Abstract
Molecular chaperones and co-chaperones are crucial for cellular development and maintenance as they assist in protein folding and stabilization of unfolded or misfolded proteins. Prefoldin (PFDN), a ubiquitously expressed heterohexameric co-chaperone, is necessary for proper folding of nascent proteins, in particular, tubulin and actin. Here we show that a genetic disruption in the murine Pfdn5 gene, a subunit of prefoldin, causes a syndrome characterized by photoreceptor degeneration, central nervous system abnormalities, and male infertility. Our data indicate that a missense mutation in Pfdn5, may cause these phenotypes through a reduction in formation of microtubules and microfilaments, which are necessary for the development of cilia and cytoskeletal structures, respectively. The diversity of phenotypes demonstrated by models carrying mutations in different PFDN subunits suggests that each PFDN subunit must confer a distinct substrate specificity to the prefoldin holocomplex.
Keywords: Chaperone Chaperonin, Mouse Genetics, Neurodevelopment, Photoreceptors, Protein Folding
Introduction
For essential cellular functions such as cell division, motility, molecular transport, cytoskeletal stability, and biological signal transduction, correct and timely synthesis of actins and tubulins is critical. This fundamental process is carried out by the chaperone proteins, prefoldin (PFDN)2 and chaperonin containing TCP1 (CCT), a eukaryotic group II chaperonin. In eukaryotes, prefoldin is a heterohexameric co-chaperone consisting of two α-like (PFDN3 and -5) and four β-like (PFDN1, -2, -4, and -6) subunits (1, 2) that form a jellyfish like structure with coiled-coil tentacles that participate in substrate binding. This unique conformational structure is thought to confer selective substrate specificity for nonnative target proteins. PFDN stabilizes nascent proteins as they are translated and delivers them to the CCT (3). The CCT consists of eight subunits that form a cylindrical pocket that creates an environment conducive for substrate folding. In yeast, the CCT folding rate increases by 5-fold in the presence of PFDN, in part, by preventing the premature release of nonnative proteins from the CCT (4). It is not surprising, therefore, that mutations within the subunits of prefoldin and of the CCT lead to similar phenotypes that affect cellular development and growth (1, 5–7). For example, CCT subunit mutations in Saccharomyces cerevisiae cause cytoskeletal disruption, temperature sensitivity, and growth retardation resulting from disrupted actin and tubulin synthesis (5, 7, 8). Similarly, deletion of single or multiple subunits of prefoldin in yeast results in increased sensitivity to microtubule destabilizing agents and to low temperature stress, and retarded growth with disruption in actin/tubulin-based cytoskeletal structures (1, 3, 6, 9, 10). In Caenorhabditis elegans, reduction of functional prefoldin subunits 1, 2, 3, and 6 by RNAi result in developmental defects such as improper cell division that can lead to embryonic lethality (11–13). In Arabidopsis, prefoldin 6 is necessary for proper microtubule organization and dynamics (14, 15).
Whereas the studies above provide important information regarding the function of prefoldin in lower eukaryotes, the effects of prefoldin mutations in mammals are only now coming to light. Sakono et al. (16) suggested that improper function of prefoldin may be involved in the formation of toxic soluble β-amyloid oligomers, a common pathology observed in neurodegenerative diseases. In support of this notion, a number of human neurodegenerative diseases associated with genetic mutations in actin, tubulin, tubulin-associated proteins, and chaperones have been described (15, 17–21). Recently, a prefoldin subunit-1-null mouse model demonstrating a disruption in cytoskeletal structure has been reported (22). Loss of nerve bundles, hydrocephaly, mucus clearance defects, abnormal lymphocyte development, and function, and a shortened life span were observed in Pfdn1−/− mice (22).
In this report, we describe a mouse model bearing a missense mutation in prefoldin subunit-5 (Pfdn5) generated by N-ethyl-N-nitrosurea (ENU) mutagenesis. Despite its broad tissue expression, Pfdn5nmf5a homozygotes, surprisingly, show fairly restricted neuronal defects, which result in progressive neurodegeneration and hydrocephalus, and in reproductive abnormalities. Although there are some similarities in disease phenotypes between Pfdn1-null (22) and Pfdn5 mice with a missense mutation, the differences provide important insight into the unique function of prefoldin subunits. Reduced availability of microfilament and microtubules, necessary for development of cellular structures, is likely to contribute to the cytoskeletal and ciliary abnormalities that underlie the disease pathologies observed in Pfdn5nmf5a mutants.
MATERIALS AND METHODS
Animals
Mice were bred and housed in standardized conditions in the Research Animal Facility at The Jackson Laboratory and monitored regularly to maintain a pathogen-free environment. Mice were provided free access to NIH 6% fat chow and acidified water in a vivarium with a 12:12 h dark-light cycle. Experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee. C57BL/6 (B6)-Scn8a5J;nmf5a/+ and DBA2/J mice were used in the initial mapping and subsequent high-resolution mapping cross. B6-Pfdn5nmf5a mice were used for molecular and cellular phenotypic studies. Homozygous C57BL/6- Pfdn5nmf5a/Pfdn5nmf5a mice are referred to as Pfdn5nmf5a or nmf5a mutants throughout the text.
Fundus Photography
Mouse pupils were dilated with atropine and fundi examined by indirect ophthalmoscopy with a 60 diopter aspheric lens. Fundus photographs were taken with a Kowa fundus camera using a Volk superfield lens held 2 inches from the eye.
Light Microscopy
Enucleated eyes were fixed in a methanol/acetic acid (3:1) solution overnight, embedded in paraffin, and sectioned in a plane to include the ora serrata and optic nerve, and stained with hematoxylin and eosin (H&E). Mice were perfused with 4% paraformaldehyde prior to dissection of brain tissue. Brains were embedded in paraffin, and sectioned in a sagittal plane and stained with H&E or with cresol luxol fast blue.
Immunohistochemistry
Mice homozygous for the Pfdn5nmf5a mutation and wild-type, age-matched controls were sacrificed by carbon dioxide asphyxiation. For paraffin sections, eyes were enucleated and placed in methanol acetic acid (3:1) fixative overnight and embedded in paraffin. In longitudinal studies, a minimum of three eyes from different animals for each genotype was used per time point. Sections of 6-μm thickness were cut and mounted on slides pretreated with Vectabond (Vector Laboratories). After blocking with 5% normal horse serum in PBS, sections were incubated overnight with the following antibodies: anti-acetylated-α tubulin (Sigma), anti-β-actin (Sigma), anti-β-tubulin (Abcam), anti-β-calbindin (Sigma), anti-caspase 3 (Cell Signaling Technology), anti-PFDN5 (Santa Cruz Biotechnology), anti-rhodopsin (Leinco Technology), anti-ROM1 (a kind gift from R. McInnes, Toronto, Canada), and anti-ZO-1 (Zymed Laboratories Inc.) at a dilution of 1:200. Secondary antibodies were Cy3-conjugated anti-rabbit IgG (1:200, Vector Laboratories) for β-tubulin, ROM1, ZO-1, caspase 3, and PFDN5, and Cy3-conjugated anti-mouse IgG (1:250, Vector Laboratories) for acetylated α-tubulin, β-actin, calbindin, and rhodopsin. Nuclear counterstaining was performed with 4,6 diamidine 2-phenylindoledihydrochloride (DAPI) at a final concentration of 5 μg/ml. Images were collected on a Leica DMRXE fluorescent microscope (Leica, Deerfield, IL) equipped with a SPOTTM CCD camera (Diagnostic Instruments, Sterling Heights, MI) using appropriate bandpass filters for each fluorochrome.
