Abstract
cAMP is an ubiquitous second messenger. Localized areas with high cAMP concentration, i.e. cAMP microdomains, provide an elegant mechanism to generate signaling specificity and transduction efficiency. However, the mechanisms underlying cAMP effector targeting into these compartments is still unclear. Here we report the identification of radixin as a scaffolding unit for both cAMP effectors, Epac and PKA. This complex localizes in a submembrane compartment where cAMP synthesis occurs. Compartment disruption by shRNA and dominant negative approaches negatively affects cAMP action. Inhibition can be rescued by expression of Rap1b, a substrate for both Epac1 and PKA, but only in its GTP-bound and phosphorylated state. We propose that radixin scaffolds both cAMP effectors in a functional cAMP-sensing compartment for efficient signal transduction, using Rap1 as a downstream signal integrator.
Keywords: Cyclic AMP (cAMP), Cytoskeleton, Protein Assembly, Protein Kinase A (PKA), Protein Phosphorylation, Protein-Protein Interactions, Thyroid, ERM, Epac, Radixin
Introduction
Contrary to the original concept of a freely diffusible second messenger homogenously distributed in the cell (1), cAMP diffusion is rather restricted (2–7). The concept of cAMP microdomains was originally proposed to explain how in a single cell hormones acting via cAMP were able to trigger distinct biological responses (8). More recently, modeling studies indicate that restricted diffusion is also necessary to attain cAMP levels matching effector activation constants (9). Thus, compartmentalization in a defined spatially restricted unit has the potential to explain signaling specificity and transduction efficiency. Although the mechanisms involved in microdomain establishment and maintenance are unknown (10), implicit in the model is the co-localization of second messenger and cAMP targets. Although a central role of A-kinase-anchoring protein (AKAP)2 proteins (11, 12) was clearly established for PKA (13), the mechanisms involved in anchoring Epac are still ill-defined (14, 15). In the present work, we report the identification of the ERM family member radixin as a scaffolding unit for Epac and PKA into a functional compartment required for cAMP-mediated cell proliferation.
EXPERIMENTAL PROCEDURES
Materials
TSH, forskolin, and H89 were obtained from Sigma. GSH-agarose was from Amersham Biosciences. Antibodies against HA (HA.11) and Myc (9E10) were from Covance. Anti-Epac antibody (H70, 1:250) was from Santa Cruz Biotechnology. Anti-GFP antibody (A11122, 1:1500) was from Invitrogen. Affinity-purified anti-radixin antibody (215R+) was a generous gift of Dr. Crouch (Scotland). Secondary antibodies (1:2000, anti-mouse Alexa 488/594, anti-rabbit Alexa 594) were purchased from Invitrogen. Bodipy-forskolin was from Invitrogen.
DNA Constructs
pCGN-HA-Rap constructs, pCMV-myc-N-Epac, pCMV myc-Epac, and pCMV myc-Δ(1–148)-Epac plasmids were already described (16). pCEFL GST-Epac was subcloned as a EcoRI/Klenow-NotI fragment. Full-length and Δ(1–148)-Epac constructs for in vitro translation were subcloned into pET28b (Novagen), as HindIII-SalI fragments. pEF1 myc/His FL-, N-, and C-radixin constructs were generated by PCR with BamHI-NotI. pGEX-2T radixin, N-radixin, C-radixin, and pCDNA HA-radixin were kindly provided by Dr. Voyno-Yasenetskaya. pEGFP-radixin was kindly provided by Dr. Ravichandran. pEF1-N-Rad-dsRed and pEF1-C-Rad-dsRed were generated upon replacement of the myc/His cassette with a dsRed express by PCR using BstBI-PmeI enzymes. (1–52)-Epac-dsRed was generated by replacing the BamHI-NotI fragment from pEF1-N-Rad-dsRed with (1–52)-Epac generated by PCR.
Radixin Knock-down
The radixin target sequence was designed with PSICOLIGOMAKER1.5 software (rat/mouse, 5′-GCACCTCGTCTGAGAATCA-3′) and subcloned into pLL3.7 vector. To generate an off-target control (shRNAR), silent mutations were introduced (5′-GCACCTCGTCTGAGAATCA-3′) into pCDNA3.1-radixin-HA with QuikChange mutagenesis kit (Stratagene). Radixin L421P (CTT to CCT), was generated with the same kit using radixin-HA shRNAR as template.
Statistical Analysis
Cluster analysis upon Radixin knock-down, were performed by one-tailed t test, using Graphpad Prism software.
Cell Lines and Transfections
PCCL3 cells were grown in Coon's modified F-12 medium (Sigma), supplemented with 5% FBS and the combination of four hormones: TSH (1 mIU/ml; IU is international unit), insulin (1 μg/ml), apo-transferrin (5 μg/ml), and hydrocortisone (1 nm). HEK-293T cells were maintained in Dulbecco's modified Eagle's medium (Cambrex) supplemented with 10% FBS. Cells were kept at 37 °C in a 5% CO2/95% humidified air environment. Transfections were performed in 6-well plates with polyethylenimine (PEI, Polysciences, Inc), with 6 μg of PEI/2 μg DNA/well.
