Abstract
SRPK2 belongs to a family of serine/arginine (SR) protein-specific kinases (SRPKs), which phosphorylate SR domain-containing proteins in the nuclear speckles and mediate the pre-mRNA splicing. Previous studies have shown that SRPK2 plays a pivotal role in cell proliferation and apoptosis. However, how SRPK2 is regulated during the apoptosis is unclear. Here, we show that SRPK2 is cleaved by caspases at Asp-139 and -403 residues. Its N terminus cleaved product translocates into the nucleus and promotes VP16-induced apoptosis. Akt phosphorylation of SRPK2 prevents its apoptotic cleavage by caspases. 14-3-3β, the binding partner of Akt-phosphorylated SRPK2, further protects it from degradation. Hence, our results suggest that the N-terminal domain of SRPK2 cleaved by caspases translocates into the nucleus, where it promotes chromatin condensation and apoptotic cell death.
Keywords: Akt PKB, Apoptosis, Caspase, Nuclear Translocation, Protein-Protein Interactions, 14-3-3, SRPK2
Introduction
Serine/arginine (SR)2 proteins are a family of RNA-binding proteins that contain a marker SR domain enriched with serine/arginine repeats. Several prototypical SR proteins are essential splicing factors, but the majority of SR domain-containing factors are implicated in altering splice site selection in vitro or in transfected cells. SR proteins and the related proteins are generally believed to modulate splice site selection via RNA recognition motif (RRM)-mediated binding to exonic splicing enhancers and SR domain-mediated protein-protein and protein-RNA interactions during spliceosome assembly (1). RNA-binding SR proteins play critical roles in multiple steps in gene expression, from transcriptional elongation, mRNA splicing, RNA export to translation. The integration of these activities by single SR proteins may constitute the requirement of SR proteins for cell viability and proliferation. Recent findings also suggest some unexpected roles of SR proteins in organizing gene networks in the nucleus, maintaining genome stability, and facilitating cell-cycle progression (2). Presumably, all SR domain-containing proteins are post-translationally modified by phosphorylation, and reversible phosphorylation has been shown to play an important role in splicing.
Two families of kinases, SR protein-specific kinase (SRPK), and Clk/Sty, have been identified to phosphorylate SR domain-containing splicing factors. SRPKs, a family of cell cycle-regulated protein kinases, phosphorylate SR domain-containing proteins in the nuclear speckles and mediate the pre-mRNA splicing. SRPK1 and SRPK2 are highly specific kinases for the SR family of splicing factors. SRPK1 is predominantly expressed in pancreas, whereas SRPK2 is highly expressed in brain, although both are coexpressed in other human tissues and in many experimental cell lines (3). The SRPK family of kinases, containing bipartite kinase domains separated by a unique spacer, is mainly localized in the cytoplasm, which is critical for nuclear import of SR proteins in a phosphorylation-dependent manner. Removal of the spacer in SRPK1 has little effect on the kinase activity, but triggers the nuclear translocation of kinases and consequently induces aggregation of splicing factors in the nucleus (4). Fu et al. (5) identify and clone human SRPK1 in the pursuit of an activity that mediates splicing factor redistribution in the cell cycle. SRPK2 that is discovered based on its sequence similarity to SRPK1. A series of biochemical experiments demonstrate that SRPK1 and -2 are very similar with respect to their enzymatic activity and substrate specificity. Both kinases promote specific protein-protein interactions between SR domain-containing splicing factors and their overexpression induced the redistribution of splicing factors from the nuclear speckles to the nucleoplasm, indicating that both kinases may be involved in the regulation of spliceosome assembly in vivo (6).
In addition to phosphorylating SR proteins and regulating pre-mRNA splicing, SRPKs also play an important role in cell proliferation and apoptosis. For instance, SRPK1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas (7). Kamachi et al., report that SRPK1 is associated with the U1-snRNP autoantigen complex in healthy and apoptotic cells. SRPK1 is activated early during apoptosis, followed by caspase-mediated proteolytic inactivation at later time points. SRPKs are cleaved in Jurkat cells after multiple apoptotic stimuli, and the cleavage can be inhibited by overexpression of bcl-2 and bcl-x(L), and by exposure to soluble peptide caspase inhibitors. Incubation of recombinant caspases with in vitro-translated SRPKs demonstrates that they are in vitro substrates for caspases-8 and -9 (8). Recently, we have shown that SRPK2 triggers cell cycle progression in post-mitotic neurons and induces apoptosis through up-regulation of nuclear cyclin D1 (9). Ablation of SRPK2 abrogates cyclin A1 expression in leukemia cells and arrest cells at G1 phase. Knocking down of SRPK2 induces caspase-3 activation in cortical neurons (9). SRPK2 overexpression increases leukemia cell proliferation and elicits primary cortical neuronal cell death (9, 10), indicating that SRPK2 is a critical player in regulating cell survival. Too much or too little may tilt the balance, leading to programmed cell death.
The 14-3-3 proteins are a family of phosphoserine/phosphothreonine-binding molecules that control the functions of a wide array of cellular proteins and promote cell survival. 14-3-3 binds the client proteins through an amphipathic binding cleft that preferentially recognizes the phosphorylated motifs: RSXpSXP or RXXXpSXP (11), which share a common region in the consensus Akt phosphorylation elements preserved in numerous Akt substrates. Our previous study has shown that Akt phosphorylates SRPK2 on Thr-492 and promotes its nuclear translocation leading to cyclin D1 up-regulation, cell cycle re-entry and neuronal apoptosis. However, overexpression of 14-3-3β prevents this event and promotes neuronal survival (9).
Caspases (cysteine proteases with aspartate specificity) are a family of proteases specifically recognizes an Asp residue at the substrate P1 site (12). Based on their substrate preferences, three caspases subgroups were identified. The first subgroup (caspases-1, -4, and -5) prefers bulky hydrophobic residues in the P4 site and has the optimal substrate cleavage sequence WEXD. The second subgroup (caspases-2, -3, -7, and CED-3) favors an Asp in P4 and preferentially cleaves targets C-terminal to a DEXD motif. The third subgroup (caspases-6, -8, and -9) are less discriminating in their P4 preferences; their optimal substrate cleavage sequence is (L/V)EXD (12). In this report, we show that SRPK2 is cleaved in vitro by recombinant caspase-3. Asp-139 and Asp-403 residues are two major cutting sites. Apoptotically cleaved SRPK2 promotes VP16-induced apoptosis. Caspase cleavage of SRPK2 releases its N-terminal fragment that translocates to the nucleus and further promotes apoptotic cell death.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
HEK293 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mg/ml glutamine, and 100 units of penicillin-streptomycin at 37 °C in a humidified incubator containing 5% CO2. Anti-Myc, anti-caspase-3, anti-PARP, and anti-p473AKT antibodies were purchased from Cell Signaling. Anti-tubulin antibody, 4,6-diamidino-2-phenylindole (DAPI), and Etoposide (VP16) were from Sigma. Anti-acinus and anti-SRPK2 antibodies were from BD Bioscience. Glutathione-Sepharose 4B was supplied by Pharmacia Biotech. The purified active caspase-3 and Akt proteins were purchased from Strategene.