Chromosomal Localization and Fine Mapping
DNAs of 32 affected backcross (BC) offspring between (C57BL/6J x DBA/2J)F1-Scna85J;nmf5a/+ and C57BL/6J- Scna85J;nmf5a/+ or C57BL/6-nmf5a/+ were subjected to a genome-wide scan using simple sequence length polymorphic (SSLP) markers to confirm the original map position on Chr 15. DNAs from 245 BC progeny were subsequently tested to identify additional recombinants to narrow the critical interval of the disease locus. BC mice that were recombinant between the flanking markers but unaffected were progeny tested to determine if they were heterozygous for the nmf5a allele. A minimum of 20 offspring from each progeny test was genotyped and phenotyped. DNAs from 59 informative, recombinant mice were subjected to further genetic marker analyses to generate a high-resolution map and to narrow the critical interval around the disease locus. For PCR amplification, 25 ng of DNA was used in a 10-μl volume containing 50 mm KCl, 10 mm Tris-Cl, pH 8.3, 2.5 mm MgCl2, 0.2 mm oligonucleotides, 200 μm dNTP, and 0.02 units of AmpliTaq DNA polymerase. The reactions which were initially denatured for 2 min at 95 °C were subjected to 49 cycles of 20 s at 94 °C, 20 s at 50 °C, 30 s at 72 °C, and a 7-min extension at 72 °C. PCR products were separated by electrophoresis on a 4% MetaPhor (FMC, ME)-agarose gel and visualized under UV light after staining with ethidium bromide. Mutant and wild-type mRNA was obtained from whole-eye tissues using TRIzol, according to the manufacturer's directions (Invitrogen, Grand Island, NY), and template cDNA was generated with oligo(dT) primers. We designed exon-spanning primer pairs and used them to amplify the coding region of Pfdn5. RT-PCR was performed in an MJ Research PTC-200 using the following program: initial denaturation at 94 °C for 2 min; then 35 cycles of 94 °C for 30 s, 56 °C for 30 s, and 68 °C for 2 min. The resulting product was electrophoresed on a 1% agarose gel and isolated using a NucleoSpin Extraction kit (Clontech, Palo Alto, CA). The purified DNA was sequenced using Big Dye Terminator Cycle Sequencing Chemistry on a PE Applied Biosystems 3600 using the amplification primers.
Electron Microscopy
For transmission electron microscopy (TEM), eyes were fixed for 3 h in a cold, phosphate-buffered glutaraldehyde-paraformaldehyde fixative. For the eye, the anterior segment was removed and the posterior segment cut into 1- × 2-mm blocks. The dissected tissue was placed in fresh fixative for an additional 2–8 h and was post-fixed in 1% osmium tetroxide, dehydrated, and embedded in plastic. Thick sections were first cut for orientation, and then thin sections were cut and stained with uranyl acetate and lead citrate and examined using a transmission electron microscope. For scanning electron microscopy (SE), brains cut longitudinally were fixed in 2.5% glutaraldehyde in cacodylate buffer overnight. Tissues were rinsed twice in cacodylate buffer, dehydrated, and freeze fractured with liquid nitrogen. Tissues were then rehydrated, treated with osmium tetroxide, dehydrated, and sputter-coated. Specimens were mounted and examined by SE.
Electroretinography
After dark-adaptation for at least 2 h, mice were anesthetized with ketamine (80 mg/kg) and xylazine (16 mg/kg). The pupils were dilated by administration of 1% cyclopentolate. Body temperature was maintained at 37 °C using a heating pad. Retinal responses were recorded as described previously (23).
Videomicroscopic Analysis of Ependymal Cilia Function
Brains were removed from the skull cavity immediately after euthanasia by decapitation and kept in prewarmed sterile L-15 oocyte medium (Specialty Media, Phillipsburg, NJ) at room temperature. The cerebellum was removed, as were small (3 mm) edge regions of the frontal cortex and the left and right cortical hemispheres. A mid-sagittal cut along the longitudinal fissure was made to reveal the ventricles. The ependymal cells, which make up the epithelial membrane lining the ventricles, were exposed by making a parallel cut along the ventricle and these were flat mounted onto a glass slide. 50 μl of a 0.01% suspension of green fluorescent latex beads (0.046 μm, Sigma) were placed on the ependymal cells of the brain explants. The movement of green fluorescent beads was video recorded at a rate of 3–10 frames per second in a continuous stream mode. Images were collected on a Princeton Instrument MicroMax-cooled CCD mounted on a Zeiss Axiovert 200 microscope. Recordings were done using autoscale to maximize brightness and a bin of 2 in the MetaMorph software (Universal Imaging, Ypsilanti, MI). The software traced the movement of six selected beads per sample. The distance that the beads traveled was divided by the time elapsed to obtain the rate of ciliary beating. Because movement of the beads was slower in mutants than in controls, the time intervals assessed were ∼3 and 11 s, respectively. Six mutant and age matched controls between postnatal day (P) 84 to P112 of age were used in these analyses.
Atomic Model of the Complex between Eukaryotic PFDN and Unfolded Actin
The atomic model of eukaryotic PFDN was generated using as a template the atomic structure of the archaeal PFDN from M. thermoautotrophicum (24) with the sequences from the six PFDN subunits from mouse and the subunit topology established by Torrey-Simons et al. (25). To model the atomic structure of the complex between eukaryotic PFDN and the unfolded actin, the atomic structure of actin (26) opened to fit the three-dimensional reconstruction of actin bound to nucleotide-free CCT (27) and the atomic model of the eukaryotic PFDN were docked visually using ‘”O” software (28) into the corresponding part of the three-dimensional reconstruction of the PFDN:actin complex (3).
Yeast Growth Studies
Wild-type and mutant (T1362G) Pfdn5 PCR products with blunt ends were prepared using a 5′-primer containing a Kozak translation initiation sequence and a 3′-primer: Forward primer: (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCGCAGTCGATTAAC-3′), and reverse primer: (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGGCTTTGACCGTGGCCTGCG-3′). Each product was cloned into the pENTR TOPO vector using a pENTR directional TOPO cloning kit (Invitrogen) and transformed into One Shot competent Escherichia coli (Invitrogen) for a miniprep (Qiagen). The purified entry vector was used to insert the targeted genes into pDEST32 using the LR Recombination Reaction Method (Invitrogen). Each recombination reaction was transformed into competent TOPO10 cells (Invitrogen). Each pDEST32 vector containing wild-type or mutant Pfdn5 was transformed into MaV203 yeast cells. Transformed yeast cells were grown on SC-L agar plates for selection. For the growth assay, the initial concentration at A600 of each yeast sample was 0.1 optical density (OD). Growth curves were obtained at 30 °C by recording the OD of each sample at 600 nm (Spectronic 601, Bausch & Lomb, Rochester, NY) every hour until growth reached a plateau. For growth under stress conditions, yeast cells were exposed to cold temperature at 20 °C or chemicals such as 1.2 m KCl, 10 mm Nocodazole, or 1.8 sorbitol in YPAD medium, or in SC-L plates.
