci. 27, 303–307 [DOI] [PubMed] [Google Scholar]
- 5. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 6. Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. (2003) Science 300, 1922–1926 [DOI] [PubMed] [Google Scholar]
- 7. Grabe M., Lai H. C., Jain M., Jan Y. N., Jan L. Y. (2007) Nature 445, 550–553 [DOI] [PubMed] [Google Scholar]
- 8. Prole D. L., Yellen G. (2006) J. Gen. Physiol. 128, 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrer T., Rupp J., Piper D. R., Tristani-Firouzi M. (2006) J. Biol. Chem. 281, 12858–12864 [DOI] [PubMed] [Google Scholar]
- 10. Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) Nature 384, 78–80 [DOI] [PubMed] [Google Scholar]
- 11. Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Nature 384, 80–83 [DOI] [PubMed] [Google Scholar]
- 12. Boulet I. R., Labro A. J., Raes A. L., Snyders D. J. (2007) J. Physiol. 585, 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. John S. A., Weiss J. N., Xie L. H., Ribalet B. (2003) J. Physiol. 552, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilchrist A., Li A., Hamm H. E. (2002) Sci. STKE. 2002, PL1 [DOI] [PubMed] [Google Scholar]
- 15. Le Maout S., Welling P. A., Brejon M., Olsen O., Merot J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10475–10480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bellocq C., Wilders R., Schott J. J., Louérat-Oriou B., Boisseau P., Le Marec H., Escande D., Baró I. (2004) Mol. Pharmacol. 66, 1093–1102 [DOI] [PubMed] [Google Scholar]
- 17. Cole K. S., Moore J. W. (1960) Biophys. J. 1, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silva J., Rudy Y. (2005) Circulation 112, 1384–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu Z., Klem A. M., Ramu Y. (2001) Nature 413, 809–813 [DOI] [PubMed] [Google Scholar]
- 20. Lu Z., Klem A. M., Ramu Y. (2002) J. Gen. Physiol. 120, 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang W., Xia J., Kass R. S. (1998) J. Biol. Chem. 273, 34069–34074 [DOI] [PubMed] [Google Scholar]
- 22. Park K. H., Piron J., Dahimene S., Mérot J., Baró I., Escande D., Loussouarn G. (2005) Circ. Res. 96, 730–739 [DOI] [PubMed] [Google Scholar]
- 23. Loussouarn G., Baró I., Escande D. (2006) Methods Mol. Biol. 337, 167–183 [DOI] [PubMed] [Google Scholar]
- 24. Kang C., Tian C., Sönnichsen F. D., Smith J. A., Meiler J., George A. L., Jr., Vanoye C. G., Kim H. J., Sanders C. R. (2008) Biochemistry 47, 7999–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng R., Thompson K., Obeng-Gyimah E., Alessi D. M., Chen J., Cheng H., McDonald T. V. (2010) Biochem. J. 428, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y. H., Xu S. J., Bendahhou S., Wang X. L., Wang Y., Xu W. Y., Jin H. W., Sun H., Su X. Y., Zhuang Q. N., Yang Y. Q., Li Y. B., Liu Y., Xu H. J., Li X. F., Ma N., Mou C. P., Chen Z., Barhanin J., Huang W. (2003) Science 299, 251–254 [DOI] [PubMed] [Google Scholar]
- 27. Lin P., Luby-Phelps K., Stull J. T. (1999) J. Biol. Chem. 274, 5987–5994 [DOI] [PubMed] [Google Scholar]
- 28. Rex T. S., Peet J. A., Surace E. M., Calvert P. D., Nikonov S. S., Lyubarsky A. L., Bendo E., Hughes T., Pugh E. N., Jr., Bennett J. (2005) Mol. Vis. 11, 1236–1245 [PubMed] [Google Scholar]
- 29. Chen J., Mitcheson J. S., Tristani-Firouzi M., Lin M., Sanguinetti M. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11277–11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soler-Llavina G. J., Chang T. H., Swartz K. J. (2006) Neuron 52, 623–634 [DOI] [PubMed] [Google Scholar]
- 31. Labro A. J., Boulet I. R., Choveau F., Mayeur E., Bruyns T., Loussouarn G., Raes A. L., Snyders D. J. (2010) J. Biol. Chem. 285, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.