Plasmids
Full-length, truncated, and site-mutated SRPK2 were inserted into pEGFP-C2 between BglII and SalI cutting sites, or into pEGX-4T-2 between SalI and NotI cutting sites. 14-3-3β-wt and K50E were inserted into pEGFP-C2 between BglII and EcoR I cutting sites. The site mutations were constructed using overlap elongation PCR.
Preparation of Cytosolic Fraction (S-100) from HEK293 Cells
The procedures were performed as described previously (13). Briefly, the pellets of 293 cells were washed once with ice-cold PBS and resuspended in 5 volumes of buffer C (20 mm HEPES-KOH, pH 7.5), 10 mm KCl, 1.5 mm MgCl2, 1 mm sodium EDTA, 1 mm sodium EGTA, 1 mm dithiothreitol (DTT), and 0.1 mm PMSF, supplemented with protease inhibitors mixture). After sitting on ice for 15 min, the cells were broken by passing 15 times through a G22 needle. After centrifugation 1000 × g for 10 min at 4 °C., the supernatants were further centrifuged at 105 × g for 30 min in an ultracentrifuge (Beckman). The resulting supernatants were used for in vitro apoptotic assay. Cytochrome c and dATP were added into S-100 extract to initiate caspase cascade.
TnT Quick-coupled Transcription/Translation Reaction and Cell-free Apoptotic Cleavage Assay of SRPK2
SRPK2 was subcloned into TNT vector (Promega, Madison, WI), and in vitro transcripted and translated in rabbit reticulocyte lysate in the presence of [35S]methionine. The reaction components T7 Quick Master Mix (16 ml), [35S]methionine (1 μl), PCR enhancer (1 μl), and plasmid DNA template (1 μg) were assembled in a 1.5-ml microcentrifuge tube with a total volume of 20 μl. The reaction mixture was incubated at 25 °C for 90 min. To conduct in vitro apoptotic cleavage, 2 μl of translation product was incubated with 200 μg (60 μl in volume) inactive or active S-100 at 37 °C for 3 h. The reaction mixture was then analyzed by SDS-PAGE, and then the gel was dried and exposed to the film.
In Vitro Kinase Assay
GST- or GFP-tagged SRPK2 and its mutants were immunoprecipitated with glutathione beads or anti-GFP antibody. After extensive washing, the precipitants were employed in the kinase assay. The precipitated SRPK2 were incubated with 2 μg of acinus in 20 μl of kinase reaction buffer (20 mm Tris, pH 7.5 with 10 mm MgCl2) containing 20 μm ATP and 1 μCi of [γ-32P]ATP for 30 min at 30 °C. Reactions were terminated by adding 5 μl of 5× sample buffer and boiling for 5 min. The sample was separated on a SDS-PAGE gel and autoradiographed.
Apoptotic Analysis of GFP-positive Cells
24 h after transfection, the cells were treated VP16 for another 16 h. Morphological changes in the nuclear chromatin of cells undergoing apoptosis were detected by staining with DAPI. Normal nuclei and apoptotic nuclei (condensed new moon-type or fragmented chromatin) were easily distinguished. A minimum of 300 cells from 3–5 different microscopic fields were counted to obtain reliable estimates of cell apoptosis. GFP-positive cells were counted, and the percentage of apoptosis was quantified.
TUNEL Assay
TUNEL staining was performed with an in situ cell death detection kit (Roche Diagnostics). Briefly, HEK293 cells were transfected with various SRPK2 mutants for 24 h and then treated with 50μm VP16 for another 16 h. Then, the adherent and non-adherent cells were collected, mounted onto the slides, and stained with the TUNEL reagents according to the manufacturer's instruction. The cells were also counterstained with DAPI, and the ratio of TUNEL/DAPI was calculated as apoptotic rate (%). A minimum of 300 cells from 3–5 different microscopic fields was counted.
Statistical Analysis
The data were analyzed with one-way ANOVA. p < 0.05 was considered as statistically significant.
RESULTS
SRPK2 Is a Substrate of Caspases
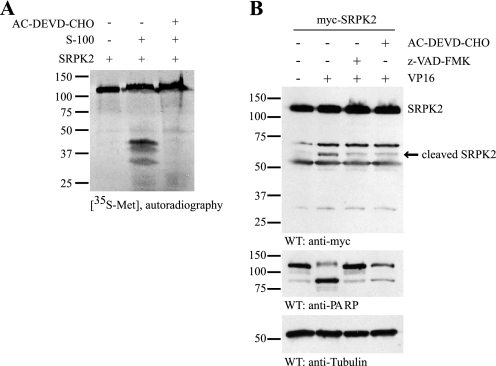
In our previous study, we show that SRPK2 promotes cell cycle progression and cell death in neurons (9). Moreover, SRPKs have been implicated in apoptosis (8). To determine whether SRPK2 is cleaved during apoptotic cells death and to study the molecular mechanism of its proteolytic cleavage, we performed a cell-free apoptotic assay. [35S]methionine-labeled SRPK2 was incubated with the activated cell-free apoptotic solution (S-100), prepared from cytosolic fraction of HEK293 cells, in the presence or absence of caspase-3/7 inhibitor. As shown in Fig. 1A, SRPK2 was cleaved in the activated S-100, which was mostly abolished in the presence of caspase-3/7 inhibitor AC-DEVD-CHO. To further study the cleavage of SRPK2 in the intact cells, HEK293 cells were transfected with the Myc-tagged SRPK2, and treated with VP16 (50 μm) in the presence or absence of caspase-3/7 or pan-caspase inhibitor. Immunoblotting with anti-Myc antibody demonstrated that VP16 treatment induced the cleavage of SRPK2 in intact cells. The two major fragmented bands were ∼60 and 30 kDa in size and were partially blocked by pan-caspase inhibitor z-VAD-FMK and caspase-3/7 inhibitor AC-DEVD-CHO (Fig. 1B), fitting with the observation in Fig. 1A. These data indicate that caspase 3 and 7 might be the major enzymes that contribute to its degradation. These results suggest that SRPK2 can be cleaved during apoptotic cell death in caspase-dependent manner, and SRPK2 may be the substrate of caspases. The slightly different cleavage patterns between in vitro cleavage assay and VP16-induced apoptosis might be due to different detection approaches. In the in vitro cleavage assay, SRPK2 was labeled with 35S, while in the apoptotic cells treated with VP16, SRPK2 was detected with the Myc antibody that recognizes the N-terminal fragment of SRPK2.
