Abstract
Epithelial cells are dependent on extracellular matrix (ECM) attachment for maintenance of metabolic activity and suppression of apoptosis. Here we show that loss of ECM attachment causes down-regulation of epidermal growth factor receptor (EGFR) and β1 integrin protein and mRNA expression and that ErbB2, which is amplified in 25% of breast tumors, reverses these effects of ECM deprivation. ErbB2 rescue of β1 integrin mRNA and protein in suspended cells is dependent on EGFR, however, the rescue of EGFR expression does not require β1 integrin. We show that there is a significant decrease in the stability of EGFR in ECM-detached cells that is reversed by ErbB2 overexpression. Rescue of both EGFR and β1 integrin protein by ErbB2 is dependent on Erk activity and induction of its downstream target Sprouty2, a protein known to regulate EGFR protein stability. Interestingly, expression of EGFR and β1 integrin protein is more dependent on Erk/Sprouty2 in ECM-detached ErbB2-overexpressing cells when compared with ECM-attached cells. These results provide further insight into the ErbB2-driven anchorage independence of tumor cells and provide a new mechanism for regulation of EGFR and β1 integrin expression in ECM-detached cells.
Keywords: Apoptosis, Breast Cancer, Cell Death, ERK, Extracellular Matrix, MAP Kinases (MAPKs), Receptor Tyrosine Kinase, Signal Transduction, Epidermal Growth Factor Receptor (EGFR), ErbB2
Introduction
One of the hallmarks of tumor cells is the ability to survive without attachment to the extracellular matrix (ECM)3 (1). Detachment of normal epithelial cells from ECM leads to metabolic impairment and induction of apoptotic death (anoikis) (2–5). However, tumor cells are generally able to survive without ECM attachment, a property known as “anchorage independence” (2, 3). This is believed to allow tumor cells to invade and proliferate outside their natural ECM niches. For example, one early feature of breast cancer is the proliferation of cells into the ECM-deficient hollow lumen of the glandular epithelium (6). These premalignant cells are able to survive despite the lack of contact to the basement membrane. Additionally, it is believed that anchorage independence is an important aspect of the metastatic process, both for survival in the vasculature and lymphatic system and also for survival in distant sites with altered matrix environments (2). Thus, elucidation of the mechanisms responsible for tumor cell anchorage independence is critical for understanding of the tumorigenic processes and may lead to identification of novel drug targets.
ECM detachment of mammary epithelial cells results in a decrease of growth factor signaling through the Mek/Erk and PI3K/Akt pathways (2, 4, 5, 7). This leads to decreased cell viability, both through the induction of anoikis and through a caspase-independent metabolic impairment (2, 4). We have previously found that decreased Erk activation in ECM-detached cells leads to increased expression of the pro-apoptotic protein, Bim (7–9). In addition, the lack of Akt signaling in the ECM-detached cells results in a dramatic decrease in glucose uptake and ATP levels, causing a severe energy deficiency (4). Therefore, survival of mammary epithelial cells under ECM-deprived conditions requires evasion of both apoptotic death and metabolic impairments.
We have previously shown that loss of integrin engagement in the ECM-detached mammary epithelial cells leads to a striking decrease in expression of the epidermal growth factor receptor (EGFR) (7). Overexpression of ErbB2, an ErbB family member that is amplified and overexpressed in breast tumors (10), maintains EGFR expression and EGF-induced signaling and permits cell survival under ECM-detached conditions (4, 7, 10, 11). Several mechanisms contributing to ErbB2 regulation of survival and metabolism have been elucidated (4, 7, 10, 12–14). ErbB2 signaling through Mek/Erk maintains low Bim levels after ECM detachment and inhibits anoikis (7, 8), and signaling through PI3K/Akt activation sustains glucose uptake and ATP levels (4). The ErbB2 rescue of glucose uptake and ATP is strictly dependent upon its ability to maintain EGFR expression in ECM-detached cells. Hence, elucidation of the mechanisms whereby ErbB2 regulates EGFR under altered conditions of ECM attachment will further our understanding of processes associated with anchorage independence.
The degradation of EGFR after ligand stimulation of ECM-attached cells is in large part regulated by c-Cbl, the E3-ubiquitin ligase that binds to phosphorylated EGFR (15). The ubiquitination of EGFR by Cbl promotes receptor trafficking to lysosomes and is necessary for efficient degradation of EGFR (15, 16). We have previously shown that Cbl binds less efficiently to ErbB2-EGFR heterodimers when compared with EGFR homodimers (17); this and other mechanisms contribute to an inefficient degradation of ErbB2-EGFR heterodimers (16).
Epithelial cells cultured in suspension lose growth factor signaling in part due to the loss of engagement of integrins and activation of downstream signaling pathways (2–5, 7). Interestingly, growth factor receptors and integrins have been shown to be functionally coupled (18–20). For example, EGFR and β1 integrin physically interact and are coregulated (7, 18, 19, 21, 22). The loss of EGFR in mammary epithelial cells cultured in suspension is prevented by the addition of reconstituted basement membrane to the suspended cells (7). This rescue is dependent on integrin engagement as a β1 integrin-blocking antibody prevents the stabilization of EGFR (7). The reciprocal regulation of EGFR and β1 integrin has also been reported in T4-2 transformed mammary epithelial cells in three-dimensional culture (22). In these cells, which express high levels of both EGFR and β1 integrin, treatment with either a β1 integrin-blocking antibody or an EGFR inhibitor leads to decreased expression of both EGFR and β1 integrin.
In this study, we demonstrate that loss of ECM attachment down-regulates both EGFR and β1 integrin at the protein and mRNA level and that ErbB2 overexpression significantly attenuates the reduction of EGFR and β1 integrin in ECM-detached cells. ErbB2 maintains EGFR and β1 integrin protein expression in an Erk-dependent manner via increased Sprouty2 expression and stabilization of EGFR protein. These results reveal a novel mechanism of coordinated regulation of growth factor receptor and integrin expression that cells are more dependent on when deprived of ECM contact. The findings also highlight an additional phenotypic effect of hyperactivation of the Erk pathways that could play a role in promoting cancer progressions by increasing survival of tumor cells outside their niches. In addition, these studies suggest that tumor cells, which are often challenged to survive in altered ECM environments, may be especially reliant on these pathways.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF-10A cells were cultured as described previously (23). MCF-10A cells expressing ErbB2 or MekDD were generated as described previously (24). Human mammary epithelial cells were cultured as described previously (25). SK-BR-3 cells were cultured in RPMI medium plus 10% FBS and penicillin/streptomycin. The pMSCV-IRES-PURO and pMSCV-SPRY2-IRES-PURO constructs were kindly provided by M. Teitell (UCLA). Assays on ECM-detached cells were performed on cells grown on tissue culture plates coated with poly(2-hydroxy methacrylate) (poly-HEMA) for 24 h except where indicated.
For conditioned media experiments, MCF-10A or MCF-10A ErbB2 cells were grown for 24 h on poly-HEMA plates. These media were then collected, and new MCF-10A or MCF-10A ErbB2 cells were cultured for 24 h in the conditioned media on poly-HEMA plates.