BrdUrd Labeling
Cells were grown to 50% confluency on glass coverslips, transfected for 24 h, and made quiescent by serum starvation in Coon's/0.2% BSA for 16 h. Upon agonist stimulation (10%FBS/TSH) for 8 h, cells were labeled for 16 h with 100 μm BrdUrd (Sigma). At the end of the labeling period, cells were fixed in 4% paraformaldehyde (10 min, room temperature) and permeabilized with 0.5% Triton X-100 (20 min, room temperature). After washing, incorporated BrdUrd was detected by indirect immunofluorescence. Samples were co-stained for 1 h at room temperature with sheep anti-BrdUrd antibody (Biodesign International; diluted 1/100 in PBS/1% BSA) and the corresponding primary antibody (HA-11, 1:400; Myc, 1:400 or Flag, 1:5000) in the presence of RQ1 DNase (Promega; 10 units/ml). After extensive washes in PBS/1% BSA, samples were incubated for 1 h at room temperature with a combination of FITC-conjugated goat-antisheep (Sigma, dilution, 1/150 in PBS/2% BSA) and the appropriate secondary anti-mouse, containing 0.2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI, Sigma). After extensive washes in PBS-0.1% Tween20, samples were mounted in PermaFluor (Thermo) and viewed by epifluorescence (60×).
Yeast Two-hybrid
A bait construct expressing hEpac1 residues 1–200 fused to the C terminus of Gal4 DBD (residues 1–147) was transformed into yeast strain PNY200 (MATα trp1-901 leu2-3,112 ura3-52 his3-200 ade2gal4Δ gal80). A human brain cDNA library (Clontech) constructed as a fusion to the C terminus of the Gal4 AD (residues 768–881) was transformed into yeast strain BK100 (MATa trp1–901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80 LYS2::GAL-HIS3 GAL2-ADE2 met2::GAL7-lacZ). Upon mating, ∼5 million diploid yeast cells were plated and selected in the presence of 3 mm 3-amino-1,2,4-triazole in medium lacking Trp, Leu, His, and Ade. A fragment of radixin (residues 6–511) was isolated, and the interaction confirmed by liquid β-galactosidase assays.
Immunoprecipitation
pCMVmyc-Epac or the Δ(1–148) mutants were co-transfected with HA-radixin-expressing plasmids. After 48 h, cells were lysed with a buffer containing 0.3% CHAPS, 150 mm NaCl, 50 mm Tris/HCl pH 7.5, 1 mm benzamidine, 10% glycerol, and protease inhibitors. Lysates were incubated for 1 h at 4 °C with 9E10 Myc antibody and 1 h with A/G-Sepharose beads (Amersham Biosciences) followed by five washes with lysis buffer.
GST Pull-downs
Cells were co-transfected with GST-fusion and Myc-tagged mammalian expression vectors. Upon lysis in immunoprecipitation buffer, GST proteins were pulled down with glutathione-Sepharose beads and associated proteins resolved on SDS-PAGE and blot with anti-Myc antibodies.
Protein Purification and in Vitro Binding
BL21/DE3 Escherichia coli cells transformed with the appropriate pGEX plasmids were grown until 0.8 OD and induced for 3 h at 37 °C with 0.5 mm IPTG. After centrifugation, cells were lysed, and aliquots of the lysates were frozen with 10% glycerol. Proteins were isolated on GSH-Sepharose (Amersham Biosciences) just before use. Purified C-radixin was obtained from immobilized GST-C-radixin after overnight incubation with thrombin. pET28b-Epac mutants, containing the T7 promoter, were in vitro translated using TnT T7 quick (Promega) and Redivue [35S]methionine (Amersham Biosciences), following the manufacturer's directions. Binding assays were performed in 50 mm Tris/Cl pH 7.5, 50 mm NaCl, and 0.1% Tween-20 for 1 h at room temperature, and complexes were washed four times with binding buffer. Competition assays included the appropriate soluble protein competitors purified after thrombin cleavage. Samples were run on 10% SDS-PAGE, fixed with 45% methanol-15% acetic acid solution, soaked with Amplify (Amersham Biosciences), dried, and exposed.
Confocal Microscopy
Radixin and Epac co-localization was assayed by confocal microscopy (Olympus Fluoview FV1000), utilizing the appropriate filters. Images were analyzed with the built-in software and ImageJ. Quantitative co-localization was performed on background subtracted images using Pearson and Spearman (ranked) correlation coefficients utilizing a plug-in available from ImageJ (17). These dimensionless values range from −1 to +1, with 0 indicating random co-localization.
Bodipy-Forskolin Labeling
Cells transfected with (1–52)-Epac-dsRed were washed in 1× PBS and incubated with BODIPY-FL forskolin (0.5 μm, Invitrogen) for 10 min at room temperature. After washing with 1× PBS, samples were directly mounted (Permafluor) for confocal microscopy analysis.
RESULTS AND DISCUSSION
Epac-mediated Rap1 activation is required, in synergy with PKA, to transduce the TSH mitogenic response in thyroid cells (16). Expression of the Epac N-terminal regulatory fragment including its DEP domain (amino acids 72–148) acted as dominant negative (16). The ability of Epac to act as a cAMP-dependent Rap GEF in vitro, however, does not require the presence of its DEP domain (18). Thus, we hypothesized that the DEP domain was required in vivo to transduce the signal because of its effects on Epac localization. This implies the existence of Epac DEP-binding proteins; radixin, a member of the ERM family (19), was identified from a yeast two-hybrid screen using the first 200 residues of Epac as bait.