Quantitative Western Analyses of Retinal Microtubules
The protocol for isolation of microtubules in Microtubule Protocols (29) was followed with slight modification. In brief, harvested retinas were homogenized in 250 μl of paclitaxel microtubule stabilization buffer (100 mm PIPES buffer, pH 6.9, 5 mm MgCl2, 1 mm EGTA, 30%(v/v) glycerol, 0.1% Nonidet P40, 0.1% Triton X-100, 0.1% Tween 20, 0.1% β-mercaptoethanol, 0.001% Antifoam, 0.2% ethanethiol (BME), 1 μg/ml pepstatin, 1 μg/ml leupeptin, 10 μg/ml benzamidine, 500 μg/ml tosylargininemethylester, and 10 mm paclitaxel] for 30 s using a homogenizer (PowerGen 125, Fisher Scientific, Hanover Park, IL). Microtubules were separated by centrifugation of the lysate at 150,000 × g for 10 min using a TL100 Tabletop Centrifuge (Beckman). All procedures were done at 4 °C. The supernatant was removed into a new tube. 50 μl of 4× SDS sample buffer and an additional 150 μl of paclitaxel microtubule stabilization buffer was added to the supernatant, and boiled for 2 min before loading onto a 10% gel. The pellet was also resuspended in 400 μl hot loading buffer (350 μl of paclitaxel microtubule stabilization buffer and 50 μl of 4× SDS sample buffer) and boiled 2 min prior to electrophoresis. Electrophoresis was performed in MOPS buffer (Invitrogen) at 200 V for 1 h, and protein bands were transferred to a nitrocellulose membrane (Millipore, Temecula, CA) at 100 V for 1 h. Membranes were probed with anti-α-acetylated tubulin (Sigma), anti-β-actin (Sigma), and anti-synaptotagmin (Synetic Systems Goettengen, Germany) antibodies as primary antibodies with horseradish peroxidase anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA) as the secondary antibody. Density of bands of tubulin and actin was analyzed using ImageJ and normalized against the band density of synaptotagmin.
Connecting Cilia (CC) and Photoreceptor Cell Body Counts
After immunohistochemistry with acetylated α-tubulin, all CC stained in a 500-pixel area along the RPE were counted. For photoreceptors, all DAPI-positive nuclei within the same area were counted. Single sections in which the diameter of the optic nerve head was at its widest from at least three different mice were assessed for mutant and control groups. ImageJ was used to assist in the analyses.
Tunel Assay
TUNEL assays were performed according to manufacturer's directions (Chemicon, Temecula, CA). Briefly, after de-paraffination, tissue sections were washed once with 1× PBS and digested with 20 μg/ml proteinase K for 12 min. After two washes with 1× PBS, 75 μl/5 cm2 equilibration buffer (Chemicon) was applied directly on the sections and incubated for 15 s. After removing excess liquid, 55 μl/5 cm2 of working strength TdT enzyme was applied and the sections incubated in a humidified chamber at 37 °C for 1 h. The reaction was stopped by incubating the specimens in Working Strength Stop/Wash Buffer (Chemicon) for 10 min at room temperature. After washing three times with 1× PBS, 65 μl/5 cm2 of anti-digoxigenin conjugate was applied to the sections, and sections were incubated in a humidified chamber for 30 min at room temperature. Sections were washed three times with 1× PBS and 4′-6′ diamino-2-phenylindole (DAPI) was applied for nuclear staining.
RESULTS
A Point Mutation in the Prefoldin 5 (Pfdn5) Gene Leads to the nmf5a Mouse Phenotypes
The Neuroscience Mutagenesis Facility (NMF) at The Jackson Laboratory identified an ENU (N-ethyl-N-nitrosourea)-derived mouse model, nmf5, which was severely runted, presented with a retinal degenerative phenotype, and died shortly after weaning age. Buchner et al. (30) determined that the nmf5 mutant mice carried a mutation in Scn8a, now designated C57BL/6J-Scn8a5J, and that the retinal degeneration phenotype did not fully co-segregate with the Scn8a mutation, indicating the potential of an independent, but closely linked second mutation (30). By a series of backcrosses to strain C57BL/6J and intercrosses, we were able to segregate the retinal degenerative phenotype away from the Scn8a5J mutation, confirming the existence of the second mutation, which we named nmf5a.
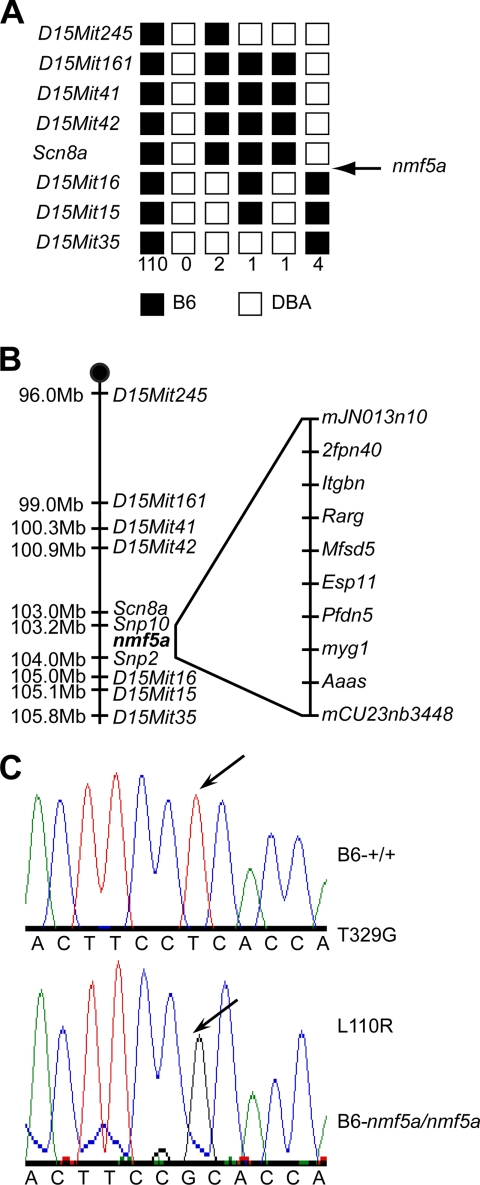
nmf5a was initially localized to an interval between D15Mit42 and D15Mit16 on Chromosome 15 (Fig. 1A). The fine structure mapping of the disease locus was carried out by backcrossing (C57BL/6J x DBA/2J)F1-Scn8a;nmf5a/+ to C57BL/6J-Scn8a;nmf5a/+, as homozygous male mice were infertile and female mice had difficulty in reproduction. A total of 245 backcross progeny were phenotyped and genotyped for the flanking markers, D15Mit245 and D15Mit35. All unaffected mice with a recombination between these two markers were progeny tested with C57BL/6J-nmf5a/+ mice to determine whether the recombinants carried the nmf5a allele. The region containing nmf5a was narrowed to 476 kb between SNP markers mJN013710 and mCV23763448 that included 21 transcripts (Fig. 1B). Seven of these genes, which were either expressed in the eye and/or had biologically relevant predicted functions, were analyzed by direct sequence comparison. A missense mutation, T329G, was found in the Pfdn5 gene, which is predicted to change amino acid 110 from leucine to arginine (Fig. 1C). The leucine residue is highly conserved across species, and in the 7 divergent mouse strains in which Pfdn5 was sequenced, none showed a guanosine at nucleotide 329 (data not shown).
FIGURE 1.
Molecular genetic analyses of the Pfdn5nmf5a mouse. A, haploytpes. 59 informative, recombinant BC mice from (D2.B6-Scn8a;nmf5a/+) F1 X B6-Scn8a;nmf5a/+ or (D2.B6-Scn8a;nmf5a/+) F1 X B6-nmf5a/+ backcrosses were phenotyped for the retinal photoreceptor degeneration and genotyped for the indicated SSLP markers. Black boxes represent the B6-derived allele, and white boxes represent the DBA-derived allele. The number of chromosomes sharing the corresponding haplotype is indicated below each column of squares. The order of marker loci was determined by minimizing the number of crossovers. B, schematic of chromosome 5 with markers (depicted on the right of the vertical line) and the physical distances between them (left of the vertical line). The physical map was assembled using data from Ensembl 50, NCBI m37 mouse assembly. C, point mutation (T329G) in the Pfdn5 gene. Chromatographs of genomic DNA sequence of Pfdn5nmf5a mouse versus control and corresponding amino acids are shown. The arrow in the upper panel indicates the original nucleotide, thymidine at nt 329 in the Pfdn5 gene of WT mice. The arrow in the lower panel indicates the missense mutation to a guanosine in the Pfdn5 gene of the nmf5a mutant.