FIGURE 1.
SRPK2 is a substrate of caspases. A, SRPK2 is cleaved in the cell-free apoptotic solution in caspase-3/7-dependent manner. The active or inactive cell-free apoptotic S-100 was preincubated with or without 10 μm AC-DEVD-CHO for 30 min, followed by addition of [35S]methionine-labeled SRPK2. The reaction mixture was incubated at 37 °C for another 2 h, resolved in SDS-PAGE, and visualized by autography. B, VP16 treatment induces apoptotic SRPK2 cleavage in cells. HEK293 cells were transfected with Myc-tagged SRPK2, pretreated with 30 μm AC-DEVD-CHO or z-VAD-FMK for 0.5 h, and then treated with or without 50 μm VP16 for another 16 h. The cell lysates were analyzed by Western blot with the anti-Myc antibody.
Identification of Asp-139 and Asp-403 Residues as the Cleavage Sites in SRPK2
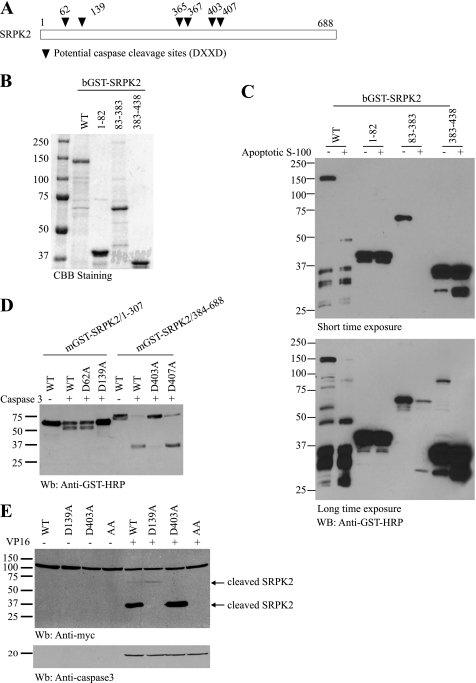
To locate the caspase cleavage sites in SRPK2, we examined the amino acid sequence of SRPK2, and identified six putative caspase cleavage sites (DXXD, Fig. 2A). According to the distribution of these potential cutting sites, various SRPK2 fragments were constructed into pGEX4T2 vector. GST-SRPK2 recombinant proteins were expressed and purified from bacteria and analyzed in the cell-free apoptotic cleavage assay (Fig. 2B). The wild-type SRPK2 full-length, and fragments SRPK2 (amino acids 83–383) and SRPK2 (amino acids 383–438) were cleaved in the active S-100, whereas the fragment SRPK2-(1–83) was not (Fig. 2C). Based on the molecular weight of the fragments and the putative caspase cleavage sites, we prepared numerous single point mutants. GST-tagged SRPK2 fragments with D62A, D139A, D403A, or D407A mutants were expressed in HEK293 cells, and the cell lysates were incubated with or without active caspase-3 at 37 °C for 2 h. While the mutants of 1–307-D62A and 1–307-D407A were markedly cleaved by caspase-3, the mutants 1–307-D139A and 384–688-D403A were not (Fig. 2D). These results indicate that Asp-139 and Asp-403 residues might be the major caspase cleavage sites in SRPK2. Therefore, we constructed D139A, D403A, and D139/403A mutants in the full-length of SRPK2. Upon VP16 treatment, wide-type SRPK2 showed two distinct bands with a molecular mass of about 60∼70 and 37 kDa, with the latter much stronger than the former, suggesting that the latter might be the predominant cutting product. However, mutants D139A and D403A only gave one band at 60∼70 and 37 kDa, respectively, indicating that Asp-139 might be the major cutting site. The apoptotic cleavage of SRPK2 was predominantly abolished in double mutant D139A/D403A (Fig. 2E). Therefore, Asp-139 and Asp-403 residues are the main cutting sites by the caspases, with Asp-139 residue the most principal cleavage position.
FIGURE 2.
Identification of Asp-139 and Asp-403 residues as the cleavage sites in SRPK2. A, schematic representation of potential caspase cleavage sites (DXXD) within SRPK2. B and C, various bGST-tagged SRPK2 fragments harboring the potential cleavage sites were constructed, and the recombinant proteins were bacterially expressed and analyzed in the in vitro cleavage assay. D, aspartate 139 and 403 residues are the caspase cleavage sites in SRPK2. GST-tagged SRPK2 fragments with D62A, D139A, D403A, or D407A mutation was expressed and incubated with or without active caspase-3 at 37 °C for 2 h. Mutants mGST-SRPK2/1–307-D139A and mGST-SRPK2/384–688-D403A were not cleaved by caspase-3. E, double mutant D139/403A totally abolished the apoptotic cleavage of SRPK2 in cells. HEK293 cells were transfected with wild-type and point mutant SRPK2 plasmids, followed by DMSO or VP16 treatment. The cell lysates were analyzed by immunoblotting with anti-Myc and caspase-3 antibodies. Mutation of Asp-139 or Asp-403 into A blocked its apoptotic cleavage (upper panel). Caspase-3 was selectively activated in VP16-treated cells (lower panel).
