Abstract
The BolA homologue Fra2 and the cytosolic monothiol glutaredoxins Grx3 and Grx4 together play a key role in regulating iron homeostasis in Saccharomyces cerevisiae. Genetic studies indicate that Grx3/4 and Fra2 regulate activity of the iron-responsive transcription factors Aft1 and Aft2 in response to mitochondrial Fe-S cluster biosynthesis. We have previously shown that Fra2 and Grx3/4 form a [2Fe-2S]2+-bridged heterodimeric complex with iron ligands provided by the active site cysteine of Grx3/4, glutathione, and a histidine residue. To further characterize this unusual Fe-S-binding complex, site-directed mutagenesis was used to identify specific residues in Fra2 that influence Fe-S cluster binding and regulation of Aft1 activity in vivo. Here, we present spectroscopic evidence that His-103 in Fra2 is an Fe-S cluster ligand in the Fra2-Grx3 complex. Replacement of this residue does not abolish Fe-S cluster binding, but it does lead to a change in cluster coordination and destabilization of the [2Fe-2S] cluster. In vivo genetic studies further confirm that Fra2 His-103 is critical for control of Aft1 activity in response to the cellular iron status. Using CD spectroscopy, we find that ∼1 mol eq of apo-Fra2 binds tightly to the [2Fe-2S] Grx3 homodimer to form the [2Fe-2S] Fra2-Grx3 heterodimer, suggesting a mechanism for formation of the [2Fe-2S] Fra2-Grx3 heterodimer in vivo. Taken together, these results demonstrate that the histidine coordination and stability of the [2Fe-2S] cluster in the Fra2-Grx3 complex are essential for iron regulation in yeast.
Keywords: Circular Dichroism (CD), Gene Regulation, Iron Metabolism, Iron-Sulfur Protein, Protein Metal Ion Interaction, Raman Spectroscopy, Site-directed Mutagenesis, Yeast, BolA-like Protein, Monothiol Glutaredoxin
Introduction
Although iron is a cofactor for many essential proteins, an excess of this redox-active metal ion is toxic to cells. To maintain optimal intracellular iron levels, iron homeostasis is tightly regulated in all eukaryotic cells ranging from yeast to humans. Transcription of iron uptake and storage genes in the model eukaryote Saccharomyces cerevisiae is regulated by the iron-responsive transcriptional activator Aft1 and its paralogue Aft2 (1–3). Under iron-replete conditions, Aft1 is mainly cytosolic, and under iron-deplete conditions, Aft1 accumulates in the nucleus where it activates genes involved in iron uptake and transport, collectively known as the iron regulon (4). Interestingly, activation of the iron regulon by Aft1 and Aft2 does not respond directly to cytosolic iron but rather to mitochondrial iron-sulfur cluster biogenesis via a signaling pathway that includes the cytosolic proteins Fra1, Fra2, Grx3, and Grx4 (5–8). Fra2 is a homologue of the protein encoded by the Escherichia coli stationary phase morphogene bolA. The monothiol glutaredoxins Grx3 and Grx4 also interact with Fra2 in addition to Aft1 in vivo (5–7). In the absence of the Fra2-Grx2 signaling complex, Aft1 exhibits constitutive activation in both low and high iron growth conditions. Thus, the Fra-Grx complex inhibits Aft1 (and presumably Aft2) activation of the iron regulon when intracellular iron levels are sufficient and mitochondrial Fe-S cluster biogenesis is functional. However, the specific role of the Fra-Grx complex and the precise mechanism of iron-dependent inhibition of Aft1/2 activity are still unclear.
To better understand this signaling pathway, we recently characterized the molecular interactions between Fra2 and Grx3/4 (9). Fra2 forms heterodimers with Grx3 or Grx4 with a [2Fe-2S]2+ cluster bound at the interface ligated by both cysteine and histidine residues. The spectroscopic characteristics of these complexes are distinct from [2Fe-2S]2+-bridged Grx3 or Grx4 homodimers, which bind the Fe-S cluster with all Cys residues provided by Grx3/4 and two glutathione (GSH) molecules, similar to other members of the monothiol Grx family (10–12). Grx3 mutagenesis studies demonstrate that Cys-176, located in the conserved Grx-like domain active site (CGFS), and residues in the GSH binding pocket (Trp-214/Pro-215) are also required for heterodimer formation and cluster binding. These same residues are indispensable for iron-dependent inhibition of Aft1 activity in vivo (6), suggesting that there is a direct link between the formation of the [2Fe-2S] Fra2-Grx3/4 complex and their in vivo role in iron signaling.