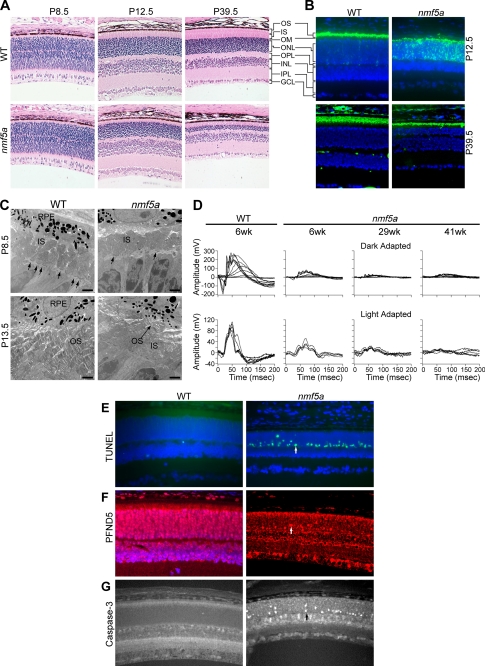
Pfdn5 is expressed ubiquitously in mice (GNF SymAtlas v 1.2.4, Genomics Institute of the Novartis Research Foundation), including most regions of the brain, and is highly expressed in testes and ovary (GNF SymAtlas v 1.2.4). Within the retina, the protein is found in the inner segments (IS), outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL) (Fig. 2F).
FIGURE 2.
Retinal phenotypes of Pfdn5nmf5a homozygotes. A, histological images (×200) of retina show delayed outer limiting membrane and photoreceptor development in Pfdn5nmf5a (nmf5a) homozygotes compared with unaffected C57BL/6J WT littermate controls at postnatal days P8.5, 12.5, and 39.5. B, rhodopsin (green) is mislocalized to the ONL and IS of photoreceptors in Pfdn5nmf5a retinas at P12.5 compared with WT (upper panel). Rhodopsin localization in retinas of Pfdn5nmf5a homozygotes appears to normalize by P42 (lower panel). C, electron micrographs of the outer retina of P8.5 (upper panel) and P13.5 (lower panel) old WT and mutant mice show normal OLM (arrows), IS, and OS development in WT mice, and retarded development of the OLM (arrows), IS, and OS in Pfdn5nmf5a mice. At P13.5, the OS of Pfdn5nmf5a mice are not properly developed and disrupted (arrow indicates shortened OS). In all micrographs; scale bar, 2 microns and nuclei are stained with DAPI (blue). D, comparison of ERG of dark- (upper panel) and light-adapted (lower panel) WT and Pfdn5nmf5a mice shows significantly reduced a- and b-wave responses at P42, 203, 287 in mutants, indicating disrupted function of photoreceptor and/or secondary neurons within the retina. At least three sets of mutant and control mice were examined for each time point. E, TUNEL assay (green) of photoreceptor cells (arrow pointing at an apoptotic cell nucleus in the ONL) Mag. 200×. F, anti-PFDN5 (red). Intense nuclear staining of cells under apoptotic stress (arrow indicating an apoptotic cell in the ONL) Mag. 200×. G, anti-caspase-3 stain (arrow indicating a caspase-3-positive nucleus in the ONL) Mag. 200×. OPL, outer plexiform layer; IPL, inner plexiform layer.
The Pfdn5nmf5a Mutation Causes Retinal Abnormalities in Mutants
At P28.5, Pfdn5nmf5a mutants exhibit a very subtle depigmented, yellowish, granular fundus appearance (data not shown). By gross morphological assessment, photoreceptor inner and outer segments (OS) do not reach their full length. In addition, the outer limiting membrane (OLM) is attenuated as early as P8.5 in mutants when compared with controls (Fig. 2A). Between P12.5 and P39.5, photoreceptors rapidly degenerate panretinally (Fig. 2A). Thereafter, the thickness of the central retina remains stationary. In contrast, the peripheral retina continues to degenerate with a marked reduction in both inner and outer nuclear layers (data not shown).
Immunohistochemical studies indicate that rhodopsin, which is normally found in the outer segments (31) at P12.5, is mislocalized to the IS and ONL in mutants (Fig. 2B). However, by P42.5, rhodopsin is found mainly in the residual OS (Fig. 2B).
Ultrastructural studies using transmission electron microscopy revealed a reduction in the number of adherens junction complexes in the OLM and confirmed the observed thinning of the IS of mutants at P8.5 (Fig. 2C). At P14.5, OS were severely fragmented, disorganized, and diminished in length and number (Fig. 2C). However, at P42, although the mutant OS were shorter than those of littermate controls, OS had a normal laminar appearance indicating a recovery from the earlier developmental defects (data not shown). This may explain the immunohistochemical results in which rhodopsin localization is partially restored in mutant mice at P42.5.
To determine how the clinical, histological, and immunohistochemical defects affected retinal signal transduction, electroretinographic analyses were conducted. Fig. 2D shows representative electroretinograms (ERGs) obtained from dark and light adapted P42, 203, and 287 day old mutant mice and littermate control mice at P42. The ERGs indicate that there is a significant reduction of both a- and b-waves in dark and light adapted mutant mice. This functional defect becomes progressively worse as mutants age despite the apparent normalization of OS morphology at P42.
TUNEL-positive photoreceptor cells were observed in mutant retinas but not controls at P13.5 (Fig. 2E). Interestingly, PFDN5 levels appeared to be up-regulated in the photoreceptor cells under apoptotic stress (Fig. 2F). Apoptosis appears to be induced through the caspase-3-dependent pathway as many photoreceptor cells in the ONL of mutant mice (Fig. 2G) stained positively with anti-caspase-3. However, after P42, the point at which the photoreceptors appear to stabilize and normalize, no TUNEL positive cells were observed in mutant retinas (data not shown).
The nmf5a Mutation Causes Central Nervous System Abnormalities
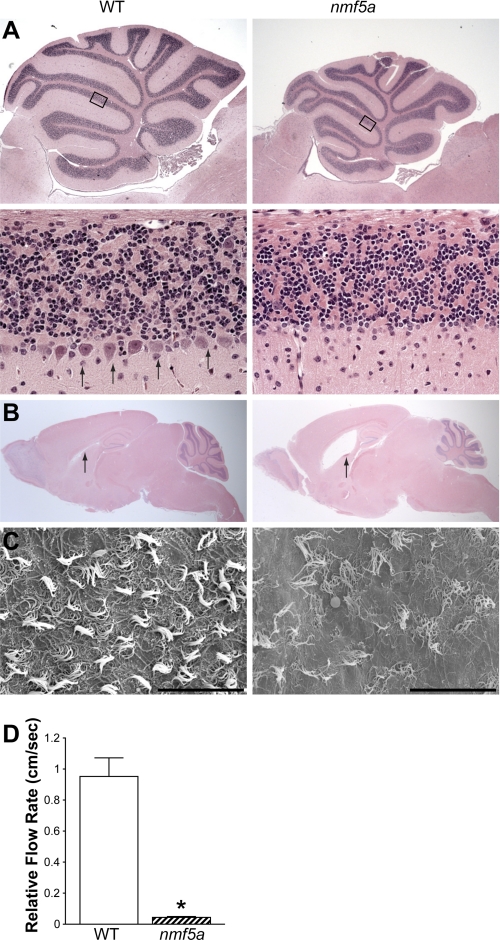
5–10% of Pfdn5nmf5a homozygotes develop hydrocephalus between birth and P112 with varying degrees of severity resulting in premature death. At approximately five months of age, Pfdn5nmf5a mutants become ataxic, the earliest clinical indication of potential defects in the cerebellum. Histologically, the cerebellum of Pfdn5nmf5a homozygotes appears to be smaller than in control littermates. Purkinje cell death is readily apparent at 5 months (Fig. 3A), with the rostral region of the cerebellum most profoundly affected (data not shown). At the same time point, a trend toward increased apoptotic profiles was observed in the granule cell layer but not the molecular layer (data not shown). At P56, overall brain weight of mutants was 19% less than that of controls (0.48 ± 0.02 g versus 0.39 ± 0.04 g).