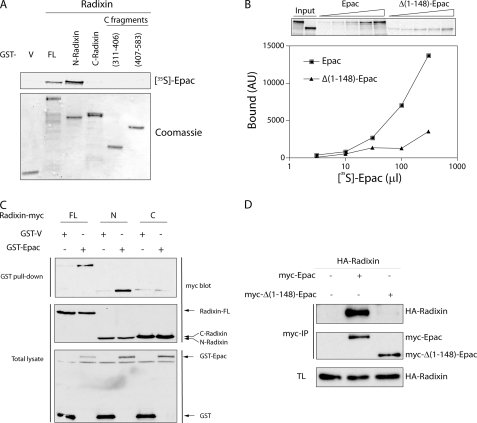
In vitro binding and in vivo pull-down and co-immunoprecipitation assays were then performed to validate Epac-radixin interaction.3 To confirm direct binding, full-length Epac was in vitro translated with [35S]methionine, and its ability to bind to purified GST-radixin full-length or deletion mutants was assayed. Full-length and N-terminal (1–318) radixin, but not the C-terminal fragments were able to interact with Epac (Fig. 1A). Based on the two-hybrid assay, the first 200 residues of Epac are sufficient for radixin interaction. Binding assays were repeated with bacterially expressed GST-N-radixin and 35S-labeled in vitro translated Epac or the Δ(1–148)-Epac mutant. N-radixin specifically interacted with full-length Epac but not Δ(1–148)-Epac (Fig. 1B). Similar results were obtained in vivo upon co-transfection with GST-Epac and Myc-tagged radixin constructs (Fig. 1C). This interaction was also confirmed by co-immunoprecipitation assays in 293T cells co-transfected with HA-radixin and full-length or Δ(1–148)-Epac constructs. Despite similar expression levels for both Epac constructs, radixin only associated with full-length Epac (Fig. 1D). These results validate the two-hybrid data and furthermore support a direct interaction between Epac and radixin, involving the first 148 residues of Epac and the radixin N-terminal (1–318) FERM (Four.1, Ezrin, Radixin, Moesin) domain, respectively.
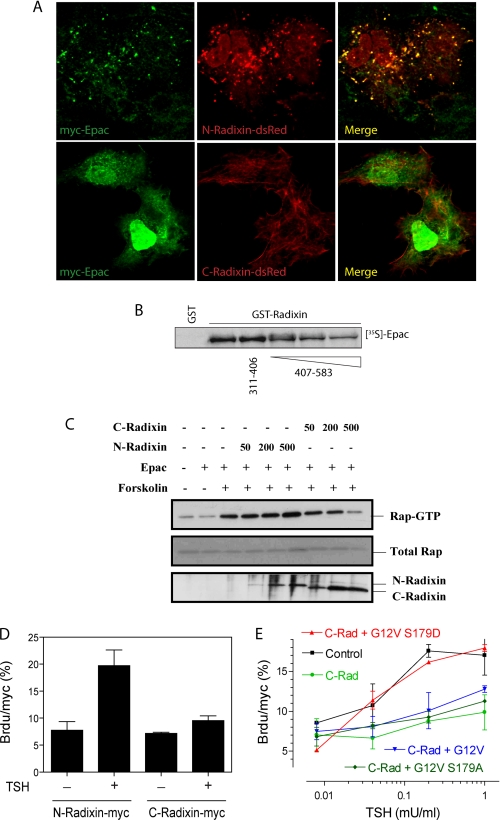
FIGURE 1.
Specific Epac-radixin interaction. A, direct Epac-radixin interaction via the radixin N-terminal domain. Immobilized GST-radixin proteins (Coomassie stain shown on lower panel) were incubated with 35S in vitro translated Epac. After extensive washes, samples were run on SDS-PAGE. Once dried, radioactivity was visualized by fluorography using a phosphorimager (FL, Epac full-length; N-radixin, 1–318; C-radixin, 319–583). B, direct Epac-radixin interaction via the Epac N-terminal domain. Immobilized GST-N-radixin was incubated with different amounts of in vitro translated [35S]Epac or Δ(1–148)-Epac. After extensive washes, bound Epac proteins were resolved on SDS-PAGE, and quantified using a phosphorimager (AU, arbitrary units). C, in vivo Epac-radixin interaction. PCCL3 cells were transfected with radixin-myc constructs along with GST-V or GST-Epac mammalian expression plasmids. After 48 h, cells were lysed, and GST fusion proteins were pulled-down. The presence of radixin in the complex was assessed with a Myc-specific antibody (GST-pull-down/Myc blot). Expression level was assessed from total lysate by GST and Myc blots before the pull-down step (lower panels). D, in vivo Epac-radixin interaction via the Epac N-terminal domain. PCCL3 cells were co-transfected with Myc-Epac and HA-radixin expression vectors. After 48 h, cells lysates were immunoprecipitated with an anti-Myc antibody (9E10), and the presence of radixin in the immunocomplex was assessed by Western blots with an HA-specific antibody.