Immunoblotting
Cells were grown either on control plates (attached, indicated as A in Figs. 1–3 and 5) or on poly-HEMA coated plates (detached, indicated as D in Figs. 1–3 and 5). After 24 h (except where indicated), cells were lysed in 1% Nonidet P-40 containing 5 mm sodium fluoride, 1 mm sodium orthovanadate, 5 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride, and 5 μg/ml leupeptin on ice for 20 min. Lysates were spun at 16,000 × g at 4 °C for 30 min and normalized for protein concentration. Lysates were then subjected to SDS-PAGE, and transfer/blotting were performed as described previously (26). Blots were imaged either by chemiluminescence or by fluorescent imaging using the Odyssey infrared imaging system (LI-COR Biosciences; Millennium Science, Surrey Hills, Australia). Immunoblots were quantified using the NIH ImageJ software. The figures including Western blots show representative blots from three or more independent experiments. The figures including quantification are presented as the average of three or more independent experiments. The following antibodies were used: EGFR (Cell Signaling Technology, 2232), phospho-Akt Ser-473 (Cell Signaling Technology, 4060), phospho-Erk1/2 Tyr-185/187 (Invitrogen, 44-680G), β-actin (Sigma-Aldrich), β1 integrin (BD Biosciences, 610468), β-tubulin (Abcam), Erk1/2 (Cell Signaling Technology, 9107), and Sprouty2 (Abcam, ab60719).
FIGURE 1.
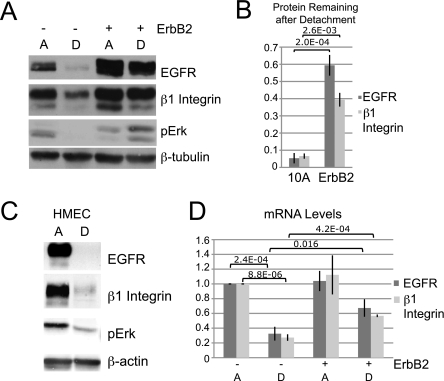
ErbB2 prevents the decrease of EGFR and β1 integrin protein and mRNA in ECM-detached cells. A, MCF-10A or MCF-10A ErbB2 cells were plated under ECM-attached (lane A) or ECM-detached (lane D) conditions for 24 h. Expression of the indicated proteins was determined by immunoblotting. pErk, phosphorylated Erk. B, quantification from three experiments performed as described in A. Data are presented as ratios of protein in detached cells to protein in attached cells. C, human mammary epithelial cells (HMEC) were plated and analyzed as described in A. D, MCF-10A or MCF-10A ErbB2 cells were processed as in A, and mRNA levels were measured by qPCR.
FIGURE 2.
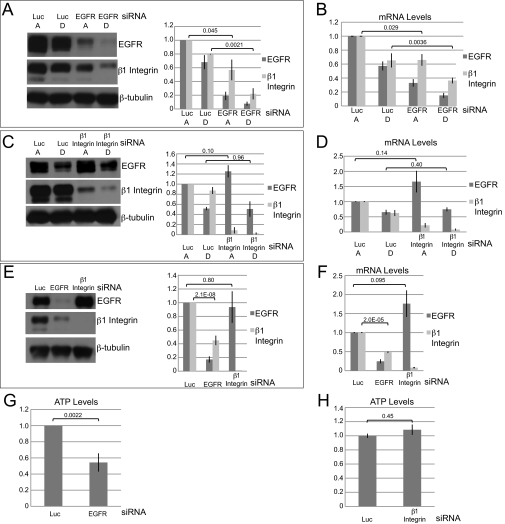
ErbB2 rescue of β1 integrin protein and mRNA levels is dependent on EGFR, but β1 integrin is not required for rescue of EGFR. A and B, MCF-10A ErbB2 cells were transfected with siRNAs targeting luciferase (Luc) or EGFR. 24 h later, cells were plated under attached or detached conditions, and after an additional 24 h, protein levels were assessed by immunoblotting (A), and mRNA levels were measured by qPCR (B). Quantification of average protein levels from three independent experiments is also shown (A, right panel). Lane A, ECM-attached; lane D, ECM-detached. C and D, MCF-10A ErbB2 cells were transfected with siRNAs targeting luciferase or β1 integrin. 24 h later, cells were plated under attached or detached conditions, and after an additional 24 h, protein levels were assessed by immunoblotting (C), and mRNA levels were measured by qPCR (D). Quantification of average protein levels from three independent experiments is also shown (C, right panel). E and F, MCF-10A cells were transfected with siRNAs targeting luciferase, EGFR, or β1 integrin. 48 h later, protein levels were analyzed by immunoblotting (E), and mRNA levels were measured by qPCR (F). Quantification of average protein levels from three independent experiments is also shown (E, right panel). G and H, MCF-10A ErbB2 cells were transfected with siRNAs targeting luciferase, EGFR (G), or β1 integrin (H). Cells were then plated under attached and detached conditions, and ATP levels were measured using the ATPlite assay.
FIGURE 3.
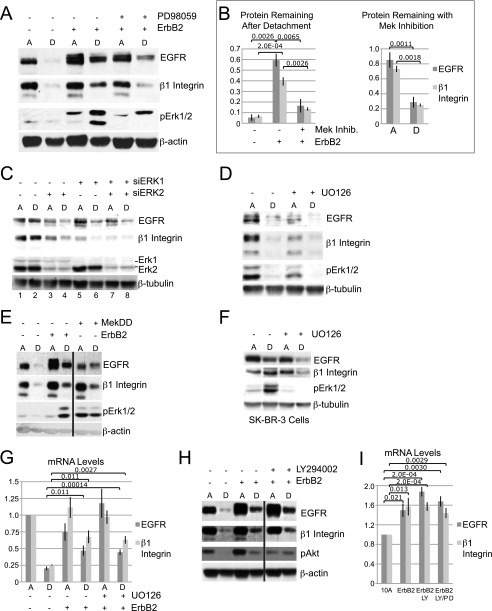
Erk signaling is necessary for the ErbB2 maintenance of EGFR and β1 integrin protein expression in ECM-detached cells. A, MCF-10A cells or MCF-10A ErbB2 cells treated with 20 μm PD98059 (+) or vehicle (−) were plated and analyzed as described in the legend for Fig. 1. Lane A, ECM-attached; lane D, ECM-detached. B, quantification from three experiments as described in A. Data are presented as a ratio of protein in detached cells to protein in attached cells (left panel) or ratio of protein in Erk inhibitor-treated cells to control cells (right panel). Mek Inhib., Mek inhibitor. C, MCF-10A ErbB2 cells were transfected with siRNA targeting luciferase (−), ERK1, ERK2, or ERK1 and ERK2. Cells were plated under attached and detached conditions and analyzed as in A. D, MCF-10A cells treated with 10 μm UO126 (+) or vehicle (−) were plated under attached or detached conditions for 24 h, and protein levels were analyzed by immunoblotting. E, MCF-10A, MCF-10A ErbB2, or MCF-10A MekDD was plated and analyzed as in A. F, SK-BR-3 cells were treated with 10 μm UO126 (+) or vehicle (−) and plated under attached or detached conditions for 24 h. Protein levels were analyzed by immunoblotting. G, MCF-10A or MCF-10A ErbB2 cells were treated with 10 μm UO126 (+) or vehicle (−) and plated under attached or detached conditions for 24 h. mRNA levels were then measured by qPCR. H, MCF-10A cells, or MCF-10A ErbB2 cells treated with 20 μm LY29402 (+) or vehicle (−) were plated and analyzed as in A. I, MCF-10A cells or MCF-10A ErbB2 cells treated with 10 μm LY29402 or 10 μm LY29402 and 20 μm PD98059 (+) or vehicle (−) were plated under detached conditions for 24 h, and mRNA levels were measured by qPCR.
FIGURE 5.