Phosphorylation of SRPK2 by Akt Prevents Its Proteolytic Cleavage
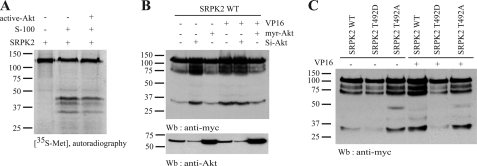
Our previous study showed that Akt phosphorylates SRPK2 at Thr-492 (9). To explore whether Akt phosphorylation can protect SRPK2 from proteolytic cleavage, we labeled SRPK2 with [35S]methionine, followed by incubation with active Akt prior to the in vitro caspase cleavage assay. We observed that SRPK2 incubation with active Akt decreased its degradation (Fig. 3A), suggesting that Akt phosphorylation may protect SRPK2 from caspase cleavage. To further explore the notion that Akt phosphorylation of SRPK2 might prevent it from apoptotic cleavage, we transfected Myc-SRPK2 into HEK293 cells, which were transfected with Myr-Akt, a plasma membrane resident constitutively active Akt, and siRNA of Akt, respectively. The transfected cells were treated with vehicle control or VP16 (50 mm) overnight. In the vehicle control-treated cells, knocking down of Akt elicited spontaneous SRPK2 apoptotic degradation, which was not detected in control or active Akt-transfected cells. On the other hand, VP16 treatment triggered SRPK2 apoptotic cleavage in both control and si-RNA Akt-treated cells, which was evidently inhibited in active Akt-expressed cells. Hence, down-regulation of Akt with it si-RNA promoted SRPK2 cleavage even in the absence of VP16, whereas the constitutively active Akt (myr-Akt) inhibited SRPK2 cleavage upon VP16 treatment (Fig. 3B). To further confirm the effect of Akt phosphorylation on SRPK2 cleavage, we employed two mutants, T492D and T492A, which mimic the constitutively phosphorylated state and non-phosphorylated state of SRPK2 by Akt, respectively. The transfected cells were subjected to VP16 treatment overnight. As shown in Fig. 3C, spontaneous degradation occurred in wild-type SRPK2 even in the absence of VP16 stimulation. The fragmentation of wild-type SRPK2 was further enhanced by VP16. By contrast, T492D robustly prevented SRPK2 from cleavage. SRPK2 T492A exhibited evident fragmentation regardless of VP16 treatment. These results indicate that Akt phosphorylation plays a protective role in SRPK2 cleavage.
FIGURE 3.
Activation and phosphorylation of SRPK2 by Akt prevent its cleavage. A, active AKT alleviates SRPK2 cleavage in S-100. [35S]methionine-labeled SRPK2 was phosphorylated by active Akt prior to the in vitro caspase cleavage assay. After being incubated with or without S-100 at 37 °C for another 2 h, the reaction mixtures were resolved in SDS-PAGE and visualized by autography. B, Akt plays an essential role in preventing SRPK2 apoptotic cleavage. HEK293 cells were transfected with Myc-tagged SRPK2 and myr-Akt or Si-Akt for 24 h, and treated with DMSO or VP16 for 16 h. The cell lysates were subjected to Western blotting analysis with anti-Myc and anti-Akt antibodies. Active Akt protected SRPK2 from degradation, whereas depletion of endogenous Akt enhanced SRPK2 cleavage (upper panel). Verification of Akt expression by immunoblotting (lower panel). C, Akt phosphorylation mimetic mutant prevents SRPK from apoptotic degradation. HEK293 cells were transfected with Myc-tagged SRPK2WT, T492A, or T492D. At 24 h after transfection, the cells were treated with vehicle or VP16 for another 16 h, and the cell lysates were subjected to Western blot analysis with anti-Myc antibody.
14-3-3β Binding Prevent Apoptotic SRPK2 Cleavage
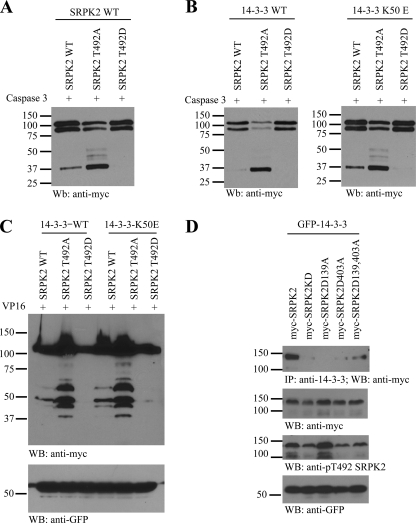
Accumulative evidence demonstrates that 14-3-3 proteins bind to numerous Akt substrates and prevent their apoptotic cleavage. Our previous study showed that Akt phosphorylation of SRPK2 facilitates its binding to 14-3-3β (9). We wonder whether 14-3-3β binding to SRPK2 would affect its proteolytic cleavage. To test this possibility, we cotransfected HEK293 cells with Myc-tagged SRPK2 WT, T492A, or T492D in a combination with empty vector, 14-3-3β-WT or 14-3-3β-K50E for 24 h, and then the cell lysates were incubated in vitro with active caspase-3 at 37 °C for 2 h. Immunoblotting analysis revealed that wild-type SRPK2 was cleaved by caspase-3, and the apoptotic cleavage was substantially elevated in SRPK2 T492A; in contrast, no fragmentation occurred in SRPK2 T492D (Fig. 4A). As expected, wild-type 14-3-3β, which strongly binds wild type and T492D SRPK2, inhibited SRPK2 cleavage; however, T492A mutant was markedly cut, fitting with its incapability of interacting with 14-3-3 (9). Nonetheless, 14-3-3β-K50E, which weakly binds SRPK2, was unable to protect wild-type SRPK2 from cleavage in vitro (Fig. 4B). To examine whether these observations also occur in the intact cells, we cotransfected SRPK2 and 14-3-3β wild-type or K50E into HEK293 cells and treated the transfected cells with VP16. In consistent with the in vitro experiment, 14-3-3β-WT but not K50E inhibited the fragmentation of SRPK-WT, and T492D did not show apparent cleavage bands under any circumstances. On the contrary, T492A was significantly cleaved in 14-3-3β-WT or K50E-transfected cells (Fig. 4C). Because D139A and D403A mutants prevented SRPK2 cleavage during apoptosis, we wondered whether they would affect its interaction with 14-3-3. The binding assay showed that the single point mutation decreased the interaction between 14-3-3β and SRPK2. Interestingly, the double mutant (D139A, D403A) possessed partial binding affinity to 14-3-3. The remnant binding activity might result from the conformational change by the double mutation. Nonetheless, the mutation did not substantially alter SRPK2 phosphorylation by the endogenous Akt (Fig. 4D).
FIGURE 4.
14-3-3β binding prevents apoptotic SRPK2 cleavage. A, SRPK2 mutant apoptotic cleavage assay. HEK293 cells were transfected with Myc-tagged SRPK2WT, T492A, or T492D for 24 h, and then the cell lysates were incubated with active caspase-3 at 37 °C for 2 h. B, 14-3-3β wild-type but not K50E mutant protects SRPK2 from apoptotic cleavage. HEK293 cells were transfected with Myc-tagged SRPK2WT, T492A, or T492D and 14-3-3WT or 14-3-3K50E for 24 h, and then the cell lysates were incubated with active caspase-3 at 37 °C for 2 h. C, 14-3-3β protects SRPK2 from VP16-triggered apoptotic cleavage. HEK293 cells were transfected with Myc-tagged SRPK2WT, T492A, or T492D and 14-3-3WT or 14-3-3β-K50E for 24 h, and then treated with DMSO or VP16 for another 16 h. The cell lysates were analyzed by immunoblotting. D, D139A mutation affects SRPK2 binding to 14-3-3β. HEK293 cells were co-transfected with Myc-tagged SRPK2WT, T492A, or T492D and GFP-14-3-3β-WT for 48 h, and then the cell lysates were subjected to immunoprecipitation with anti-14-3-3β antibody. The coprecipitated proteins were analyzed by immunoblotting with Myc antibody.