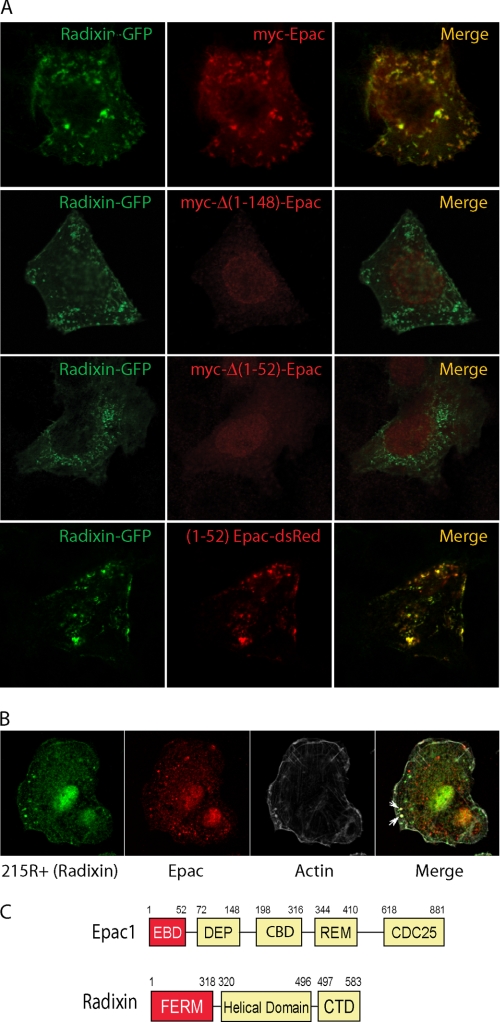
Epac-radixin co-localization was studied by confocal microscopy. Radixin-GFP was co-transfected along with myc-Epac constructs in thyroid PCCL3 cells. Radixin localized in clusters near the plasma membrane and in actin-containing cortical structures (i.e. microvilli, filopodia, lamellipodia), and a strong Epac-radixin co-localization was observed, mostly in the submembrane clusters (Fig. 2A). Deletion of the N-terminal domain (Δ1–148) dramatically changed Epac distribution in the cell, increasing its steady-state accumulation in the nuclear compartment, and decreasing accordingly its degree of co-localization with radixin-GFP. To confirm DEP dependence (amino acids 72–148) in Epac-radixin interactions, further deletion mutants were tested. Surprisingly, deletion of the first 52 residues of Epac (Δ1–52) behave exactly as Epac Δ1–148 (i.e. nuclear localization), indicating that either the N-terminal domain (1–52) is required to maintain DEP(72–148) integrity, or alternatively, that the radixin-binding domain is contained within the N-terminal 52 amino acids. Consistent with this idea, addition of the radixin first 52 residues was sufficient to target dsRed to the radixin clusters. Most importantly, Epac-radixin interaction was also observed on native proteins by immunostaining, confirming that an endogenous pool of intracellular Epac complexes with radixin and cortical actin cytoskeleton (Fig. 2B). Thus, the combined in vivo and in vitro results reveal a DEP-independent Epac-radixin interaction and the identification of a new ERM-binding domain (EBD, amino acids 1–52) in the Epac N terminus (Fig. 2C).
FIGURE 2.
A new EBD targets Epac to clusters. A, Epac-(1–52) is sufficient for radixin binding and cluster localization. PCCL3 cells were co-transfected with radixin-GFP and Epac constructs. After 36 h, cells were fixed, stained with an anti-Myc antibody (9E10) and analyzed by confocal microscopy. Representative cells are shown; radixin-GFP (green channel), myc-Epac/dsRed (red channel), and merged channels. B, endogenous Epac-radixin complex. Fixed PCCL3 cells were co-stained with 215R+ (rabbit anti-radixin, 1:20), C17 (goat anti-Epac, 1:50) and phalloidin-Alexa 647 (F-actin probe) followed by a mix of donkey anti-rabbit Alexa 488 (1:1000), and bovine anti-goat Alexa 594 (1:1000) secondary antibodies. Single (radixin, green; Epac, red; F-actin, gray), and merged images are shown. Clear Epac-radixin-actin co-localization in clusters is marked by white arrows. C, interacting domains. Schematic domain representation of the human Epac1 and mouse radixin proteins used in this study. Boxes shown in red represent the binding domains (EBD, ERM binding domain and FERM, Four.1, Ezrin, Radixin, Moesin, in Epac and radixin, respectively). Other domains in Epac: CBD, cAMP binding domain; DEP, Disheveled, Egl-10, Pleckstrin; REM, Ras exchange motif; CDC25, catalytic GEF. Other domains in radixin: helical domain, including the AKAP-like domain; CTD, C-terminal domain, including Thr-564 phosphorylation site, the F-actin, and FERM binding domains.
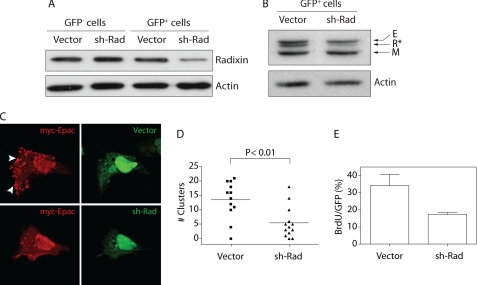
To determine the biological significance of Epac-radixin clusters, experimental maneuvers were devised for complex disruption. The use of shRNA to suppress radixin expression was evaluated in the context of cluster formation, Epac localization, and proliferative action of TSH in thyroid cells. Expression of endogenous radixin was efficiently (Fig. 3A) and specifically (Fig. 3B) down-regulated as evaluated with anti-radixin and anti-panERM antibodies, respectively. Radixin down-regulation led to an increased nuclear localization of Epac (Fig. 3C) accompanying the significant decrease in Epac-containing clusters (Fig. 3, C and D). Most importantly, when assessed by BrDU labeling, radixin down-regulation blocked TSH-mediated cell proliferation (Fig. 3E). Off-target control experiments were performed with a radixin mutant insensitive to the shRNA (radixinR), whose levels did not change upon radixin shRNA-mediated down-regulation (Fig. 4A). Like wt-radixin, radixinR localized in clusters (Fig. 4B) and completely rescued the inhibitory effect of shRNA on TSH action (Fig. 4C). These results confirmed that, despite the presence of wild-type levels of the other ERM members, i.e. ezrin and moesin (Fig. 3B), radixin expression is required to maintain the integrity of the cluster compartment for proper Epac localization and efficient TSH action.