Biophysical and biochemical analyses of [2Fe-2S] Fra2-Grx3/4 complexes also demonstrate the presence of a single histidine ligand for the cluster, which is presumably provided by Fra2. Because there have been no published studies identifying specific Fra2 residues required for interaction with Grx3/4 and regulation of Aft1/2 function in vivo, this study is aimed at identifying potential Fe-S-binding ligands on Fra2 and gauging their role in regulating Aft1/2 activity. Here, we present mutagenesis, spectroscopic, genetic, and biochemical evidence that His-103 in Fra2 is an Fe-S cluster ligand and plays an indispensable role in Aft1 regulation in vivo. Furthermore, we demonstrate that apo-Fra2 binds tightly to [2Fe-2S] Grx3 to form the [2Fe-2S] Fra2-Grx3 heterodimer, suggesting a possible mechanism for formation of this iron regulatory complex in vivo. Mutation of His-103 does not interfere in Fra2 binding to the [2Fe-2S] Grx3 homodimer, suggesting that this residue is critical for a different step in the yeast iron signaling pathway. Overall, these results provide a more detailed picture of the Fra2-Grx3 interaction and support a model in which Fra2 acts as an adaptor protein converting Grx3 from an Fe-S scaffold/delivery protein with a relatively labile cluster to an iron sensor with a stable cluster.
EXPERIMENTAL PROCEDURES
E. coli Plasmid Construction
Construction of recombinant Fra2 plasmids pRSFDuet-1-Fra1-His6/Fra2 and pET21a-Fra2 for overexpression in E. coli was described previously (9). The Fra2 ORF without the N-terminal 35 amino acids, Fra2Δ1–35, was amplified from pET21a-Fra2 by PCR using the primers shown in supplemental Table 1 and subcloned at the NdeI/KpnI sites of pRSFDuet-1-Fra1-His6 to generate pRSFDuet-1-Fra1-His6/Fra2Δ1–35. Recombinant Fra2 mutants were created by site-directed mutagenesis of pRSFDuet-1-Fra1-His6/Fra2 or pRSFDuet-1-Fra1-His6/Fra2Δ1–35 (QuikChange mutagenesis kit, Agilent Technologies, Santa Clara, CA) using primers listed in supplemental Table 1. The mutated pRSFDuet-1-Fra1-His6/Fra2 plasmids were used for coexpression and purification of Fra2-Grx3 heterodimers, whereas the mutated pRSFDuet-1-Fra1-His6/Fra2Δ1–35 plasmids were used for expression and purification of apo-Fra2. As noted previously, Fra1-His6 is not expressed at detectable levels from pRSFDuet-1-Fra1-His6/Fra2 plasmids (9). Nevertheless, these coexpression plasmids were used for these studies because the presence of the FRA1 gene does not interfere in WT or mutant Fra2 overexpression.
Protein Expression and Purification
Recombinant Grx3 was purified as described previously (13). Coexpression and copurification of Grx3 with Fra2Δ1–35 or full-length Fra2 His or Cys mutants were accomplished using the same procedures described for the WT Grx3-Fra2 complex (9). Overexpression of recombinant apo-Fra2Δ1–35 was performed in 1 liter of LB media at 30 °C with shaking until the A600 = 0.6–0.8 followed by induction with 1 mm isopropyl β-d-thiogalactoside. The cells were collected after an 18-h induction, resuspended in 50 mm Tris/MES, pH 8.0, sonicated, and centrifuged to remove cell debris. The cell-free extract was subsequently loaded onto a DEAE anion-exchange column (GE Healthcare) pre-equilibrated with 50 mm Tris/MES, pH 8.0. The protein was eluted with a 0–1 m NaCl gradient using 50 mm Tris/MES, pH 8.0, 1 m NaCl. Fra2Δ1–35-containing fractions were concentrated and loaded onto a HiLoad Superdex 75 gel filtration column (GE Healthcare) equilibrated with 50 mm Tris/MES, pH 8.0, 150 mm NaCl. The purest fractions of Fra2Δ1–35 as judged by SDS-PAGE were collected and concentrated to ∼500 μl with the addition of 5% glycerol (v/v) and stored at −80 °C.
