Abstract
Surfactant protein A (SP-A), a C-type lectin, plays an important role in innate lung host defense against inhaled pathogens. Crystallographic SP-A·ligand complexes have not been reported to date, limiting available molecular information about SP-A interactions with microbial surface components. This study describes crystal structures of calcium-dependent complexes of the C-terminal neck and carbohydrate recognition domain of SP-A with d-mannose, d-α-methylmannose, and glycerol, which represent subdomains of glycans on pathogen surfaces. Comparison of these complexes with the unliganded SP-A neck and carbohydrate recognition domain revealed an unexpected ligand-associated conformational change in the loop region surrounding the lectin site, one not previously reported for the lectin homologs SP-D and mannan-binding lectin. The net result of the conformational change is that the SP-A lectin site and the surrounding loop region become more compact. The Glu-202 side chain of unliganded SP-A extends out into the solvent and away from the calcium ion; however, in the complexes, the Glu-202 side chain translocates 12.8 Å to bind the calcium. The availability of Glu-202, together with positional changes involving water molecules, creates a more favorable hydrogen bonding environment for carbohydrate ligands. The Lys-203 side chain reorients as well, extending outward into the solvent in the complexes, thereby opening up a small cation-friendly cavity occupied by a sodium ion. Binding of this cation brings the large loop, which forms one wall of the lectin site, and the adjacent small loop closer together. The ability to undergo conformational changes may help SP-A adapt to different ligand classes, including microbial glycolipids and surfactant lipids.
Keywords: Calcium-binding Proteins, Carbohydrate Complex, Crystal Structure, Innate Immunity, Lectin, Pattern Recognition Receptor, Conformational Change, SP-A, Surfactant Proteins, Collectins
Introduction
Surfactant protein A (SP-A)2 is an abundant protein associated with pulmonary surfactant. Together with the lung protein homolog SP-D, SP-A plays an important role in pulmonary innate immunity by recognizing canonical patterns on microbial surfaces (1–8). These host defense proteins protect the lung from infection by recognizing the carbohydrate and/or lipid component on pathogens, including bacteria, virus, and fungi, and by helping to initiate various clearance mechanisms (9).
SP-A and SP-D are members of the collectin family, a subgroup of C-type (calcium-dependent) lectins (10), which also includes serum proteins, such as human mannose-binding protein (MBP) or lectin and bovine conglutinin (11). SP-A and SP-D have distinct preferences among the mannose-type saccharides and other glycans, which may enable these proteins to interact differentially with respiratory pathogens (12). Where the two collectins share a common microbial target, such as LPS from the outer membrane of Gram-negative bacteria, the microbial features recognized may be dissimilar. For instance, SP-D interacts with the LPS core saccharides, whereas SP-A prefers the lipid A domain (13, 14). Distinct mechanisms, which may not require lectin interactions, may underlie such divergent interactions. For example, SP-D binds to influenza A virus through lectin-type interactions (15, 16), whereas influenza A virus binds to SP-A via interactions involving the sialylated N-linked carbohydrate attached to the C-terminal domain of SP-A (17, 18). SP-A, the more abundant of the two lung collectins, thus plays an important and complementary role in pathogen clearance by interacting with microbial patterns that are not well recognized by SP-D and vice versa.
The carbohydrate recognition domain (CRD) confers the calcium-dependent lectin activity and is responsible primarily for pathogen recognition. Structural and other types of studies show that the CRD of C-type lectins characteristically folds into a canonical double-loop structure. The two loops, the long loop (residues 181–204) and the short loop (residues 171–177), are stabilized by disulfide bonds at the ends of the loops and bound calcium ions. These lectins recognize their carbohydrate ligands through extensive interactions between two pyranose hydroxyl groups of the sugar, the calcium ion, and four to five residues of the lectin site (10). Fragments corresponding to the CRD or the neck and CRD (NCRD) of these large multidomain proteins have provided useful tools for studies of collectin-ligand recognition and binding interactions.
SP-A has features in its loop sequences that are rare in the C-type lectin family, making it difficult to predict how this surfactant protein interacts with its ligands (10). The substitution in SP-A of arginine (rat) or alanine (human) for asparagine in the consensus lectin site of related collectins results in the loss of one calcium ligand in the SP-A loop. The side chain of another conserved calcium ligand is positioned outside the calcium coordination shell, as demonstrated crystallographically. Furthermore, residues that bind a structural non-lectin calcium ion found in homologous lectins are lost to substitution in SP-A. The effects of these lost functionalities on ligand recognition have been unclear but have suggested some deviation of SP-A from prototypical CRD-mediated recognition properties. This study provides the first structural information regarding SP-A NCRD complexes with carbohydrates and reveals an unexpected feature: a significant conformational change associated with ligand binding. This property, which has not been reported in other collectins, could facilitate interactions between SP-A and non-carbohydrate ligands.
EXPERIMENTAL PROCEDURES
Protein Preparation
A recombinant rat SP-A construct consisting of the CRD and neck region and lacking the consensus residue for the asparagine-linked glycosylation site (ΔN1-P80, N187S) was synthesized and purified as described previously (19). Protein concentrations were determined with protein assay reagent (Pierce) using bovine serum albumin as the standard. d-Mannose, α-methyl-d-mannoside, and l-glycerol were obtained from Sigma and used without further purification.