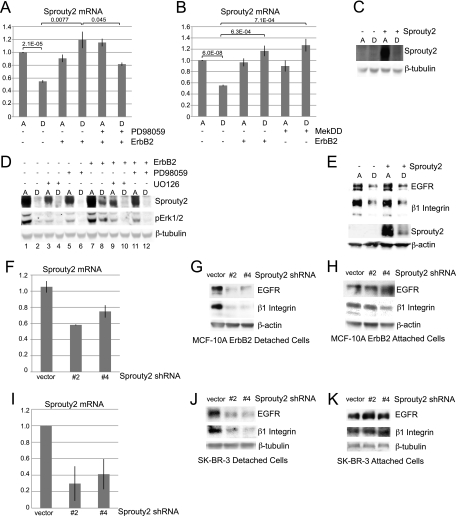
ErbB2 regulates EGFR and β1 integrin via Erk-dependent regulation of Sprouty2. A, MCF-10A or MCF-10A ErbB2 cells treated with 20 μm PD98059(+) or vehicle (−) were plated under attached or detached conditions for 24 h. mRNA levels were measured by qPCR. Column A, ECM-attached; column D, ECM-detached. B, MCF-10A, MCF-10A ErbB2, or MCF-10A MekDD cells were plated under attached or detached conditions for 24 h, and mRNA levels were measured by qPCR. C, MCF-10A cells infected with pMSCV-IRES-PURO or pMSCV-SPRY2-IRES-PURO were plated under attached or detached conditions for 24 h, and protein levels were analyzed by immunoblotting. D, MCF-10A or MCF-10A ErbB2 cells infected with pMSCV-SPRY2-IRES-PURO were plated under attached or detached conditions with 10 μm UO126 or 20 μm PD98059 (+) or vehicle (−). 24 h later, protein levels were analyzed by immunoblotting. pErk1/2, phosphorylated Erk1/2. E, cells were plated and analyzed as described in C. F, MCF-10A ErbB2 cells were infected with shRNA vectors targeting Sprouty2, and mRNA levels were measured by qPCR to confirm knockdown. G and H, MCF-10A ErbB2 cells infected with shRNAs targeting Sprouty2 were plated under detached (G) or attached (H) conditions for 24 h, and protein levels were measured by immunoblotting. I, SK-BR-3 cells were infected with shRNA vectors targeting Sprouty2, and mRNA levels were measured by qPCR to confirm knockdown. J and K, SK-BR-3 cells infected with shRNA vectors targeting Sprouty2 were plated under detached (J) or attached (K) conditions for 24 h, and protein levels were measured by immunoblotting.
Reagents
The following reagents were used at the doses indicated and as described under “Results” and in the figure legends 1–5: poly-HEMA (Sigma-Aldrich), LY294002 (EMD Biosciences), U0126 (EMD Biosciences), PD98059 (Calbiochem), cell dissociation buffer (Invitrogen), cycloheximide (Sigma-Aldrich), chloroquine diphosphate salt (Sigma-Aldrich), bafilomycin A1 (Sigma-Aldrich), and concanamycin A (Sigma-Aldrich).
RNA Interference
The following small interfering RNA (siRNA) SMARTpool reagents (Dharmacon, Lafayette, CO) were used: EGFR (M-003114-03), β1 integrin (L-004506-00), MAPK1 (L-003555-00), MAPK3 (L-003592-00), and luciferase GL2 duplex (D-00110-01-20).
For each transfection, 25 nmol of siRNA were transfected into cells using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Knockdown efficiency was examined after 48 h by immunoblotting or quantitative real-time PCR (qPCR). For experiments involving siRNA in detached cells, cells were plated on poly-HEMA-coated plates 24 h after siRNA transfection, and then assays were conducted 48 h after siRNA transfection. The figures including data using siRNAs show representative experiments from three or more independent experiments. Spry2 short hairpin RNAs (shRNAs) TRCN0000007522 and TRCN0000007524 were obtained from the RNAi Consortium, and lentiviruses were generated according to standard protocol (11).
EGFR Protein Stability
For EGFR stability experiments, MCF-10A or MCF-10A ErbB2 cells were plated on poly-HEMA-coated plates. After 3 h, half of the cells per well were lysed as described above. To the other half, 10 μg/ml cycloheximide with or without 100 nm concanamycin A was added. After an additional 3 h, the remainder of the cells were lysed. EGFR levels were then quantified by immunoblotting as described above. Data are presented as an average of three or more independent experiments.
For experiments with chloroquine or bafilomycin A1 treatment, MCF10A cells were plated on poly-HEMA plates, and 3 h after plating vehicle control, 20 μm chloroquine diphosphate or 20 nm bafilomycin A1 was added. Cells were lysed 24 h after plating, and EGFR levels were then quantified by immunoblotting as described above. Data are presented as an average of three independent experiments.
Quantitative Real-time PCR
Total RNA was prepared from cells using TRIzol (Invitrogen). For real-time PCR, cDNA was synthesized from 2 μg of RNA using the SuperScript first-strand synthesis system (Invitrogen). Real-time PCR was performed using an ABI PRISM 7900HT fast real-time PCR system with gene-specific primers and Power SYBR Green PCR master mix (Applied Biosystems). Quantification of relative mRNA expression levels was determined and normalized to RPLP0. The data are presented as an average of three or more independent experiments.
ATP Assay
For the comparison of ATP levels, the ATPlite assay (PerkinElmer Life Sciences) was used. Cells were plated in triplicate in 96-well control or poly-HEMA-coated plates at a density of 13,000 cells/well. After 24 h, ATP assay was conducted according to the manufacturer's protocol. The data shown from these assays are averages of three or more independent experiments.
Oncomine Data Mining
Normalized Sprouty2 expression data and p values were derived from the Wooster cell line (array.nci.nih.gov) (28) and the Hoeflich Cell Line 2 (27) datasets in Oncomine. Detailed descriptions of data collection and analysis for the Hoeflich Cell Line 2 are in Hoeflich et al. (27). Briefly, cells were treated at 10 different concentrations of PD0325901, at 3-fold dilutions, and the CellTiter-Glo luminescent cell viability assay (Promega) was used to measure viability at 72 h of treatment. The EC50 was then calculated using the XLfit software from IDBS, and cell lines for which an EC50 could not be calculated were defined as resistant. RNA expression was measured using the Affymetrix HG-U133-Plus 2.0 chips.
Detailed descriptions of data collection and analysis for the Wooster Line 2 (from GlaxoSmithKline cancer cell line genomic profiling data) are in Greshock et al. (28). Briefly, cells (obtained from ATCC) were treated at 10 different concentrations of GSK1120212, at 3-fold dilutions. Nuclei staining with 4′,6-diamidino-2-phenylindole (DAPI) and an IN Cell Analyzer 1000 high content analyzer was used to quantify cell numbers at 72 h of treatment. The cell growth IC50 was then calculated using model 205 of XLfit; cell lines with an IC50 greater than 1000 nm were classified as resistant, and those with an IC50 below 10 nm were classified as sensitive. RNA expression was measured using the Affymetrix HG-U133-Plus 2.0 chips.
Box plots were generated in the JMP 7.0 software. The p values were calculated using a Student's t test comparing the means of Sprouty2 expression values in resistant and sensitive cell lines.
Statistics
All average results are presented as mean ± S.E. p values were calculated using a Student's two-tailed t test.