Biochemical Methods
In vitro Fe-S reconstitution of Grx3 was done anaerobically in the presence of GSH, ferrous ammonium sulfate, l-cysteine, and Azobacter vinelandii NifS as described previously (9). Protein concentrations were determined by the Bradford assay (Bio-Rad) using BSA as the standard. Iron concentrations were determined using the colorimetric ferrozine assay (14). Acid-labile sulfur concentrations were determined using published methods (15, 16). For GSH measurements, the purified Fe-S protein complexes were denatured and precipitated with 1% (w/v) 5-sulfosalicylic acid, and GSH in the supernatant was measured using 5,5′-dithiobis(2-nitrobenzoic acid) and GSSG reductase as described previously (17). Analytical gel filtration analyses were performed on a Superdex 75 10/300 GL column (GE Healthcare) as described previously (9).
Spectroscopic Methods
UV-visible absorption spectra were recorded under anaerobic conditions at room temperature using a Shimadzu UV-3101 or a Beckman DU-800 spectrophotometer. CD spectra were also recorded under anaerobic conditions on identical samples using a Jasco J-715 or J-800 spectropolarimeter (Jasco, Easton, MD). Resonance Raman spectra were recorded as described previously (18), using a Ramanor U1000 spectrometer (Instruments SA, Edison, NJ) coupled with a Sabre argon ion laser (Coherent, Santa Clara, CA) with 20-μl frozen droplets of 1.8–2.6 mm sample mounted on the cold finger of a Displex model CSA-202E closed cycle refrigerator (Air Products, Allentown, PA). X-band (9.6 GHz) EPR spectra were recorded using an ESP-300D spectrometer (Bruker, Billerica, MA), equipped with an ESR 900 helium flow cryostat (Oxford Instruments, Concord, MA), and quantified under nonsaturating conditions by double integration against a 1.0 mm CuEDTA standard.
X-ray Absorption Spectroscopy
X-ray absorption experiments were completed at the Stanford Synchrotron Radiation Laboratory on beamlines 9-3 and 7-3. An Oxford liquid helium cryostat maintained the sample temperature at 10 K, and the iron Kα fluorescence was detected using a Canberra 30 element solid-state detector. Individual scans were calibrated by simultaneously measuring the spectrum of an iron foil and by assigning the inflection point of the rising foil edge to 7111.2 eV. As a precaution against photoreduction, for most samples, the beam position was moved after 2–3 scans to a fresh area on the sample surface. Samples were between 1 and 2 mm iron and were flash-frozen in liquid nitrogen in 30% (v/v) glycerol. All other data collection and analysis details were described previously (9).
Yeast Strains and Plasmid Construction
The yeast strains used in this study are the wild-type strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the fra2 deletion strain BY4741 fra2Δ::kanMX4. Yeast were maintained at 30 °C on YPD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose) or in complete synthetic medium (CM). Iron-replete media were made by supplementing with 100 μm FeSO4, and iron-depleted media included 100 μm bathophenanthroline sulfonate as a ferrous iron chelator. A FRA2 yeast expression vector was constructed by inserting a DNA fragment containing 700 bp of 5′ sequences and the entire Fra2 open reading frame followed by a His6 epitope into a YCp vector containing the CYC1 terminator. Single amino acid mutations in the Fra2 coding sequence were made using the QuikChange site-directed mutagenesis kit (Agilent Technologies). The Fra2 C66A/H103A double mutant was constructed using overlapping PCR. All yeast transformations were performed using the lithium acetate procedure, and all plasmid sequences were confirmed by DNA sequencing.
S1 Nuclease Protection Assays
RNA was extracted from cells grown to mid-log phase using the hot acid phenol method, and S1 analysis was performed as described previously (19). In each reaction, 12 μg of total RNA were hybridized to a 32P end-labeled DNA oligonucleotide probe before digestion with S1 nuclease and separation on an 8% polyacrylamide, 8 m urea polyacrylamide gel. Dried gels were imaged using a Bio-Rad FX phosphorimager and quantified using Quantity One software, prior to autoradiography.
Immunodetection
Cellular lysates were prepared by glass bead lysis in PBS, 150 mm NaCl. Following centrifugation, the supernatant was treated with SDS sample buffer, resolved by 12.5% SDS-PAGE, and transferred to nitrocellulose. The membranes were probed with mouse anti-Pgk1 (Invitrogen) or rabbit anti-Fra2 (kindly provided by Jerry Kaplan) and detected by chemiluminescence (SuperSignal West Dura, Pierce).
