Abstract
Gephyrin mediates the postsynaptic clustering of glycine receptors (GlyRs) and GABAA receptors at inhibitory synapses and molybdenum-dependent enzyme (molybdoenzyme) activity in non-neuronal tissues. Gephyrin knock-out mice show a phenotype resembling both defective glycinergic transmission and molybdenum cofactor (Moco) deficiency and die within 1 day of birth due to starvation and dyspnea resulting from deficits in motor and respiratory networks, respectively. To address whether gephyrin function is conserved among vertebrates and whether gephyrin deficiency affects molybdoenzyme activity and motor development, we cloned and characterized zebrafish gephyrin genes. We report here that zebrafish have two gephyrin genes, gphna and gphnb. The former is expressed in all tissues and has both C3 and C4 cassette exons, and the latter is expressed predominantly in the brain and spinal cord and harbors only C4 cassette exons. We confirmed that all of the gphna and gphnb splicing isoforms have Moco synthetic activity. Antisense morpholino knockdown of either gphna or gphnb alone did not disturb synaptic clusters of GlyRs in the spinal cord and did not affect touch-evoked escape behaviors. However, on knockdown of both gphna and gphnb, embryos showed impairments in GlyR clustering in the spinal cord and, as a consequence, demonstrated touch-evoked startle response behavior by contracting antagonistic muscles simultaneously, instead of displaying early coiling and late swimming behaviors, which are executed by side-to-side muscle contractions. These data indicate that duplicated gephyrin genes mediate Moco biosynthesis and control postsynaptic clustering of GlyRs, thereby mediating key escape behaviors in zebrafish.
Keywords: Development, Neurobiology, Neuron, Synapses, Zebra Fish
Introduction
Chemical synaptic transmission requires presynaptic release of neurotransmitters that trigger the opening of ligand-gated ion channels at postsynaptic sites. The accumulation or clustering of neurotransmitter receptors at postsynaptic specializations is critical for rapid and efficient synaptic transmission. Gephyrin, a major postsynaptic protein at inhibitory synapses, was initially identified as a tubulin-binding protein that co-purified with glycine receptors (GlyRs)3 (1–3). Gephyrin was later characterized as an essential scaffold protein, which forms postsynaptic aggregations of GlyRs or GABAA receptors at glycinergic or GABAergic synapses, respectively (4–7). Gephyrin consists of three distinct domains, an N-terminal G domain and a C-terminal E domain that are linked by a central C domain. Gephyrin directly binds to the large M3-M4 intracellular loop of the GlyR β subunit via the E domain (8–12), whereas the association to the M3-M4 intracellular loop of the GABAA receptor subunits is mediated by the boundary motif of the C and E domains (13). The G domain mediates trimer formation in vitro, whereas the E domain forms a dimer, thus potentially enabling the accumulation of GlyRs and GABAA receptors at postsynaptic sites (10, 14, 15). Gephyrin also interacts with Pin1 (peptidylprolyl isomerase) (16), Dlc1/2 (cargo-binding components of cytoplasmic dynein and myosin Va complexes) (17, 18), and collybistin (a RhoGEF for Cdc42) (19) via the C domain. In addition to their roles as aggregation frameworks, the E and G domains of gephyrin are crucial components for the biosynthesis of molybdenum cofactor (Moco) from molybdopterin (MPT) (5, 20). The G domain binds to MPT and catalyzes an adenylyl transfer to MPT (68). Adenylylated MPT is subsequently transferred to the E domain, where adenylylated MPT is hydrolyzed, thereby yielding Moco (21, 69). Because molybdenum-dependent enzymes (molybdoenzymes) such as nitrate reductase and sulfite oxidase are required for basic metabolism, it is not surprising that Moco biosynthesis is highly conserved in animals as well as in plants and bacteria (22). In fact, Moco deficiency in humans usually results in death in early childhood due to toxic sulfite levels (22, 23).
Antisense oligonucleotide-mediated knockdown of gephyrin in cultured spinal neurons eliminated GlyR clustering (4). Similarly, antisense oligonucleotides or shRNA-mediated depletion of gephyrin in cultured neurons blocked postsynaptic aggregation of GABAA receptors (6, 24). Neonatal gephyrin knock-out mice appeared externally normal but exhibited severe apnea (5). They did not suckle and thus their stomach never turned the normal white color. Because of these respiratory and nutritional defects, gephyrin-deficient newborns died in early neonatal stages. In addition, they displayed a progressive startle reflex in response to tactile stimuli. These symptoms were more severe than those reported for the phenotypes of mice carrying mutations in the GlyR α1 or β subunit genes, which display startle responses following sudden acoustic or tactile stimuli (25–31).
Mammals and birds are known to have a single gephyrin gene (3, 32–35), but gephyrin has not yet been studied in fish. Using the latest release of the zebrafish genome, we identified and characterized two duplicated gephyrin genes (gphna and gphnb) and examined alternative splicing, transcript distribution, and molybdoenzyme activity to determine whether these genes have evolved distinct structural and functional properties. Taking advantage of the ease of performing gene knockdown in zebrafish, we investigated whether one or both gephyrin proteins contribute to postsynaptic GlyR clustering, thereby mediating escape behaviors. Our findings suggest that the zebrafish gphna and gphnb have undergone functional partitioning by evolving distinct gene expression, while retaining the ability to synthesize Moco and to cluster GlyRs at synaptic sites.
EXPERIMENTAL PROCEDURES
Animals
Zebrafish (Danio rerio) were maintained on a 14-h light and 10-h dark cycle at 28.5 °C and fed twice daily according to the established procedures (36, 37). Embryos were obtained by natural crossing. Fertilized eggs were raised in system water at 28.5 °C and staged according to the standard developmental method (38). All experiments were conducted in accordance with the guidelines approved at National Institute of Genetics.
Database Searches
We searched the zebrafish genome database Zv8 using full-length peptide sequence of the human gephyrin P1 isoform (NP_001019389) as a query in BLAT/BLAST searches. We also identified multiple distinct zebrafish partial cDNAs using the same query in expression sequence tag databases (www.ncbi.nlm.nih.gov). Data were aligned and analyzed by Genetyx software (Genetyx Corp.).
Molecular Cloning
Total RNA was extracted from whole zebrafish embryos (48 h post-fertilization (hpf)) with TRIzol reagent (Invitrogen). After DNase treatment, total RNA (500 ng) was reverse-transcribed to single strand cDNA using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT)12–18 primer (Invitrogen) according to the manufacturer's instructions. To clone full-length gephyrin cDNAs, two sets of primers were used for PCR as follows: gphna forward primer, 5′-ATGGCGTCGGACGGGATGATTTTAAC-3′, and reverse primer, 5′-TCATAGCCGTCCTATGACCATGACG-3′; gphnb forward primer, 5′-ATGGCGTCAGACGGGATGATTTTAACAAAC-3′, and reverse primer, 5′-GTCATAGACGCCCGATGACCATG-3′.
Physical Mapping
The LN54 radiation hybrid panel (39) was used for physical mapping with the following primer sets: gphna RH forward primer, 5′-GTTGAGATTGCTCTGGCCTGACTCCTCCCC-3′, and reverse primer, 5′-GACAAACCACAGCAGGCAGCAGCGACAACG-3′; gphnb RH forward primer 1, 5′-CACACACGCTTGGGGAGGAG-3′, and reverse primer 1, 5′-CTCTCCCCCTCCAGAGTTCG-3′; gphnb RH forward primer 2, 5′-CCCATTTCAGATCTGCAAAGATGTGCCCTC-3′, and reverse primer 2, 5′-CATGCACCGTGAACCACCCGTCCATCTCTG-3′.