RESULTS
ErbB2 Prevents the Decrease of EGFR and β1 Integrin after ECM Detachment
We have previously shown that EGFR expression as well as activation of its downstream targets Erk and Akt are dramatically decreased when mammary epithelial cells are plated in suspension and that this decrease can be blocked by overexpression of ErbB2 (4, 7). Because modulation of EGFR expression can reciprocally affect β1 integrin expression (22), we examined how ECM detachment and ErbB2 overexpression alter β1 integrin expression (Fig. 1, A and B). Matrix detachment caused on average more than a 90% reduction of β1 integrin expression in control MCF-10A cells. Overexpression of ErbB2 (MCF-10A ErbB2) significantly rescued β1 integrin expression in suspended cultures (Fig. 1, A and B). The effects of matrix detachment and ErbB2 on β1 integrin closely paralleled the effect on EGFR expression, suggesting that these events may be coupled. These findings were not limited to MCF-10A cells; human mammary epithelial cells also display decreased growth factor signaling (as measured by phosphorylated Erk (pErk) levels) as well as a reduction in both EGFR and β1 integrin under ECM-detached conditions (Fig. 1C). Thus, detachment from matrix results in a significant down-regulation of β1 integrin as well as EGFR.
The expression of EGFR and β1 integrin has previously been reported to be regulated at both the transcriptional and the post-transcriptional level (29–33). We found that the mRNA levels of EGFR and β1 integrin paralleled those of the protein as they were decreased in ECM-detached MCF-10A cells, and this was partially rescued by ErbB2 overexpression (Fig. 1D). Thus, ECM detachment of mammary epithelial cells leads to a decrease in both protein and mRNA levels of EGFR and β1 integrin, and ErbB2 overexpression partially prevents these reductions.
The ErbB2 Rescue of EGFR, β1 Integrin, and ATP in ECM-detached Cells Is Dependent on EGFR, but Not β1 Integrin
The evidence that β1 integrin and EGFR expression is regulated in parallel by ECM detachment and ErbB2 overexpression raised the question whether one of these proteins regulates the expression of the other. To address this question, we examined whether knockdown of EGFR or β1 integrin in MCF-10A ErbB2 cells affects ErbB2 rescue in suspended cells. Knockdown of EGFR using an siRNA SMARTpool decreased both β1 integrin protein (Fig. 2A) and mRNA (Fig. 2B) in MCF-10A ErbB2 cells under ECM-attached and ECM-detached conditions. However, although we were able to achieve robust knockdown of β1 integrin with an siRNA SMARTpool, neither EGFR protein (Fig. 2C) nor mRNA levels (Fig. 2D) were decreased by β1 integrin knockdown in ECM-attached or -detached conditions. We found that this unidirectional regulatory relationship exists in the control MCF-10A ECM-attached cells as well; knockdown of EGFR in MCF-10A cells plated in attached conditions resulted in reduced β1 integrin protein and mRNA, whereas knockdown of β1 integrin did not decrease either EGFR protein (Fig. 2E) or mRNA (Fig. 2F). These results clearly demonstrate that reduction of EGFR suppresses β1 integrin expression, but the reciprocal regulation is not observed.
We have previously shown that the ErbB2 rescue of ATP in matrix-detached cells is dependent on EGFR expression (4). As reported previously, knockdown of EGFR in ECM-detached MCF-10A ErbB2 cells resulted in a dramatic decrease in ATP levels (Fig. 2G) (4); however, β1 integrin knockdown did not affect ATP levels (Fig. 2H). These data demonstrate that EGFR is the critical effector for the ErbB2 rescue of both β1 integrin expression and ATP levels in ECM-detached cells.
The ErbB2 Maintenance of EGFR and β1 Integrin Protein Levels Is Erk-dependent in ECM-detached Cells
To investigate the mechanism involved in ErbB2 regulation of EGFR and β1 integrin expression in ECM-detached cells, we examined whether ErbB2 induction of Erk or PI3K is required for the maintenance of EGFR and β1 integrin. Treatment of MCF-10A ErbB2 cells with either of two Mek inhibitors, PD98059 (Fig. 3A) or UO126 (data not shown), caused a dramatic decrease in EGFR and β1 integrin protein in the ECM-detached MCF-10A ErbB2 cells (Fig. 3, A and B, left panel). The dependence of EGFR and β1 integrin expression on Mek/Erk is weaker in ECM-attached MCF-10A ErbB2 cells; Mek inhibition in attached cells resulted in only a 15 and 25% decrease of EGFR and β1 integrin, respectively, whereas in ECM-detached cells, there was over a 70% reduction of both EGFR and β1 integrin (Fig. 3B, right panel).
To further examine the involvement of Erk in ErbB2 regulation of EGFR and β1 integrin, we utilized RNA interference to knock down ERK1 and/or ERK2 in the MCF-10A ErbB2 cells. As with the Erk inhibitors, we found that knockdown of either ERK1 or ERK2 alone or in combination resulted in decreased expression of EGFR and β1 integrin (Fig. 3C), and again the reduction of EGFR was more pronounced in ECM-detached cells. Although β1 integrin expression was decreased by Erk knockdown in both ECM-attached and ECM-detached conditions, there was a proportionally more significant decrease in the ECM-detached cells (Fig. 3C, compare lanes 3 and 4 and lanes 5 and 6). Therefore, ErbB2 maintains EGFR and β1 integrin via Erk signaling, an effect that is most apparent upon ECM detachment. As shown in Fig. 3D, control MCF-10A cells show a much stronger dependence on Erk activation for expression of EGFR and β1 integrin expression in ECM-attached conditions as there is a dramatic reduction in EGFR and β1 integrin in cells treated with a Mek inhibitor (Fig. 3D). These results suggest that under ECM-detached conditions (where ErbB2 is required to maintain EGFR and β1 integrin expression), Mek/Erk signaling is critical in maintenance of these proteins. However, in ECM-attached MCF-10A ErbB2 cells, where ErbB2 is not required for EGFR and β1 integrin expression, ErbB2 overexpression results in an Erk-independent regulation of these proteins, which is not observed in control, ECM-attached cells.
Overexpression of a constitutively activated form of Mek (MekDD) results in activation of the Erk pathway independent of growth factor signaling. Consistent with our results, we found that overexpression of MekDD in the ECM-detached MCF-10A cells is sufficient to rescue the expression of EGFR and β1 protein (Fig. 3E). The dependence of EGFR and β1 integrin protein expression on Erk signaling is not specific to MCF-10A ErbB2 cells; we also found that in the ErbB2-expressing breast cancer cell line SK-BR-3, maintenance of EGFR and β1 integrin expression after ECM detachment is dependent on Erk signaling (Fig. 3F). Thus, ECM-detached cells overexpressing ErbB2 are highly dependent upon Erk signaling for maintenance of EGFR and β1 integrin protein expression, a contrast to the relative lesser dependence upon the Erk pathway for maintenance of these proteins in ECM-attached cells.
We also examined the dependence on Erk for ErbB2 rescue of EGFR and β1 integrin mRNA. Surprisingly, unlike for protein expression, inhibition of Erk signaling with either of two inhibitors (Fig. 3G and data not shown) did not suppress the ability of ErbB2 to maintain EGFR and β1 integrin mRNA levels in ECM-detached cells. Therefore, ErbB2 maintains EGFR and β1 integrin protein expression in ECM-detached cells via Erk-dependent pathways but regulates mRNA levels through Erk-independent signaling.
In contrast, ErbB2 rescue of EGFR and β1 integrin protein in ECM-detached cells is not dependent on PI3K signaling as treatment of the MCF-10A ErbB2 cells with the PI3K/mTor (mammalian target of rapamycin) inhibitor LY294002 did not affect the ability of ErbB2 to maintain EGFR or β1 integrin protein (Fig. 3H). Consistent with these results, we have previously shown that overexpression of the insulin-like growth factor 1 receptor in ECM-detached MCF-10A cells is sufficient to maintain PI3K activation but not EGFR protein levels (4). In addition, PI3K inhibition in MCF-10A ErbB2 cells did not affect EGFR and β1 integrin mRNA levels, and dual inhibition with both an Erk inhibitor and a PI3K inhibitor did not reduce the ErbB2 rescue of EGFR or β1 integrin mRNA expression (Fig. 3I). These results indicate that Erk (and not PI3K) signaling downstream of ErbB2 maintains EGFR and β1 integrin protein levels after ECM detachment and that neither PI3K nor Erk signaling is required for the regulation of the mRNA levels in ECM-detached MCF-10A ErbB2 cells.