VP16 Treatment Down-regulates SRPK2 Kinase Activity
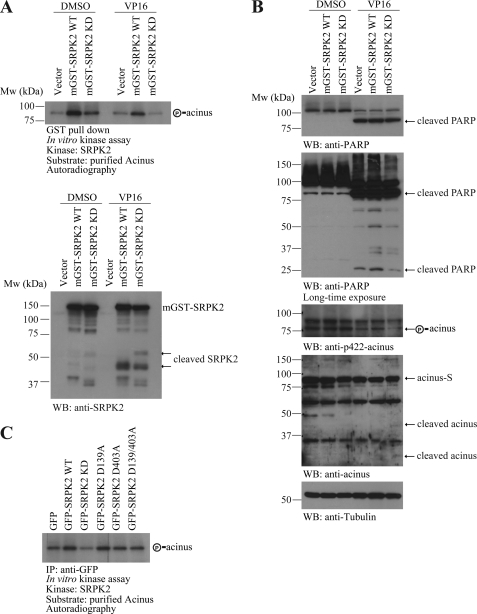
Previous study shows that apoptotic cleavage of SRPK1 and SRPK2 abrogates their in vitro kinase activity on ASF/SF2 in Fas-treated Jurkat cells (8). Recently, we have shown that SRPK2 phosphorylates acinus on Ser-422 and triggers its redistribution from the nuclear speckles to the nucleoplasm (10). Next, we sought to determine whether SRPK2 kinase activity was affected during apoptotic cell death by VP16. We transfected HEK293 cells with GST-tagged SRPK2-WT or SRPK2-KD, a kinase-dead mutant with K110A, and then treated the cells with DMSO or VP16 for another 16 h. The cell lysates were subjected to GST pull-down. After extensive wash of the precipitates, in vitro kinase assay was conducted using bacterial expressed acinus as a substrate in the presence of [γ-32P]ATP. In DMSO-treated cells, wild-type SRPK2 robustly phosphorylated acinus, and SRPK2-KD weakly phosphorylated acinus. VP16 treatment evidently triggered the proteolytic cleavage of SRPK2 wild-type and KD (Fig. 5A, lower panel), and down-regulated SRPK2 kinase activity on acinus (Fig. 5A, upper panel).
FIGURE 5.
VP16 treatment down-regulates SRPK2 kinase activity. A, VP16 treatment down-regulates SRPK2 kinase activity on acinus. HEK293 cells were transfected with GST-tagged SRPK2WT or SRPK2KD for 24 h, and then treated with DMSO or VP16 for another 16 h. The cell lysates were subjected to GST pull-down with GSH beads. The precipitated proteins were analyzed in the in vitro kinase assay using bacterial expressed acinus as the substrate. B, wild-type but not KD SRPK2 escalates VP16-provoked apoptosis. The cell lysates from above cells were subjected to Western blot analysis with indicated antibodies. VP16 treatment provoked PARP cleavage, and SRPK2-WT overexpression increased in VP16-induced PRAP cleavage (top and 2nd panel). Acinus Ser-422 phosphorylation was decreased in VP16-treated cells (3rd panel). C, in vitro kinase assay with acinus. GFP-tagged SRPK2-WT, KD, D139A, D403A, and D139/403A were transfected into the HEK293 cells, and then the cell lysates were subjected to immunoprecipitation with anti-GFP antibody. The precipitated kinase was incubated with purified acinus in the kinase reaction buffer in the presence of [γ-32P]ATP.
To ensure that SRPK2 kinase activity was indeed reduced in the apoptotic cells, we monitored acinus S422 phosphorylation in the cells. Immunoblotting analysis showed that acinus phosphorylation was substantially diminished upon VP16 treatment. The p-acinus in SRPK2-KD-transfected cells was decreased more than SRPK2 wild-type-transfected cells (Fig. 5B, 3rd panel). In VP16-treated cells, acinus was cleaved with slightly more apoptotic degradation occurred in the cells transfected with SRPK2 KD than SRPK2 wild-type (Fig. 5B, 4th panel). VP16 treatment elevated PARP fragmentation in all cells, but SRPK2 wild-type transfected cells revealed stronger PARP cleavage (Fig. 5B, 1st and 2nd panel). This finding fits with our previous report that overexpression of SRPK2 wild-type but not KD triggers spontaneous neuronal apoptosis in primary cortical neurons (9). To study whether the point mutation of D139A and D403A would influence the kinase activity of SRPK2, we conducted an in vitro kinase assay with acinus and found that the mutation did not affect the kinase activity of SRPK2 (Fig. 5C).
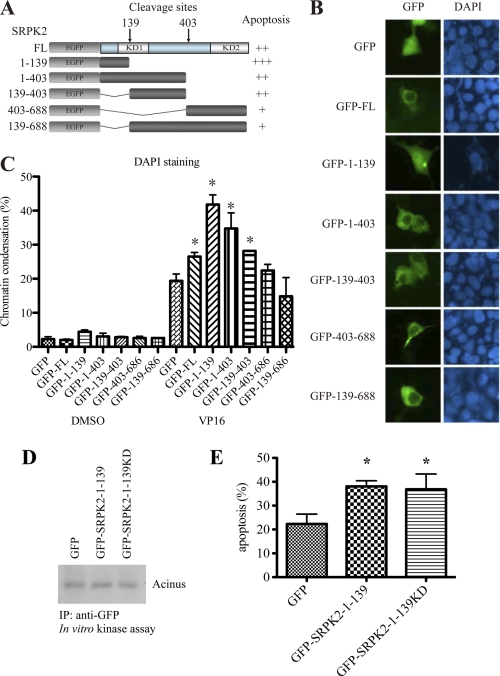
N Terminus of SRPK2 Localizes to the Nucleus and Promotes Apoptotic Cell Death
Caspases-mediated proteolytic cleavage may exert different impacts on different proteins. Some proteins, such as PARP, are inactivated after cleavage; however, others, like pro-caspases, are activated and further magnify the apoptotic signal. To clarify the consequence of SRPK2 cleavage, we constructed five SRPK2 truncations that reflect the cleaved products (Fig. 6A). While the fragment SRPK2-(1–139) showed a prominent nuclear localization, other fragments acted like the full-length SRPK2 by residing in the cytoplasm (Fig. 6B). The cytoplasmic residency of GFP-SRPK2-(139–688) indicates that this fragment might bind to other cellular partners that tether SRPK2 in the cytoplasm. To investigate each fragments role in provoking apoptosis, we transfected HEK293 cells with different GFP-tagged SRPK2 fragments. In the vehicle control treated cells, no any significant apoptosis was triggered by any of the fragments. VP16 induced marked apoptosis in GFP-transfected control cells, and overexpression of SRPK2 full-length and a few of the fragments further elevated the programmed cell death. The most prominent apoptosis occurred in the N-terminal fragment SRPK2-(1–139)-transfected cells. By contrast, the C-terminal fragment GFP-SRPK2-(403–688) or the N-terminal 1–139-deleted fragment (GFP-SRPK2-(139–688)) did not escalate apoptotic cell death as compared with GFP control (Fig. 6C). This finding suggests that the N-terminal 1–139 fragment is indispensable for SRPK2 to increase apoptosis. To study whether the 1–139 fragment possesses any kinase activity that contributes to the pro-apoptotic effect, we constructed SRPK2-(1–139)K110A. However, neither SRPK2-(1–139) nor SRPK2-(1–139)KD showed any substantial kinase activity, and they both display similar pro-apoptotic effects in VP16-treated cells (Fig. 6, D and E).