RT-PCR
Total RNA was isolated from 1, 2, or 5 days post-fertilization (dpf) zebrafish whole embryos or dissected heads (2 dpf). To compare transcript levels of alternative splicing isoforms, RT-PCR was performed with the following primers: gphna splicing forward primer, 5′-GGTGTGGCGTCCACCGAGGACAGCGGGTC-3′, and reverse primer, 5′-ATGTCTACAGCGCTGTGACTTGCTCTCAGG-3′; gphnb splicing forward primer, 5′-CACCGCCGCATCCATCGCTGCCAAGATTCC-3′, and reverse primer, 5′-CCTGAGCCAGAACTCGACCCATGCCGTCTC-3′. The PCR products were separated in a 2% (w/v) agarose gel and imaged by Printgraph (ATTO, AE-6933FXCF).
Determination of Moco Content in Escherichia coli mogA and moeA Mutant Cells
Zebrafish gephyrin constructs pQE80-gephyrin a (P1), -gephyrin a (C3), -gephyrin a (C4), -gephyrin a (C3 and C4), -gephyrin b (P1) and -gephyrin b (C4) as well as empty pQE80 vector were expressed in the E. coli mogA mutant RK5206 (40) and the moeA mutant SE1581 (41), respectively. Moco content was determined using the nit-1 reconstitution assay (42). In brief, 50-ml cultures were inoculated to an initial A600 of 0.25, induced with 50 μm isopropyl 1-thio-β-d-galactopyranoside, grown overnight anaerobically at 25 °C, harvested, washed with nit-1 buffer (50 mm sodium phosphate, 200 mm NaCl, 5 mm EDTA), solubilized in 1 ml of nit-1 buffer, and homogenized by sonication at 4 °C. The expression levels of each sample were analyzed by Western blotting. Aliquots of equally expressed proteins were used for the determination of Moco activity by nit-1 reconstitution using 1–10 μl of crude protein extract and 25 μl of Neurospora crassa nit-1 extract supplemented with 2 mm reduced glutathione. After 2 h of anaerobic reconstitution, nitrate reductase activity was determined as described previously (23). Expression levels of the different gephyrin splice variants in E. coli cells and zebrafish embryos were analyzed by Western blotting using either mouse monoclonal anti-gephyrin antibodies mAb3B11 (1:20) (42) or polyclonal rabbit anti-gephyrin antisera pAb-GepG (1:2000, Puszta Serum) (43) as primary antibodies.
Determination of Moco Content in Zebrafish Embryos
Control and gphna/gphnb and gphna/gphnb antisense morpholino-oligonucleotide (MO)-injected zebrafish embryos were solubilized in 250 μl of nit-1 buffer, homogenized with Potter S homogenizer (Sartorius), sonicated, and centrifuged for 20 min at 4 °C. For the determination of Moco content, 5–20 μl of crude extract were incubated with 30 μl of nit-1 extract. After 2 h or overnight anaerobic reconstitution, nitrate reductase activity was determined.
In Situ Hybridization
Plasmids encoding zebrafish full-length gephyrins were linearized by restriction enzyme digestion, followed by in vitro transcription reactions with T3 RNA polymerase (Stratagene) to synthesize digoxigenin-labeled antisense RNA probes. Whole-mount in situ hybridization using digoxigenin-labeled RNA probes was carried out as reported previously (44). In brief, embryos were raised in system water supplemented with 0.003% (5 mm) 2-phenylthiourea (Sigma) to prevent pigmentation after 24 hpf. At 24 and 48 hpf, embryos were fixed in 4% (w/v) paraformaldehyde at 28 °C for 8 h and treated with methanol. After proteinase K treatment, embryos were refixed and hybridized with the appropriate RNA probes at 65 °C, followed by incubation with anti-digoxigenin antibody conjugated with alkaline phosphatase and stained with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrates (Roche Applied Science) to produce purple insoluble precipitates. For sectioning after color development, 10-μm sections were cut with a cryostat (Leica, CM3050S).
Western Blotting
Whole-cell extracts were prepared from zebrafish embryos (48 hpf). 50 μg of protein was separated by 10% (w/v) SDS-PAGE and blotted onto polyvinylidene difluoride membrane (Bio-Rad). Anti-gephyrin (clone 45, mouse IgG1, 1:1000, BD Transduction Laboratories) and anti-mouse IgG, HRP-linked antibody (1:1000, Cell Signaling Technology) were used as primary and secondary antibodies, respectively.
Immunostaining
Zebrafish embryos were embedded in OCT compound and gradually frozen in liquid nitrogen. Cryosections (20 μm) were fixed in 4% (w/v) paraformaldehyde at room temperature for 10 min and subjected to immunostaining with anti-GlyRα (mAb4a, mouse IgG1, 1:1000, Synaptic Systems (45)), anti-gephyrin, anti-Islet1 (39.4D5, mouse IgG2, 1:200, Developmental Studies Hybridoma Bank), anti-GluR2/3 (EP929Y, rabbit monoclonal IgG, 1:1000, Epitomics), and anti-synaptic vesicles (SV2, mouse IgG1, 1:200, Developmental Studies Hybridoma Bank). Alexa 488-conjugated anti-mouse IgG1, Alexa 555-conjugated anti-mouse IgG1, Alexa 555-conjugated anti-mouse IgG2, Alexa 568-conjugated anti-mouse IgG, and Alexa 568-conjugated anti-rabbit IgG were used as secondary antibodies (1:1000, Invitrogen). Double labeling with anti-GlyR and anti-gephyrin was performed sequentially. Fluorescent images were captured by a confocal microscopy (Olympus, FV300).
Antisense Morpholino Injection
MO synthesized by Gene Tools were resuspended in sterile water at a concentration of 0.8 mm and delivered into zebrafish embryos at the 1–4-cell stage by microinjection (46). The injection apparatus consisted of a heat-pulled glass capillary (GD1.5, Narishige) positioned by a micromanipulator (UM-3C, Narishige), through which the MO was pressure-injected using a nitrogen air supply controlled by a microinjector (Picospritzer III, Parker). The MOs were designed against 15 bases of the 5′-UTR and 10 bases of the coding sequence on gphna and gphnb mRNAs. The CAT sequence in boldface corresponds to the start codon (ATG). The sequences of MO were as follows: gphna MO, 5′-CCGACGCCATGTTTAGCAGCGCTCT-3′; gphnb MO, 5′-CTGACGCCATATTCAGCACCGCTGT-3′; and control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′. Wild-type embryos were injected with 5–10 ng of MO. At these doses, control MOs produced no discernible phenotypes.
Video Recording of Zebrafish Touch Responses
Embryonic behaviors were observed and video recorded at 24 and 48 hpf using a dissection microscope (M165FC, Leica) and a high speed CCD camera (Fastcam-Ultima 1024, Photron). Mechanosensory stimulation was delivered to the tail with a forceps.