Crystallization, Soaking, and Data Collection
SP-A crystals are grown at 17 °C from hanging drops after equilibration of a 1–2-μl droplet containing 10 mg/ml protein with 0.5 ml of mother liquor containing 1.2–1.6 m Li2SO4, 50 mm sodium cacodylate (pH 6.5), and 10 mm CaCl2. For monosaccharide soaking, the crystal-containing droplet was diluted with crystallization buffer (50 mm sodium cacodylate (pH 6.5) and 10 mm CaCl2) to reduce the salt concentration to 0.6–0.8 m. Simultaneously, monosaccharide in powder form was added to the diluted crystal soaking solution and dissolved by gentle stirring with a wire loop. Crystals were soaked in this solution for 30 min to several hours. Just prior to data collection, additional buffer was added to further reduce the salt concentration, and more monosaccharide powder was dissolved in the crystal solution quickly to a concentration high enough to prevent ice formation under a nitrogen vapor stream. The final crystal solution contained 0.3–0.5 m Li2SO4, 50 mm sodium cacodylate (pH 6.5), 10 mm CaCl2, and monosaccharide concentrations above 1 m for cryoprotection. For the unliganded form, crystals were prepared similarly except that maltose (which does not bind to SP-A under these conditions) was used as a cryoprotectant. For the glycerol complex, 5% (v/v) glycerol was present in the crystallization conditions, and its concentration was increased to cryoprotectant levels before data collection. Crystallographic data sets of unliganded SP-A and the saccharide complexes were collected using an R-AXIS IV imaging plate detector mounted on a Rigaku RU-300 rotating anode generator. The SP-A·glycerol complex data set was collected at the National Synchrotron Light Source at Brookhaven National Laboratory. All crystals were cooled to 80–100 K in a vapor nitrogen stream prior to data collection.
Data Processing and Refinement
Data sets from the Boston University facility were indexed and integrated using DENZO and scaled and merged using SCALEPACK software (both part of the HKL suite of programs). The SP-A complex structures were solved by difference Fourier from the previously reported structure of the SP-A NCRD (Protein Data Bank code 1R13) (19). The SP-A NCRD forms a symmetry (P63)-related trimer with one monomer per asymmetric unit. The N-terminal first three residues (residues 81–83) of the neck region are invisible in the electron density maps for all structures, presumably because of their high mobility. Structure refinements were carried out using the CNS software package (20) with iterative cycles of model rebuilding in program O (21) or Coot (22). Initially, each ligand was manually fit into simulated annealing-omit Fo − Fc electron density maps and then refined to acceptable Rfree and Rcryst values. Protein geometry was analyzed by PROCHECK. Data collection and refinement statistics are presented in Table 1. The statistics show that the data are of comparably good quality and have been refined to good geometry. Figures were prepared using PyMOL (23), Swiss-PdbViewer, and POV-Ray (24). Alignments were performed in Swiss-PdbViewer (25).
TABLE 1.
Data collection and refinement statistics of SP-A structures
| Protein·ligand |
||||
|---|---|---|---|---|
| SP-A·glycerol | SP-A·d-Man | SP-A·a-MMA | SP-A | |
| Data collection | ||||
| Space group | P6 (3) | P6 (3) | P6 (3) | P6 (3) |
| Resolution (Å) | 1.8 | 1.9 | 2.1 | 2.3 |
| Rmerge | 4.4 (22.9) | 5.0 (28.7) | 3.8 (37.9) | 3.9 (26.6) |
| I/σI | 14.8 (5.6) | 24.5 (4.5) | 21.5 (3.9) | 21.9 (4.3) |
| Completeness (%) | 99.6 (98.2) | 99.4 (98.9) | 99.6 (99.8) | 98.2 (91.3) |
| Redundancy | 5.0 (3.0) | 5.0 (3.9) | 4.0 (3.9) | 4.0 (2.9) |
| Wavelength (Å) | 0.9795 | 1.5418 | 1.5418 | 1.5418 |
| No. of reflections | 22,680 (2206) | 19,500 (1279) | 14,325 (946) | 10,984 (683) |
| Refinement | ||||
| Rwork/Rfree | 20.3/22.0 | 21.8/24.2 | 22.4/24.4 | 22.6/24.0 |
| Ca2+ ions/subunit | 1 | 1 | 1 | 1 |
| Ligands/subunit | 3 | 1 | 1 | 0 |
| Average B-factor (Å2) | 27.8 | 36.4 | 40.5 | 45.4 |
| Protein | 26.5 | 35.7 | 39.9 | 45.0 |
| Calcium ions | 23.0 | 37.7 | 44.2 | 43.2 |
| Water | 36.8 | 41.6 | 42.7 | 44.3 |
| Ligands | 31.7 | 54.8 | 64.4 | |
| r.m.s.d. bonds (Å) | 0.005 | 0.004 | 0.005 | 0.006 |
| r.m.s.d. angles | 1.1° | 1.1° | 1.1° | 1.2° |
| Ramachandran plot | ||||
| Core/allowed (%) | 94.4/5.6 | 95.2/4.8 | 92.7/7.3 | 88.6/11.4 |
| General/disallowed (%) | 0/0 | 0/0 | 0/0 | 0/0 |
RESULTS
d-Mannose Complex with SP-A
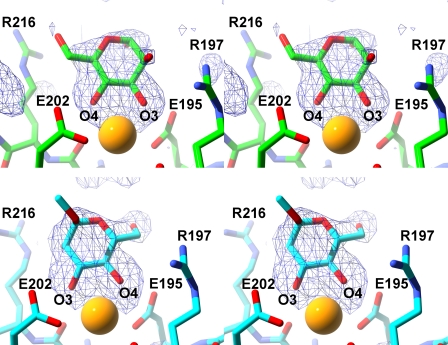