ErbB2 Increases EGFR Protein Stability in ECM-detached Cells
Because conditions that maintain EGFR mRNA levels in ECM-detached MCF-10A ErbB2 cells did not allow maintenance of EGFR protein expression (Fig. 3, A and G) and because ErbB2 rescue of EGFR but not β1 integrin is critical for rescuing ATP levels (Fig. 2, G and H) and β1 integrin expression (Fig. 2, A and B) in ECM-detached cells, we focused our efforts on elucidating the Erk-dependent mechanism by which ErbB2 regulates EGFR protein levels in ECM-detached cells. Lysosomal degradation is the primary mechanism regulating EGFR turnover (15, 16). To determine the effects of ECM detachment and ErbB2 on EGFR protein stability, we first examined the kinetics of EGFR expression following ECM detachment (Fig. 4A). EGFR protein was low at 1 h after detachment, transiently increased at 3–6 h, and then decreased again from 12–24 h. MCF-10A ErbB2 cells also displayed decreased levels of EGFR 1 h after ECM detachment but then showed a steady increase in EGFR expression over the following 24 h. This low expression at the 1-h time point is not due to cleavage by trypsin as the same expression levels were seen in both cell types when cells were detached using non-enzymatic methods (cell dissociation buffer, Invitrogen) (Fig. 4B).
FIGURE 4.
ErbB2 increases EGFR protein stability in ECM-detached cells. A, MCF-10A or MCF-10A ErbB2 cells were plated under detached conditions and lysed after the indicated amount of time. Protein expression was analyzed by immunoblotting. B, MCF-10A or MCF-10A ErbB2 cells were detached from tissue culture plates using either trypsin or cell dissociation buffer (CDB, Invitrogen). Cells were then plated under detached conditions for 1 h, and protein levels were analyzed by immunoblotting. C, MCF-10A or MCF-10A ErbB2 cells were plated under detached conditions. After 3 h, cells were either lysed or treated with 10 μg/ml cycloheximide (CHX) for an additional 3 h. Protein levels were measured by immunoblotting, and data are presented as the percentage of EGFR remaining after the 3-h cycloheximide treatment. D, MCF-10A cells were plated under detached conditions. After 3 h, cells were either lysed or treated with 10 μg/ml cycloheximide or 10 μg/ml cycloheximide and 100 nm concanamycin A (CHX & CCA) for an additional 3 h. Protein levels were measured by immunoblotting, and data are presented as the amount of EGFR remaining after the 3-h cycloheximide treatment. E and F, MCF-10A cells were plated under detached conditions. After 3 h, 20 μm chloroquine diphosphate salt (E), 20 nm bafilomycin A1 (F), or vehicle was added. Cells were lysed 24 h after plating, and EGFR protein levels were measured by immunoblotting.
Based on these studies, we examined EGFR protein stability by treating cells with the translation inhibitor cycloheximide 3 h after ECM detachment and then measured the level of protein after an additional 3 h. ErbB2 overexpression caused a dramatic increase in EGFR protein stability in ECM-detached cells; although only ∼40% of the EGFR remained in the control cells after 3 h of cycloheximide treatment, the ErbB2-overexpressing cells retained ∼80% (Fig. 4C). EGFR degradation has been shown to occur predominantly through lysosomal degradation (15, 34); to ensure that the above results are not due to unintended effects of cycloheximide treatment, we examined whether we could reverse the loss of EGFR in the MCF-10A detached cells with the lysosomal inhibitor concanamycin A. Indeed, the combined treatment of concanamycin A together with cycloheximide restored EGFR expression in the ECM-detached MCF-10A cells (Fig. 4D). We also found that treatment with either of two other lysosomal inhibitors (chloroquine and bafilomycin A1) could increase EGFR expression in the ECM-detached MCF-10A cells (Fig. 4, E and F). Thus, inhibition of EGFR degradation contributes significantly to the maintenance of EGFR expression in ECM-detached cells, and ErbB2 rescues EGFR expression by increasing its protein stability.
ErbB2 Maintains EGFR Expression after ECM Detachment through Erk-dependent Sprouty2 Expression
Lysosomal degradation of EGFR is in large part regulated by the E3-ubiquitin ligase c-Cbl that targets EGFR to lysosomes (15, 16). One mechanism of Cbl regulation is through Erk inhibition via expression of Sprouty2. Erk signaling leads to increased transcription of Sprouty2, which binds to and inhibits Cbl, thereby decreasing its ability to promote the degradation of EGFR (35–40). Thus, through negative regulation of Cbl, Sprouty2 functions to increase EGFR protein stability. In certain contexts, Erk signaling, via induction of Sprouty2 mRNA and protein, negatively regulates Cbl activity (35–37, 40–44). To address whether Sprouty2 may be involved in regulation of EGFR expression by ECM, ErbB2, or Erk, we first examined whether Sprouty2 expression is affected by ECM detachment or ErbB2 overexpression. ECM detachment caused a 50% decrease in Sprouty2 mRNA levels (Fig. 5A), and ErbB2 overexpression completely prevented the reduction of Sprouty2 mRNA in ECM-detached cells in an Erk-dependent manner (Fig. 5A). In addition, expression of the activated variant of Mek, MekDD, rescued Sprouty2 mRNA in ECM-detached cells (Fig. 5B). These results demonstrate that expression of Sprouty2 mRNA correlates with Erk activation and EGFR stabilization.
To address whether ectopic expression of Sprouty2 could rescue EGFR expression in MCF-10A cells, we overexpressed Sprouty2 in these cells. Remarkably, although we were able to express high levels of Sprouty2 protein in MCF-10A cells, there was a dramatic decrease upon ECM detachment (Fig. 5C). Additionally, inhibition of Erk signaling with either PD98059 or UO126 also reduced Sprouty2 protein, even under ECM-attached conditions (Fig. 5D, lanes 1, 3, and 5). Consistent with the ability of ErbB2 to maintain EGFR expression and Erk signaling in ECM-detached cells, ErbB2 overexpression partially prevented the reduction of Sprouty2 protein in detached cells in an Erk-dependent manner (Fig. 5D, lanes 2 and 8, and lanes 8, 10, and 12). Sprouty2 expression in the MCF-10A ErbB2 attached cells was also dependent on Erk activation (Fig. 5D, lanes 7, 9, and 11). Thus, in MCF-10A and MCF-10A ErbB2 cells, Sprouty2 is regulated by Erk signaling both at the transcriptional and at the post-transcriptional level. We saw no rescue of EGFR or β1 integrin expression in the ECM-detached MCF-10A cells overexpressing Sprouty2 (Fig. 5E), but this is not surprising given the inability to maintain Sprouty2 overexpression under these conditions.
To address whether Sprouty2 is required for the ErbB2 rescue of EGFR and β1 integrin, we stably expressed two shRNAs targeting Sprouty2 in the MCF-10A ErbB2 cells (Fig. 5F). Down-regulation of Sprouty2 resulted in a decrease of both EGFR and β1 integrin in the ECM-detached MCF-10A ErbB2 cells (Fig. 5G) but not in ECM-attached cells (Fig. 5H).