RESULTS
Identification of Two Gephyrin Genes in Zebrafish
We searched the zebrafish genome database using full-length peptide sequence of the human gephyrin P1 isoform (NP_001019389), which is known to be the standard isoform among many splicing products, and we found at least two genomic fragments each covering a gephyrin gene. We also identified multiple distinct zebrafish partial cDNAs using the same query in expression sequence tag databases. These in silico searches resulted in the identification of two distinct gephyrin genes located in Scaffold2082/Chromosome17 (designated gphna) and in Scaffold2347/Chromosome20 (designated gphnb). However, a match to C-terminal gphnb exons was found in Scaffold1318/Chromosome11. Given the current ambiguity of some gene locations in Zv8 due to incorrect contig assemblies, we physically mapped the chromosomal locations of gphna and gphnb using the LN54 radiation hybrid panel (39). This confirmed that gphna was located on chromosome 17 (logarithm of odds score, 17.6) as predicted by the genome database. By contrast, both the N and C termini of gphnb were physically mapped to chromosome 20 (logarithm of odds score, 11.2). No PCR matches were observed to chromosome 11. Thus, zebrafish appear to have two distinct gephyrin genes that are likely to have been generated by the ancestral genomic duplication during teleost evolution. We then amplified full-length cDNAs from zebrafish total RNA by RT-PCR using forward and reverse primers matching gphna and gphnb N and C termini. For gphna, we amplified four distinct gephyrin cDNAs, which were generated by alternative splicing. The shortest gephyrin a isoform was designated as gphna (P1) (GenBankTM accession number AB546096), which corresponds to the P1 construct in mammals (Fig. 1) (3, 35). The others were named gphna (C3) (AB546097), gphna (C4) (AB546098), and gphna (C3,C4) (AB546099), according to the inclusion of C3 and/or C4 cassette exons (see below). By contrast, fewer isoforms were identified for gphnb. We designated the shortest isoform as gphnb (P1) (AB546100), which corresponds to the P1 in mammals (3, 35), and the longer as gphnb (C4) (AB546101) due to the presence of C4 cassette exons. The P1 isoforms of gephyrin a and b were 735 and 736 amino acids long, respectively, and showed 95.9% sequence identity. Residues in the E domain required for binding to the GlyR β subunit (11) or in the G and E domains required for Moco biosynthesis (14, 47) were completely conserved in both zebrafish gephyrins. The boundary region of the C and E domains, which is responsible for GABAA receptor binding (13), was also conserved. Furthermore, in the C domain, the collybistin- and Dlc1/2-binding sites (17–19) were highly conserved, although the Pin1-binding site (16) is divergent in gephyrin b. These findings suggest that both zebrafish gephyrins are predicted to have equivalent functional properties to mammalian gephyrin in terms of Moco biosynthesis and GlyR clustering.
FIGURE 1.
Sequence alignment of zebrafish gephyrin proteins with frog, avian, and mammalian counterparts. Sequence alignment of human (GenBankTM accession number NP_001019389), rat (NP_074056), mouse (modified from NP_766540), chick (NP_001026720), and frog (modified from NP_001090459) gephyrin protein (P1 isoform) with zebrafish gephyrin a (BAI68421) and gephyrin b (BAI68425). The G, C, and E domains are represented as colored boxes (G domain, green; C domain, blue; E domain, orange). The positions of C3 and C4 cassettes are indicated by open triangles. The binding sites for Pin1, Dlc1/2, and Collybistin are highlighted by yellow boxes. The GABAAR-binding site is highlighted by a magenta box. Residues essential for GlyR β interactions and Moco biosynthesis are highlighted by red and blue circles below the residues, respectively.
Moco Synthetic Activity of Gephyrin Isoforms
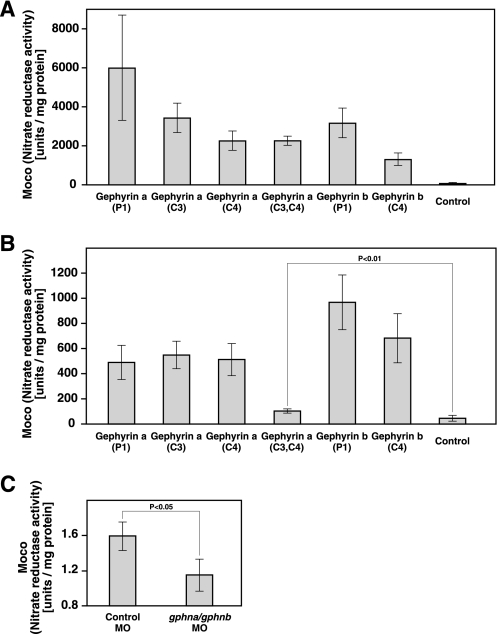
To demonstrate Moco synthetic activity, gephyrin isoforms were analyzed in Escherichia coli by reconstituting Moco synthesis in mutants of mogA and moeA, which have biosynthetic activity corresponding to the gephyrin G and E domains, respectively. Six recombinant gephyrin isoforms were expressed in both E. coli mutants, and Moco contents of crude extracts were determined by nit-1 reconstitution. All gephyrin a and b isoforms were able to restore Moco synthesis in mogA and moeA mutants (Fig. 2, A and B), confirming that Moco synthetic activity is maintained in gephyrin a and b regardless of inclusion of C3 and/or C4 cassette exons.
FIGURE 2.
Moco synthetic activity of zebrafish gephyrin. A, nitrate reductase activity in E. coli mogA RK5206 mutants carrying pQE80-gephyrin a (P1), -gephyrin a (C3), -gephyrin a (C4), -gephyrin a (C3,C4), -gephyrin b (P1), or -gephyrin b (C4) expression vector or control vector (pQE80). Error bars represent S.D. of three measurements. B, nitrate reductase activity in E. coli moeA SE1581 mutants expressing zebrafish gephyrin isoforms. Error bars represent S.D. of five measurements. Note that gephyrin a (C3 and C4) shows reduced but significant Moco synthetic activity. C, nitrate reductase activity in zebrafish embryos injected with a control MO or a mixture of gphna and gphnb MOs. Nit activity was significantly reduced in gephyrin morphants compared with controls, consistent with a large reduction in Moco biosynthesis. Although we were not able to completely abolish Moco biosynthesis, this is likely due to persistent maternal gephyrin or Moco. Error bars represent S.D. of four measurements.
To further examine whether zebrafish gephyrin products have Moco synthetic activity in zebrafish embryos, we used antisense morpholino-oligonucleotides (MO) to block gephyrin protein synthesis. We injected a control MO or a mixture of gphna and gphnb MOs into wild-type zebrafish embryos at the 1–4-cell stage. Moco contents of whole zebrafish embryos at 72 hpf were determined by nit-1 reconstitution. Moco synthetic activity was significantly reduced in gphna/gphnb MO-injected embryos compared with control MO-injected embryos (Fig. 2C). These successful complementation and knockdown experiments indicate that zebrafish gephyrin a and gephyrin b isoforms have Moco synthetic activity in vitro and in vivo.