d-Mannose is capable of assuming either α- or β-anomeric forms in solution, and the sugar binds here as the α-anomer, as indicated by simulated annealing-omit and other difference electron density maps (Fig. 1). The structural data cannot rule out binding of the β-anomer by SP-A in the non-crystalline state because a close contact between the side chain of Glu-149 from a crystallographically related molecule and the mannose C1 may preclude binding of the β-anomer in the crystal. For d-mannose, the 3-OH and 4-OH groups are the only vicinal equatorial hydroxyl groups and therefore the only possible oxygen ligands for the lectin calcium. In the complex, the sugar forms two calcium coordination bonds and three hydrogen bonds with the protein through the ring 3-OH and 4-OH groups at the lectin site, which is partially formed by the long loop. When the hydroxyl oxygens bind to the calcium ion, they replace the two water molecules in the coordination shell observed in ligand-free SP-A. In this complex, the calcium coordination shell comprises five protein oxygen ligands from four residues: side chain oxygens of Glu-195, Glu-202, Asn-214, and Asp-215 plus one main chain oxygen from Asp-215. One water molecule is shared between the calcium and the main chain oxygen of Arg-197 (Table 2). The sugar ring 3-OH and 4-OH groups also form hydrogen bonds with side chain oxygens of Glu-195 and Glu-202, respectively. The 4-OH group additionally forms a hydrogen bond with the Asn-214 side chain amide moiety. There is some evidence in the electron density maps for a conformation of mannose where the O3 and O4 hydroxyl groups have switched positions in the calcium coordination sphere, rotating the sugar ring 180° about an axis bisecting the mannose ring; however, the density was not strong enough to fit an alternate conformation.
FIGURE 1.
SP-A complex with d-mannose and α-methyl-d-mannose. The upper and lower panels show stereoviews of the d-mannose and α-methylmannose complexes, respectively. All maps are omit (Fo − Fc) at the 3σ level, generated in CNS. The calcium ion in the lectin site is depicted as a yellow sphere.
TABLE 2.
Distances between calcium ions and coordinating oxygens in unliganded and complex SP-A structures
SC, side chain; MC, main chain; NA, not applicable.
| Asp-215 SC | Asp-215 MC | Asn-214 SC | Glu-195 SC | Glu-202 SC | Arg-197 MC | Water | Ligand 1-OH to Glu-195 SC | Ligand 2-OH to Glu-202 SC | |
|---|---|---|---|---|---|---|---|---|---|
| Unliganded | 2.8 | 2.3 | 2.5 | 2.4 | a | 2.9 | NA | 2.6 | 2.7 |
| Glycerol | 2.5 | 2.6 | 2.6 | 2.6 | 2.9 | a | 2.8 | 2.8 | 2.7 |
| Mannose | 2.3 | 2.4 | 2.3 | 2.4 | 2.7 | a | 2.5 | 2.3 | 2.4 |
| α-MMA | 2.5 | 2.5 | 2.3 | 2.4 | 3.0 | a | 2.6 | 2.4 | 2.3 |
a No direct interaction.
Other interactions between d-mannose and the protein include contacts between mannose and Arg-197 and Arg-216, on either side of the mannose ring. The O6 atom of mannose is 3.0 Å from the Nϵ side chain atom of Arg-216. Similarly, the axial 2-OH group may interact with NH1 or NH2 of Arg-197, sitting 3.2 Å from the guanidinium group.
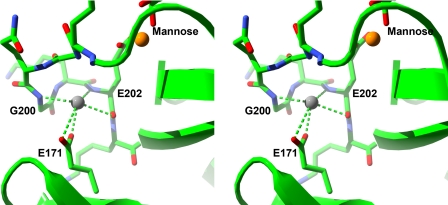
In the mannose complex, the main chain of the long loop is closer to the adjacent short loop (residues 171–177) than in the starting unliganded model structure (e.g. 3.5 Å closer from the Gly-200 nitrogen to Pro-175 Cβ). This difference is accompanied in the complex by a strong density peak in a site formed by the backbone nitrogens of Gly-200 and Glu-202 (2.8 and 3.0 Å, respectively), the backbone oxygen of Glu-202 (2.9 Å), and the side chain carboxylate group of Glu-171 (2.5 Å). The geometry of the interactions in the site is not tetrahedral (Fig. 2), which precludes interpretation of this density as water, and the site appears to be suitable for a metal ion. The only cations present in the crystal soaking solutions are calcium, lithium, and sodium ions. Lithium would be expected to have shorter coordination distances than those observed and a smaller peak height in the electron density map, if it would even be visible in maps at this resolution. A search of calcium- and sodium-containing structures in the Protein Data Bank (26) for four-coordinate metal sites interacting with backbone nitrogens revealed 29 structures for sodium and only three for calcium. Because calcium very rarely interacts with backbone nitrogen atoms and the density peak is not strong enough to suggest a calcium ion, this peak is more likely to correspond to a sodium ion than calcium. It is interesting to note that previous studies with SP-A showed an effect of both sodium and calcium concentrations on binding of SP-A to rough LPS-coated beads (14).
