Abstract
We applied fluorescence lifetime imaging microscopy to map the microenvironment of the myosin essential light chain (ELC) in permeabilized skeletal muscle fibers. Four ELC mutants containing a single cysteine residue at different positions in the C-terminal half of the protein (ELC-127, ELC-142, ELC-160, and ELC-180) were generated by site-directed mutagenesis, labeled with 7-diethylamino-3-((((2-iodoacetamido)ethyl)amino)carbonyl)coumarin, and introduced into permeabilized rabbit psoas fibers. Binding to the myosin heavy chain was associated with a large conformational change in the ELC. When the fibers were moved from relaxation to rigor, the fluorescence lifetime increased for all label positions. However, when 1% stretch was applied to the rigor fibers, the lifetime decreased for ELC-127 and ELC-180 but did not change for ELC-142 and ELC-160. The differential change of fluorescence lifetime demonstrates the shift in position of the C-terminal domain of ELC with respect to the heavy chain and reveals specific locations in the lever arm region sensitive to the mechanical strain propagating from the actin-binding site to the lever arm.
Keywords: Fluorescence, Microscopic Imaging, Molecular Motors, Myosin, Protein Conformation, Protein-Protein Interactions, Skeletal Muscle, Fluorescence Lifetime Imaging Microscopy
Introduction
The function of striated muscles is brought about by the relative sliding of actin and myosin filaments. Muscle myosin is a hexameric protein, assembled from two heavy chains, each bearing a regulatory and an essential light chain (RLC3 and ELC, respectively). The heavy chains consist of a long tail region responsible for the formation of a filament structure and a globular head containing the catalytic domain and the lever arm, which binds the light chains. The interaction of the globular head with actin filament facilitates the release of ATP hydrolysis products from myosin. The accompanying structural changes result in rotation of the lever arm (1–3).
The molecular details of the change in position of the lever arm with respect to the catalytic domain were revealed by crystallographic studies of myosin head in the presence of nucleotide analogues in the ATPase pocket (2). Although there is still no crystal structure of actomyosin available, x-ray diffraction (4) and fluorescence polarization studies of both RLC (5–7) and ELC (8) confirmed rotation of the lever arm when comparing active, relaxed, and rigor muscle fibers. However, the angular changes were substantially different from those expected by crystallography. This discrepancy could be due to the crystal packing or the use of a truncated form of myosin head, which lacked most of the lever arm. In addition, the fluorescence polarization studies were focused on the distal part of the lever arm, which is remote from the catalytic domain. Therefore, the proximal part of the lever arm, which forms an interface with the catalytic domain, is of particular interest. The fluorescence polarization study of this region (9) showed its reorientation when muscle fibers were moved from relaxation to rigor. However, only a single labeling site was used, and the details of this reorientation are still not clear.
The aim of this work is to investigate the response of cross-bridges to the chemo-mechanical transitions in muscle fibers by probing the microenvironment of the interface region of the myosin lever arm domain. Fluorescence lifetime imaging microscopy (FLIM) is a method specifically measuring changes in local environment of a probe while also providing spatial information. Although local environmental changes can be detected by other methods, particularly fluorescence polarization, their interpretation is often complicated by other contributing factors, such as changes in orientation and protein mobility. For this purpose, we utilized the fluorescence lifetime of coumarin covalently bound to the interface. Coumarin is known to be particularly sensitive to the microenvironmental changes, and it was previously used as a phosphate and ADP probe in muscle fibers (10, 11). Dependence of the fluorescence lifetime of coumarin-labeled ATP analogue on the location in the sarcomere was shown (12).
The interface region of the lever arm is occupied by the ELC, a calmodulin family protein consisting of two halves, each comprised of two helix-loop-helix motifs (EF-hands). The C-terminal half adopts a semi-open conformation, with the pairs of EF-hands relatively separated from each other when bound to the heavy chain (13). This half is nearest to the catalytic domain. To investigate it, we prepared a number of mutants of recombinant ELC, each containing a single cysteine residue in a different position. The mutation sites were placed in the loops connecting helices of the C-terminal domain so that the conformational changes caused by the interhelical movements could be detected. The cysteines were covalently bonded with coumarin, and the labeled ELC mutants were exchanged with the native ELC in muscle fibers. We investigated the fluorescence lifetime properties of these fibers in relaxed and rigor conditions and following a short stretch of rigor fibers. The results showed a response of the C terminus of the ELC to the stretch, with the amplitude of response depending on location of the probe.
EXPERIMENTAL PROCEDURES
Reagents and Solutions
7-Diethylamino-3-((((2-iodoacetamido)ethyl)amino)carbonyl)coumarin (IDCC) and tetramethylrhodamine-5-iodoacetamide dihydroiodide (5IATR) dyes were obtained from Invitrogen. All other chemicals and reagents, unless otherwise stated, were from Sigma-Aldrich or VWR.