Alternative Splicing of Zebrafish Gephyrin mRNAs and Temporal Expression
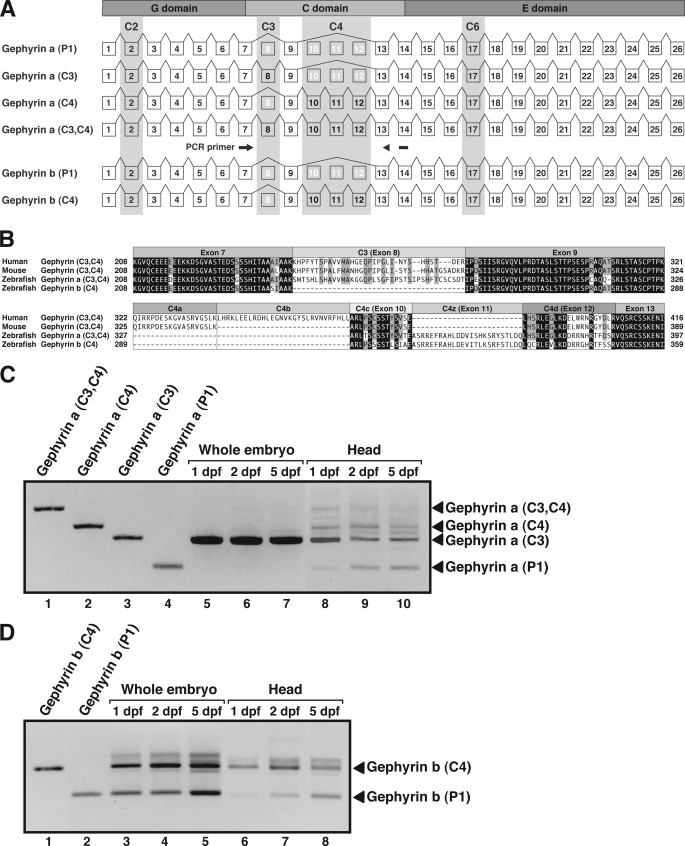
In mammals, the gephyrin gene consists of up to 30 exons (35, 48). Ten of these are so-called cassette exons, which are alternatively spliced into gephyrin mRNAs, giving rise to numerous gephyrin isoforms. These cassettes were originally named C1–C7 (3, 48). However, gephyrin isoforms appear to be expressed in a tissue-specific manner with species-specific differences (33, 35, 49–51). Different cassettes have been discovered by multiple groups and named independently; thus, the nomenclature of gephyrin isoforms is confusing. In this paper, we employed the cassette definition proposed by a recent comprehensive review on gephyrin (52). In zebrafish, gphna includes 26 exons, whereas gphnb has 25 exons (Fig. 3A). The C2 and C6 cassettes, which are constitutively spliced into the majority of mammalian and avian gephyrin isoforms, are now considered to be the common exons rather than cassette exons (52). In accordance with this notion, the corresponding exons (2 and 17) were present in all of the gphna and gphnb isoforms. By contrast, the C3 cassette, which is constitutively spliced into gephyrin transcripts in peripheral tissues and glial cells (35, 42, 49, 51, 53), was found in zebrafish gphna but not in gphnb, suggesting that gphna but not gphnb can generate non-neuronal gephyrin isoforms. In humans, four C4 cassette exons (C4a, C4b C4c, and C4d) have been characterized (Fig. 3B) (35, 52). In zebrafish, C4c and C4d as well as a zebrafish-specific C4 exon (designated C4z) located between exons C4c and C4d were found in both gphna and gphnb. No other cassettes were found in either gphna or gphnb genes.
FIGURE 3.
Alternative splicing of zebrafish gephyrin mRNAs. A, schematic diagram showing genomic structure of zebrafish gphna and gphnb. Exons are shown as boxes and introns as lines. Shaded exons 2, 8, 10–12, and 17 represent splicing cassettes C2, C3, C4, and C6, respectively. Positions of PCR primers to amplify gephyrin cDNAs isoforms are shown by arrows. B, amino acid sequence alignment of human, mouse, and zebrafish gephyrin isoforms containing C3 and/or C4 cassettes. C, temporal expression of gphna isoforms. Lanes 1–4, cloned gphna cDNAs were used as templates to represent the sizes of corresponding PCR products. Lanes 5–7, total RNA was extracted from 1, 2, or 5 dpf whole embryos and subjected to RT-PCR. Lanes 8–10, total RNA extracted from dissected heads of 1, 2, or 5 dpf embryos was subjected to RT-PCR. D, temporal expression of gphnb isoforms. Lanes 1 and 2, cloned gphnb cDNAs were used as templates. Lanes 3–5, RT-PCR using total RNA extracted from 1, 2, or 5 dpf whole zebrafish embryos. Lanes 6–8, RT-PCR using total RNA extracted from 1, 2, or 5 dpf zebrafish heads.
To examine temporal expression of alternative splicing isoforms of gphna and gphnb, we used semi-quantitative RT-PCR with total RNA extracted from whole embryos or dissected heads. Primers were designed in exons 7–14, which amplify cDNAs containing both C3 and C4 cassettes, for both gphna and gphnb. For gphna, we found that the C3 isoform was the major transcript detected in whole embryos, whereas P1, C3, C4, and C3/C4 isoforms could be detected in dissected heads (Fig. 3C). By contrast, for gphnb, both P1 and C4 were the major transcripts in whole embryos and in dissected heads (Fig. 3D). Note that no predicted C3 isoforms were detected for gphnb.
Spatial Expression Pattern of Gephyrin mRNA
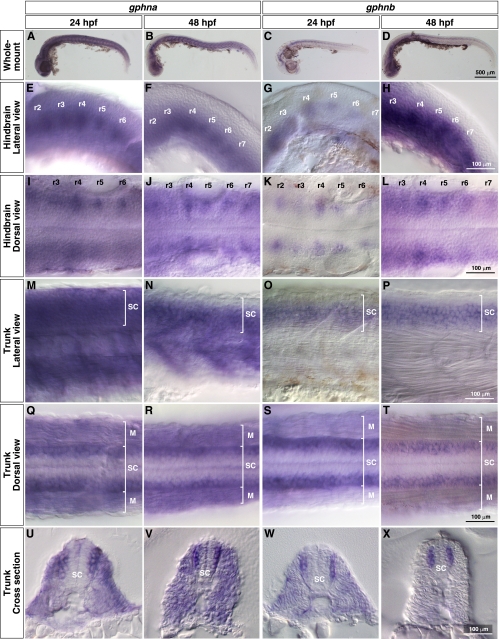
In mammals, the C3 cassette is excluded from neuronal gephyrins by a splicing factor called Nova (53). Thus, whereas C3 is constitutively spliced into gephyrin transcripts in peripheral tissues and glial cells, it is excluded in neurons (35, 42). If this assumption is correct, gphna, which includes the C3 cassette, should be expressed in many tissues, whereas gphnb, which does not harbor a C3 cassette exon, might only be expressed in neurons. Expression of gphna and gphnb was examined by whole-mount in situ hybridization using full-length cDNA probes. As predicted, gphna mRNA was detected in the whole embryo at 24 and 48 hpf (Fig. 4, A and B). In the hindbrain, gphna was predominantly expressed in repeating bilateral clusters of cells, which appeared to be reticulospinal neurons (Fig. 4, E, F, I, and J) (54). In the trunk region, gphna was expressed in both the spinal cord and muscle (Fig. 4, m, N, Q, and R). Cross-sections clearly demonstrated that gphna mRNA was predominantly found in the lateral spinal cord (Fig. 4, U and V). By contrast, gphnb expression was restricted to neurons at 24 and 48 hpf (Fig. 4, C and D). In the hindbrain, gphnb was expressed in reticulospinal neurons (Fig. 4, G, H, K, and L). In the trunk, expression of gphnb was observed in the spinal cord at 24 and 48 hpf (Fig. 4, O, P, S, and T). Cross-sections of the trunk revealed that gphnb was exclusively expressed in the lateral spinal cord (Fig. 4, W and X). These spatial patterns indicate that expression of gphna is ubiquitous (CNS and peripheral tissues), whereas gphnb encodes a neuronal gephyrin. However, the expression patterns of both genes overlap in the CNS, suggesting that both gphna and gphnb are important for the function of hindbrain reticulospinal neurons and lateral spinal neurons.