FIGURE 6.
N terminus of SRPK2 localizes to the nucleus and promotes apoptotic cell death. A, schematic representation of various SRPK2 fragments that mimic the cleaved products by caspase-3. B, subcellular localization of SRPK2 fragments. HEK293 cells were transfected with GFP-tagged SRPK2 fragments for 24 h. The cell nuclei were stained with DAPI and visualized under a fluorescent microscope. C, apoptosis quantification. HEK293 cells were transfected with GFP-tagged SRPK2 fragments for 24 h, and then treated with DMSO or VP 16 for another 16 h. The cells were stained with DAPI- and GFP-positive cells were counted for chromosome condensation. *, p < 0.05. A minimum of 300 cells were counted from 3 different fields. D, SRPK2-(1–139) and SRPK2-(1–139)KD possess no kinase activity. GFP, GFP-SRPK2-(1–139) or GFP-SRPK2-(1–139)KD was transfected into HEK293 cells and then pulled-down with anti-GFP antibody. The precipitated proteins were incubated with purified Acinus as a substrate and analyzed by in vitro kinase assay. E, quantification of apoptosis. HEK293 cells were transfected with GFP, GFP-SRPK2-(1–139) or GFP-SRPK2-(1–139)KD for 24 h and then treated with VP16 for another 16 h. The adherent and non-adherent cells were collected, mounted on the slides, and stained with DAPI. The percentages of cells with chromosome condensation were counted. *, p < 0.05. A minimum of 300 cells were counted from 3–5 different fields.
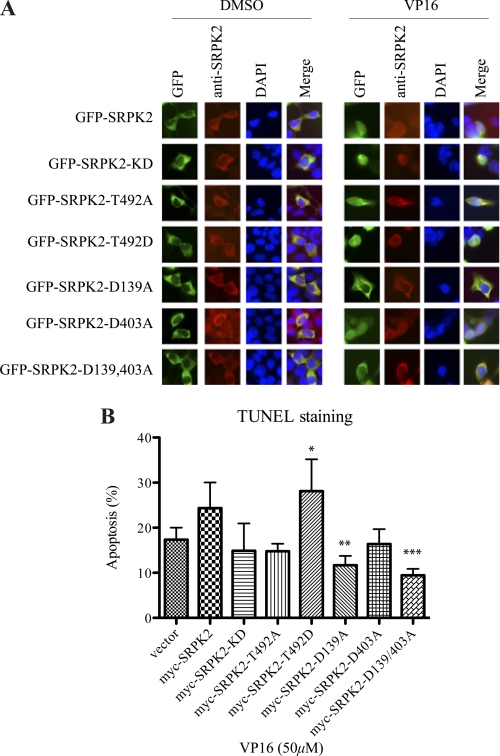
D139A Mutation Inhibits the Pro-apoptotic Effect of SRPK2
Because D139A/D403A double mutation reduces the proteolytic cleavage of SRPK2, we wondered whether this mutation could inhibit the pro-apoptotic effect of SRPK2. To test this possibility, we transfected HEK293 cells with various GFP-tagged SRPK2 constructs and examined their subcellular localization and apoptosis upon VP16 treatment. The cells were stained with anti-SRPK2 to show the C terminus of SRPK2. GFP-tagged SRPK2-WT, -KD, D139A, D403A, and D139A/D403A all distributed in the cytoplasm before stimulation. Except for D139A and D139A/D403A double mutant, other constructs displayed nuclear residency upon VP16 treatment as visualized by GFP fluorescent. However, anti-SRPK2 staining revealed the cytoplasmic localization for these constructs, indicating that VP16 treatment provoked the cleavage of SRPK2 in the N terminus and elicited the nuclear translocation of GFP-tagged N-terminal fragment (Fig. 7A). These results demonstrate that inhibition of Asp-139 cleavage prevents SRPK2 nuclear translocation. Presumably, the nuclear translocation observed in GFP-SRPK2 WT, KD, and D403A cells reflects the cleaved N-terminal GFP-SRPK2-(1–139) fragment. Apoptotic analysis with TUNEL staining showed that both SRPK2 WT and T492D enhanced VP16-triggered programmed cell death, while D139A and D139A/D403A significantly blocked VP16 provoked apoptosis, underscoring that apoptotic cleavage of SRPK2 might be required for its pro-apoptotic actions (Fig. 7B). Hence, prevention of D139 cleavage decreases the pro-apoptotic effect of SRPK2.
FIGURE 7.
D139A mutation inhibits the pro-apoptotic effect of SRPK2. A, subcellular localization of GFP-SRPK2 and its mutants upon VP16 treatment. HEK293 cells were transfected with GFP, GFP-tagged SRPK2WT, KD, T492A, T492D, D139A, D403A, or D139/403A for 24 h, and then treated with DMSO or VP16 for another 24 h. The cells were stained with DAPI and visualized under the fluorescent microscope. B, quantification of apoptosis. HEK293 cells were transfected with empty vector, Myc-tagged SRPK2WT, KD, T492A, T492D, D139A, D403A, or D139/403A for 24 h, and then treated with DMSO or VP16 for another 16 h. Then, the adherent and non-adherent cells were collected, mounted on the slides and stained with the TUNEL reagents and DAPI. The percentages of TUNEL-positive cells were counted. *, vector versus T492D; **, SRPK2 versus D139A; ***, SRPK2 versus D139/403A, p < 0.05. A minimum of 300 cells were counted from three different fields.