FIGURE 3.
Epac-radixin clusters are required for TSH-mediated G1/S progression. A, efficient down-regulation of radixin by shRNA. PCCL3 cells were transfected with radixin shRNA (sh-Rad) or empty vector (GFP cassette in vector). After 72 h, GFP-positive cells were sorted and endogenous radixin expression was tested by Western blot using a radixin specific antibody (Sigma). Sorted GFP-negative cells were used as controls. An actin antibody was used for loading control. B, specific down-regulation by radixin shRNA. GFP-positive cells sorted from empty vector or sh-Rad-transfected samples were analyzed by Western blot with a pan-ERM antibody (Santa Cruz Biotechnology) to visualize ezrin (E), radixin (R*), and moesin (M) proteins. An actin antibody was used for loading control. C, radixin is necessary for Epac localization in clusters. PCCL3 cells were co-transfected with empty vector or radixin shRNA (sh-Rad) along with myc-Epac expression vectors. After 72 h, cells were fixed and stained with an anti-Myc antibody (9E10) to visualize Epac. Representative cells are shown; GFP (green channel), Myc-Epac (red channel). D, radixin down-regulation affects cluster number. Quantification of clusters in cells expressing GFP control (vector) and Epac (n = 12), or GFP-shRNA (sh-Rad) and Epac (n = 13) proteins. E, radixin down-regulation blocks TSH-mediated G1/S progression. PCCL3 cells were transfected with GFP control or GFP shRNA expression vectors. BrDU labeling was used to monitor TSH-dependent G1/S progression in the presence of 1 mU/ml TSH and 10% FBS as co-mitogen. BrDU/GFP results are expressed as mean ± S.E. (n = 3, >500 cells/experiment).
FIGURE 4.
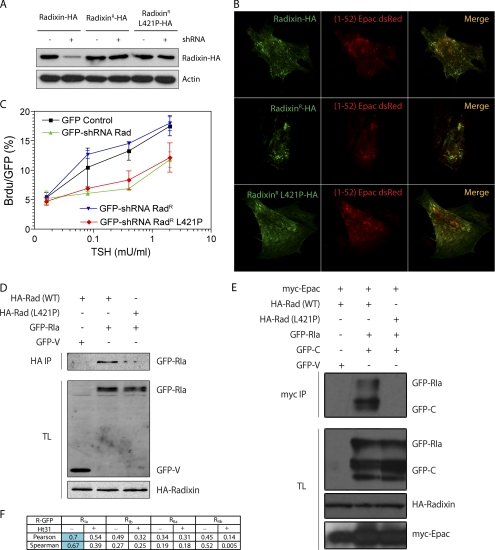
Radixin is a functional AKAP required for TSH-mediated G1/S progression. A, expression of Radixin shRNAR proteins. 500 ng of Radixin WT, Radixin shRNAR, or Radixin shRNAR L421P were co-transfected with 1.5 μg of GFP-V (−) or GFP-shRNA (+). After 48 h, cells were lysed, and radixin was detected using an HA-specific antibody. An actin antibody was used for loading control. B, radixin shRNAR localize in clusters. PCCL3 cells were co-transfected with radixin-HA along with Epac-(1–52)-dsRed to visualize clusters. After 36 h, cells were fixed and stained for radixin-HA detection and analyzed by confocal microscopy. Representative cells are shown; radixin-HA (green channel), Epac (1–52)-dsRed (red channel) and merged channels. C, rescue with Radixin shRNAR proteins. PCCL3 cells were transfected with 500 ng of empty vector (GFP control) or radixin-shRNA (GFP-shRNA) and Brdu labeling was used to monitor TSH-dependent G1/S progression in the presence of 10% FBS as co-mitogen. For rescue experiments, 1.5 μg of the corresponding shRNAR expression plasmids were co-transfected. BrDU/GFP results are expressed as mean ± S.E. (n = 3, >500 cells/experiment). D, radixin interacts with RIa. Cells were co-transfected with HA-radixin WT or AKAP-deficient L421P mutant, and RIa-GFP or empty GFP-V as control. After 36 h, cells were lysed, immunoprecipitated with anti-HA antibody (HA.11) and associated proteins revealed by Western blot (TL, total lysate). E, ternary complex Epac-Radixin-PKA. Cells were co-transfected with myc-Epac, HA-radixin (WT or L421P), and RIa-GFP or GFP-V as control. Lysates were immunoprecipitated with an anti-Myc antibody (9E10) and Epac-associated proteins revealed by Western blot (TL, total lysate; GFP is not observed since it ran-off the gel). F, radixin binds RIa. PCCL3 cells were co-transfected with Radixin-HA along with GFP-tagged R subunits. After 36 h, cells were fixed and stained for Radixin-HA detection and analyzed by confocal microscopy. Background subtracted images were subjected to co-localization assays using correlation analysis.