FIGURE 4.
Spatial expression of zebrafish gphna and gphnb. In situ hybridization with gphna and gphnb antisense probes. A–D, whole-mount images. Lateral views (E–H) and dorsal views (I–L) of hindbrains show gphna and gphnb expression in bilaterally located reticulospinal neurons of rhombomere segments (r2–r7). Lateral views (M–P) and dorsal views (Q–T) of trunks show gphna expression in muscles (M) and spinal cords (SC) and gphnb expression in spinal cord at 24 and 48 hpf. Cross-sections of trunks (U–X) reveal that both gphna and gphnb are expressed in lateral spinal neurons.
GlyR Clustering in Spinal Neurons
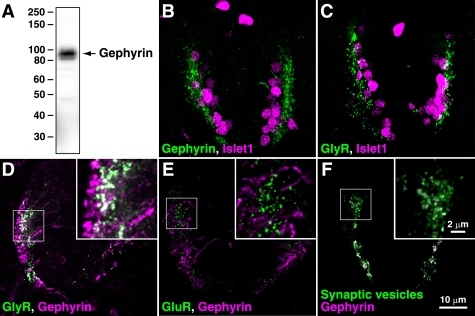
In mammals, monoclonal antibodies mAb7a (45) (Synaptic Systems) have been successfully used to demonstrate synaptic localization of gephyrin in vivo. However, in our hands, this antibody did not function in immunostaining experiments in zebrafish, perhaps due to subtle differences in zebrafish and mammalian gephyrin sequences. Instead, we found that another anti-gephyrin monoclonal antibody, clone 45 (BD Biosciences), reacts with zebrafish gephyrin proteins. Western blotting of whole embryo extracts with this antibody yielded a major band at ∼93 kDa, which is identical to the molecular weight of mammalian gephyrin (Fig. 5A). Immunolabeling of the spinal cord sections with this antibody revealed gephyrin puncta in the lateral area of the spinal cord, where both gphna and gphnb mRNA are expressed (Fig. 5B). Double labeling with anti-Islet1 (a marker for motor neuron nuclei and primary sensory Rohon-Beard neuron nuclei) revealed that gephyrin proteins are likely to be expressed in motor neurons and some inter-neurons. GlyR aggregates labeled using the anti-GlyRα (mAb4a) were also present in lateral spinal neurons, including motor neurons (Fig. 5C). In fact, co-labeling of GlyR and gephyrin demonstrated that GlyR and gephyrin were co-localized in lateral spinal neurons (Fig. 5D). By contrast, gephyrin did not co-localize with excitatory glutamate receptors (Fig. 5E), corroborating the notion that gephyrin functions as a scaffolding protein at inhibitory but not excitatory synapses. We also confirmed co-labeling of gephyrin and synaptic vesicles, indicating that, as expected, gephyrin is located at synapses. Along with the conservation of GlyR α and β proteins in vertebrates (44, 55, 56), these results strongly suggest that zebrafish gephyrins mediate GlyR clustering.
FIGURE 5.
Gephyrin co-localizes with GlyRs in the spinal cord. A, protein extracts from whole zebrafish embryos (48 hpf) were separated by SDS-PAGE and probed with an anti-gephyrin antibody, resulting in the detection of a major ∼93-kDa protein. B–F, immunostaining of zebrafish spinal cord sections (48 hpf). Cross-sections were co-labeled with anti-gephyrin and anti-Islet1 (B), with anti-GlyRα and anti-Islet1 (C), with anti-GlyRα and anti-gephyrin (D), with anti-GluR and anti-gephyrin (E), or with anti-synaptic vesicles and anti-gephyrin (F). Islet1-positive cells are motor neurons (ventral side) and mechanosensory Rohon-Beard neurons (dorsal side). Note that gephyrin is co-localized with GlyRs but not with GluRs.
Gephyrin Is Necessary for GlyR Clustering and Escape Behavior
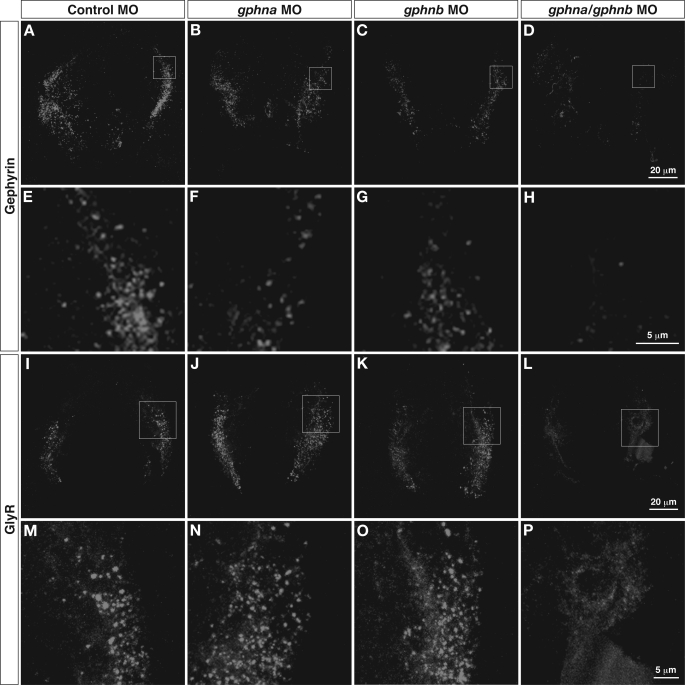
To assess whether gephyrin is essential for postsynaptic accumulation of GlyRs in zebrafish, we used MO to block gephyrin protein synthesis. We injected control MO, gphna MO, gphnb MO, or a mixture of both MOs into wild-type embryos at the 1–4-cell stage. Cross-sections of morphants were labeled with anti-gephyrin or anti-GlyRα antibodies at 48 hpf. Following gphna or gphnb MO injection, punctate gephyrin immunostaining was similar to that seen following the control MO injection (Fig. 6, A–C and E–G), whereas gephyrin puncta were not observed when a mixture of both MOs was injected (Fig. 6, D and H). These immunolabelings confirm that the anti-gephyrin antibody recognizes both gephyrin a and b proteins in zebrafish and that gephyrin immunoreactivity is eliminated only when translation of both mRNA species is blocked. Labeling with an anti-GlyRα antibody revealed GlyR clusters in the lateral spinal cord in the control and gphna and gphnb morphants (Fig. 6, I–K and M–O). However, in the double morphants, sparse diffuse signals rather than puncta were seen in the lateral spinal neurons (Fig. 6, L and P). These results strongly suggest that gephyrin is necessary for the synaptic clustering of GlyRs in developing zebrafish embryos.
FIGURE 6.