FIGURE 2.
Cation-binding site in ligand complexes. Shown is a stereo diagram of the cation-binding site found in liganded SP-A complexes. SP-A is represented as a green ribbon with the exception of residues 197–203 (long loop; upper) and the side chain of Glu-171 (short loop; lower), which are depicted in stick representation. Interactions between the bound sodium ion (gray sphere) and the protein are shown. The ligand-binding site is in the upper right-hand corner, with the calcium ion shown as a yellow sphere.
α-Methyl-d-mannoside Complex with SP-A
α-Methyl-d-mannoside (α-MMA), a nonreducing derivative of d-mannose with a 1-OH linkage to the methyl group, binds to SP-A in a different orientation from d-mannose. Although both sugars bind in the crystals as α-anomers, the mannoside binds in an orientation that is flipped 180° around a local symmetry axis, reversing the 3,4-order of the oxygen–calcium coordination bonds in the mannose complex to 4,3 for the mannoside (Fig. 1). This reversed orientation also leads to hydrogen bonds between 4-OH and 3-OH groups and Glu-195 and Glu-202, respectively, which are opposite pairs compared with the mannose complex. The 3-OH is potentially close enough to form a hydrogen bond with the side chain nitrogen of Asn-214. Alignment of the structures of the mannose and α-MMA complexes to compare the pyranose ring positions gives a root mean square deviation (r.m.s.d.) of 0.73 Å calculated using the ring atoms only. Other contacts between α-MMA and the two arginine side chains nearby include the sugar 2-OH with Arg-216 Nϵ (2.9 Å) and C6 with Arg-197 NH2 (3.0 Å). Analogous to the mannose complex, there is a sodium ion modeled into density between the long and short loops making very similar interactions.
There are some contacts between the ligand and a neighboring molecule in the crystal that might influence the observed binding orientation. First, in the current orientation, the C6 of the mannose ring makes close contacts to a neighboring carboxyl oxygen of Glu-149 of an adjacent subunit related by crystal symmetry. This contact may have altered the observed conformation of the mannose ring. Second, the mannose-like conformation of α-MMA could potentially fit into the crystal binding site; however, the O-methyl group would need to displace a conserved water molecule held in place by the backbone amide of Glu-149. It is possible that both orientations would be observed in the crystal structure were it not for this contact.
Glycerol Complex with SP-A
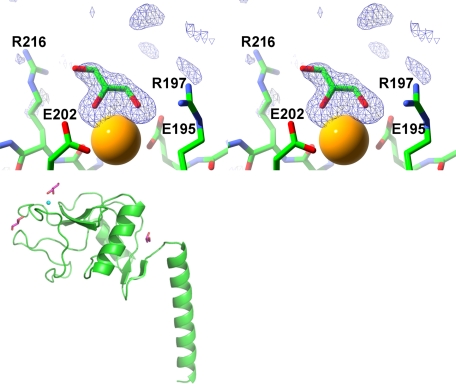
The crystal structure of the SP-A NCRD containing glycerol (Fig. 3) as cryoprotectant shows a glycerol molecule bound at the lectin site. As seen in the sugar complexes, the glycerol bound to the lectin site donates two calcium coordination bonds through vicinal hydroxyl groups. The third hydroxyl group approaches the side chain of Arg-216 with a distance of 3.5 Å from the glycerol O3 to the Nϵ of Arg-216. The remainder of the calcium coordination shell is the same as in the other complexes. Two additional glycerol molecules are observed, one near the lectin site and another at the linkage region between the neck domain and the CRD. The first glycerol molecule makes hydrogen bonds with the backbone carbonyl group of Pro-196 and the backbone nitrogen of Gln-199 and van der Waals contacts with the backbone atoms of residues 175 and 176 of the short loop and residues 196–199 of the long loop. The other non-lectin glycerol molecule is almost totally buried in a pocket formed at the linker region between the CRD and neck domain. It forms hydrogen bonds with the side chain of Lys-160 and the backbone nitrogen of Thr-121 as well as several waters. Additional van der Waals contacts are made by residues 109–112, Ser-120, and Ile-157. The crystal structure of a crystal grown similarly with glycerol present but subsequently soaked in the same buffer but without glycerol or another sugar present was consistent with the unliganded conformation described below (data not shown).
FIGURE 3.
SP-A·glycerol complex structure. Shown is a stereo diagram of an electron density map calculated with Fourier coefficients Fo − Fc of the final refined SP-A·glycerol complex at 1.9 Å resolution, omitting the glycerol atoms from the calculation (upper). The lectin-binding site is shown with the calcium ion as a yellow sphere. The other two binding sites are shown in the lower drawing of the SP-A NCRD monomer. The protein backbone is shown in green in ribbon representation, and glycerol molecules are in magenta. The lectin calcium ion shown as a small cyan sphere in the upper left of the drawing.