The relaxing solution contained 7 mm ATP, 5 mm EGTA, 8 mm magnesium acetate, 6 mm imidazole, 70 mm potassium propionate, pH 7.1. The 2,3-butanedione 2-monoxime rigor solution contained 60 mm TES, 30 mm EGTA, 4.5 mm MgCl2, 20 mm glutathione, 10 mm 2,3-butanedione 2-monoxime, 20.64 mm potassium propionate, pH 7.1. The rigor solution contained 60 mm TES, 0.25 mm EGTA, 29.75 mm Ca-EGTA, 2.02 mm MgCl2, 20 mm glutathione, 23.28 mm potassium propionate, pH 7.1. The activating solution contained 60 mm TES, 0.21 mm EGTA, 29.79 mm Ca-EGTA, 5 mm ATP, 6.63 mm MgCl2, 20 mm glutathione, 1.73 mm potassium propionate, pH 7.1.
The solution used for the exchange of labeled ELC with endogenous light chains of myosin in muscle fibers was a minor modification of that described in Refs. 9 and 14 and contained 2 mg/ml ELC in 10 mm imidazole, 5 mm ATP, 10 mm KH2PO4, 5 mm EDTA, 5 mm EGTA, 150 mm potassium propionate, 5 mm DTT, 0.5 mm trifluoperazine, pH 6.5.
Proteins
The previously described (9) DNA construct of human ventricular ELC (ELC-180) was used to generate several mutants containing single cysteine residues at different positions in the C-terminal half of the protein (ELC-127, ELC-142, and ELC-160) with QuikChange site-directed mutagenesis kit (Stratagene). First, a cysteine-free sequence was produced in which the Cys-180 was mutated to alanine. Using this sequence, three constructs were obtained by mutating Thr-127, Gly-142, and Glu-160 to cysteines. The ELC was then overexpressed in M15(pREP4) Escherichia coli cells and purified using nickel-nitrilotriacetic acid-agarose affinity chromatography (Qiagen) (9). Troponin C was a gift from Dr. Yin-Biao Sun (King's College, London).
Protein Labeling
Each mutant ELC was labeled with IDCC. The protein was mixed with the fluorophore at 1:3 molar ratio in solution containing 10 mm potassium phosphate, 100 mm KCl, pH 7.1, and incubated at room temperature for 3 h. The reaction was terminated by adding 1 mm 2-mercaptoethanesulfonate. The protein and the unreacted dye were separated on a 5-ml HiTrap desalting column (GE Healthcare) equilibrated in the exchange solution without trifluoperazine and stored at −80 °C freeze-dried or in solution. The labeling efficiency was determined by spectrophotometry using the extinction coefficient of coumarin E430 = 50 000 m−1 and calculating the protein concentration as CELC = A280 − A430/R, where R = 4.13 is the experimentally determined ratio of coumarin absorption at 430 and 280 nm. Typically, 60–80% of the ELC was labeled. The reactive cysteine SH1 of myosin in muscle fibers was labeled by incubating the fibers in relaxing solution with 100 μm 5IATR at 15 °C for 20 min. Fluorescence spectra were acquired using a FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon Inc.).
ELC Exchange
Single permeabilized fibers, prepared as described previously from psoas muscle of adult rabbits (15, 16), were suspended via aluminum T-clips at the fiber ends on two hooks in a trough, with one of the fiber ends attached to a force transducer (AE801 Sensor One Technologies) and the other end attached to a micrometer drive (Mitutoyo). The sarcomere size was adjusted to 2.4 μm using a diffraction pattern generated by a 532 nm He-Ne laser shone through the fiber. The exchange solution containing 2 mg/ml labeled ELC was injected into the trough, and the temperature was raised to 37 °C for 30 min. After the incubation, the temperature was lowered to 15 °C, and the fiber was extensively washed with relaxing solution and further incubated for 30 min at 15 °C in relaxing solution containing 1 mg/ml troponin C. The fibers used in FLIM experiments were dissolved in SDS-PAGE sample buffer, and their protein composition was analyzed on 8% gels to estimate the level of the ELC exchange. The gels were scanned on an Ettan DIGE imager 1.0 (GE Healthcare) using the Cy2 filter set for coumarin fluorescence, stained with Coomassie Blue, and scanned again using the Cy5 filter set to estimate the staining signal.
Isometric Force Measurements
The temperature jump method was used to measure the isometric force. The fibers were suspended via T-clips at 2.4-μm sarcomere length on a home-built rotating stage in a trough containing relaxing solution at 0 °C, with one fiber end attached to the force transducer. The fibers were moved to activating solution at 0 °C, where no force was developed during 2-s observation period. The temperature was raised by a jump to 20 °C to initiate force development. After 3 s, the fibers were moved back to relaxing solution.
Fluorescence Microscopy
Following the exchange with coumarin-labeled ELC, the setup was placed under the upright Leica TCS SP5 confocal microscope (Fig. 1) equipped with an SPC-830 time-correlated single photon counting module (Becker & Hickl GmbH). Fluorescence lifetime images were acquired at 21 °C in a time-domain mode using two-photon excitation at 850 nm by a picosecond Mai Tai laser (Spectra-Physics Inc.) pulsing at 80 MHz rate. A Leica HCS APO L63/0.9 U-V-I lens was used to acquire 256 × 256-pixel images at the zoom 4 setting with 64 time bins during 3-min periods. This provided several thousand photon counts per pixel with at least 100 photons in the peak of the fluorescence decays in the A-band regions. No photobleaching was observed during the acquisition period. In the case of stretched fibers, the acquisition was initiated 10–20 s after the application of stretch. Confocal images were acquired on the same microscope as two-photon, except that the image resolution was increased to 512 × 512 pixels. Fluorescence lifetime measurements in solution were done either using the same microscope or in a cuvette system as described previously (17) in relaxing solution at 21 °C. No effect on the lifetime was detected when relaxing or rigor solutions were used.