Inhibition of zebrafish gephyrin translation. Control, gphna, or gphnb MO or a mixture of gphna/gphnb MOs were injected into wild-type embryos. Cross-sections of these morphants (48 hpf) were immunolabeled with anti-gephyrin (A–H) or anti-GlyR (I–P). Expression of gephyrin in the lateral spinal cord was observed in control, gphna, and gphnb morphants (A–C) but not in gphna/gphnb double morphants (D). High magnification images of the lateral spinal cord revealed that gephyrin immunoreactivities are puncta in control, gphna, and gphnb morphants (E–G). Gephyrin protein was eliminated by knocking down both gphna and gphnb (H). GlyR clusters were observed in control, gphna, and gphnb morphants (I–K) but not in gphna/gphnb double morphants (L). High magnification images (M–P) confirm that unlike control or gphna and gphnb morphants (M–O), GlyR immunoreactivity in gphna/gphnb double morphants is weak and diffuse, presumably representing GlyRs trapped in intracellular compartments (P).
Gephyrin knock-out mice assume a rigid, hyperextended posture in response to tactile stimuli (5). To address whether gephyrin morphants in zebrafish also exhibit abnormalities in motility and touch-elicited escape behaviors, gphna and gphnb morphants as well as double gphna/gphnb morphants were examined at 24 and 48 hpf using high speed video recording. At 24 hpf, control, gphna and gphnb morphants responded to touch with normal side-to-side alternating contractions of the trunk muscles that resulted in typical coiling behaviors (supplemental Movies 1–3). However, double morphants demonstrated apparent simultaneous bilateral contractions of trunk muscles and showed dorsal flexure and shortening of the body (supplemental Movie 4). Likewise at 48 hpf, control and gphna and gphnb morphants swam away following touch (supplemental Movies 5–7), whereas double knockdown embryos displayed a startle response by contracting antagonistic muscles simultaneously (supplemental Movie 8). These abnormal behaviors of gphna/gphnb double morphants are highly reminiscent of zebrafish bandoneon mutants that carry mutations in glrbb, encoding one of two duplicated GlyR β subunits (44). These results, along with the GlyR clustering defects in double knockdown embryos, indicate that both zebrafish gephyrins are critical for GlyR clustering in vivo and as a consequence for proper touch-evoked escape behavior.
DISCUSSION
In this study, we identified gphna and gphnb, encoding two zebrafish orthologs of vertebrate gephyrin. Both genes were expressed in the CNS, including reticulospinal neurons and motor neurons. However, only gphna was expressed in non-neural tissues in addition to the CNS and carried the C3 cassette that is the exon constitutively spliced into peripheral and glial gephyrin isoforms in mammals. Antisense-mediated single knockdown of either gphna or gphnb did not cause any apparent defects. However, double knockdown embryos showed impairments in synaptic GlyR clustering and, as a consequence, exhibited simultaneous contraction of bilateral muscles, resulting in abnormal startle reflexes in response to touch. Therefore, it appears that these duplicated gephyrin genes redundantly regulate synaptic GlyR aggregation and, thus, escape behavior.
Mammals, birds, and amphibians each have a single gephyrin gene, whereas zebrafish have two gephyrin orthologs. The existence of two distinct counterparts of a mammalian gene is not uncommon in zebrafish due to suspected duplication of the whole genome during fish evolution (57, 58). Zebrafish gphna and gphnb were located on chromosome 17 and 20, respectively, whereas human gephyrin gene is located on chromosome 14 (32). In agreement with the suspected genome duplication, comparative genomic studies have suggested that zebrafish chromosome 17 and 20 and human chromosome 14 are derived from the same ancestral chromosome (59, 60).
The interesting question raised by such duplicated orthologs is as follows. How are the functions of the genes divided in the case of duplicated genes? In some cases, one ortholog plays a dominant role and the other plays a minor role (61). In the other cases, the two orthologs serve as backups for each other, thereby working redundantly (62). But if the two orthologs alter their spatial or temporal expression patterns, each of the genes can acquire separate essential functions (63). In zebrafish, gphna was expressed ubiquitously as shown previously for mammalian gephyrin (3, 35, 48, 51, 53), whereas gphnb was expressed only in the CNS. Because the gphna/gphnb double morphants but neither of the single morphants showed abnormal neuronal functions, it appears that gphna and gphnb redundantly operate GlyR clustering in the CNS.
Because the C3 cassette is excluded in mouse neurons by the splicing factor Nova (53), C3 has been thought of as an important exon in non-neuronal functions such as Moco biosynthesis. Similarly in zebrafish, none of the isoforms of gphnb, which is expressed exclusively in neurons, contained C3, corroborating the notion that the C3 cassette is repressed in neurons. In addition, gphna without C3 was enriched in the head, whereas most of the gphna mRNA in whole zebrafish embryos consisted of the C3 isoform. Taken together along with previous results showing a glial and peripheral expression of gephyrin C3 (35, 42), our results suggest that C3 is somehow linked to non-neuronal roles of gephyrin. However, regardless of the presence of C3, all of the zebrafish gephyrin a and gephyrin b isoforms showed Moco synthetic activity as in the case of rat gephyrin isoforms (42).
C4 cassettes are generated by four distinct exons (C4a, C4b, C4c, and C4d) in humans (35), three exons (C4a, C4c, and C4d) in mice (48), and three exons (C4c, C4z, and C4d) in zebrafish. Thus, it appears that the number and types of C4 exons are variable between species. In this study, we found novel C4z exons between C4c and C4d that are spliced into both gphna and gphnb mRNAs. Although we could not find the corresponding potential exon sequence between C4c and C4d in the human and mouse gephyrin genes, additional gephyrin isoforms may remain to be uncovered in other species. Gephyrin containing both the C4 and the G2 cassette (formerly C5 or C5′ cassette) blocks postsynaptic aggregation of gephyrin. However, this inhibitory effect is attributable to the insertion of the G2 cassette into the N-terminal G domain, which interferes with gephyrin trimerization and consequently higher order oligomerization (12, 64, 65). Likewise, Moco synthetic activity is affected by inclusion of G2 but not of C4 cassettes (42). We confirmed that Moco synthetic activity of zebrafish gephyrin is not affected by the presence or absence of the C4 cassette. Interestingly, however, we noted that inclusion of both C3 and C4 cassettes reduces the activity compared with the P1, C3, or C4 isoforms. Thus, function of the C4 cassettes remain elusive but must serve some important function, because they are conserved in mammals and zebrafish.
Future experiments with gene or splice cassette-specific morpholinos in zebrafish have the potential to reveal additional specialized roles of gphna and gphnb. These experiments will require antibodies that recognize gephyrin a and b to confirm selective knockdown, because both gephyrin proteins exhibit Moco synthetic activity in recombinant expression systems (Fig. 2). Even then, it may not be possible to completely eliminate Moco activity, because zebrafish embryos have a yolk, which contains maternally supplied mRNA and proteins.