Unliganded SP-A
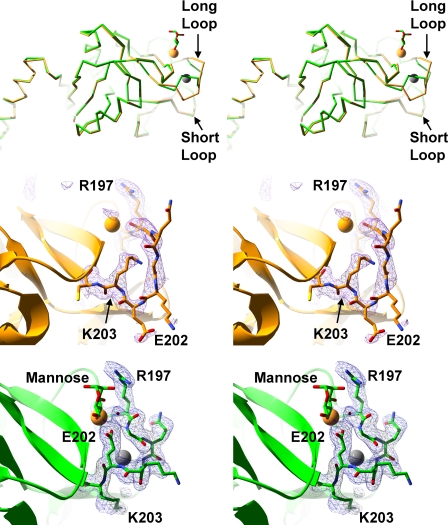
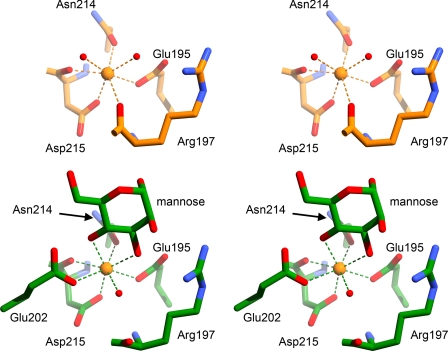
Crystals prepared similarly as the complexes but without sugar added were analyzed to provide a comparison between liganded and unliganded SP-A. The calcium coordination at the lectin site is similar to that reported previously for the unliganded SP-A NCRD, although that structure was solved at a higher salt concentration (19). However, there are minor differences between the two unliganded structures in the C-terminal end of the long loop, and we used the present one as the more accurate comparison with liganded SP-A. The residues of the long loop are significantly different between the unliganded and liganded complexes (Fig. 4). In the present unliganded structure, calcium coordination is achieved by the side chain oxygens of Glu-195, Asn-214, and Asp-215 and the backbone carbonyl oxygens of Arg-197 and Asp-215. Two water molecules also coordinate the lectin site calcium, bringing the total coordination sphere to seven atoms (Fig. 5). Glu-202 is in a solvent-exposed conformation such that its side chain points away from the lectin site calcium, unlike the liganded structure, where this residue coordinates the lectin calcium. The side chain of Lys-203 occupies space in between the long and short loops, where it makes a salt bridge with the side chain of Glu-171. In the liganded structures, this site is filled by a sodium ion. The overall effect of the conformation of the long loop in the unliganded form is a larger gap between the long and short loops than that observed with liganded complexes.
FIGURE 4.
Comparison of SP-A complex and native structures. Upper, stereo diagram of the Cα trace of the SP-A NCRD·mannose complex (green) superimposed on the unliganded SP-A NCRD (orange). Middle and lower, stereo diagrams of an electron density map calculated with Fourier coefficients Fo − Fc of unliganded SP-A (middle) at 2.3 Å resolution and the SP-A·mannose complex (lower), omitting atoms from residues 197–203 from the calculation. The lectin calcium and sodium ions are shown as yellow and gray spheres, respectively.
FIGURE 5.
Comparison of the lectin site interactions of the complex and unliganded SP-A structures. Upper, unliganded SP-A shown with carbons colored orange. Lower, SP-A·mannose shown with carbons colored green. Calcium ions are orange spheres, and water molecules are red spheres. Calcium coordination interactions are shown with dashed lines.
DISCUSSION
SP-A Complexes with Carbohydrates
In all SP-A complexes, consistent with the calcium dependence of the interactions, cis-diol hydroxyl oxygens from the ligands coordinate the calcium ion bound at the lectin site. The variable ring orientations in d-mannose and α-MMA demonstrate that SP-A is capable of binding sugars in more than one orientation. This “ambidexterity” in sugar binding is a feature that was initially described for this family of proteins in MBP-A structures (27, 28). SP-D complexes subsequently have shown that additional sugar residues in oligosaccharides can influence the ring orientation of the residue bound at the lectin site (29). Other SP-D studies have shown that both l-glycero-d-mannose and d-glycero-d-mannose, heptoses found in bacterial lipopolysaccharides, can coordinate calcium via either the glycerol or mannose moieties (29, 30). Such versatile accommodation of broadly similar sugar ligands may be a general feature in this family of pattern recognition proteins and may serve to broaden the range of potential targets.
Unlike the SP-A·monosaccharide complexes, which show only one bound sugar per CRD, the SP-A complex with glycerol reveals two bound glycerol molecules in addition to the lectin-bound ligand. One of the two non-lectin glycerol sites is located in between the long and short loops. The other is located in a pocket between the head and neck domains. Given that a glycerol molecule structurally mimics one-half of a hexose monosaccharide, these additional glycerol sites may be structural markers for non-lectin monosaccharide-binding sites. If so, their distal locations suggest that they are unlikely to bind the same oligosaccharide as that bound by the lectin site. Such additional sites could, however, reflect further points of contact for the polyvalent microbial targets favored by SP-A and other lectins.