FIGURE 1.
Experimental setup. The isolated permeabilized muscle fibers were suspended on hooks in a trough chamber (bottom left) and incubated at 37 °C in exchange solution to introduce fluorescent ELC. The chamber was moved under an upright Leica SP5 microscope equipped with a 63×/0.9 water immersion lens. The fluorescence was excited by a pulsed Mai Tai laser at 850 nm, and the fluorescence lifetime images were recorded using a time-correlated single photon counting module. A typical fluorescence lifetime image is shown (top right). The force developed after fiber stretch was measured using a force transducer (bottom right).
Data Analysis
Fluorescence decay in each pixel of an image was fitted with a single exponential function using the SPCImage software (Becker & Hickl GmbH) to obtain a fluorescence lifetime image. ImageJ software (National Institutes of Health) was used to extract the fitted lifetime values from the A-band regions. First, a threshold was applied to an intensity image, and a selection mask was created that included all A-bands in the image. The mask was applied to a fluorescence lifetime image, and a profile of lifetime values distribution in the mask with 5-ps bins was generated. The lifetime distributions were fitted with a Gaussian function in SigmaPlot (SPSS Inc.) to obtain the mean lifetime and the half-maximal width of the distribution. Several images were acquired in each condition (relaxation, rigor, and stretch in rigor) applied to the fibers. The data in the results section represent the measurements from 5–6 fibers with standard errors for each mutant ELC. A one-tailed paired Student's t test analysis was used to assess the significance of the lifetime differences.
RESULTS
Exchange of the ELC in Permeabilized Muscle Fibers
To investigate the interface region of the lever arm, we developed a protocol to substitute the native ELC with a fluorescent recombinant ELC in isolated skeletal muscle fibers. Human LC1 isoform of the ELC was used, which is identical to the rabbit LC1, except for two extra amino acid residues in the N-terminal half. The LC1 isoform is prevalent in rabbit psoas muscle; thereby the replacement of the endogenous ELC with the recombinant protein did not drastically alter the fiber isoform composition. To estimate the efficiency of ELC exchange, the fiber protein composition was analyzed using 8% SDS-PAGE. First, from an isolated labeled ELC used for exchange, a ratio of fluorescence and Coomassie Blue staining signals was found (supplemental Fig. 1A). Then from the band corresponding to the ELC in the treated muscle fiber, the proportion of the exchanged ELC was found as the difference between the isolated ELC and the treated fiber staining ratios. Based on these calculations, 66 ± 7% (n = 3) of the native ELC was exchanged with recombinant protein. Because the recombinant ELC was fluorescently labeled to 80%, the degree of ELC labeling in fibers was about 50%. This level of labeling produced a sufficiently high number of photons for FLIM, and as the fluorescence lifetime value is independent of fluorophore concentration, the partial labeling did not affect the measurements. The functional integrity of the fibers following exchange was confirmed by their unimpaired ability to undergo transitions from relaxed to rigor states and by development of 144 ± 51 kilonewtons/m2 (n = 21) force caused by a 1% stretch when in rigor. The fibers were also able to develop and maintain the isometric force in the presence of calcium and ATP, although it was reduced in comparison with untreated fibers (Fig. 2). The fibers carrying ELC-142, ELC-160, and ELC-180 produced 65 ± 13, 55 ± 12, and 71 ± 3% isometric force, whereas the force development after ELC-127 exchange was reduced to 41 ± 5%. Importantly, all fibers were able to relax to the initial level. The ability of fibers to undergo transition from relaxation to rigor and from rigor to relaxation was monitored immediately before and/or after the fluorescence lifetime data acquisition. All fibers used in FLIM experiments were able to switch between relax and rigor, as well as to develop force after activation when these measurements were carried out.
FIGURE 2.
Effects of ELC exchange and cysteine mutations on isometric force development. The force was measured using a temperature jump protocol. The fibers suspended in a trough of a rotating stage in relaxing solution were moved to activating solution at 0 °C. The temperature was raised after 2 s by a jump to 20 °C to initiate force development. After 3 s, the fibers were moved back to relaxing solution. Shown are average force records of fibers normalized to untreated fibers. n = 4 in each case. Following ELC-180 exchange, about 71 ± 3% force was developed. The effect of cysteine mutation was greatest in the case of ELC-127 (41 ± 5% force).
Although the ELC binds specifically to the heavy chain in solution, nonspecific binding could occur during exchange in muscle fibers. To check the specificity of exchange, we conducted a fluorescence microscopy study. Confinement of the labeled ELC to the A-band is confirmed by colocalization imaging. In this experiment, the reactive myosin cysteine SH1 was labeled with 5IATR in a muscle fiber following coumarin-ELC exchange. The confocal microscopy analysis showed a complete overlap of the rhodamine and coumarin fluorescence signals (supplemental Fig. 1B), thus confirming that the ELC binds only in the region containing myosin heads. Moreover, when the fibers were stretched to a sarcomere length of 3 μm or more in relaxation condition, all label remained in the A-band. The point mutations introduced in the ELC did not affect the exchange specificity.