Titration of [2Fe-2S]2+ Cluster-bound Grx3 with Fra2
The titration of [2Fe-2S]2+ cluster-bound Grx3 with apo-Fra2 was monitored under anaerobic conditions at room temperature using UV-visible CD spectroscopy. N-terminal truncated forms (Δ1–35) of WT, H103A, and H103C Fra2 were used for these studies because removal of the first 35 residues from Fra2 greatly improves its solubility when expressed in the absence of Grx3. Reactions were carried out in 100 mm Tris-HCl buffer, pH 7.8, with the [2Fe-2S]2+ cluster concentration of Grx3 held constant at 50 or 200 μm and Fra2:Grx3[2Fe-2S] ratios varying from 0 to 10. Samples were equilibrated for 5 min at room temperature after addition of Fra2 prior to recording CD spectra. To confirm that removal of residues 1–35 in Fra2 does not influence apo-Fra2 binding to [2Fe-2S]2+ Grx3, full-length WT Fra2 was incubated with [2Fe-2S]2+ Grx3 and produced identical CD changes as Fra2Δ1–35 (data not shown).
RESULTS
Identifying Potential Fe-S Cluster Ligands in Fra2
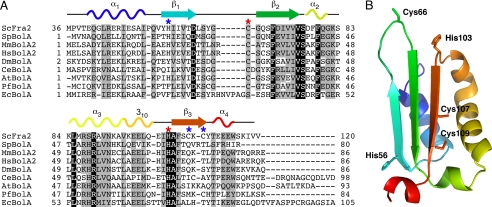
To identify potential Fra2 Fe-S cluster ligand(s) in the [2Fe-2S] Fra2-Grx3 complex, the Fra2 amino acid sequence was aligned with other BolA family members to highlight conserved residues (Fig. 1A). Available structural information for both prokaryotic and eukaryotic BolA-like proteins indicates that this protein family adopts an α1β1β2α2α3β3α4 topology (Fig. 1B) (20, 21). Because the spectroscopic data suggest that both His and Cys residues play a role in cluster binding (9), yeast Fra2 was modeled on the structure of its mouse homologue (BolA2) to identify well conserved Cys/His residues located within close proximity to each other. Cys-66 and His-103 located on the β1/β2 loop and α3/β3 loop, respectively, were identified as good candidates for Fe-S binding (Fig. 1). In addition, Fra2 has several other nonconserved histidines (His-19, His-33, and His-56) and cysteines (Cys-107 and Cys-109). Both the conserved and nonconserved Cys/His residues were individually mutated or deleted and overexpressed with WT Grx3 to assess their role in Fra2-Grx3 complex formation and Fe-S cluster binding. We verified by SDS-PAGE that the expression levels of both mutant Fra2 and WT Grx3 were similar to the coexpression results for the WT forms (data not shown). Because the conserved BolA-like domain of Fra2 only includes amino acids 36–120, we also tested whether removal of the 1st 35 amino acids at the Fra2 N terminus influenced the interaction between Fra2 and Grx3. This shortened form of Fra2 (Fra2Δ1–35) was able to form an Fe-S complex with Grx3 with spectroscopic signatures identical to the WT complex as determined by UV-visible absorption, CD, EPR, and resonance Raman spectroscopy (data not shown). Therefore, the additional N-terminal sequence of Fra2 (which includes His-19 and His-33) is not critical for heterodimer formation. All the Fra2 Cys mutants (C66S, C107S, and C109S) and the Fra2 H56A mutant also copurified with Grx3 and formed [2Fe-2S]-bridged heterodimers that were spectroscopically identical to the WT complex (data not shown). These results suggest that the Cys residues in Fra2 and His-56 are not cluster ligands.
FIGURE 1.