FIGURE 3.
The Pfdn5nmf5a mutation causes Purkinje cell degeneration, reduced ciliary function, and hydrocephalus. A, H&E staining of the cerebellum of C57BL/6J-Pfdn5nmf5a mice and unaffected C57BL/6J littermate controls (n = 10). A reduced number of Purkinje cells are observed in mutants compared with controls (lower panel, arrows). The lower panels show a magnification of the respective rectangular area shown above. Upper panel, Mag. 25×; lower panel, Mag. 400×. B, representative histological sections of brains reveal hydrocephalus in Pfdn5nmf5a mutants (arrow) (n = 5), Mag. 10×. C, electron microscopic analysis of ventricular cilia in WT and Pfdn5nmf5a mice (n = 3) shows disrupted and reduced number of cilia in mutant mice, Mag. 1.5k×; scale bar, 30 microns. D, relative flow rate (cm/s) was measured. *, p < 0.001.
With the exception of the hydrocephalus, other neuronal phenotypes examined were fully penetrant in homozygous Pfdn5nmf5a mice. Therefore, to determine whether subclinical signs of hydrocephalus were observable in Pfdn5nmf5a homozygotes, sagittal sections of brains from mutants and WT were examined. All mutants examined that were greater than one year of age showed variable but increased dilatation of the third ventricle when compared with age matched WT controls (Fig. 3B). Because hydrocephalus is associated with structural or functional defects of the choroid plexus of the brain which generates the cerebrospinal fluid (CSF) (32) and/or of the cilia in the ventricles of the brain that maintain proper circulation of the CSF, these structures were examined in Pfdn5nmf5a mutants. Immunohistochemical examination of the choroid plexus from homozygous Pfdn5nmf5a mice showed TUNEL positive ependymal cells undergoing apoptosis (data not shown). Scanning electron microscopy (SE) revealed disorganization and a significant reduction in the number of cilia in the ventricles of Pfdn5nmf5a mutants compared with controls (Fig. 3C).
To determine whether these morphological alterations affected cilia function, the ability of the ependymal cilia to beat and move fluid was examined by videomicroscopic analyses. Ventricular ependymal cell tissue explants were incubated with green fluorescent latex beads (0.046 μm, Sigma) in L-15 oocyte medium and the movement of the beads was recorded (data not shown). The distances beads moved during a 2.7 or a 10.7 s interval in control and in Pfdn5nmf5a mutant samples, respectively, were compared. A significant reduction in ciliary beating was observed in Pfdn5nmf5a mutants, with a relative flow rate of 0.044 ± 0.005 cm/sec compared with wild-type controls at 0.95 ± 0.12 cm/sec (Fig. 3D, p < 0.005).
Hypogonadism, a Primary Defect in Pfdn5nmf5a Mutants
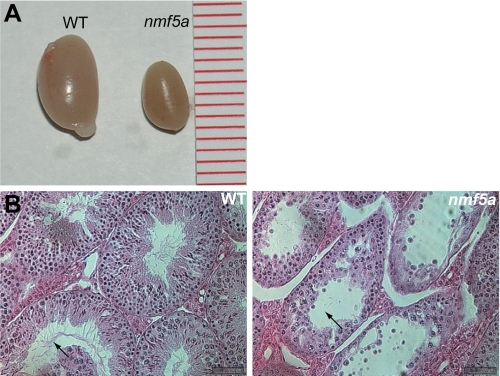
Male Pfdn5nmf5a mutant mice are infertile, with testes size and weight half those of littermate controls (Fig. 4A). Testicular development was independent of overall growth as mutants were similar in total size and weight compared with littermate controls until early adulthood. The ratio of mutant body weight over control was 0.923 ± 0.10, while the ratio of testis weight of the same animals was 0.494 ± 0.05. Mutant animals were oligospermic with 1/10 the normal number of mature sperm (data not shown). Most seminiferous tubules appeared abnormal and contained sloughed off spermatocytes and spermatids (Fig. 4B). High incidence of immature, round spermatids was observed in seminal fluid (data not shown).
FIGURE 4.
The Pfdn5nmf5a mutation causes hypogonadism. A, comparison of WT and nmf5a/nmf5a mutant testis (n = 5). One unit of the scale bar is equivalent to 1 mm. B, in the WT seminiferous tubules, normally developing spermatogonia (SA), spermatocyte (SE), spermatid (SD) cells, and spermatid tails (arrow) are observed. In the Pfdn5nmf5a mutant seminiferous tubules, these cells (arrow) were atrophied and reduced in number, n = 5, Mag. 1000×.
Unlike male mutant mice, females are fertile. However, they often experienced difficulty in the birthing process and died during labor, a complication rarely observed in B6 WT controls. Also, Pfdn5nmf5a mutant dams have a shorter reproductive life, about half that of WT controls.
Examination of the Effects of the Pfdn5nmf5a Mutation by Modeling
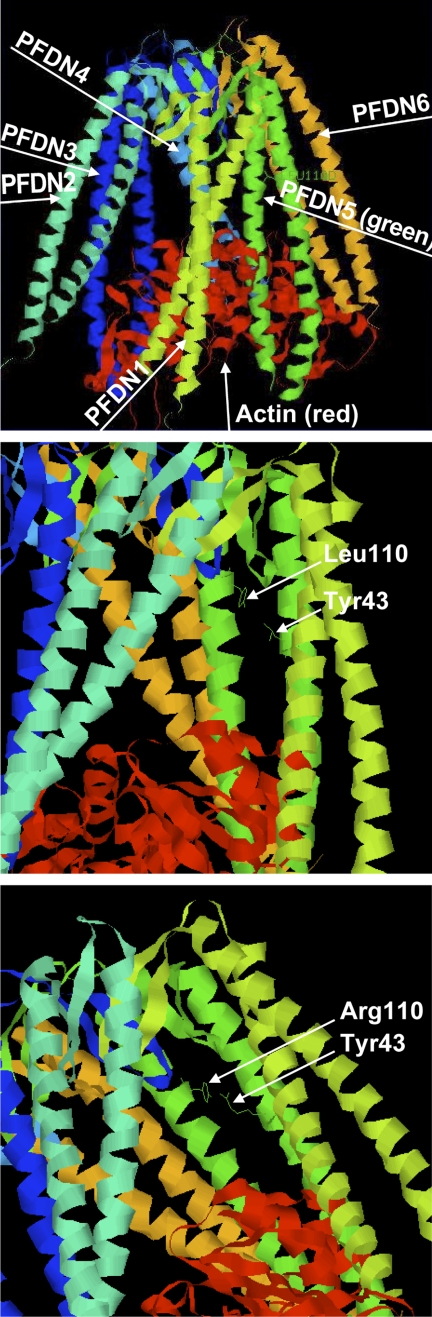
To assess the potential effects of the missense mutation on the structural conformation of the protein, we performed modeling based on the reported structural conformation of PFDN5 (3). Unlike the wild-type leucine (L) residue, an arginine (R) at amino acid 110 is predicted to create a favorable interaction environment with the tyrosine at amino acid residue 43 in PFDN5 (Fig. 5). Since substrate binding takes place at the tips of the prefoldin tentacles (33) and the mutation is located further away from them, the L110R mutation may not have a direct effect on its ability to bind to substrates. The mutation is more likely to affect the flexibility of the tentacles of prefoldin through which substrate accommodation in the interior of the cavity is mediated (33), thereby causing an abnormal interaction with its substrates.