Fluorescence Lifetime Dependence on Probe Location
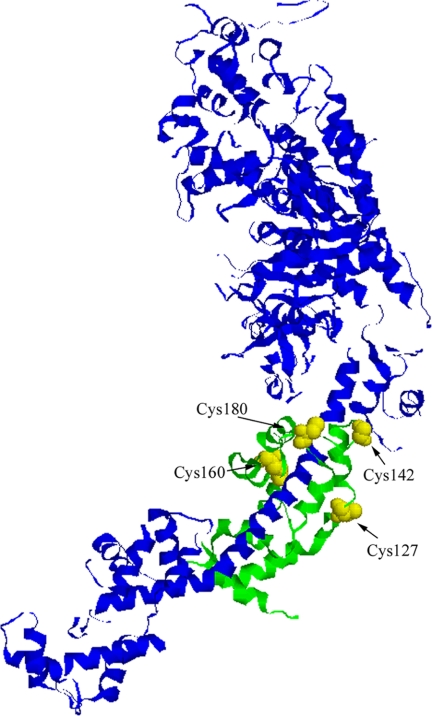
To obtain a detailed view of the ELC conformation, we prepared by site-directed mutagenesis four mutants of the ELC, each bearing a single cysteine residue: ELC-127, ELC-142, ELC-160, and ELC-180, respectively (Fig. 3). Three of the locations were in the loops connecting helices E, F, G, and H in the C-terminal half of the protein, with the fourth (ELC-127) in the linker between the N- and C-terminal halves. The cysteines were labeled with a thiol-reactive coumarin fluorophore IDCC. The sensitivity of coumarin to location on the protein can be immediately seen from the emission spectra of free coumarin dye and labeled ELC mutants (supplemental Fig. 2). The fluorescence emission peak shifted from 476 nm for the free IDCC to 466 nm for ELC-142, 464 nm for ELC-127 and ELC-160, and 463 nm for ELC-180.
FIGURE 3.
Location of mutated residues in the structure of myosin subfragment-1 (after PDB entry 2mys (27)). The ELC and the mutated residues are highlighted in green and yellow, respectively. The Cys-180 of the wild-type ELC was removed, and three ELC constructs were generated by introducing Cys-127, Cys-142, and Cys-160. The heavy chain and the RLC are shown in blue.
Fluorescence lifetime is a suitable parameter to characterize the microenvironment of a fluorophore (18, 19). It is believed to be modified by interaction with the solvent and by the flexibility and conformation of the fluorophore-bearing region. To assess the differences between coumarin-labeled ELC mutants in greater detail, their fluorescence lifetimes were measured in solution using fluorescence lifetime imaging microscopy. The fluorescence decays in each pixel of an image were fitted with a single exponential function, and a Gaussian distribution of fluorescence lifetime was found in the A-bands (Fig. 4). Analysis of the mean fluorescence lifetimes showed that ELC-142 had a shorter lifetime at 1674 ± 1 ps (n = 3), consistent with location of the label in a highly exposed loop between helices E and F (Fig. 3). ELC-127 had a significantly longer fluorescence lifetime at 1831 ± 1 ps (n = 3). The N and C-terminal halves of the ELC are likely to interact in solution, forming a compact globule and thus protecting the linker region, where the Cys-127 is located, from the solvent, which can be related to the longer fluorescence lifetime of the probe attached to this residue. ELC-160, located in a relatively exposed loop between helices F and G, displayed a fluorescence lifetime in between the values of ELC-127 and ELC-142, namely 1777 ± 4 ps (n = 3). The ELC-180, located in a loop between helices G and H, which is likely to be shielded from the solvent, had the longest lifetime at 1936 ± 9 ps (n = 3). Thus, it can be concluded that the fluorescence lifetime of coumarin-labeled ELC is strongly affected by the location of the probe.
FIGURE 4.
Fluorescence lifetime of coumarin-labeled ELC in solution. 20 μm of each labeled ELC mutant was placed on a microscope slide in relaxing solution, and the fluorescence lifetime was acquired with 64 time bins in a 256 × 256-pixel image. A minimum of 1000 photons/pixel was collected. The solid lines represent Gaussian fits of the lifetime distributions. n = 3 for each ELC label position.
Effect of ELC Binding to the Myosin Heavy Chain
To study ELC conformation in situ, we applied FLIM to map the microenvironment of the ELC mutants, initially in relaxed muscle fibers. An example fit to a single pixel decay is shown in supplemental Fig. 3. The mean fluorescence lifetimes obtained from the fluorescence lifetime histograms for ELC-127, ELC-142, ELC-160, and ELC-180 (Fig. 5A) were 1643 ± 21 (n = 17), 1442 ± 46 (n = 12), 1728 ± 52 (n = 16), and 1836 ± 19 ps (n = 16), respectively.
FIGURE 5.
The effect of ELC binding to the myosin heavy chain in muscle fibers. A, fluorescence lifetime distributions obtained from FLIM images in the A-bands of relaxed fibers. The fluorescence lifetime strongly depends on the location of the probe. The solid lines represent Gaussian fits of the lifetime distributions. B, difference between mean fluorescence lifetimes of ELC mutants measured in solution and in relaxed fibers. Note that although all ELC labels have a fluorescence lifetime decrease, ELC-127 and ELC-142 show a much greater change, suggesting a complex rearrangement after ELC binding to myosin heavy chain. n = 17, 12, 16, and 16 for each ELC label position, p < 0.05 in each case.