Ligand-induced Conformational Change in SP-A
The differences between the unliganded and complex SP-A structures correspond to a significant ligand-associated conformational change. Such a feature has not been described for other collectins, such as MBP and SP-D. In SP-A, the changes take place primarily in the long loop. Whereas the tertiary structures in the unliganded protein or ligand complexes are indistinguishable from one another when averaged over all backbone atoms (r.m.s.d. < 0.9 Å), the complexes differ substantially (r.m.s.d. > 3 Å) from the unliganded structure in a portion of the long loop (Table 3). This portion corresponds to residues 197–203 (sequence RGQGKEK), which form one wall of the lectin site. In the complex structures, the long loop shows evidence of conformational stabilization with well defined and continuous electron density in the region. In the unliganded SP-A structure, however, the long loop suggests greater flexibility, as evidenced by high B-factors and broken electron density locally. These alternate loop conformations may be interconvertible depending upon whether ligand is present because soaking glycerol out of crystals returns the structure to the unliganded conformation (data not shown). Furthermore, in the SP-A·sugar complexes, the main chain of the long loop is closer to the adjacent short loop (residues 171–177) than in the unliganded structure (e.g. 3.5 Å closer from the Gly-200 nitrogen to Pro-175 Cβ). This closing of the space between the two loops produces a more compact protein structure in the complexes relative to unliganded SP-A.
TABLE 3.
r.m.s.d. values of superimpositions of unliganded and complex SP-A structures
| r.m.s.d. of backbone atoms (all residues/loop 197–203) |
|||
|---|---|---|---|
| Unliganded | Glycerol | Mannose | |
| Å | |||
| Glycerol | 0.82/3.39 | ||
| Mannose | 0.83/3.32 | 0.21/0.22 | |
| α-Methylmannose | 0.82/3.33 | 0.20/0.32 | 0.16/0.25 |
Lectin Site Changes with Carbohydrate Binding
The lectin calcium ion coordination undergoes two significant changes when carbohydrate binds at the site. The more striking effect involves the Glu-202 side chain, in which the carboxylate group translocates 12.8 Å to fill a previously unfilled calcium coordination position in the complexes. As part of this translocation, the side chains of Glu-202 and Lys-203 effectively swap positions. In the complexes, Glu-202 bends inward to coordinate the lectin calcium, whereas Lys-203 extends out into the solvent. However in the unliganded structure, Glu-202 extends outward toward the solvent, whereas Lys-203 folds back underneath the long loop and forms a salt bridge with Glu-171. This salt bridge likely stabilizes the loop in the absence of carbohydrate binding. It is notable that although Glu-202 coordinates calcium in the SP-A complexes, the calcium–oxygen distances are relatively long. The average distance for calcium–glutamate coordination in 1112 published eight-coordinate calcium is 2.5 Å (26). For the SP-A complexes, the coordination distances are 2.7–3.0 Å, respectively. It is possible that binding of longer or more structurally complex ligands may shorten this distance. However, this longer bond length also may be an intrinsic feature of SP-A, which may facilitate a distinct mode of ligand interactions.
A second difference between unliganded and liganded SP-A involves Arg-197. Its backbone oxygen coordinates calcium in the unliganded form. In the complexes, however, a water molecule inserts itself between the calcium and the Arg-197 backbone, replacing the Arg-197 backbone oxygen in the calcium coordination shell. This substitution increases favorable interactions in the complex through additional hydrogen bonds provided by the water. The insertion of this water molecule moves the Arg-197 backbone oxygen away from the calcium (e.g. from 2.6 to 5.3 Å in the unliganded and mannose complex structures, respectively). The displacement is facilitated by a change in the backbone conformation of Arg-197 and Gly-198. In the unliganded structure, these two residues adopt backbone dihedral angles characteristic of extended conformation (ϕ/ψ = −173.1°/135.3° and 179.6°/165.8°, respectively). In the complexes, however, ϕ/ψ angles for those same residues are characteristic of α-helical conformation (ϕ/ψ = −146.4°/36.6° and −80.6°/−9.0°, respectively, in the SP-A·mannose complex). In all, the displacement of the Arg-197 oxygen is 2.7 Å, comparing its position in the liganded and complex structures.
Crystallographic data of other collectins show that carbohydrate-induced conformational changes are minimal in these proteins and involve mostly side chains (31–33). This relative conformational insensitivity to sugar binding is in contrast to the significant transition caused by calcium binding to the lectin site. In MBP, this event has been shown crystallographically to correspond to a proline isomerization and associated structural changes (34). Although no crystal structure has been determined for calcium-free SP-A, calcium-dependent changes at the lectin site have been observed spectroscopically (35). Moreover, the key residues that effect the transition in MBP are conserved in SP-A. Therefore, it is likely that SP-A undergoes a similar calcium-dependent conformational transition. However, unlike MBP, SP-A additionally undergoes a second, major transition when a carbohydrate substrate binds at the lectin site.
Human CD23, a low affinity IgE receptor and C-type lectin of the asialoglycoprotein class, does show a similar conformation to that of the SP-A long loop in the unliganded state. It remains to be seen, if crystallographic complexes become available for CD23, whether a similar conformational change occurs upon carbohydrate binding at the lectin site (36). Interestingly, it has been suggested that in CD23, the conformational form that corresponds to an unoccupied lectin site may be the one that binds to IgE through sites on the protein that do not contain carbohydrate (36).