EGFR and β1 integrin expression is maintained in an Erk-dependent manner in SK-BR-3 cells as well (Fig. 3F). Sprouty2 expression has also been found to be Erk-dependent in these cells (45). Knockdown of Sprouty2 in SK-BR-3 cells (Fig. 5I) also decreased EGFR and β1 integrin expression in ECM-detached cells (Fig. 5J) but not ECM-attached cells (Fig. 5K). These results indicate that Sprouty2 is required for the ErbB2 rescue of EGFR and β1 integrin in ECM-detached cells but not ECM-attached cells.
DISCUSSION
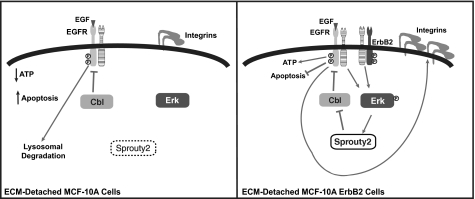
In this study, we provide new insights into the mechanisms that contribute to the decrease in growth factor receptor and integrin expression in mammary epithelial cells in ECM-detached conditions. We demonstrate that ECM regulates the expression of EGFR and β1 integrin protein and mRNA. Interestingly, down-regulation of β1 integrin protein and mRNA is a secondary consequence of loss of EGFR protein. An understanding of the basis for EGFR down-regulation was derived from analysis of the mechanism by which ErbB2 sustains the expression of EGFR in ECM-detached cells. ErbB2 was found to maintain EGFR expression through Erk-dependent transcriptional and post-transcriptional regulation of Sprouty2 (Fig. 6). In control, ECM-attached cells, the combined effects of EGFR and integrin engagement activate Erk signaling, which increases Sprouty2 expression, thereby contributing to the maintenance of EGFR protein expression. ECM-detached cells display decreased Erk activation and Sprouty2 expression, and these alterations likely contribute to the decreased EGFR (and β1 integrin) in ECM-detached cells. ECM-attached cells that overexpress ErbB2 maintain EGFR and β1 integrin expression through Erk-independent pathways, whereas the ECM-detached cells require Erk signaling and Sprouty2.
FIGURE 6.
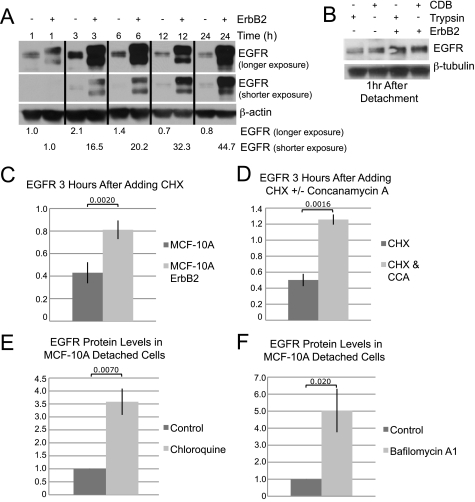
Model for regulation of EGFR and β1 integrin expression by ErbB2 and extracellular matrix contact. Detachment of MCF-10A cells from ECM prevents integrin engagement, leading to decreased Erk signaling and decreased Sprouty2; Cbl is thus active, and EGFR is degraded via the lysosome and can no longer maintain β1 integrin expression or ATP levels, or prevent apoptosis (left panel). Overexpression of ErbB2 maintains activation of Erk under ECM-detached conditions, leading to increased Sprouty2, which inhibits Cbl-induced degradation of EGFR and maintains β1 integrin expression and ATP levels and prevents apoptosis (right panel). The striped receptor represents any of the ErbB family members. Circled P, phosphorylation.
The ability of Sprouty2 to stabilize EGFR protein has been previously described (35–41, 43, 44, 46–48); however, Sprouty2 function appears to be context- and cell type-dependent (35, 44, 49–51). One of the most confounding aspects of Sprouty2 function is that it has been found to act as both a negative and a positive regulator of Erk signaling (40, 44, 47, 50, 52). These dichotomous activities may in part reflect differences in the regulation of Sprouty2 by different growth factors as Sprouty2 inhibits Erk activity induced by fibroblast growth factor (FGF) and vascular endothelial growth factor, whereas augmenting signaling from EGF by increasing EGFR protein stability (46–48, 53, 54). Tyrosine phosphorylation of Sprouty2 may contribute to these differences in the function of Sprouty2 as FGF but not EGF induces phosphorylation at Tyr-227 of Sprouty2, and this phosphorylation is required for inhibition of FGF-induced Erk signaling (55). MCF-10A cells are dependent on EGF for survival, proliferation, and growth factor signaling and express high levels of EGFR (7, 56). FGF receptor expression, however, is undetectable in MCF-10A cells (57). Thus, the role of Sprouty2 as a regulator of EGFR expression in these cells is consistent with the dependence of these cells on EGFR signaling.
Interestingly, Erk and Sprouty2 are essential for EGFR expression only in suspended MCF-10A ErbB2 cells, whereas this pathway is largely dispensable in attached conditions. In a previous study of a tumorigenic breast cell line, it was shown that EGFR and β1 integrin were coordinately regulated by Erk signaling only in matrix-attached cells grown in three-dimensional culture systems and not when cells were grown in standard monolayer culture (22). Our findings indicate that EGFR and β1 integrin are regulated by Erk both in ECM-attached MCF-10A cells and in ECM-detached MCF-10A ErbB2 cells. However, ECM-attached MCF-10A ErbB2 cells are not dependent on Erk for EGFR and β1 integrin expression; similarly, the cells used in the previous study are tumorigenic and express high levels of EGFR and β1 integrin. It is possible that transformation can induce Erk-independent regulation of these proteins in the context of ECM attachment as the Erk dependence in the MCF-10A ErbB2 cells was only evident when these cells were forced to grow without ECM contact. Therefore, the coregulation of EGFR and β1 integrin by Erk signaling can vary depending on differences in ECM interactions and cell context.
Given that the maintenance of EGFR and β1 integrin expression is more highly dependent on Mek/Erk signaling and Sprouty2 expression in ECM-detached MCF-10A ErbB2 cells, ErbB2-overexpressing cells may be more dependent upon Mek/Erk signaling and Sprouty2 for survival under ECM-detached conditions. As tumor cells are often challenged to survive in altered ECM environments, signaling pathways that are required for ECM-detached cells, but not ECM-attached cells (such as Mek/Erk and Sprouty2), may be candidate targets for cancer therapies. The increased dependence of ErbB2-overexpressing cells on Mek/Erk and Sprouty2 in ECM-detached conditions also raises important issues with respect to drug screening because these findings lead to the prediction that the sensitivity to targeted inhibitors will vary greatly depending upon different ECM conditions (attached versus detached and two-dimensional versus three-dimensional cell culture) (58–62). These results highlight the importance of ECM context in control of intracellular signaling and illustrate that the dependence of epithelial cells on attachment to ECM may serve a tumor suppressive function that could be exploited pharmacologically.
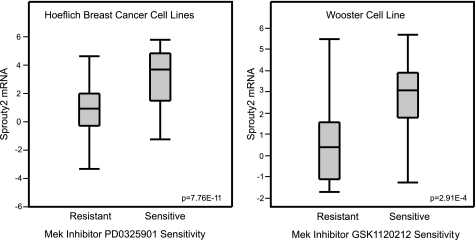
Although the dependence on Erk for ErbB2 stabilization of EGFR and β1 integrin has yet to be evaluated in vivo, it is possible that ErbB2 tumor cells would be sensitive to inhibition of the Mek/Erk pathway and Sprouty2, especially cells that reside in ECM-altered or -deprived conditions. In support of this, mining of two datasets (27, 28) that compare cell lines with differing sensitivities to Mek inhibitors revealed that Sprouty2 mRNA expression is significantly higher (p = 7.76 × 10–11 and p = 2.91 × 10–4) in cell lines sensitive to Mek inhibitors (Fig. 7).