Comparing the mean lifetimes of the ELC mutants in muscle fibers with the mean lifetimes obtained in solution (Fig. 5B), a change in the microenvironments of the fluorophores is inferred. In both solution and fibers, the ELC-142 exhibited the shortest fluorescence lifetime. In the case of all mutants, there is a 50–230-ps decrease in fluorescence lifetime in fibers when compared with values in solution, which can be attributed to a conformational change in the C-terminal half of the ELC upon binding to the heavy chain. ELC-127, where coumarin is attached in the linker connecting the N- and C-terminal halves of the ELC, and ELC-142 show a strikingly greater change in lifetime (Fig. 5B). These residues located at the opposite ends of the helix E could be more affected as a result of the extended binding of ELC along the heavy chain.
Response of ELC to the Length Change in Rigor
When the muscle fibers were moved from relaxing solution to the rigor state, the fluorescence lifetime increased by 78 ± 16 (n = 13), 91 ± 28 (n = 14), 84 ± 22 (n = 14), and 58 ± 13 ps (n = 16) for ELC-127, ELC-142, ELC-160, and ELC-180, respectively (Fig. 6A). This uniform increase suggests a gross conformational shift in the myosin cross-bridge following its attachment to the actin filament and could result both from the change in the microenvironment and from the lower mobility of the probe-bearing region, which was previously shown for rhodamine-labeled ELC (8, 9).
FIGURE 6.
Fluorescence lifetime change following transition from relaxation to rigor. A, difference between the mean fluorescence lifetimes of ELC in relaxation and rigor. All ELC label positions have a longer fluorescence lifetime in rigor. n = 13, 14, 14 and 16, p < 0.05 in each case. B, the effect of partial thick and thin filament overlap on the fluorescence lifetime profile of A-bands in relaxed and rigor fibers. The relaxed fiber was stretched to 2.6 μm and then moved to rigor. Shown are the half-sarcomere fluorescence intensity (black) and fluorescence lifetime profiles in relaxed (green) and rigor (red) fibers. Two populations of fluorescence lifetime are observed in rigor at the center and periphery of the A-band, which can be attributed to the presence of unattached and attached cross-bridges, respectively. arb., arbitrary units.
Because the transition from relaxation to rigor was done at a full overlap of thick and thin filaments (2.4-μm sarcomere length), a more complex distribution of cross-bridges can be expected if the fiber is stretched to partial overlap before the application of strain. Accordingly, relaxed fibers bearing ELC-180 were stretched to 2.6 μm and then moved to rigor. The fluorescence lifetime profile of sarcomeres was measured. The half-sarcomere profiles (Fig. 6B) show that although in relaxed fibers the profile is uniform through the A-band, in rigor two populations can be distinguished. The observation of two populations can be explained by the presence of unattached cross-bridges at the center of the A-band, in the non-overlap H-zone.
To test the relation between the actin-binding site and the lever arm of cross-bridges, rigor fibers were stretched by 1% of the fiber length. This length change is comparable with the movement of each myosin head during contraction achieved by tilting of the lever arm while remaining attached to actin. As shown in Fig. 7A, the length changes were followed by a left-hand shift in the fluorescence lifetime histogram of the ELC-180. When a series of stretches and releases was applied to fibers with ELC-180, the fluorescence lifetime followed the mechanical movements (Fig. 7B). It is notable, however, that the amplitude of stretch response decreased after several stretch-release cycles. Because the fluorescence lifetime value in the released state does not change significantly after each cycle, these results suggest that stretch releases return the fiber to the state prior to stretch and that the process is reversible (Fig. 7B). However, the rigor fibers tend to pull out of the T-clips, so the amplitude of the effective stretch gradually decreases after each cycle, resulting in loss of fluorescence lifetime difference.
FIGURE 7.
Fluorescence lifetime response to stretch. Muscle fibers bearing fluorescent ELC were stretched by 1% length in rigor. A, fluorescence lifetime distribution of ELC-180 in relaxation, in rigor, and after short stretch in rigor. The distribution shifts to longer fluorescence lifetime after transition from relaxation to rigor, but the shift is partially reversed when a short stretch is applied to rigor fibers. The solid lines represent Gaussian fits of the lifetime distributions. B, response of fluorescence lifetime of ELC-180 to a series of stretches and releases. The fiber is initially in the rigor state. The fiber was stretched by 1% and then returned to initial state by 1% stretch release. The cycle was repeated four times. Fluorescence lifetime images were acquired after each step in each of the two states, stretched, and released. The bars represent the mean fluorescence lifetime from all pixels in the A-bands in an image. C, the effect of a short stretch of rigor fibers on the mean fluorescence lifetime of ELC mutants. A significant decrease in the lifetime is detected for ELC-127 and ELC-180 (n = 18 and 19, respectively, p < 0.05), whereas ELC-142 and ELC-160 showed no stretch effect (n = 12 and 19, respectively, p > 0.05). D, half-maximal widths of fluorescence lifetime distributions. The parameter remained unchanged between ELC mutants during relaxation, rigor, and stretch, suggesting single population behavior under all experimental conditions for all ELC mutants.