FIGURE 5.
Prefoldin protein modeling. Whole view of wild-type human prefoldin:actin (red) complex model (upper panel). Leu-110 and Tyr-43 in PFDN5 are labeled (middle panel). Mutated human prefoldin:actin complex with L110R in PFDN5 (lower panel). A rotomer for L110R allows for the guanido group of the arginine side chain to potentially interact favorably with the side chain of Tyr-43, and may act to stabilize the two helices. All other rotamers create steric clashes with other amino acids.
Overexpression of Mutant Pfdn5 in Yeast Causes Abnormal Cell Growth and Viability
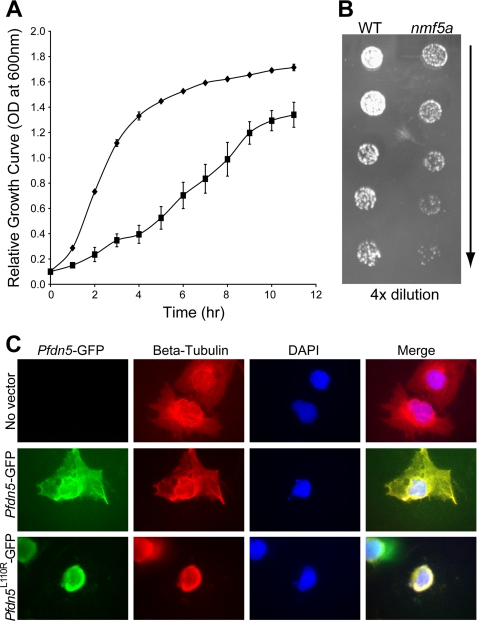
To further assess the effects of the Pfdn5nmf5a mutation, we over-expressed WT and mutant Pfdn5nmf5a in yeast cells. In YPAD, a complete growth medium, mutant Pfdn5nmf5a transformed yeast cells showed a 4-fold slower growth during the exponential growth phase when compared with WT transformed cells (Fig. 6A). Further, yeast cells transformed with Pfdn5nmf5a were more sensitive to 10 mm nocodazol, a microtubule polymerization inhibitor, than were WT controls (Fig. 6B).
FIGURE 6.
Overexpression of mutant Pfdn5 causes growth retardation of yeast cells. A, growth curve of MaV203 yeast cells overexpressing wild-type (diamonds, ♦) and mutant (squares, ■) Pfdn5 shows that there is about a 4-fold difference in growth between wild-type and mutant Pfdn5 expressing yeast cells at the peak of the exponential proliferation period. OD was measured at 600 nm. Standard error was obtained from three independently performed experiments. Significance (p < 0.001). B, serial dilutions (from top to bottom) of the MaV203 yeast strain with wild-type (left panel) and mutant Pfdn5 (right panel) were spotted onto YPAD agar plates with 2 mm nocodazol, microtubule inhibitor. Yeast cells with mutant Pfdn5 expression were more sensitive to nocodazol (n = 3). C, overexpression of wild-type and Pfdn5L110R in COS-7 cells. The upper panel shows COS-7 cells (no vector expression) with normal microtubule organization. The middle and lower panels depict Pfdn5-GFP wild type and PfdnL110R-GFP expression, respectively, showing abnormal microtubule organization and lack of peripheral structure in COS-7 cells overexpressing PfdnL110R-GFP (lower panel). Magnification, 1000X.
Reduced Microtubules and Microfilament in Retina May Lead to Abnormal IS and OS Development
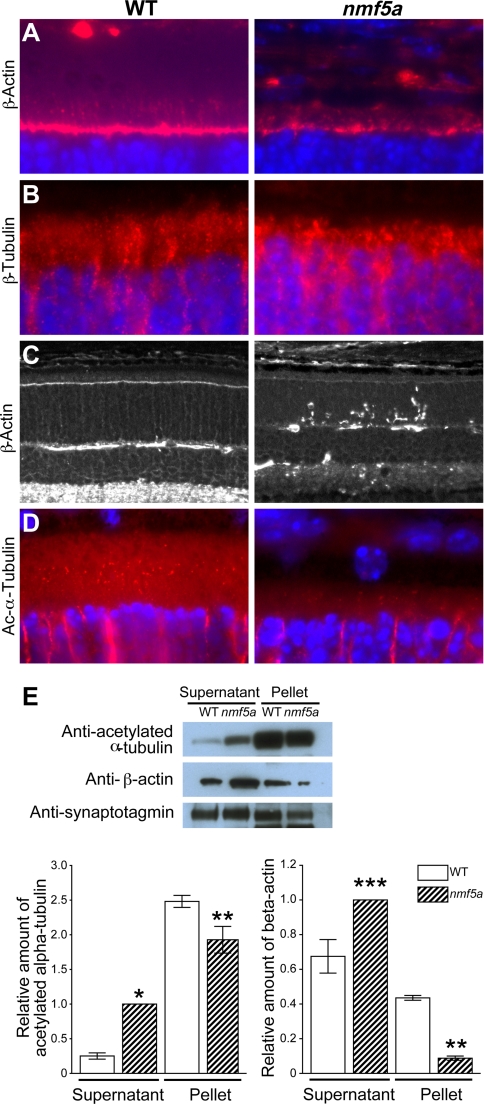
Consistent with the retarded development of the OLM observed in mutant mice at P8.5 (Fig. 2C), at P13.5, β-actin staining was reduced in the OLM, and structures in the IS stained by β-actin appear disorganized and aggregated in mutant mice compared with the wild-type controls (Fig. 7A). β-Tubulin showed a similar IS staining pattern in mutant mice as β-actin (Fig. 7B). Focal aggregates of actin were observed in the ONL at P13.5 (Fig. 7C).
FIGURE 7.
The Pfdn5 mutation leads to aggregation and a reduced level of the polymer forms of cytoskeletal components. A, anti-β-actin stain (red) of IS (vertical, red staining immediately above the OLM) and OLM (horizontal, red staining), ONL (blue), Mag. 400×. B, anti-β-tubulin (red) stain of IS. ONL (blue), Mag. 1000×. C, anti-β-actin (white) staining of aggregates in the ONL of mutant mice, Mag. 200×. D, connecting cilia stained with acetylated α-tubulin (red), Mag. 1000×. E, Western blot of tubulin and actin in the retina after microtubule and microfilament ultracentrifugal separation of the supernatant (monomers) and pellet (polymers). Loading control-synaptotagmin. Relative densitometric representation of tubulin and actin stain in the supernatant and pellet from three independent experiments. Standard error: *, p < 0.1; **, p < 0.05; ***, p < 0.025. ONL of photoreceptor cells, Mag. 400×. A minimum of three mutant and control mice were used for each analysis.
Relatively fewer acetylated α-tubulin positive staining basal bodies of the CC were found in mutant when compared with the WT control retinas (Fig. 7D), suggesting a delayed and/or disrupted CC development. Fewer or disrupted CC at a time of rapid OS development could potentially lead to the observed mislocalization and accumulation of the OS proteins such as rhodopsin (Fig. 2B) in the IS and ONL. Relative ratios of the number of positively stained CC over the number of photoreceptor cell bodies in a given section from mutant and WT mice at P12.5 to P13.5 were 0.034 ± 0.01 and 0.26 ± 0.02, respectively.