Sequence comparison and structural model for yeast Fra2. A, multiple sequence alignment of Fra2 with BolA homologues in several model organisms using Kalign. Identical residues are highlighted in black, and similar residues are shown in gray. Red and blue asterisks indicate the location of conserved and nonconserved Cys/His residues in yeast Fra2, respectively. Putative secondary structure of yeast Fra2 corresponding to the model in B is shown above the alignment. ScFra2, S. cerevisiae Fra2 (gi:6321218); SpBolA, Schizosaccharomyces pombe BolA-like protein (gi:19115194); MmBolA2, Mus musculus BolA2 (gi:29171318); HsBolA2, Homo sapiens BolA2 (gi:46577124); DmBolA, Drosophila melanogaster BolA-like protein CG33672 (gi:78707212); CeBolA, Caenorhabditis elegans BolA-like protein T12D8.10 (gi:71990814); AtBolA, Arabidopsis thaliana BolA-like protein (gi:18416103); PfBolA, Plasmodium falciparum BolA-like protein PFE0790c (gi:23504643); EcBolA, E. coli BolA (gi:78099828). B, putative structure of yeast Fra2 modeled on the mouse BolA2 structure (Protein Data Bank code 1V9J) (21) using ESyPred3D (43). Residues corresponding to the asterisks in A are highlighted.
UV-visible Absorption and CD Spectroscopy Reveal Differences in the Cluster Coordination Environment for Fra2 His-103 Mutants Coexpressed with Grx3
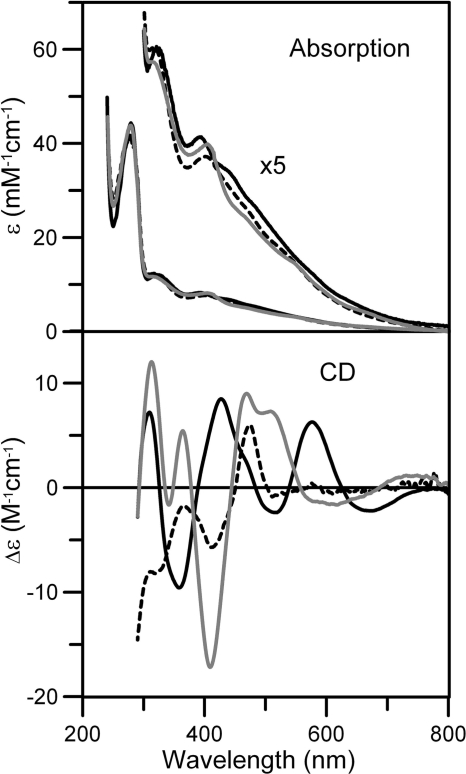
To test whether His-103 in Fra2 was important for Fe-S cluster binding, we first generated a His to Ala mutation in full-length Fra2 to gauge how elimination of this putative metal-binding residue influenced the Fra2-Grx3 interaction and cluster binding. Fra2 His-103 was also changed to a Cys to replace, rather than eliminate, this potential cluster ligand. Interestingly, the Fra2 H103A and H103C mutants also formed 1:1 [2Fe-2S] complexes with Grx3 upon coexpression with one GSH bound per cluster. However, the Grx3-Fra2(H103A) and Grx3-Fra2(H103C) absorption spectra are both significantly different from the WT Grx3-Fra2 heterodimer spectrum in the visible region between 350 and 600 nm that is associated with S → Fe(III) charge transfer transitions (Fig. 2). More dramatic changes are apparent upon comparison of the CD spectra in this region, indicating differences in cluster ligation and/or chirality of the cluster environments in each of the three samples (Fig. 2).
FIGURE 2.
Comparison of the UV-visible absorption and CD spectra of [2Fe-2S] Fra2-Grx3 (black line), [2Fe-2S] Fra2(H103C)-Grx3 (gray line), and [2Fe-2S] Fra2(H103A)-Grx3 (broken line). Spectra were recorded under anaerobic conditions in sealed 0.1-cm cuvettes in 50 mm Tris/MES buffer, pH 8.0. ϵ and Δϵ are based on concentrations of the cluster-bound heterodimer concentrations ([2Fe-2S] Fra2-Grx3 (1.30 mm), [2Fe-2S] Fra2(H103A)-Grx3 (0.17 mm), and [2Fe-2S] Fra2(H103C)-Grx3 complex (0.66 mm)).
Interestingly, we found that a Fra2(C66S/H103A) double mutant did not form an Fe-S cluster complex with Grx3 even though the corresponding single mutants were able to form the Fra2-Grx3 heterodimer. Coexpression of Grx3 with Fra2(C66S/H103A) resulted in purification of the [2Fe-2S] Grx3 homodimer, with analogous spectroscopic signatures to the reconstituted [2Fe-2S] Grx3 homodimer reported previously (9). Fra2(C66S/H103A) was expressed and soluble under these conditions but did not coelute with Grx3. Apparently Fra2 Cys-66 plays a role in Grx3 binding when His-103 is absent even though it does not appear to be a cluster ligand.