The detailed analysis of the ELC mutants (Fig. 7C) showed that the short stretch applied to rigor fibers induced a decrease in fluorescence lifetime by about 32 ± 13 ps (n = 18) for ELC-127 and 37 ± 11 ps (n = 19) for ELC-180. No significant change was observed for ELC-142 (4 ± 2 ps, n = 12) and ELC-160 (17 ± 8 ps, n = 19). These data indicate a partial reversal of the lifetime in comparison with the change induced by the relaxation to rigor transition. Moreover, a lack of change in cases of ELC-142 and ELC-160 lifetimes suggests a variable response in different parts of the protein.
Another parameter that could be affected by the perturbations of muscle fibers is the width of the lifetime distribution. However, as evident from Fig. 7D, there is no significant change in the width between ELC mutants in relaxation, rigor, and after stretch, suggesting that the lifetime changes in response to chemical or mechanical perturbation affect the whole light chain population.
Although fluorescence lifetime profiles shift uniformly between different experimental conditions, a mean fluorescence lifetime is likely to represent a set of subpopulations of myosin conformations and the determined mean lifetime reports about a dominant state. Within each image pixel, it is likely that the fluorophores will exist in a range of different populations, where each population has a slightly different local environment. This will result in a complex multi-exponential fluorescence decay. However, the relatively low number of photons acquired in each image pixel means that it is not feasible to carry out multi-exponential decay analysis on the data presented here. However, in the future, a more sophisticated global-fitting FLIM analysis could allow the distribution and relative concentration of multiple fluorophore populations to be studied in more detail.
DISCUSSION
The ELC binds to a specific IQ sequence of the myosin heavy chain, as demonstrated by crystallographic studies of myosin light chain-binding region (13). The ability of the ELC to dissociate and rebind to the myosin heavy chain in vitro is well known (20). We used this property to exchange the native ELC with a recombinant fluorescently labeled protein. Characterization of the protocol by SDS-PAGE and fluorescence microscopy confirmed the high efficiency and specificity of the exchange, whereas the functionality of muscle fibers was retained. This is consistent with the previously reported work, where the ability of muscle fibers to activate following ELC exchange was found to be largely preserved (8, 9, 14). The development of the exchange protocol allows a thorough investigation of the proximal end of the lever arm, where the ELC is located.
A useful feature of the striated muscle ELC is that its sequence contains only one cysteine residue near the C terminus. This residue has been exploited previously to attach fluorescent or spin probes (9, 21–24). Observing a reduced fluorescence of fluorescein attached to the cysteine in chicken myosin subfragment-1 (S1) in the presence of ATP, Marsh et al. (21) first proposed a change in the microenvironment of this residue. Further fluorescence resonance energy transfer studies produced contradictory results for the distance between the ELC and the catalytic domain (22, 24). When energy transfer was measured in myosin S1, no significant change in distance was detected (24). However, a large change in distance between ELC and the catalytic domain, as well as between ELC and RLC, was reported when reconstituted myosin filaments were used (22). This contradiction could arise from differences of the in vitro systems used. Fluorescence polarization investigation of both S1 and muscle fibers showed a nucleotide- and actin-dependent change in the orientation of the ELC-180 (9, 23). The electron paramagnetic resonance measurements detected a change in the mobility of a spin probe attached in reconstituted myosin filaments (25). However, despite the accumulated information, the interpretation of these findings is limited by the use of fragmentary protein systems and a single labeling site in the ELC.
Using a site-directed mutagenesis approach, we designed a number of ELC mutants, each bearing a single cysteine residue (Fig. 3). The labeling sites were placed in the linker between N- and C-terminal domains (ELC-127) and in the loops connecting helices E and F (ELC-142), F and G (ELC-160), and G and H (ELC-180). The point mutations are separated by 15–20 amino acid residues, thus evenly covering the entire 60-residue long C-terminal half of the ELC. Characterization of the labeled mutants in solution showed that the location of the probe strongly affected their fluorescent properties, with the value of mean fluorescence lifetime varying by up to 260 ps. It was previously shown by electron paramagnetic resonance spectroscopy that in solution the ELC adopts a tight configuration formed by the hydrophobic cores in its N- and C-terminal lobes (26). Crystallographic studies demonstrated that the C-terminal domain of ELC is characterized by a particular semi-open conformation when bound to the heavy chain (13, 27), and it was proposed that this domain switches to a closed conformation when separated from the heavy chain (28). We applied fluorescence lifetime imaging microscopy to map the microenvironment of coumarin attached to ELC mutants introduced in permeabilized skeletal muscle fibers. All mutants with the probe located in the C-terminal domain displayed a decrease in fluorescence lifetime upon binding to the heavy chain. This finding points to a conformational switch in the C-terminal domain associated with the adaptation of the semi-open conformation.