P-selectin, a platelet cell adhesion protein that is structurally unrelated to SP-A except for a common CRD, has been shown to undergo a similar ligand-induced conformational change to that observed for SP-A. For P-selectin in the liganded form, the structural equivalent of Glu-202 in SP-A coordinates the lectin calcium, whereas this residue is solvent-exposed in the unliganded form. The conformational change in P-selectin is localized to a similar region of the long loop as observed for SP-A, although their unliganded structures differ somewhat. Furthermore, the Glu-202 side chain equivalent translocates only 4.8 Å in P-selectin, a modest difference compared with the distance of 12.8 Å seen in SP-A. It has been proposed that for P-selectin, this ligand-induced conformational change converts P-selectin from a low to a high affinity binding state (37).
Other Cation-binding Sites in SP-A
In several SP-A structures, variable binding of non-lectin cations is observed at a common site between the long and short loops. This site, which involves the Glu-171 side chain on the short loop, is occupied variously by the Lys-203 amino group (in the unliganded structure), a samarium ion (where samarium replaces calcium), or a sodium ion (in the carbohydrate complexes). Curiously, calcium binding in this site, which would be expected, has not been observed in any structures to date. In the previously reported samarium complex (19), calcium was soaked out and samarium soaked into crystals in preparation for anomalous phasing. Two samarium ions were located in the Patterson maps: one bound at the lectin site and the other at a site formed by the side chain of Glu-171 and the backbone oxygen of Lys-201. In the sugar complexes, a sodium ion is bound by the backbone nitrogens of Gly-200 and Glu-202, the backbone oxygen of Glu-202, and the side chain of Glu-171. Electron density, coordination geometry and ligands, and the B-factor strongly suggested the identity of this ion as sodium rather than calcium. In the presence of calcium but without carbohydrate (i.e. unliganded structure), the Lys-203–Glu-171 salt bridge precludes metal ion binding at this site, whereas in the complexes, the metal ion occupies the space vacated by the Lys-203 amino group. These observations suggest that SP-A requires at least some stabilization of the two-loop structure; however, its requirements are relatively loose and opportunistic compared with the more structured calcium sites of the other collectins. The data also suggest that Glu-171, which is invariant in SP-A across species, may be an important stabilizing residue. This conclusion is supported by the greater susceptibility of a Glu-171 → Ala mutant to trypsin digestion compared with wild-type SP-A.3 Even with the stabilization afforded by residues such as Glu-171 and available cations, the data collectively suggest that the long and short loops of SP-A are inherently more flexible than lectins that contain structural calcium ions in the CRD.
Conclusion
SP-A is unusual among members of its class, the collectin subfamily of C-type lectins, in that its long and short loops lack aspartate and glutamate side chains to bind one or both non-lectin calcium ions, and there are no other aspartate or glutamate residues in the vicinity of the expected calcium sites that could replace the missing conserved residues. In MBP and SP-D CRDs, the structural calcium sites form multiple interrelated interactions that effectively cross-link the two loops in a highly stable conformation. Relatively rigid preformed sites tend to effect highly selective higher affinity binding of ligands, whereas more flexible open sites tend in the opposite direction. The structural data presented herein suggest that SP-A belongs to the latter category.
The ability to undergo a significant ligand-induced conformational change may allow SP-A to bind not only to carbohydrates but also to certain non-carbohydrate substrates, such as dipalmitoylphosphatidylcholine and lipid A of Gram-negative bacteria. In the airways, SP-A is abundantly associated with the major surfactant lipid, dipalmitoylphosphatidylcholine, on the surface of tubular myelin. Attachment of SP-A to dipalmitoylphosphatidylcholine membrane surfaces potentially could occur proximal to the lectin site assisted by an alternate conformation of the long loop. It is also possible that the SP-A conformational change could assist transfer of SP-A, e.g. from the dipalmitoylphosphatidylcholine surface to glycolipids or carbohydrate-rich pathogen surfaces. We suggest that such notable properties of SP-A as the ligand-induced conformational change, flexible loop structure, and ability to utilize varied cations for stabilization may allow SP-A to broaden its range of ligand classes and ultimately increase the scope of innate immune surveillance in the lung.
This work was supported, in whole or in part, by National Institutes of Health Grant AI083222 from NIAID (to B. A. S.). This work was also supported by a Department of Veterans Affairs merit award (to F. X. M.).
The atomic coordinates and structure factors (codes 3PAK, 3PAQ, 3PAR, and 3PBF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