The purported co-chaperone function of prefoldin in tubulin and actin folding, and the observed aggregation of tubulin and actin in Pfdn5nmf5a mutants, suggested that the mutation in Pfdn5 might lead to an inefficient transport of these nascent proteins to the CCT with the net result being a decrease in microfilament and microtubule formation. Because in vitro expression of the six prefoldin subunits to form a functional prefoldin complex is inherently difficult and we were unable to directly assess the rate of binding, transport and release of these proteins in vivo, an indirect method was used to assess the effects of the Pfdn5 mutation on its substrates. We determined the levels of monomeric tubulin and actin and their respective polymers, microtubules, and microfilaments, in retina by Western analyses after separation by high-speed centrifugation. The relative quantity of α-acetylated tubulin and β-actin monomer in the supernatant isolated from mutant retina was 3.99 ± 0.62- and 1.48 ± 0.07-fold higher, respectively, when compared with normal controls. The relative amount of microtubules and microfilament in mutant retina contained in the pellet was 0.78 ± 0.27- and 0.20±.03-fold lower, respectively, when compared with controls (Fig. 7E). The latter result is consistent with the observation of the disruption in cytoskeletal development and of the reduction in the number of CC in the retina (Fig. 7, A, B, and D, respectively). The observation that the relative amount of actin monomer is higher and microfilament is lower than that of tubulin and microtubules, respectively, may indicate that the L110R mutation in prefoldin 5 affects actin processing to a greater degree than tubulin. To confirm this hypothesis, further experiments are required. However, differential effects of the truncation of the coiled-coil tips of the various prefoldin subunits have been shown to affect substrate specificity in yeast (25).
DISCUSSION
The importance of prefoldin as a co-chaperone has been demonstrated in previous studies in yeast cells in which the deletion of GIM5, a homolog of Pfdn5 or of PAC10, a homolog of Pfdn3, resulted in growth retardation and a disruption of actin- and tubulin-based cytoskeletal development (1, 6). Additionally, loss of GIM5 in yeast cells was shown to cause increased sensitivity to microtubule polymerization inhibitors and low temperature stress (1). While loss of individual prefoldin subunits does not affect the survival of yeast, loss of Pfdn1 in Caenorhabditis elegans leads to embryonic lethality (1, 4, 6). Deletion of Pfdn1 in mice causes a loss of nerve bundles, hydrocephaly, mucus clearance defects, and abnormal lymphocyte development and function (22). The prefoldin 1 null mutant suggests an important role of prefoldin in neuronal development (22). Although in vivo relevance has yet to be established, in humans, PFDN3 is reported to interact with Von Hippel-Lindau syndrome (VHL) protein, a tumor suppressor, which acts as a molecular chaperone that transports PFDN3 between perinuclear granules to the nucleus or cytoplasm (34). It has also been reported that PFDN3 is involved in protein degradation (35). Additionally, Mori et al. (16) have suggested that PFDN5 (MM-1) modulates c-Myc expression in relation to tumor suppression. The in vivo data in yeast, C. elegans and mouse taken together with the in vitro data suggest that the prefoldin co-chaperone complex plays a central role in cellular development.
In this report, we show that a recessive point mutation in murine Pfdn5 primarily causes neurodegeneration and hypogonadism. Early photoreceptor degeneration, which has not been reported in the Pfdn1-null mutants, Purkinje cell degeneration, and hydrocephalus result in severely diminished ERG responses, ataxia and premature death, respectively. Our data also indicate that abnormal actin and/or tubulin cytoskeleton assembly and microtubule formation, respectively, may underlie the Pfdn5nmf5a disease phenotypes, and cellular stress appears to lead to apoptosis.
Although PFDN5 is expressed in a wide variety of tissues, defects in Pfdn5 homozygotes appear to be restricted to neuronal cells such as the ependymal cells of the third ventricle, Purkinje and photoreceptor cells, and to sperm. These cell-types share a common feature of having motile cilia or modified cilia as a structural component, e.g. connecting cilium in the photoreceptors. However, kidney disease reported in human patients with ciliopathies (36–38) is not observed in Pfdn5nmf5a mutant mice (data not shown). This may suggest a redundancy of the protein-folding machinery in non-affected tissues. Alternatively, it is possible that substrates of prefoldin 5 may be only expressed in the affected cell types. Previous studies have shown that certain isoforms of actin, are predominantly expressed in neuronal cells, and even within or between neuronal cells they are differentially distributed (39). For example, γ-actin polymers are found in the soma and proximal regions of extended neurites, whereas β-actin is enriched in the terminal tip of the neurites (39).
The differences in disease phenotypes between the Pfdn1-null and Pfdn5nmf5a mutant mice may be due to the nature of the respective mutations. The Pfdn1-targeted mutation, which leads to a total deficiency of PFDN1, may affect the proper assembly of the PFDN holocomplex while the hypomorphic PFDN5 allele may lead to conformational changes that affect substrate binding affinity. Alternatively, each subunit may have unique binding partner(s) within different cell-types, such that the loss or impaired function of either subunit might affect different downstream pathways and lead to a different disease phenotype.
Ohtaki et al. (40) predict from their electron microscopy and biochemical studies that the β-subunits (PFDN1, -2, -4, and -6) are crucial for nonnative protein binding (40). They also suggest that the α subunits (PFDN3 and -5) may be involved in substrate specificity and stabilizing the complex of prefoldin/substrate by changing their conformation and position according to substrate size and mode of binding (40). The main function of prefoldin is to transport non-native substrates in a linear state to the CCT for proper folding and thereby prevent aggregation. The predicted effect of the Pfdn5nmf5a mutation by computer modeling, and the aggregates of actin and tubulin observed in mutant retinas (Fig. 7, A, B, and C) suggests that the Pfdn5nmf5a mutation may result in a less than optimum binding of prefoldin to its substrates. If the demand for and generation of actin and tubulin are high, for example, during photoreceptor IS and OS development, the reduced affinity could potentially exceed the capacity of the mutant prefoldin complex to carry out its necessary function and lead to aggregates of unfolded protein. When the accumulation of protein exceeds the ability of the cell to remove unfolded or misfolded proteins, apoptosis occurs (41). The accumulation of OS proteins, such as rhodopsin and ROM1, in the IS and ONL could add additional stress to the photoreceptor cells that are already perturbed, thereby causing a rapid photoreceptor degeneration. Perhaps, the OS development and proper localization of OS proteins observed in mutant photoreceptors at P42 (Fig. 2B) may indicate that the generation of substrates for synthesis of microfilaments and microtubules are at a level at which the mutant prefoldin complex and the normal cytosolic mechanisms that remove aberrantly formed protein can accommodate, and thus, allow for some OS to be formed.
The Pfdn5nmf5a mutant mouse will be a valuable model to further examine these possibilities and elucidate the functions of the co-chaperone, prefoldin, and its disease mechanisms related to protein transport and folding, and induction of apoptosis. Because single nucleotide substitutions are the most common cause of human disease, the nmf5a allele of Pfdn5 described here may be particularly relevant to understanding diseases caused by mutations within human chaperones (42). Further, disruption of other subunits of prefoldin may assist in determining tissue/substrate specificity of the prefoldin complex.
Acknowledgments
We thank Drs. Susan Ackerman and Gregory Cox for critical review of the manuscript, Adrienne Mehalow for initiating the mapping crosses, Dr. Robert Braun for testis analysis, Jeanie Hansen for mouse colony care, and Douglas Howell for ERG measurement. Shared scientific services were supported by an Institutional Cancer Core grant (CA-24190).
This work was supported, in whole or in part, by NEI, National Institutes of Health Grants EY016501 and EY11996.
- PFDN
- prefoldin
- CCT
- chaperonin containing TCP1
- TEM
- transmission electron microscopy
- SE
- scanning electron microscopy
- CC
- connecting cilia
- IS
- inner segments
- ONL
- outer nuclear layer
- INL
- inner nuclear layer
- GCL
- ganglion cell layer
- OLM
- outer limiting membrane
- OS
- outer segment
- ERG
- electroretinogram.