DISCUSSION
In the present study, we show that SRPK2 is cleaved both in vitro and in vivo in a caspase-dependent manner. The pan-caspase inhibitor z-VAD-FMK and the caspase-3/7 inhibitor AC-DEVD-CHO both partially inhibit SRPK2 cleavage in apoptotic cells. Presumably, higher concentration of pan-caspase inhibitor is needed to completely block the apoptotic cleavage of SRPK2. We also show the direct evidence that SRPK2 is cleaved in vitro by recombinant caspase-3. These results imply that SRPK2 is the substrate of caspase-3, and other caspases may also be involved in the apoptotic cleavage of SRPK2. Consistent with our finding, Kamachi et al. (8) suggests that SRPK1 and SRPK2 are cleaved during apoptotic cell death and are in vitro substrates for caspase-8 and -9, respectively. Further, in the present study, we identified the caspase-cleavage sites within SRPK2 as Asp-139 and -403 residues. Double mutant of D139A/D403A evidently abolished SRPK2 cleavage in vivo. In alignment with the observation that SRPK2 is a substrate of caspases, we found that Akt phosphorylation of SRPK2 strongly protects it from apoptotic cleavage. Binding to 14-3-3 proteins by SRPK2 further protects it from proteolytic cleavage by the active caspases. Recently, we show that SRPK2 wild-type but not KD elicits spontaneous apoptosis in the primary neurons, indicating that the kinase catalytic activity is implicated in this event. This finding is further supported by the observation that SRPK2 T492D, a kinase hyperactive SRPK2 mutant, substantially elevates VP16-provoked apoptosis (Fig. 7). Although Akt phosphorylates SRPK2 and escalates its kinase activity, 14-3-3 proteins associate with the phosphorylated SRPK2 and anchor it from translocation into the nucleus. Because SRPK2 triggers apoptosis dependent on its catalytic activity, indicating some of its kinase substrates can provoke apoptosis in the nucleus. Among the numerous SRPK2 substrates, one of the major SR proteins that implicate in apoptosis is acinus. Acinus, predominantly located in the nucleus, induces apoptotic chromatin condensation after cleavage by caspases (14). Acinus resides in the nuclear speckles and is cleaved by caspases on both its N and C termini, producing a p17 active form (amino acids 987–1093), which triggers chromatin condensation in the absence of caspase-3. Acinus contains a region similar to the RNA recognition motif (RRM) of Drosophila splicing regulator Sxl, suggesting that it is implicated in RNA metabolism. Indeed, acinus is a component of functional splicesomes (15, 16). It consists of three SR dipeptide repeat domains in the C terminus. Moreover, different acinus isoforms are found in the apoptosis-associated and splicing-associated protein (ASAP) complex (17). Conceivably, nuclear translocated SRPK2 phosphorylates acinus, and promotes its degradation, leading to generation of an active form p17 and elevation of chromatin condensation and apoptosis.
One of the major functions proposed for SRPKs is to phosphorylate SR proteins, which in turn regulates alternative splice site selection of a subset of mRNA molecules, including the apoptosis regulatory protein Ich-1 (18–20). Over 30 different mRNAs have been identified that exist as two or more different splice variants, each with opposing apoptotic functions (20). SR proteins such as ASF/SF2 alter splice site selection of several RNAs, including Ich-1, and this correlates with resetting of the apoptotic threshold of transfected cells (19). Therefore, it is possible that alternative splicing may play a critical role in the sensing phase of cellular stress including VP16. Thus, mRNA splicing may represent an unrecognized and unexplored post-transcriptional modification that determines cell fate in response to extracellular stimuli. Conceivably, SRPKs such as SRPK2 are activated at the initial stage of cell stress, resulting in alternative splicing of mRNAs such that anti-apoptotic isoforms such as bcl-xL and Ich1-α. Caspase-mediated cleavage of SRPKs at the later stage of cell stress would diminish their kinase activity, and ensure that the apoptotic program is faithfully executed in condemned cells (8).
Protein kinases are critical regulators for most of the cellular processes in a kinase-dependent manner. For example, inhibition of Akt kinase activity with a small molecule inhibitor results in suppression of cell growth and induction of apoptosis in human cancer cells that harbor constitutively activated Akt due to overexpression of Akt or other genetic alterations such as PTEN mutation (21). Previously, we found that SRPK2 provoked neuronal apoptosis through up-regulating cyclin D1/Cdk4, which was abolished in SRPK2-KD-transduced neuronal cells (9). Here, we show that abrogation of SRPK2 kinase activity inhibited its pro-apoptotic effect in HEK293 cells. These results indicate that SRPK2 kinase activity is responsible for the pro-apoptotic effect of SRPK2. However, the underlying mechanism how SRPK2 kinase activity regulates the programmed cell death needs further investigation.
Regulating the balance between survival and apoptotic signaling is a key aspect of cell fate decisions and 14-3-3 proteins contribute to this process in multiple ways. The binding of 14-3-3 has often been found to enhance the activity of proteins with proliferative or survival functions, while antagonize the activity of proteins that promote cell death and senescence (22). Our previous study demonstrates that Akt phosphorylation of SRPK2 facilitates its binding with 14-3-3, which in turn prevents the pro-apoptotic effect of SRPK2 in primary neurons (9). Our previous study shows that 14-3-3β-WT binds to SRPK2, while the mutant K50E impairs its binding ability. Also, SRPK2T492D strongly binds to 14-3-3β, where T492A inhibits the interaction between SRPK2 and 14-3-3β. The present study shows that T492A is robustly cleaved despite the presence of 14-3-3β, whereas T492D is not cut under any circumstances. The fragmentation of SRPK2-WT is inhibited when 14-3-3β-WT is overexpressed, and the cleavage is up-regulated in the presence of K50E. Collectively, we provide further evidence that 14-3-3 interaction with SRPK2 suppresses its cleavage by caspases, and inhibits the release of the N-terminal pro-apoptotic fragment of SRPK2 (Fig. 4). Therefore, these findings suggest a new cue that how 14-3-3 proteins regulate cell death and survival.