Although the lever arm is assumed to be largely rigid, comparing crystal structures (PDB entries 2MYS, 1BR1, and 1BR4) of myosin head in pre- and post-power stroke states (1, 2) shows a potential interaction between the C-terminal domain of ELC and the N-terminal part of the heavy chain (9, 29). To test this, we investigated the effect of the lever arm rotation on the fluorescence lifetime properties of ELC mutants in relaxed and rigor fibers. The fluorescence lifetime of all ELC mutants in rigor fibers increased by 60–90 ps when compared with relaxation, suggesting a conformational shift transmitted from the ATPase and actin-binding site. This shift implies some degree of intramolecular flexibility of the ELC and can be related to a change in its position with respect to the heavy chain, which would produce a different microenvironment of the probe (Fig. 8). The fluorescence lifetime data presented here are in parallel with the fluorescence polarization study of ELC labeled at the N-terminal half with bifunctional rhodamine probes, which showed a broad polarization distribution in relaxation and narrow distribution in rigor, suggesting a greater order of ELC caused by the binding of myosin to actin (8). On the contrary, fluorescence polarization and rotational correlation time measurements of ELC-180 showed greater degree of order in relaxation (9). It was suggested that the ELC-180 becomes immobilized in relaxation due to its interaction with the heavy chain of myosin. Therefore, it is necessary to distinguish between global and local disorder, which can be affected by specific interactions.
FIGURE 8.
Diagram of the proposed mechanism of the fluorescence lifetime changes in ELC. The binding to actin (not shown) changes the position of the C-terminal domain of ELC (green) with respect to the N-terminal subdomain (red) of the heavy chain. Tilting of the lever arm by stretch partially reverses the effect of actin binding, with position relative to the converter domain (blue) unchanged. The locations of the probe in ELC-127, ELC-142, ELC-160, and ELC-180 are shown as red circles, and the α-helices of the ELC C-terminal domain are also shown, with the helix F highlighted in dark blue. This representation suggests why ELC-142 and ELC-160 are insensitive to stretch, whereas ELC-180 is sensitive to stretch. Stretch sensitivity of ELC-127 may reflect a change in the interdomain conformation of ELC.
We further tested the relation between the catalytic domain and the ELC by stretching rigor fibers by a distance comparable with the myosin step size. In these conditions, the tilting of the lever arm is expected, whereas myosin heads remain attached to actin filaments. A significant (30–40-ps) decrease of fluorescence lifetime was detected for the ELC-127 and ELC-180. However, in comparison with the lifetime increase caused by the transition from relaxation to rigor, the change is only 41 and 68% for the two mutants, respectively. Moreover, no change was detected for the ELC-142 and ELC-160. On the other hand, the difference between rigor and relaxation was greatest for these mutants. This rules out the strain-induced local conformational change for ELC-142 and ELC-160 defined as a rearrangement of an area in the vicinity of a probe leading to a change in local environment. Nevertheless, it does not rule out a change in orientation of that area caused by rotation of the lever arm because the environment of the probe may not be affected in this case. The probes in these cases are located at the opposite ends of the helix F (Fig. 8). At the same time, the ELC-142 probe is nearest to the converter domain. The converter is thought to rotate following conformational change in the relay helix, which leads to the tilting of the lever arm (1). The insensitivity of ELC-142 and ELC-160 to the tilting after stretch indicates that their position with respect to the converter domain does not change, suggesting that this region of the ELC rotates in unison with the converter (Fig. 8). On the other hand, the ELC-180 is the most sensitive of the four mutants. As described above, the labeling site in this case is located in the interface area between the N-terminal subdomain of the heavy chain and this region of the ELC. A change in the interface could explain greater lifetime sensitivity as even a minor movement would lead to a change in the microenvironment, consistent with the previous findings (9, 21).
Fluorescence polarization studies previously showed a 5° change in the orientation of rhodamine attached to Cys-180 of the ELC between relaxation and rigor (9, 23). However, linking to the protein via a single covalent bond results in a wobble of the fluorophore around the bond and makes the orientation of the dipole uncertain. Therefore, it was difficult to relate these data directly to the orientation of the lever arm in the available myosin crystal structures. A recent study using bifunctional rhodamine attached around the ELC N-terminal domain also detected a 4–15° rotation depending on the location (8). These data were related to the myosin head structure, but the interpretation is complicated by two orientations observed in relaxation. An alternative approach of fluorescence lifetime microscopy applied in the present work reports a conformational shift in the C-terminal domain of the ELC. The differential reduction of lifetime following a short stretch demonstrates the sensitive points in the ELC structure, which might be associated with the pathway of the mechanical strain propagating from the actin-binding site to the lever arm. Further investigation using complementary techniques, such as fluorescence polarization and Förster resonance energy transfer, will provide additional structural information.
The behavior of the ELC C-terminal domain is likely to be substantially different in an active muscle. The effect of fiber activation on the fluorescence lifetime could not be investigated in the present work due to the long acquisition time necessary to collect a sufficient number of photons in a lifetime image. Future development of methodology for the fast lifetime acquisition in muscle fibers and fluorescence polarization studies of the C-terminal domain of ELC using bifunctional fluorophores, in particular labeling the helix F, can provide further insights in the molecular mechanism of muscle contraction.
Supplementary Material
Acknowledgments
We thank Dr. Yin-Biao Sun (King's College, London) who kindly provided troponin C, Prof. Nancy Curtin (Imperial College London) for the review of the manuscript, and the Facility for Imaging by Light Microscopy at Imperial College London.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BB/E02/573/1), by a Royal Society Wolfson Research Merit Award (to P. M. F.) and by the UK Technology Strategy Board (Technology Award CHBT/007/00030).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- RLC
- regulatory light chain
- ELC
- essential light chain
- FLIM
- fluorescence lifetime imaging microscopy
- IDCC
- 7-diethylamino-3-((((2-iodoacetamido)ethyl)amino)carbonyl)coumarin
- 5IATR
- tetramethylrhodamine-5-iodoacetamide dihydroiodide
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid.