All ERM proteins contain an amphipathic helical domain able to act as an AKAP and recruit PKA. Both PKAI (20) and PKAII (21) isoforms bind ERM, however functional consequences of radixin-PKA interactions are unknown. To test if the AKAP domain within radixin is required for TSH-mediated cell proliferation, a mutation was introduced in the radixin AKAP domain (L421P) that impaired its binding to PKA (22). Despite similar levels of expression and localization (Fig. 4, A and B), the double mutant L421P-radixinR failed to rescue the shRNA inhibitory effect (Fig. 4C), indicating that PKA binding to radixin is required for a full TSH proliferative response. To directly assess radixin-PKA interaction co-immunoprecipitation and confocal co-localization studies were performed. When HA-radixin and RIa-GFP were co-transfected, an interaction radixin-RIa was observed (Fig. 4D). Moreover, co-transfection of Myc-Epac with HA-radixin and PKA subunits (GFP-C and GFP-RIa) revealed that Epac associates with PKA in a manner dependent on a functional (i.e. AKAP) radixin protein (Fig. 4E), indicative of the presence of a ternary Epac1-radixin-PKA complex. Quantitative confocal co-localization studies between HA-radixin and GFP-R subunits indicated that PKAIa is the most likely isoform that interacts with radixin (Fig. 4F). These results demonstrate a role for radixin in scaffolding both cAMP effectors Epac1 and PKAI in a specific signaling complex marking a functional cAMP compartment required for TSH action.
ERM proteins alternate between an open and a closed conformation regulated by interaction of the N and C termini (23, 24), and independent expression of these fragments interferes with ERM function (25, 26). N- or C-radixin-dsRed fusion plasmids were co-transfected along with Myc-tagged Epac, and their respective locations were monitored by confocal microscopy. Contrary to N-radixin-dsRed that localized in clusters, C-radixin-dsRed inhibited cluster formation, accompanied by the displacement of Epac to the nuclear compartment (Fig. 5A), similar to the results observed with the deletion construct Epac-(Δ1–52) (Fig. 2A) and shRNA-mediated radixin down-regulation (Fig. 3C). As expected, C-radixin-dsRed showed a localization pattern similar to actin filaments, confirmed by co-staining with phalloidin (not shown). This suggests that C-radixin displaces Epac from radixin-containing clusters by binding to the radixin FERM domain, as previously suggested (27). Expression of soluble C-radixin-(407–583) that by itself does not bind Epac (Fig. 1A) inhibited in a dose-dependent manner the ability of 35S-labeled Epac to bind to immobilized GST-radixin in vitro (Fig. 5B). Hence, Epac displacement by C-radixin blocked Epac-mediated Rap1 activation in vivo (Fig. 5C). Consistent with a model where Epac binds to radixin in its open/active conformation, these results validated the use of a C-radixin construct as an Epac-radixin disruptor tool. As described above for radixin shRNA (Fig. 3E), expression of C-radixin blocked TSH-mediated proliferation (Fig. 5D).
FIGURE 5.
Rap1 integrates cAMP effector pathways. A, C-radixin disrupts cluster formation. PCCL3 cells were transfected with 500 ng of plasmids expressing N- or C-Radixin dsRed along with 500 ng of myc-Epac. After 36 h, cells were fixed and stained for myc-Epac detection and analyzed by confocal microscopy. Representative cells are shown; myc-Epac (green channel), dsRed constructs (red channel), and merged channels. B, soluble C-radixin binds radixin and displaces Epac from the complex. Immobilized GST-radixin or GST proteins (1 μg) were incubated with 35S in vitro translated Epac in the presence of soluble GST-free radixin fragments 311–406 (10 μg) and 407–583 (1, 5, and 10 μg). After extensive washes, samples were run in an SDS-PAGE gel and GST-bound 35S assessed by autoradiography/fluorography. C, soluble C-radixin blocks Epac-mediated Rap1 activation. Cells were co-transfected with HA-Rap1 (50 ng), Epac (100 ng), and the indicated amounts of either myc-N-radixin or myc-C-radixin. 36 h later, cells were left untreated or stimulated for 5 min with forskolin. Upon lysis, samples were processed for Ral-GDS-RBD pull-down assays. D, C-radixin blocks TSH-mediated G1/S progression. PCCL3 cells were transfected with 500 ng of N-radixin-myc or C-radixin-myc expression plasmids. Brdu labeling was used to monitor TSH-dependent G1/S progression in the presence of 10% FBS as co-mitogen. BrDU results are expressed as mean ± S.E. (n = 3, >500 cells/experiment). E, constitutively active and phosphorylated Rap1b rescues C-radixin inhibition. PCCL3 cells were transfected with 500 ng of myc-Rap1 (control) or C-radixin-myc plasmids. Brdu labeling was used to monitor TSH-dependent G1/S progression in the presence of 10% FBS as co-mitogen. For rescue experiments, 1.5 μg of HA-Rap1 expression plasmids (G12V, G12V-S179D, and G12V-S179A) were co-transfected as indicated. BrDU/myc results are expressed as mean ± S.E. (n = 3, >500 cells/experiment).