Resonance Raman Analysis Suggests That Fra2 His-103 Is an Fe-S Cluster Ligand
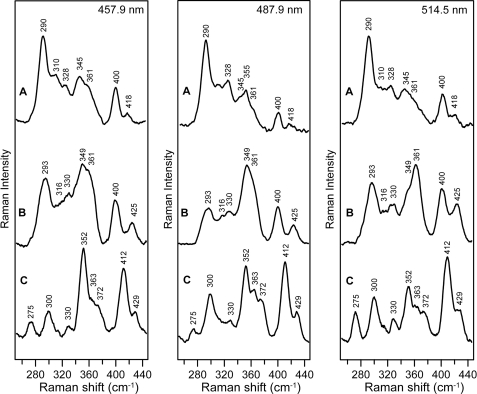
Comparison of the resonance Raman spectra of structurally characterized all-cysteinyl-ligated and mixed cysteinyl- and histidyl-ligated [2Fe-2S]2+ clusters suggests that the presence of the two low frequency modes at 275 and 300 cm−1 for the WT Grx3-Fra2 complex rather than a single broad band between 282 and 302 cm−1 is a hallmark of partial histidyl ligation (9). Hence, the replacement of these bands by a single broad band centered at 290 cm−1 in the Fra2 H103C variant and at 293 cm−1 in the Fra2 H103A variant is consistent with loss of Fra2 His-103 as a cluster ligand (Fig. 3). In the case of the H103A mutant, it is possible that the mutated His residue is replaced by a nearby ligand or by water. In fact, the differences in the vibrational frequencies for the [2Fe-2S]2+ centers in H103C and H103A are indicative of exchanging a Cys ligand in H103C with an oxygenic ligand in H103A based on resonance Raman studies of [2Fe-2S] ferredoxins with single Cys-to-Ser mutations (22). These data therefore suggest that Cys-103 in Fra2(H103C) is a cluster ligand in this mutant. Moreover, the marked differences in the resonance Raman spectra of the [2Fe-2S]2+ centers in samples with Fra2 H103A and H103C compared with that of the cluster-bound Grx3 homodimer (9) indicate that the heterodimeric complex stays intact even though His-103 is replaced as a cluster ligand. In summary, the resonance Raman data demonstrate that His-103 is clearly a ligand in the WT Grx3-Fra2 complex and that the [2Fe-2S] cluster-bound heterodimeric Grx3-Fra2 complex remains intact even when His-103 is mutated to Ala or Cys.
FIGURE 3.
Comparison of the resonance Raman spectra of [2Fe-2S] Fra2(H103C)-Grx3 (A), [2Fe-2S] Fra2(H103A)-Grx3 (B), and [2Fe-2S] Fra2-Grx3 (C) obtained with 457. 9-, 487.9-, and 514.5-nm laser excitation. Samples were ∼2 mm in the [2Fe-2S] cluster and were in the form of a frozen droplet at 17 K. Each spectrum is the sum of 100 scans, with each scan involving photon counting for 1 s at 0.5 cm−1 increments with 6 cm−1 spectral resolution. Bands due to lattice modes of ice have been subtracted from all spectra.