Gephyrin is also clearly linked to clustering of selected GABAARs containing α2 or α3 subunits in mammals (13, 66). However, although the GlyR gene family has been well characterized (55), little is known about the GABAAR gene family in zebrafish. In particular, α2 and α3 subunit homologs have not yet been identified. We have as yet been unable to find a functioning antibody for GABAAR immunolabeling in zebrafish, probably because many commercially available GABAAR subunit-specific antibodies utilize N- or C-terminal sequence as antigens that are poorly conserved between species. Identifying zebrafish GABAAR genes and development of zebrafish-specific reagents therefore remains a key challenge for the future. Although GABAergic synaptic currents have been recorded in hindbrain Mauthner neurons (67), they are barely detectable in motor neurons at 48 hpf (55). This is likely to explain why we observed a glycinergic phenotype similar to the GlyR βb mutant bandoneon in gphna/gphnb double morphants, consistent with a key role of the two zebrafish gephyrin genes in clustering GlyRs, rather than GABAARs, at this early time point.
Supplementary Material
Acknowledgments
We thank Dr. John Y. Kuwada (University of Michigan), Dr. Yoichi Oda (Nagoya University), Dr. Shin Takagi (Nagoya University), Dr. Koichi Kawakami (National Institute of Genetics), Megumi Takahashi (Nagoya University), and Yurie Matsutani (Nagoya University) for helpful discussion, support, and fish care.
This work was supported in part by grants-in-aid for young scientists (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Nakajima Foundation, Takeda Science Foundation, and Career Development Award of the Human Frontier Science Program (to H. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies 1–8.
- GlyR
- glycine receptor
- dpf
- days post-fertilization
- gphn
- gephyrin
- hpf
- hours post-fertilization
- MO
- morpholino-oligonucleotide
- Moco
- molybdenum cofactor
- MPT
- molybdopterin.
REFERENCES
- 1. Pfeiffer F., Graham D., Betz H. (1982) J. Biol. Chem. 257, 9389–9393 [PubMed] [Google Scholar]
- 2. Kirsch J., Langosch D., Prior P., Littauer U. Z., Schmitt B., Betz H. (1991) J. Biol. Chem. 266, 22242–22245 [PubMed] [Google Scholar]
- 3. Prior P., Schmitt B., Grenningloh G., Pribilla I., Multhaup G., Beyreuther K., Maulet Y., Werner P., Langosch D., Kirsch J., et al. (1992) Neuron 8, 1161–1170 [DOI] [PubMed] [Google Scholar]
- 4. Kirsch J., Wolters I., Triller A., Betz H. (1993) Nature 366, 745–748 [DOI] [PubMed] [Google Scholar]
- 5. Feng G., Tintrup H., Kirsch J., Nichol M. C., Kuhse J., Betz H., Sanes J. R. (1998) Science 282, 1321–1324 [DOI] [PubMed] [Google Scholar]
- 6. Essrich C., Lorez M., Benson J. A., Fritschy J. M., Lüscher B. (1998) Nat. Neurosci. 1, 563–571 [DOI] [PubMed] [Google Scholar]
- 7. Kneussel M., Brandstätter J. H., Laube B., Stahl S., Müller U., Betz H. (1999) J. Neurosci. 19, 9289–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer G., Kirsch J., Betz H., Langosch D. (1995) Neuron 15, 563–572 [DOI] [PubMed] [Google Scholar]
- 9. Schrader N., Kim E. Y., Winking J., Paulukat J., Schindelin H., Schwarz G. (2004) J. Biol. Chem. 279, 18733–18741 [DOI] [PubMed] [Google Scholar]
- 10. Sola M., Bavro V. N., Timmins J., Franz T., Ricard-Blum S., Schoehn G., Ruigrok R. W., Paarmann I., Saiyed T., O'Sullivan G. A., Schmitt B., Betz H., Weissenhorn W. (2004) EMBO J. 23, 2510–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim E. Y., Schrader N., Smolinsky B., Bedet C., Vannier C., Schwarz G., Schindelin H. (2006) EMBO J. 25, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saiyed T., Paarmann I., Schmitt B., Haeger S., Sola M., Schmalzing G., Weissenhorn W., Betz H. (2007) J. Biol. Chem. 282, 5625–5632 [DOI] [PubMed] [Google Scholar]
- 13. Saiepour L., Fuchs C., Patrizi A., Sassoè-Pognetto M., Harvey R. J., Harvey K. (2010) J. Biol. Chem. 285, 29623–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz G., Schrader N., Mendel R. R., Hecht H. J., Schindelin H. (2001) J. Mol. Biol. 312, 405–418 [DOI] [PubMed] [Google Scholar]
- 15. Sola M., Kneussel M., Heck I. S., Betz H., Weissenhorn W. (2001) J. Biol. Chem. 276, 25294–25301 [DOI] [PubMed] [Google Scholar]
- 16. Zita M. M., Marchionni I., Bottos E., Righi M., Del Sal G., Cherubini E., Zacchi P. (2007) EMBO J. 26, 1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuhrmann J. C., Kins S., Rostaing P., El Far O., Kirsch J., Sheng M., Triller A., Betz H., Kneussel M. (2002) J. Neurosci. 22, 5393–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maas C., Tagnaouti N., Loebrich S., Behrend B., Lappe-Siefke C., Kneussel M. (2006) J. Cell Biol. 172, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kins S., Betz H., Kirsch J. (2000) Nat. Neurosci. 3, 22–29 [DOI] [PubMed] [Google Scholar]
- 20. Stallmeyer B., Schwarz G., Schulze J., Nerlich A., Reiss J., Kirsch J., Mendel R. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwarz G. (2005) Cell. Mol. Life Sci. 62, 2792–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwarz G., Mendel R. R., Ribbe M. W. (2009) Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 23. Reiss J., Gross-Hardt S., Christensen E., Schmidt P., Mendel R. R., Schwarz G. (2001) Am. J. Hum. Genet. 68, 208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu W., Jiang M., Miralles C. P., Li R. W., Chen G., de Blas A. L. (2007) Mol. Cell. Neurosci. 36, 484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryan S. G., Buckwalter M. S., Lynch J. W., Handford C. A., Segura L., Shiang R., Wasmuth J. J., Camper S. A., Schofield P., O'Connell P. (1994) Nat. Genet. 7, 131–135 [DOI] [PubMed] [Google Scholar]
- 26. Buckwalter M. S., Cook S. A., Davisson M. T., White W. F., Camper S. A. (1994) Hum. Mol. Genet. 3, 2025–2030 [DOI] [PubMed] [Google Scholar]
- 27. Saul B., Schmieden V., Kling C., Mülhardt C., Gass P., Kuhse J., Becker C. M. (1994) FEBS Lett. 350, 71–76 [DOI] [PubMed] [Google Scholar]
- 28. White W. F., Heller A. H. (1982) Nature 298, 655–657 [DOI] [PubMed] [Google Scholar]
- 29. Kingsmore S. F., Giros B., Suh D., Bieniarz M., Caron M. G., Seldin M. F. (1994) Nat. Genet. 7, 136–141 [DOI] [PubMed] [Google Scholar]
- 30. Mülhardt C., Fischer M., Gass P., Simon-Chazottes D., Guénet J. L., Kuhse J., Betz H., Becker C. M. (1994) Neuron 13, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 31. Harvey R. J., Topf M., Harvey K., Rees M. I. (2008) Trends Genet. 24, 439–447 [DOI] [PubMed] [Google Scholar]
- 32. David-Watine B. (2001) Gene 271, 239–245 [DOI] [PubMed] [Google Scholar]