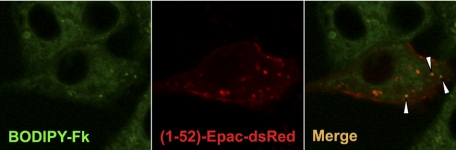
Our results indicate that radixin scaffolds cAMP effectors, i.e. Epac1 and PKA into a functional compartment required for TSH action. If Epac/PKA delocalization is responsible for the inhibition upon cluster disruption, expression of constitutively active element(s) downstream of Epac and PKA activation should bypass the requirement for an active compartment and accordingly rescue C-radixin inhibitory action. A likely candidate is Rap1, a substrate for both Epac and PKA required for TSH action (16, 28, 29). Results shown in Fig. 5E confirmed this prediction; when co-transfected with the inhibitory C-radixin construct, Rap1-G12V-S179D (but not Rap1-G12V or Rap1-G12V-S179A), was able to rescue C-radixin-mediated inhibition of TSH action. Thus, only in its activated (i.e. G12V, GTP bound) and phosphorylated form (i.e. phosphomimetic S179D), Rap1 is able to bypass the need for an active Epac-radixin-PKA complex, consistent with a model where radixin compartment acts as a cAMP sensing locale where effector activation constants are met, leading to Epac-mediated Rap activation and PKA-mediated Rap1 Ser-179 phosphorylation. Consistent with this, a pool of adenylyl cyclase is also observed into this compartment as manifested by the use of the fluorescent analog Bodipy-forskolin (Fig. 6), indicating co-compartmentalization of cAMP synthesis and cAMP effectors as predicted by a cAMP microdomain model.
FIGURE 6.
A pool of a forskolin-sensitive adenylyl cyclase co-localizes with clusters. PCCL3 cells were transfected with (1–52)-Epac-dsRed plasmid. 36 h later cells were stimulated for 10 min with Bodipy-forskolin (0.5 μm, below its EC50), washed in PBS, mounted, and visualized by confocal microscopy.
Like PKA via AKAPs, it is becoming evident that Epac can be targeted to different compartments utilizing distinct motifs (14, 30–37). The emerging picture is that different GEF pools might be subjected to distinct regulatory events, potentially leading to different biological outcomes (38). Our studies have unraveled a new compartment where cAMP synthesis and cAMP effectors reside and is required for the mitogenic effects of TSH. This compartment consists of a localized pool of cAMP that activates its effectors, Epac and PKA, via a radixin-delimited mechanism (Fig. 7). Upon cAMP elevation, activated Epac leads to the rapid recruitment and activation of Rap1, presenting it to PKA for efficient Ser-179 phosphorylation. Both events are strictly required for efficient signal transduction. However, downstream signaling seems to be independent of the integrity of this compartment because Rap1-G12V-S179D is able to rescue the inhibitory effect of compartment disruption. This indicates that this compartment is solely acting as a cAMP-sensing device matching the effector activation constant, using Rap1 as a signal integrator.
FIGURE 7.
Proposed model. Radixin, an ERM protein linking plasma membrane to the cortical actin cytoskeleton, recruits both cAMP effectors, i.e. Epac and PKA, to a compartment expressing adenylate cyclase. Epac1, via its EBD-(1–52) domain, binds to radixin FERM domain (1–318), while PKAI is recruited via an AKAP-like amphipathic α-helical region in radixin. Upon TSH stimulation, a local increase of cAMP activates (via Epac) and phosphorylates (via PKA) Rap1b and both events are strictly required to transduce the TSH/cAMP proliferative response. Disruption of this compartment blocks TSH/cAMP action and expression of constitutively active Rap1b rescues this inhibitory action in a phosphorylation-dependent manner. Our results indicate that radixin delimits a new functional cAMP compartment using Rap1 as a signaling node. (? indicates that the mechanism involved in recruiting active radixin to the plasma membrane is for the moment unknown.)
Acknowledgments
We thank Drs. Voyno-Yasenetskaya, Solomon, Ravichandran, and Tsukita for radixin reagents; Drs. Bos and Holz for Epac constructs; Dr. Zhong for GFP-R and GFP-C plasmids; Dr. Crouch for anti-radixin antiserum; Dr. N. Kotchey for her participation at early stages of this study and other laboratory members for helpful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by Public Health Service Grant CA071649 from the NCI, National Institutes of Health.
During revision of this article, a radixin-Epac interaction was reported (39).
- AKAP
- A-kinase-anchoring protein
- ERM
- ERM-binding domain.