EPR and UV-visible Absorption Studies Indicate That Replacement of Fra2 His-103 Destabilizes the [2Fe-2S]+ Cluster in Fra2-Grx3
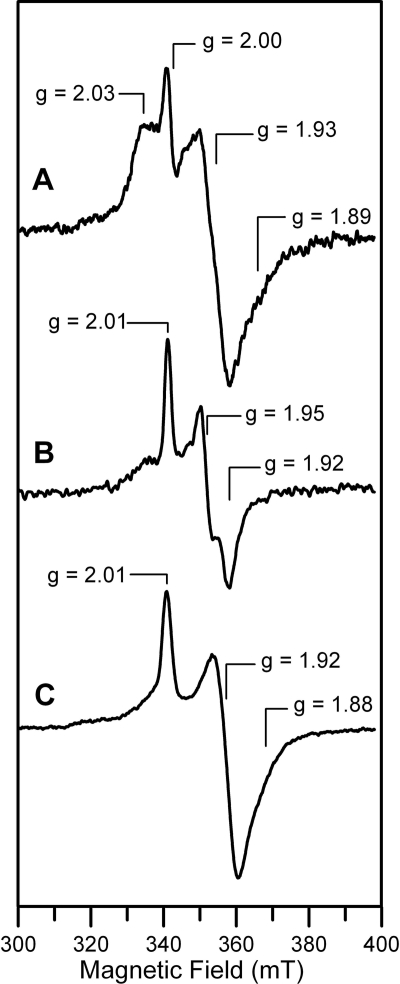
The redox properties and stability of the reduced [2Fe-2S]+ clusters in Fra2 His-103 mutant complexes were investigated by EPR and UV-visible absorption spectroscopy. As demonstrated previously (9), the [2Fe-2S]2+ center in the WT Fra2-Grx3 heterodimer is quantitatively and reversibly reduced by a 2-fold excess of dithionite to yield a stable S = 1/2 [2Fe-2S]+ center that exhibits a rhombic EPR signal, g = 2.01, 1.92, and ∼1.87 (gav ∼1.93), accounting for 1.0 spin per [2Fe-2S] cluster (Fig. 4). In contrast, the [2Fe-2S]2+ centers in the Grx3-Fra2(H103A) and Grx3-Fra2(H103C) heterodimers are relatively unstable to reduction. Both readily degrade in the presence of excess of dithionite as evidenced by irreversible bleaching of the UV-visible absorption spectra and the absence of a EPR signal corresponding to a S = 1/2 [2Fe-2S]+ center. However, addition of a 2-fold excess of dithionite coupled with rapid freezing results in weak rhombic EPR signals from metastable S = 1/2 [2Fe-2S]+ centers in both mutant complexes as follows: g = 2.03, 1.93, and ∼1.89 (gav ∼ 1.95), accounting for 0.3 spin per [2Fe-2S] cluster for Grx3-Fra2(H103C); g = 2.01, 1.95, and 1.92 (gav ∼1.96), accounting for 0.2 spin per [2Fe-2S] cluster for Grx3-Fra2(H103A) (Fig. 4). All three of the Fra2-Grx3 complexes have distinct S = 1/2 [2Fe-2S]+ EPR signals suggesting different cluster ligation in each case. Moreover, all three EPR signals are distinct from that of the metastable axial S = 1/2 [2Fe-2S]+ centers in homodimeric Grx3, g = 2.03, 1.94, and 1.94 (gav ∼1.96) accounting for 0.2 spin per [2Fe-2S] cluster, which is most likely ligated by the two active site (CGFS) Grx3 cysteines and two glutathione cysteines, based on the structure of the oxidized [2Fe-2S]2+ center in monothiol Grxs (12).
FIGURE 4.
Comparison of the X-band EPR spectra of dithionite-reduced [2Fe-2S] Fra2(H103C)-Grx3 (A), [2Fe-2S] Fra2(H103A)-Grx3 (B), and [2Fe-2S] Fra2-Grx3 (C). The samples described in Fig. 2 were reduced under anaerobic conditions by addition of stoichiometric sodium dithionite (i.e. a 2-fold excess of reducing equivalents) and frozen immediately in liquid nitrogen. High temperature EPR studies indicate that the derivative signal centered at g = 2.00 in the Fra2(H103C)-Grx3 spectrum originates from a radical degradation product of dithionite. EPR conditions are as follows: microwave frequency, 9.60 GHz; modulation frequency, 100 kHz; modulation amplitude, 0.65 millitesla; microwave power, 20 milliwatt; temperature, 26 K. Spin quantification of EPR signal indicate 1.0 spin per [2Fe-2S] cluster for Fra2-Grx3, 0.2 spin per [2Fe-2S] cluster for Fra2(H103A)-Grx3, and 0.3 spin per [2Fe-2S] cluster for Fra2(H103C)-Grx3.