F. X. McCormack and H. Wu, unpublished data.
- SP-A
- surfactant protein A
- MBP
- mannose-binding protein
- CRD
- carbohydrate recognition domain
- NCRD
- neck and carbohydrate recognition domain
- α-MMA
- α-methyl-d-mannoside
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Seaton B. A., Crouch E. C., McCormack F. X., Head J. F., Hartshorn K. L., Mendelsohn R. (2010) Innate Immun. 16, 143–150 [DOI] [PubMed] [Google Scholar]
- 2. Chroneos Z. C., Sever-Chroneos Z., Shepherd V. L. (2010) Cell. Physiol. Biochem. 25, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartshorn K. L. (2010) Front. Biosci. 2, 527–546 [DOI] [PubMed] [Google Scholar]
- 4. Sano H., Kuroki Y. (2005) Mol. Immunol. 42, 279–287 [DOI] [PubMed] [Google Scholar]
- 5. Haagsman H. P., Hogenkamp A., van Eijk M., Veldhuizen E. J. (2008) Neonatology 93, 288–294 [DOI] [PubMed] [Google Scholar]
- 6. Kingma P. S., Whitsett J. A. (2006) Curr. Opin. Pharmacol. 6, 277–283 [DOI] [PubMed] [Google Scholar]
- 7. Whitsett J. A. (2005) Biol. Neonate 88, 175–180 [DOI] [PubMed] [Google Scholar]
- 8. Wright J. R. (2004) Biol. Neonate 85, 326–332 [DOI] [PubMed] [Google Scholar]
- 9. McCormack F. X., Whitsett J. A. (2002) J. Clin. Invest. 109, 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zelensky A. N., Gready J. E. (2005) FEBS J. 272, 6179–6217 [DOI] [PubMed] [Google Scholar]
- 11. Crouch E., Hartshorn K., Ofek I. (2000) Immunol. Rev. 173, 52–65 [DOI] [PubMed] [Google Scholar]
- 12. Kishore U., Greenhough T. J., Waters P., Shrive A. K., Ghai R., Kamran M. F., Bernal A. L., Reid K. B., Madan T., Chakraborty T. (2006) Mol. Immunol. 43, 1293–1315 [DOI] [PubMed] [Google Scholar]
- 13. Kuan S. F., Rust K., Crouch E. (1992) J. Clin. Invest. 90, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Iwaarden J. F., Pikaar J. C., Storm J., Brouwer E., Verhoef J., Oosting R. S., van Golde L. M., van Strijp J. A. (1994) Biochem. J. 303, 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartshorn K. L., White M. R., Voelker D. R., Coburn J., Zaner K., Crouch E. C. (2000) Biochem. J. 351, 449–458 [PMC free article] [PubMed] [Google Scholar]
- 16. Hartshorn K. L., White M. R., Shepherd V., Reid K., Jensenius J. C., Crouch E. C. (1997) Am. J. Physiol. Lung Cell. Mol. Physiol. 17, L1156–L1166 [DOI] [PubMed] [Google Scholar]
- 17. Benne C. A., Kraaijeveld C. A., van Strijp J. A., Brouwer E., Harmsen M., Verhoef J., van Golde L. M., van Iwaarden J. F. (1995) J. Infect. Dis. 171, 335–341 [DOI] [PubMed] [Google Scholar]
- 18. Malhotra R., Haurum J. S., Thiel S., Sim R. B. (1994) Biochem. J. 304, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Head J. F., Mealy T. R., McCormack F. X., Seaton B. A. (2003) J. Biol. Chem. 278, 43254–43260 [DOI] [PubMed] [Google Scholar]
- 20. Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 21. Jones T. A., Kjeldgaard M. (1992) The Manual, Uppsala Software Factory, Uppsala, Sweden [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 24. Cason C., Froehlich T., Kopp N., Parker R. (2005) Persistence of Vision Raytracer, Version 3.6, Persistence of Vision Raytracer Pty. Ltd., Victoria Australia [Google Scholar]
- 25. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 26. Hsin K., Sheng Y., Harding M. M., Taylor P., Walkinshaw M. D. (2008) J. Appl. Crystallogr. 41, 963–968 [Google Scholar]
- 27. Weis W. I., Drickamer K., Hendrickson W. A. (1992) Nature 360, 127–134 [DOI] [PubMed] [Google Scholar]
- 28. Weis W. I., Drickamer K. (1996) Annu. Rev. Biochem. 65, 441–473 [DOI] [PubMed] [Google Scholar]
- 29. Crouch E., McDonald B., Smith K., Roberts M., Mealy T., Seaton B., Head J. (2007) Biochemistry 46, 5160–5169 [DOI] [PubMed] [Google Scholar]
- 30. Wang H., Head J., Kosma P., Brade H., Müller-Loennies S., Sheikh S., McDonald B., Smith K., Cafarella T., Seaton B., Crouch E. (2008) Biochemistry 47, 710–720 [DOI] [PubMed] [Google Scholar]
- 31. Ng K. K., Kolatkar A. R., Park-Snyder S., Feinberg H., Clark D. A., Drickamer K., Weis W. I. (2002) J. Biol. Chem. 277, 16088–16095 [DOI] [PubMed] [Google Scholar]
- 32. Shrive A. K., Tharia H. A., Strong P., Kishore U., Burns I., Rizkallah P. J., Reid K. B., Greenhough T. J. (2003) J. Mol. Biol. 331, 509–523 [DOI] [PubMed] [Google Scholar]
- 33. Shrive A. K., Martin C., Burns I., Paterson J. M., Martin J. D., Townsend J. P., Waters P., Clark H. W., Kishore U., Reid K. B., Greenhough T. J. (2009) J. Mol. Biol. 394, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ng K. K., Park-Snyder S., Weis W. I. (1998) Biochemistry 37, 17965–17976 [DOI] [PubMed] [Google Scholar]
- 35. McCormack F. X., Stewart J., Voelker D. R., Damodarasamy M. (1997) Biochemistry 36, 13963–13971 [DOI] [PubMed] [Google Scholar]
- 36. Wurzburg B. A., Tarchevskaya S. S., Jardetzky T. S. (2006) Structure 14, 1049–1058 [DOI] [PubMed] [Google Scholar]
- 37. Somers W. S., Tang J., Shaw G. D., Camphausen R. T. (2000) Cell 103, 467–479 [DOI] [PubMed] [Google Scholar]