REFERENCES
- 1. Levitzki A. (1988) Science 241, 800–806 [DOI] [PubMed] [Google Scholar]
- 2. Barnes A. P., Livera G., Huang P., Sun C., O'Neal W. K., Conti M., Stutts M. J., Milgram S. L. (2005) J. Biol. Chem. 280, 7997–8003 [DOI] [PubMed] [Google Scholar]
- 3. Jurevicius J., Fischmeister R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 295–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rich T. C., Fagan K. A., Tse T. E., Schaack J., Cooper D. M., Karpen J. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13049–13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Terrin A., Di Benedetto G., Pertegato V., Cheung Y. F., Baillie G., Lynch M. J., Elvassore N., Prinz A., Herberg F. W., Houslay M. D., Zaccolo M. (2006) J. Cell Biol. 175, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Willoughby D., Wong W., Schaack J., Scott J. D., Cooper D. M. (2006) EMBO J. 25, 2051–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zaccolo M., Pozzan T. (2002) Science 295, 1711–1715 [DOI] [PubMed] [Google Scholar]
- 8. Buxton I. L., Brunton L. L. (1983) J. Biol. Chem. 258, 10233–10239 [PubMed] [Google Scholar]
- 9. Rich T. C., Fagan K. A., Nakata H., Schaack J., Cooper D. M., Karpen J. W. (2000) J. Gen. Physiol. 116, 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaccolo M., Di Benedetto G., Lissandron V., Mancuso L., Terrin A., Zamparo I. (2006) Biochem. Soc. Trans. 34, 495–497 [DOI] [PubMed] [Google Scholar]
- 11. Dessauer C. W. (2009) Mol. Pharmacol. 76, 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dodge-Kafka K. L., Bauman A., Kapiloff M. S. (2008) Handb Exp. Pharmacol 186, 3–14 [DOI] [PubMed] [Google Scholar]
- 13. Di Benedetto G., Zoccarato A., Lissandron V., Terrin A., Li X., Houslay M. D., Baillie G. S., Zaccolo M. (2008) Circ. Res. 103, 836–844 [DOI] [PubMed] [Google Scholar]
- 14. Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., Scott J. D. (2005) Nature 437, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rampersad S. N., Ovens J. D., Huston E., Umana M. B., Wilson L. S., Netherton S. J., Lynch M. J., Baillie G. S., Houslay M. D., Maurice D. H. (2010) J. Biol. Chem. 285, 33614–33622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hochbaum D., Hong K., Barila G., Ribeiro-Neto F., Altschuler D. L. (2008) J. Biol. Chem. 283, 4464–4468 [DOI] [PubMed] [Google Scholar]
- 17. French A. P., Mills S., Swarup R., Bennett M. J., Pridmore T. P. (2008) Nat. Protoc. 3, 619–628 [DOI] [PubMed] [Google Scholar]
- 18. de Rooij J., Rehmann H., van Triest M., Cool R. H., Wittinghofer A., Bos J. L. (2000) J. Biol. Chem. 275, 20829–20836 [DOI] [PubMed] [Google Scholar]
- 19. Fehon R. G., McClatchey A. I., Bretscher A. (2010) Nat. Rev. Mol. Cell Biol. 11, 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruppelt A., Mosenden R., Grönholm M., Aandahl E. M., Tobin D., Carlson C. R., Abrahamsen H., Herberg F. W., Carpén O., Taskén K. (2007) J. Immunol. 179, 5159–5168 [DOI] [PubMed] [Google Scholar]
- 21. Dransfield D. T., Bradford A. J., Smith J., Martin M., Roy C., Mangeat P. H., Goldenring J. R. (1997) EMBO J. 16, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vijayaraghavan S., Goueli S. A., Davey M. P., Carr D. W. (1997) J. Biol. Chem. 272, 4747–4752 [DOI] [PubMed] [Google Scholar]
- 23. Gary R., Bretscher A. (1995) Mol. Biol. Cell 6, 1061–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pearson M. A., Reczek D., Bretscher A., Karplus P. A. (2000) Cell 101, 259–270 [DOI] [PubMed] [Google Scholar]
- 25. Amieva M. R., Litman P., Huang L., Ichimaru E., Furthmayr H. (1999) J. Cell Sci. 112, 111–125 [DOI] [PubMed] [Google Scholar]
- 26. Vaiskunaite R., Adarichev V., Furthmayr H., Kozasa T., Gudkov A., Voyno-Yasenetskaya T. A. (2000) J. Biol. Chem. 275, 26206–26212 [DOI] [PubMed] [Google Scholar]
- 27. Reczek D., Bretscher A. (1998) J. Biol. Chem. 273, 18452–18458 [DOI] [PubMed] [Google Scholar]
- 28. Altschuler D., Lapetina E. G. (1993) J. Biol. Chem. 268, 7527–7531 [PubMed] [Google Scholar]
- 29. Ribeiro-Neto F., Leon A., Urbani-Brocard J., Lou L., Nyska A., Altschuler D. L. (2004) J. Biol. Chem. 279, 46868–46875 [DOI] [PubMed] [Google Scholar]
- 30. Borland G., Gupta M., Magiera M. M., Rundell C. J., Fuld S., Yarwood S. J. (2006) Mol. Pharmacol. 69, 374–384 [DOI] [PubMed] [Google Scholar]
- 31. Huston E., Lynch M. J., Mohamed A., Collins D. M., Hill E. V., MacLeod R., Krause E., Baillie G. S., Houslay M. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12791–12796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Asuri S., Rebhun J. F., Castro A. F., Paranavitana N. C., Quilliam L. A. (2006) J. Biol. Chem. 281, 2506–2514 [DOI] [PubMed] [Google Scholar]
- 33. Liu C., Takahashi M., Li Y., Dillon T. J., Kaech S., Stork P. J. (2010) Mol. Cell. Biol. 30, 3956–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu C., Takahashi M., Li Y., Song S., Dillon T. J., Shinde U., Stork P. J. (2008) Mol. Cell. Biol. 28, 7109–7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Métrich M., Laurent A. C., Breckler M., Duquesnes N., Hmitou I., Courillau D., Blondeau J. P., Crozatier B., Lezoualc'h F., Morel E. (2010) Cell Signal 22, 1459–1468 [DOI] [PubMed] [Google Scholar]
- 36. Ponsioen B., Gloerich M., Ritsma L., Rehmann H., Bos J. L., Jalink K. (2009) Mol. Cell. Biol. 29, 2521–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qiao J., Mei F. C., Popov V. L., Vergara L. A., Cheng X. (2002) J. Biol. Chem. 277, 26581–26586 [DOI] [PubMed] [Google Scholar]
- 38. Wang Z., Dillon T. J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P. J. (2006) Mol. Cell. Biol. 26, 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gloerich M., Ponsioen B., Vliem M. J., Zhang Z., Zhao J., Kooistra M. R., Price L. S., Ritsma L., Zwartkruis F. J., Rehmann H., Jalink K., Bos J. L. (2010) Mol. Cell. Biol. 30, 5421–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]