REFERENCES
- 1. Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. (1993) Science 261, 58–65 [DOI] [PubMed] [Google Scholar]
- 2. Dominguez R., Freyzon Y., Trybus K. M., Cohen C. (1998) Cell 94, 559–571 [DOI] [PubMed] [Google Scholar]
- 3. Geeves M. A., Holmes K. C. (2005) Adv. Protein Chem. 71, 161–193 [DOI] [PubMed] [Google Scholar]
- 4. Dobbie I., Linari M., Piazzesi G., Reconditi M., Koubassova N., Ferenczi M. A., Lombardi V., Irving M. (1998) Nature 396, 383–387 [DOI] [PubMed] [Google Scholar]
- 5. Irving M., St Claire Allen T., Sabido-David C., Craik J. S., Brandmeier B., Kendrick-Jones J., Corrie J. E., Trentham D. R., Goldman Y. E. (1995) Nature 375, 688–691 [DOI] [PubMed] [Google Scholar]
- 6. Corrie J. E., Brandmeier B. D., Ferguson R. E., Trentham D. R., Kendrick-Jones J., Hopkins S. C., van der Heide U. A., Goldman Y. E., Sabido-David C., Dale R. E., Criddle S., Irving M. (1999) Nature 400, 425–430 [DOI] [PubMed] [Google Scholar]
- 7. Hopkins S. C., Sabido-David C., van der Heide U. A., Ferguson R. E., Brandmeier B. D., Dale R. E., Kendrick-Jones J., Corrie J. E., Trentham D. R., Irving M., Goldman Y. E. (2002) J. Mol. Biol. 318, 1275–1291 [DOI] [PubMed] [Google Scholar]
- 8. Knowles A. C., Ferguson R. E., Brandmeier B. D., Sun Y. B., Trentham D. R., Irving M. (2008) Biophys. J. 95, 3882–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borejdo J., Ushakov D. S., Moreland R., Akopova I., Reshetnyak Y., Saraswat L. D., Kamm K., Lowey S. (2001) Biochemistry 40, 3796–3803 [DOI] [PubMed] [Google Scholar]
- 10. He Z. H., Chillingworth R. K., Brune M., Corrie J. E., Webb M. R., Ferenczi M. A. (1999) J. Physiol. 517, 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. West T. G., Hild G., Siththanandan V. B., Webb M. R., Corrie J. E., Ferenczi M. A. (2009) Biophys. J. 96, 3281–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. García D. I., Lanigan P., Webb M., West T. G., Requejo-Isidro J., Auksorius E., Dunsby C., Neil M., French P., Ferenczi M. A. (2007) Biophys. J. 93, 2091–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Houdusse A., Cohen C. (1996) Structure 4, 21–32 [DOI] [PubMed] [Google Scholar]
- 14. Sweeney H. L. (1995) Biophys. J. 68, 112S–118S [PMC free article] [PubMed] [Google Scholar]
- 15. Thirlwell H., Corrie J. E., Reid G. P., Trentham D. R., Ferenczi M. A. (1994) Biophys. J. 67, 2436–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldman Y. E., Simmons R. M. (1984) J. Physiol. 350, 497–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manning H. B., Kennedy G. T., Owen D. M., Grant D. M., Magee A. I., Neil M. A., Itoh Y., Dunsby C., French P. M. (2008) J. Biophotonics 1, 494–505 [DOI] [PubMed] [Google Scholar]
- 18. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, pp. 9–12, Springer-Verlag New York Inc., New York [Google Scholar]
- 19. Dowling K., Dayel M. J., Lever M. J., French P. M., Hares J. D., Dymoke-Bradshaw A. K. (1998) Opt. Lett. 23, 810–812 [DOI] [PubMed] [Google Scholar]
- 20. Wagner P. D., Weeds A. G. (1977) J. Mol. Biol. 109, 455–470 [DOI] [PubMed] [Google Scholar]
- 21. Marsh D. J., Stein L. A., Eisenberg E., Lowey S. (1982) Biochemistry 21, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 22. Palm T., Sale K., Brown L., Li H., Hambly B., Fajer P. G. (1999) Biochemistry 38, 13026–13034 [DOI] [PubMed] [Google Scholar]
- 23. Borejdo J., Ushakov D. S., Akopova I. (2002) Biophys. J. 82, 3150–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smyczynski C., Kasprzak A. A. (1997) Biochemistry 36, 13201–13207 [DOI] [PubMed] [Google Scholar]
- 25. Baumann B. A., Hambly B. D., Hideg K., Fajer P. G. (2001) Biochemistry 40, 7868–7873 [DOI] [PubMed] [Google Scholar]
- 26. Huang W., Wilson G. J., Brown L. J., Lam H., Hambly B. D. (1998) Eur. J. Biochem. 257, 457–465 [DOI] [PubMed] [Google Scholar]
- 27. Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. (1993) Science 261, 50–58 [DOI] [PubMed] [Google Scholar]
- 28. Nelson M. R., Chazin W. J. (1998) Protein Sci. 7, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ushakov D. S. (2008) Biofizika 53, 950–955 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.