FIGURE 7.
Sprouty2 mRNA levels correlate with sensitivity to Mek inhibitors. Box plots depicting the level of Sprouty2 mRNA from two publicly available microarray study datasets are shown. Sprouty2 mRNA expression levels are in normalized expression units downloaded from Oncomine.
Further support of the pro-tumorigenic role of Sprouty2 was provided by results from a recent report that demonstrated that H-RasV12 transformation of human fibroblasts increased Sprouty2 and EGFR expression (63). Knockdown of Sprouty2 decreased EGFR levels as well as in vitro and in vivo tumorigenicity. Human fibrosarcoma cells have higher Sprouty2 expression when compared with control fibroblasts, and knockdown of Sprouty2 in these cells resulted in decreased tumor incidence in mice (63, 64). Additionally, Sprouty2 was found to be specifically up-regulated in melanomas with B-RafV600E or N-RasQ16R mutations, and these mutant proteins were not sensitive to Sprouty2 inhibition of Erk signaling (65, 66). Recently, Sprouty2 was shown to negatively regulate E-cadherin expression (67); decreased E-cadherin expression can promote tumor progression through a number of mechanisms including anoikis resistance (68), and this may be another pro-tumorigenic function of Sprouty2. Consistent with this, Sprouty2 was found to be up-regulated in high grade colon cancers and showed increased expression at the invasive front of low grade tumors in patient samples (67). Sprouty2 expression also predicted sensitivity to an EGFR kinase inhibitor in colon cancer cell lines (69).
Given these functions of Sprouty2, it is critical that this previously categorized tumor suppressor be recognized for its potential tumor-promoting functions, especially in ErbB2-driven tumors. Additionally, we found that cells without ErbB2 overexpression, but with constitutive activation of Erk signaling, maintain EGFR expression via the same mechanism, and thus, tumors with constitutive Erk activation may respond to similar drug interventions. Pharmacological targeting of Sprouty2 could potentially sensitize tumor cells that are outside their natural niches. However, given the alternative role of Sprouty2 as an inhibitor of Mek/Erk signaling, this strategy should be pursued with caution. Additionally, it will be critical to identify markers that differentiate between tumors in which Sprouty2 functions to increase growth factor signaling via stabilization of EGFR and those in which Sprouty2 inhibits growth factor signaling through Mek/Erk. These markers may include increased ErbB2 expression and mutations in Ras and Raf.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of the pMSCV-IRES-PURO and pMSCV-SPRY2-IRES-PURO constructs from Dr. Michael Teitell (David Geffen School of Medicine at UCLA, Los Angeles, CA). We give our thanks to Ekrem Emrah Er, Dr. John Blenis, Dr. Richard Wooster, Dr. Jonathon Licht, Dr. Laura Selfors, and the members of the Brugge laboratory for helpful discussions of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant from the NCI (to J. S. B.). This work was also supported by a National Science Foundation Graduate Research Fellowship (to A. R. G.).
This article was selected as a Paper of the Week.
- ECM
- extracellular matrix
- EGFR
- epidermal growth factor receptor
- qPCR
- quantitative real-time PCR
- poly-HEMA
- poly(2-hydroxy methacrylate).
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 2. Horbinski C., Mojesky C., Kyprianou N. (2010) Am. J. Pathol. 177, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frisch S. M., Screaton R. A. (2001) Curr. Opin. Cell Biol. 13, 555–562 [DOI] [PubMed] [Google Scholar]
- 4. Schafer Z. T., Grassian A. R., Song L., Jiang Z., Gerhart-Hines Z., Irie H. Y., Gao S., Puigserver P., Brugge J. S. (2009) Nature 461, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiarugi P., Giannoni E. (2008) Biochem. Pharmacol. 76, 1352–1364 [DOI] [PubMed] [Google Scholar]
- 6. Harris J., Lippman M., Morrow M., Osborne C. (1999) Diseases of the Breast, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 7. Reginato M. J., Mills K. R., Paulus J. K., Lynch D. K., Sgroi D. C., Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Nat. Cell Biol. 5, 733–740 [DOI] [PubMed] [Google Scholar]
- 8. Collins N. L., Reginato M. J., Paulus J. K., Sgroi D. C., Labaer J., Brugge J. S. (2005) Mol. Cell Biol. 25, 5282–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reginato M. J., Mills K. R., Becker E. B., Lynch D. K., Bonni A., Muthuswamy S. K., Brugge J. S. (2005) Mol. Cell Biol. 25, 4591–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 11. Haenssen K. K., Caldwell S. A., Shahriari K. S., Jackson S. R., Whelan K. A., Klein-Szanto A. J., Reginato M. J. (2010) J. Cell Sci. 123, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baselga J., Swain S. M. (2009) Nat. Rev. Cancer 9, 463–475 [DOI] [PubMed] [Google Scholar]
- 13. Workman H. C., Sweeney C., Carraway K. L., 3rd (2009) Cancer Res. 69, 2845–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carraway K. L., Theodoropoulos G., Kozloski G. A., Carothers Carraway C. A. (2009) Future Oncol. 5, 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roepstorff K., Grøvdal L., Grandal M., Lerdrup M., van Deurs B. (2008) Histochem. Cell Biol. 129, 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorkin A., Goh L. K. (2009) Exp. Cell Res. 315, 683–696 [DOI] [PubMed] [Google Scholar]
- 17. Muthuswamy S. K., Gilman M., Brugge J. S. (1999) Mol. Cell Biol. 19, 6845–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cabodi S., Di Stefano P., Leal Mdel P., Tinnirello A., Bisaro B., Morello V., Damiano L., Aramu S., Repetto D., Tornillo G., Defilippi P. (2010) Adv. Exp. Med. Biol. 674, 43–54 [DOI] [PubMed] [Google Scholar]
- 19. Ivaska J., Heino J. (2010) Cell Tissue Res. 339, 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carraway K. L., 3rd, Sweeney C. (2006) Cancer Cell 10, 93–95 [DOI] [PubMed] [Google Scholar]
- 21. Yu X., Miyamoto S., Mekada E. (2000) J. Cell Sci. 113, 2139–2147 [DOI] [PubMed] [Google Scholar]
- 22. Wang F., Weaver V. M., Petersen O. W., Larabell C. A., Dedhar S., Briand P., Lupu R., Bissell M. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14821–14826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 24. Debnath J., Mills K. R., Collins N. L., Reginato M. J., Muthuswamy S. K., Brugge J. S. (2002) Cell 111, 29–40 [DOI] [PubMed] [Google Scholar]