- 33. Heck S., Enz R., Richter-Landsberg C., Blohm D. H. (1997) Dev. Brain Res. 98, 211–220 [DOI] [PubMed] [Google Scholar]
- 34. Tsen G., Williams B., Allaire P., Zhou Y. D., Ikonomov O., Kondova I., Jacob M. H. (2000) Nat. Neurosci. 3, 126–132 [DOI] [PubMed] [Google Scholar]
- 35. Rees M. I., Harvey K., Ward H., White J. H., Evans L., Duguid I. C., Hsu C. C., Coleman S. L., Miller J., Baer K., Waldvogel H. J., Gibbon F., Smart T. G., Owen M. J., Harvey R. J., Snell R. G. (2003) J. Biol. Chem. 278, 24688–24696 [DOI] [PubMed] [Google Scholar]
- 36. Nüsslein-Volhard C., Dahm R. (eds) (2002) Zebrafish: A Practical Approach, 1st Ed., Oxford University Press, Oxford [Google Scholar]
- 37. Westerfield M. (ed) (1993) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) University of Oregon Press, Eugene, OR [Google Scholar]
- 38. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Dev Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 39. Hukriede N. A., Joly L., Tsang M., Miles J., Tellis P., Epstein J. A., Barbazuk W. B., Li F. N., Paw B., Postlethwait J. H., Hudson T. J., Zon L. I., McPherson J. D., Chevrette M., Dawid I. B., Johnson S. L., Ekker M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9745–9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stewart V., MacGregor C. H. (1982) J. Bacteriol. 151, 788–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hasona A., Ray R. M., Shanmugam K. T. (1998) J. Bacteriol. 180, 1466–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smolinsky B., Eichler S. A., Buchmeier S., Meier J. C., Schwarz G. (2008) J. Biol. Chem. 283, 17370–17379 [DOI] [PubMed] [Google Scholar]
- 43. Giesemann T., Schwarz G., Nawrotzki R., Berhörster K., Rothkegel M., Schlüter K., Schrader N., Schindelin H., Mendel R. R., Kirsch J., Jockusch B. M. (2003) J. Neurosci. 23, 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hirata H., Saint-Amant L., Downes G. B., Cui W. W., Zhou W., Granato M., Kuwada J. Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8345–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfeiffer F., Simler R., Grenningloh G., Betz H. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 7224–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nasevicius A., Ekker S. C. (2000) Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- 47. Xiang S., Nichols J., Rajagopalan K. V., Schindelin H. (2001) Structure 9, 299–310 [DOI] [PubMed] [Google Scholar]
- 48. Ramming M., Kins S., Werner N., Hermann A., Betz H., Kirsch J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Paarmann I., Schmitt B., Meyer B., Karas M., Betz H. (2006) J. Biol. Chem. 281, 34918–34925 [DOI] [PubMed] [Google Scholar]
- 50. Meier J., De Chaldée M., Triller A., Vannier C. (2000) Mol. Cell. Neurosci. 16, 566–577 [DOI] [PubMed] [Google Scholar]
- 51. Paarmann I., Saiyed T., Schmitt B., Betz H. (2006) Biochem. Soc. Trans. 34, 45–47 [DOI] [PubMed] [Google Scholar]
- 52. Fritschy J. M., Harvey R. J., Schwarz G. (2008) Trends Neurosci. 31, 257–264 [DOI] [PubMed] [Google Scholar]
- 53. Ule J., Ule A., Spencer J., Williams A., Hu J. S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., Zeeberg B. R., Kane D., Weinstein J. N., Blume J., Darnell R. B. (2005) Nat. Genet. 37, 844–852 [DOI] [PubMed] [Google Scholar]
- 54. Kimmel C. B., Powell S. L., Metcalfe W. K. (1982) J. Comp. Neurol. 205, 112–127 [DOI] [PubMed] [Google Scholar]
- 55. Hirata H., Carta E., Yamanaka I., Harvey R. J., Kuwada J. Y. (2009) Front. Mol. Neurosci. 2, 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. David-Watine B., Goblet C., de Saint Jan D., Fucile S., Devignot V., Bregestovski P., Korn H. (1999) Neuroscience 90, 303–317 [DOI] [PubMed] [Google Scholar]
- 57. Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., Westerfield M., Ekker M., Postlethwait J. H. (1998) Science 282, 1711–1714 [DOI] [PubMed] [Google Scholar]
- 58. Jaillon O., Aury J. M., Brunet F., Petit J. L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Nicaud S., Jaffe D., Fisher S., Lutfalla G., Dossat C., Segurens B., Dasilva C., Salanoubat M., Levy M., Boudet N., Castellano S., Anthouard V., Jubin C., Castelli V., Katinka M., Vacherie B., Biémont C., Skalli Z., Cattolico L., Poulain J., De Berardinis V., Cruaud C., Duprat S., Brottier P., Coutanceau J. P., Gouzy J., Parra G., Lardier G., Chapple C., McKernan K. J., McEwan P., Bosak S., Kellis M., Volff J. N., Guigó R., Zody M. C., Mesirov J., Lindblad-Toh K., Birren B., Nusbaum C., Kahn D., Robinson-Rechavi M., Laudet V., Schachter V., Quétier F., Saurin W., Scarpelli C., Wincker P., Lander E. S., Weissenbach J., Roest Crollius H. (2004) Nature 431, 946–957 [DOI] [PubMed] [Google Scholar]
- 59. Naruse K., Tanaka M., Mita K., Shima A., Postlethwait J., Mitani H. (2004) Genome Res. 14, 820–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Woods I. G., Wilson C., Friedlander B., Chang P., Reyes D. K., Nix R., Kelly P. D., Chu F., Postlethwait J. H., Talbot W. S. (2005) Genome Res. 15, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kleinjan D. A., Bancewicz R. M., Gautier P., Dahm R., Schonthaler H. B., Damante G., Seawright A., Hever A. M., Yeyati P. L., van Heyningen V., Coutinho P. (2008) PLoS Genet. 4, e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shankaran S. S., Sieger D., Schröter C., Czepe C., Pauly M. C., Laplante M. A., Becker T. S., Oates A. C., Gajewski M. (2007) Dev. Biol. 304, 615–632 [DOI] [PubMed] [Google Scholar]
- 63. Hirata H., Watanabe T., Hatakeyama J., Sprague S. M., Saint-Amant L., Nagashima A., Cui W. W., Zhou W., Kuwada J. Y. (2007) Development 134, 2771–2781 [DOI] [PubMed] [Google Scholar]
- 64. Bedet C., Bruusgaard J. C., Vergo S., Groth-Pedersen L., Eimer S., Triller A., Vannier C. (2006) J. Biol. Chem. 281, 30046–30056 [DOI] [PubMed] [Google Scholar]
- 65. Calamai M., Specht C. G., Heller J., Alcor D., Machado P., Vannier C., Triller A. (2009) J. Neurosci. 29, 7639–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tretter V., Jacob T. C., Mukherjee J., Fritschy J. M., Pangalos M. N., Moss S. J. (2008) J. Neurosci. 28, 1356–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Triller A., Rostaing P., Korn H., Legendre P. (1997) Neuroscience 80, 133–145 [DOI] [PubMed] [Google Scholar]
- 68. Llamas A., Mendel R. R., Schwarz G. (2004) J. Biol. Chem 279, 55241–55246 [DOI] [PubMed] [Google Scholar]
- 69. Llamas A., Otte T., Multhaup G., Mendel R. R., Schwarz G. (2006) J. Biol. Chem. 281, 18343–18350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.