Some of the SR proteins can shuttle between the nucleus and the cytoplasm. They are the substrates of SRPKs. Once exported to the cytoplasm, SR proteins are reimported to the nucleus by interacting with the import receptor hMtr10/Transportin-SR in a phosphorylation-dependent manner (23–27). It has been proposed that phosphorylation of shuttling SR proteins in the cytoplasm may act as a switch between mRNA unloading and SR protein reimport (28). SRPK family members share highly conserved kinase domains, which are separated by a unique spacer sequence in individual family members, indicating that specific SRPKs may be uniquely regulated by the spacer sequence. Previous studies show that the spacer in SRPK1 is able to modulate the interaction of the kinase with SR protein substrates. The spacer sequence may also interact with specific cellular proteins to anchor the kinase to the cytoplasm. Interestingly, we found that caspase-mediated proteolytic cleavage of SRPK2 released its N-terminal fragment that translocated into the nucleus and further potentiated the apoptosis. We showed the nuclear localization of N-terminal fragment SRPK2-(1–139) and the translocation of SRPK2 upon VP16 treatment, which implies that the N-terminal fragment of SRPK2 may be cleaved and translocate into the nucleus to promote VP16-induced apoptosis. However, SRPK2 D139A mutant still remained in the cytoplasm and exhibited anti-apoptotic effect, suggesting that the nuclear translocation of SRPK2 N-terminal fragment plays an essential role in promoting VP16-induced apoptosis. Though D139A or D403A mutation did not markedly alter SRPK2 Thr-492 phosphorylation capability by Akt, the mutation affected SRPK2 binding affinity to 14-3-3 (Fig. 4), indicating that these two residues might be critical for the conformation of SRPK2. Therefore, these data indicate that the site Asp-139 plays an important role in SRPK2 function during VP16-induced apoptosis. This result may unravel the molecular mechanism how SRPK2 regulates apoptotic cell death. It remains unknown how the N-terminal 139 residues escalate VP16-provoked apoptosis. Its binding partners in the nucleus are elusive. Nonetheless, the findings described in this report pave the way for the further investigation in this interesting event. Taken together, our studies show that SRPK2 is cleaved by caspase at both Asp-139 and Asp-403 residues during apoptotic cell death. Caspase cleavage of SRPK2 produces a small N-terminal fragment that translocates into nucleus and promotes VP16-induced apoptosis. This study suggests two pathways are involved in SRPK2 associated apoptotic cell death: first, the active SRPK2 can trigger apoptosis which depends on its kinase activity; second, SRPK2 is also a substrate of caspases whose action releases an N-terminal fragment from the kinase. This released fragment can also enhance apoptotic signals. 14-3-3β regulates both of these two pathways.
Acknowledgment
We thank Dr. Haian Fu (Pharmacology Dept., Emory University) for the 14-3-3β-WT plasmid.
This work was supported, in whole or in part, by Grant NS060680 from NCI, National Institutes of Health (to K. Y.).
- SR
- serine/arginine
- SRPK
- SR protein-specific kinase
- KD
- kinase-dead.
REFERENCES
- 1. Lin S., Fu X. D. (2007) Adv. Exp. Med. Biol. 623, 107–122 [DOI] [PubMed] [Google Scholar]
- 2. Zhong X. Y., Wang P., Han J., Rosenfeld M. G., Fu X. D. (2009) Mol. Cell 35, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang H. Y., Arden K. C., Bermingham J. R., Jr., Viars C. S., Lin W., Boyer A. D., Fu X. D. (1999) Genomics 57, 310–315 [DOI] [PubMed] [Google Scholar]
- 4. Ding J. H., Zhong X. Y., Hagopian J. C., Cruz M. M., Ghosh G., Feramisco J., Adams J. A., Fu X. D. (2006) Mol. Biol. Cell 17, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gui J. F., Lane W. S., Fu X. D. (1994) Nature 369, 678–682 [DOI] [PubMed] [Google Scholar]
- 6. Wang H. Y., Lin W., Dyck J. A., Yeakley J. M., Songyang Z., Cantley L. C., Fu X. D. (1998) J. Cell Biol. 140, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes G. M., Carrigan P. E., Miller L. J. (2007) Cancer Res. 67, 2072–2080 [DOI] [PubMed] [Google Scholar]
- 8. Kamachi M., Le T. M., Kim S. J., Geiger M. E., Anderson P., Utz P. J. (2002) J. Exp. Med. 196, 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jang S. W., Liu X., Fu H., Rees H., Yepes M., Levey A., Ye K. (2009) J. Biol. Chem. 284, 24512–24525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jang S. W., Yang S. J., Ehlén A., Dong S., Khoury H., Chen J., Persson J. L., Ye K. (2008) Cancer Res. 68, 4559–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 12. Cryns V., Yuan J. (1998) Genes. Dev. 12, 1551–1570 [DOI] [PubMed] [Google Scholar]
- 13. Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 14. Sahara S., Aoto M., Eguchi Y., Imamoto N., Yoneda Y., Tsujimoto Y. (1999) Nature 401, 168–173 [DOI] [PubMed] [Google Scholar]
- 15. Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002) Genome Res. 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002) Nature 419, 182–185 [DOI] [PubMed] [Google Scholar]
- 17. Schwerk C., Prasad J., Degenhardt K., Erdjument-Bromage H., White E., Tempst P., Kidd V. J., Manley J. L., Lahti J. M., Reinberg D. (2003) Mol. Cell. Biol. 23, 2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cáceres J. F., Stamm S., Helfman D. M., Krainer A. R. (1994) Science 265, 1706–1709 [DOI] [PubMed] [Google Scholar]
- 19. Jiang Z. H., Zhang W. J., Rao Y., Wu J. Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9155–9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang Z. H., Wu J. Y. (1999) Proc. Soc. Exp. Biol. Med. 220, 64–72 [DOI] [PubMed] [Google Scholar]
- 21. Yang L., Dan H. C., Sun M., Liu Q., Sun X. M., Feldman R. I., Hamilton A. D., Polokoff M., Nicosia S. V., Herlyn M., Sebti S. M., Cheng J. Q. (2004) Cancer Res. 64, 4394–4399 [DOI] [PubMed] [Google Scholar]
- 22. Morrison D. K. (2009) Trends Cell Biol. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yun C. Y., Fu X. D. (2000) J. Cell Biol. 150, 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lai M. C., Lin R. I., Huang S. Y., Tsai C. W., Tarn W. Y. (2000) J. Biol. Chem. 275, 7950–7957 [DOI] [PubMed] [Google Scholar]
- 25. Lai M. C., Lin R. I., Tarn W. Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gilbert W., Siebel C. W., Guthrie C. (2001) RNA 7, 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yun C. Y., Velazquez-Dones A. L., Lyman S. K., Fu X. D. (2003) J. Biol. Chem. 278, 18050–18055 [DOI] [PubMed] [Google Scholar]
- 28. Gilbert W., Guthrie C. (2004) Mol. Cell 13, 201–212 [DOI] [PubMed] [Google Scholar]