- 25. Overholtzer M., Zhang J., Smolen G. A., Muir B., Li W., Sgroi D. C., Deng C. X., Brugge J. S., Haber D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mailleux A. A., Overholtzer M., Schmelzle T., Bouillet P., Strasser A., Brugge J. S. (2007) Dev. Cell 12, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoeflich K. P., O'Brien C., Boyd Z., Cavet G., Guerrero S., Jung K., Januario T., Savage H., Punnoose E., Truong T., Zhou W., Berry L., Murray L., Amler L., Belvin M., Friedman L. S., Lackner M. R. (2009) Clin. Cancer Res. 15, 4649–4664 [DOI] [PubMed] [Google Scholar]
- 28. Greshock J., Bachman K. E., Degenhardt Y. Y., Jing J., Wen Y. H., Eastman S., McNeil E., Moy C., Wegrzyn R., Auger K., Hardwicke M. A., Wooster R. (2010) Cancer Res. 70, 3677–3686 [DOI] [PubMed] [Google Scholar]
- 29. Spangenberg C., Lausch E. U., Trost T. M., Prawitt D., May A., Keppler R., Fees S. A., Reutzel D., Bell C., Schmitt S., Schiffer I. B., Weber A., Brenner W., Hermes M., Sahin U., Türeci O., Koelbl H., Hengstler J. G., Zabel B. U. (2006) Cancer Res. 66, 3715–3725 [DOI] [PubMed] [Google Scholar]
- 30. Brandt B., Meyer-Staeckling S., Schmidt H., Agelopoulos K., Buerger H. (2006) Clin. Cancer Res. 12, 7252–7260 [DOI] [PubMed] [Google Scholar]
- 31. Kim L. T., Yamada K. M. (1997) Proc. Soc. Exp. Biol. Med. 214, 123–131 [DOI] [PubMed] [Google Scholar]
- 32. Burgess A. W. (2008) Growth Factors 26, 263–274 [DOI] [PubMed] [Google Scholar]
- 33. Yarden Y. (2001) Eur. J. Cancer 37, Suppl. 4, S3–S8 [DOI] [PubMed] [Google Scholar]
- 34. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 35. Wong E. S., Guy G. R. (2006) Methods Mol. Biol. 327, 61–83 [DOI] [PubMed] [Google Scholar]
- 36. Rubin C., Litvak V., Medvedovsky H., Zwang Y., Lev S., Yarden Y. (2003) Curr. Biol. 13, 297–307 [DOI] [PubMed] [Google Scholar]
- 37. Ozaki K., Kadomoto R., Asato K., Tanimura S., Itoh N., Kohno M. (2001) Biochem. Biophys. Res. Commun. 285, 1084–1088 [DOI] [PubMed] [Google Scholar]
- 38. Haglund K., Schmidt M. H., Wong E. S., Guy G. R., Dikic I. (2005) EMBO Rep. 6, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fong C. W., Leong H. F., Wong E. S., Lim J., Yusoff P., Guy G. R. (2003) J. Biol. Chem. 278, 33456–33464 [DOI] [PubMed] [Google Scholar]
- 40. Wong E. S., Fong C. W., Lim J., Yusoff P., Low B. C., Langdon W. Y., Guy G. R. (2002) EMBO J. 21, 4796–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong E. S., Lim J., Low B. C., Chen Q., Guy G. R. (2001) J. Biol. Chem. 276, 5866–5875 [DOI] [PubMed] [Google Scholar]
- 42. DaSilva J., Xu L., Kim H. J., Miller W. T., Bar-Sagi D. (2006) Mol. Cell Biol. 26, 1898–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H. J., Taylor L. J., Bar-Sagi D. (2007) Curr. Biol. 17, 455–461 [DOI] [PubMed] [Google Scholar]
- 44. Guy G. R., Jackson R. A., Yusoff P., Chow S. Y. (2009) J. Endocrinol. 203, 191–202 [DOI] [PubMed] [Google Scholar]
- 45. Pratilas C. A., Taylor B. S., Ye Q., Viale A., Sander C., Solit D. B., Rosen N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4519–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Impagnatiello M. A., Weitzer S., Gannon G., Compagni A., Cotten M., Christofori G. (2001) J. Cell Biol. 152, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yusoff P., Lao D. H., Ong S. H., Wong E. S., Lim J., Lo T. L., Leong H. F., Fong C. W., Guy G. R. (2002) J. Biol. Chem. 277, 3195–3201 [DOI] [PubMed] [Google Scholar]
- 48. Sasaki A., Taketomi T., Wakioka T., Kato R., Yoshimura A. (2001) J. Biol. Chem. 276, 36804–36808 [DOI] [PubMed] [Google Scholar]
- 49. Edwin F., Anderson K., Ying C., Patel T. B. (2009) Mol. Pharmacol. 76, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lo T. L., Fong C. W., Yusoff P., McKie A. B., Chua M. S., Leung H. Y., Guy G. R. (2006) Cancer Lett. 242, 141–150 [DOI] [PubMed] [Google Scholar]
- 51. Guy G. R., Wong E. S., Yusoff P., Chandramouli S., Lo T. L., Lim J., Fong C. W. (2003) J. Cell Sci. 116, 3061–3068 [DOI] [PubMed] [Google Scholar]
- 52. Fong C. W., Chua M. S., McKie A. B., Ling S. H., Mason V., Li R., Yusoff P., Lo T. L., Leung H. Y., So S. K., Guy G. R. (2006) Cancer Res. 66, 2048–2058 [DOI] [PubMed] [Google Scholar]
- 53. Lao D. H., Yusoff P., Chandramouli S., Philp R. J., Fong C. W., Jackson R. A., Saw T. Y., Yu C. Y., Guy G. R. (2007) J. Biol. Chem. 282, 9117–9126 [DOI] [PubMed] [Google Scholar]
- 54. Lao D. H., Chandramouli S., Yusoff P., Fong C. W., Saw T. Y., Tai L. P., Yu C. Y., Leong H. F., Guy G. R. (2006) J. Biol. Chem. 281, 29993–30000 [DOI] [PubMed] [Google Scholar]
- 55. Rubin C., Zwang Y., Vaisman N., Ron D., Yarden Y. (2005) J. Biol. Chem. 280, 9735–9744 [DOI] [PubMed] [Google Scholar]
- 56. Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., Brugge J. S. (2001) Nat. Cell Biol. 3, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koziczak M., Holbro T., Hynes N. E. (2004) Oncogene 23, 3501–3508 [DOI] [PubMed] [Google Scholar]
- 58. Nelson C. M., Bissell M. J. (2006) Annu. Rev. Cell Dev. Biol. 22, 287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weigelt B., Lo A. T., Park C. C., Gray J. W., Bissell M. J. (2010) Breast Cancer Res. Treat. 122, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meads M. B., Gatenby R. A., Dalton W. S. (2009) Nat. Rev. Cancer 9, 665–674 [DOI] [PubMed] [Google Scholar]
- 61. Xu R., Boudreau A., Bissell M. J. (2009) Cancer Metastasis Rev. 28, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nam J. M., Onodera Y., Bissell M. J., Park C. C. (2010) Cancer Res. 70, 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lito P., Mets B. D., Kleff S., O'Reilly S., Maher V. M., McCormick J. J. (2008) J. Biol. Chem. 283, 2002–2009 [DOI] [PubMed] [Google Scholar]
- 64. Lito P., Mets B. D., Appledorn D. M., Maher V. M., McCormick J. J. (2009) J. Biol. Chem. 284, 848–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bloethner S., Chen B., Hemminki K., Müller-Berghaus J., Ugurel S., Schadendorf D., Kumar R. (2005) Carcinogenesis 26, 1224–1232 [DOI] [PubMed] [Google Scholar]
- 66. Tsavachidou D., Coleman M. L., Athanasiadis G., Li S., Licht J. D., Olson M. F., Weber B. L. (2004) Cancer Res. 64, 5556–5559 [DOI] [PubMed] [Google Scholar]
- 67. Barbáchano A., Ordóñez-Morán P., García J. M., Sánchez A., Pereira F., Larriba M. J., Martínez N., Hernández J., Landolfi S., Bonilla F., Pálmer H. G., Rojas J. M., Muñoz A. (2010) Oncogene 29, 4800–4813 [DOI] [PubMed] [Google Scholar]
- 68. Jeanes A., Gottardi C. J., Yap A. S. (2008) Oncogene 27, 6920–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Feng Y. H., Tsao C. J., Wu C. L., Chang J. G., Lu P. J., Yeh K. T., Shieh G. S., Shiau A. L., Lee J. C. (2010) Cancer Sci. 101, 2033–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.