REFERENCES
- 1. Vainberg I. E., Lewis S. A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H. L., Cowan N. J. (1998) Cell 93, 863–873 [DOI] [PubMed] [Google Scholar]
- 2. Leroux M. R., Fändrich M., Klunker D., Siegers K., Lupas A. N., Brown J. R., Schiebel E., Dobson C. M., Hartl F. U. (1999) EMBO J. 18, 6730–6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martín-Benito J., Boskovic J., Gómez-Puertas P., Carrascosa J. L., Simons C. T., Lewis S. A., Bartolini F., Cowan N. J., Valpuesta J. M. (2002) EMBO J. 21, 6377–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siegers K., Waldmann T., Leroux M. R., Grein K., Shevchenko A., Schiebel E., Hartl F. U. (1999) EMBO J. 18, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vinh D. B., Drubin D. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9116–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geissler S., Siegers K., Schiebel E. (1998) EMBO J. 17, 952–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ursic D., Culbertson M. R. (1991) Mol. Cell Biol. 11, 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ursic D., Sedbrook J. C., Himmel K. L., Culbertson M. R. (1994) Mol. Biol. Cell 5, 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lacefield S., Magendantz M., Solomon F. (2006) Genetics 173, 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lacefield S., Solomon F. (2003) Genetics 165, 531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zipperlen P., Fraser A. G., Kamath R. S., Martinez-Campos M., Ahringer J. (2001) EMBO J. 20, 3984–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lundin V. F., Srayko M., Hyman A. A., Leroux M. R. (2008) Dev. Biol. 313, 320–334 [DOI] [PubMed] [Google Scholar]
- 13. Le Bot N., Tsai M. C., Andrews R. K., Ahringer J. (2003) Curr. Biol. 13, 1499–1505 [DOI] [PubMed] [Google Scholar]
- 14. Gu Y., Deng Z., Paredez A. R., DeBolt S., Wang Z. Y., Somerville C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18064–18069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vicart P., Caron A., Guicheney P., Li Z., Prévost M. C., Faure A., Chateau D., Chapon F., Tomé F., Dupret J. M., Paulin D., Fardeau M. (1998) Nat. Genet. 20, 92–95 [DOI] [PubMed] [Google Scholar]
- 16. Sakono M., Zako T., Ueda H., Yohda M., Maeda M. (2008) Febs J. 275, 5982–5993 [DOI] [PubMed] [Google Scholar]
- 17. Bouhouche A., Benomar A., Bouslam N., Chkili T., Yahyaoui M. (2006) J. Med. Genet. 43, 441–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Badano J. L., Kim J. C., Hoskins B. E., Lewis R. A., Ansley S. J., Cutler D. J., Castellan C., Beales P. L., Leroux M. R., Katsanis N. (2003) Hum. Mol. Genet. 12, 1651–1659 [DOI] [PubMed] [Google Scholar]
- 19. Eriksen J. L., Dawson T. M., Dickson D. W., Petrucelli L. (2003) Neuron 40, 453–456 [DOI] [PubMed] [Google Scholar]
- 20. Engert J. C., Bérubé P., Mercier J., Doré C., Lepage P., Ge B., Bouchard J. P., Mathieu J., Melancon S. B., Schalling M., Lander E. S., Morgan K., Hudson T. J., Richter A. (2000) Nat. Genet. 24, 120–125 [DOI] [PubMed] [Google Scholar]
- 21. Katsanis N., Beales P. L., Woods M. O., Lewis R. A., Green J. S., Parfrey P. S., Ansley S. J., Davidson W. S., Lupski J. R. (2000) Nat. Genet. 26, 67–70 [DOI] [PubMed] [Google Scholar]
- 22. Cao S., Carlesso G., Osipovich A. B., Llanes J., Lin Q., Hoek K. L., Khan W. N., Ruley H. E. (2008) J. Immunol. 181, 476–484 [DOI] [PubMed] [Google Scholar]
- 23. Lee Y., Kameya S., Cox G. A., Hsu J., Hicks W., Maddatu T. P., Smith R. S., Naggert J. K., Peachey N. S., Nishina P. M. (2005) Mol. Cell Neurosci. 30, 160–172 [DOI] [PubMed] [Google Scholar]
- 24. Siegert R., Leroux M. R., Scheufler C., Hartl F. U., Moarefi I. (2000) Cell 103, 621–632 [DOI] [PubMed] [Google Scholar]
- 25. Simons C. T., Staes A., Rommelaere H., Ampe C., Lewis S. A., Cowan N. J. (2004) J. Biol. Chem. 279, 4196–4203 [DOI] [PubMed] [Google Scholar]
- 26. Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. (1990) Nature 347, 37–44 [DOI] [PubMed] [Google Scholar]
- 27. Llorca O., Martín-Benito J., Ritco-Vonsovici M., Grantham J., Hynes G. M., Willison K. R., Carrascosa J. L., Valpuesta J. M. (2000) EMBO J. 19, 5971–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A. 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 29. Andreu J. M. (2007) in Microtubule Protocols (Zhou J. ed), Springer-Verlag, New York [Google Scholar]
- 30. Buchner D. A., Seburn K. L., Frankel W. N., Meisler M. H. (2004) Mamm. Genome 15, 344–351 [DOI] [PubMed] [Google Scholar]
- 31. Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., MacDougald O. A. (2000) Science 289, 950–953 [DOI] [PubMed] [Google Scholar]
- 32. Zhang J., Williams M. A., Rigamonti D. (2006) J. Neurol. 253, 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martín-Benito J., Gómez-Reino J., Stirling P. C., Lundin V. F., Gómez-Puertas P., Boskovic J., Chacón P., Fernández J. J., Berenguer J., Leroux M. R., Valpuesta J. M. (2007) Structure 15, 101–110 [DOI] [PubMed] [Google Scholar]
- 34. Tsuchiya H., Iseda T., Hino O. (1996) Cancer Res. 56, 2881–2885 [PubMed] [Google Scholar]
- 35. Mousnier A., Kubat N., Massias-Simon A., Ségéral E., Rain J. C., Benarous R., Emiliani S., Dargemont C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13615–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moyer J. H., Lee-Tischler M. J., Kwon H. Y., Schrick J. J., Avner E. D., Sweeney W. E., Godfrey V. L., Cacheiro N. L., Wilkinson J. E., Woychik R. P. (1994) Science 264, 1329–1333 [DOI] [PubMed] [Google Scholar]
- 37. Yoder B. K., Hou X., Guay-Woodford L. M. (2002) J. Am. Soc. Nephrol. 13, 2508–2516 [DOI] [PubMed] [Google Scholar]
- 38. Otto E. A., Schermer B., Obara T., O'Toole J. F., Hiller K. S., Mueller A. M., Ruf R. G., Hoefele J., Beekmann F., Landau D., Foreman J. W., Goodship J. A., Strachan T., Kispert A., Wolf M. T., Gagnadoux M. F., Nivet H., Antignac C., Walz G., Drummond I. A., Benzing T., Hildebrandt F. (2003) Nat. Genet. 34, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Micheva K. D., Vallée A., Beaulieu C., Herman I. M., Leclerc N. (1998) Eur. J. Neurosci. 10, 3785–3798 [DOI] [PubMed] [Google Scholar]
- 40. Ohtaki A., Kida H., Miyata Y., Ide N., Yonezawa A., Arakawa T., Iizuka R., Noguchi K., Kita A., Odaka M., Miki K., Yohda M. (2008) J. Mol. Biol. 376, 1130–1141 [DOI] [PubMed] [Google Scholar]
- 41. Schröder M., Kaufman R. J. (2005) Mutat. Res. 569, 29–63 [DOI] [PubMed] [Google Scholar]
- 42. Stenson P. D., Ball E., Howells K., Phillips A., Mort M., Cooper D. N. (2008) J. Med. Genet. 45, 124–126 [DOI] [PubMed] [Google Scholar]