Although the differences in the EPR signals for WT Grx3-Fra2, Grx3-Fra2(H103A), Grx3-Fra2(H103C), and homodimeric Grx3 suggest different cluster ligation for each of the four [2Fe-2S]+ centers, the EPR results do not provide significant insight into the nature of the fourth ligand in the Grx3-Fra2 complex. In general, the g value anisotropy and gav values of S = 1/2 [2Fe-2S]+ centers are largely determined by the nature of the ligands and the geometry at the localized valence Fe(II) site (23–26). Replacement of a cysteinate ligand by a histidine or an oxygenic (aspartate, serinate, or hydroxide) ligand is expected to change the site potential at the ligated iron atom, resulting in an increase for histidine and a decrease for oxygenic ligands (25, 27). Hence, the histidine-ligated iron site is generally expected to be the reducible site, and the decrease in the gav values from ∼1.97 in homodimeric Grx3 to ∼1.93 in WT Grx3-Fra2 and to ∼1.90 in Rieske proteins is in accord with changing the ligation at the reducible iron site from two cysteinates to one histidine and one cysteinate and to two histidines, respectively (9). In contrast, changing a histidine ligand to an oxygenic ligand in Grx3-Fra2(H103A) or to a cysteinate ligand in Grx3-Fra2(H103C) has the potential to change the reducible iron site and thereby minimize the effect of the change of ligation on the g value anisotropy and gav value compared with homodimeric Grx3. More detailed analysis of the differences in the EPR spectra in WT and mutant Grx3-Fra2 complexes is not possible without knowledge of the nature of the fourth ligand. However, the EPR results for the [2Fe-2S]+ centers are consistent with the resonance Raman data for the [2Fe-2S]2+ centers in suggesting that histidine is replaced as a cluster ligand by cysteine in Grx3-Fra2(H103C) and an oxygenic ligand in Grx3-Fra2(H103A). Moreover, the EPR and UV-visible absorption data for the WT and mutant Grx3-Fra2 complexes clearly demonstrate that ligation by Fra2 His-103 is required to obtain a stable redox-active [2Fe-2S]2+,+ center, a result that may have functional significance in terms of the regulation mechanism.
Grx3-Fra2 H103 Mutant Complexes Have Longer Fe-S Cluster Bonds
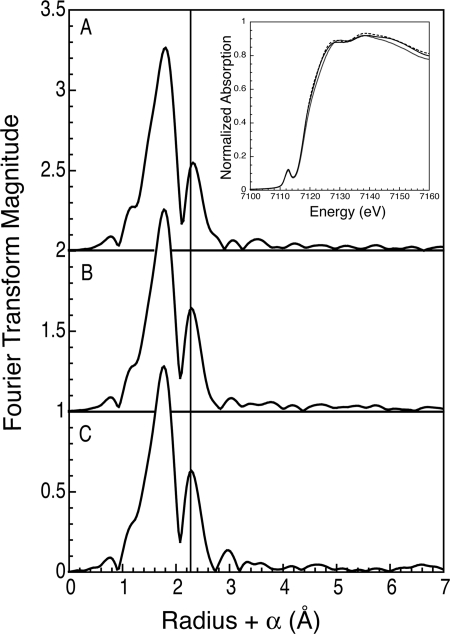
X-ray absorption spectroscopy was used to further probe the [2Fe-2S] Fra2-Grx3 complex and evaluate the effects of Fra2 mutations on the cluster coordination. The extended x-ray absorption fine structure (EXAFS) data recently reported for the WT Fra2-Grx3 Fe-S complex was best fit with a mixture of low Z (oxygen or nitrogen) and sulfur ligands with Fe-Fe and Fe-S distances of 2.67 and 2.26 Å, respectively (9), which is similar to other published values for Cys- and His-ligated [2Fe-2S] clusters (28, 29). The contribution from the single His in the WT Fra2-Grx3/4 complex is expected to be small because EXAFS reports on the average coordination for both iron atoms in the cluster, and strong scattering by the nearest neighbor sulfur ligands dominates the EXAFS data. Thus, substituting Fra2 His-103 with Cys or Ala did not have dramatic effects on the overall EXAFS or the x-ray absorption near edge spectra (Fig. 5). Nonetheless, small perturbations are observed with both substitutions. Most notably, the Fra2(H103C) mutant has longer refined Fe-Fe and Fe-S distances at 2.70 and 2.28 Å, respectively, which may contribute to the cluster instability of this mutant (Table 1). The error in distance determination with EXAFS is frequently reported to be ± 0.02 Å, in part due to the inaccuracy of the value of ΔE0. However, the error for these structurally similar clusters should be the same in both magnitude and direction for all three samples. Thus, these small changes are reliable indicators of structural perturbations to the cluster resulting from replacing the His-103 ligand. A similar expansion of the cluster core dimensions and increased cluster instability was reported in a Rieske-type [2Fe-2S] ferredoxin upon substitution of a His ligand with a Cys residue (30). The Fra2(H103A) mutant also has a longer Fe-Fe distance, but it is intermediate between the WT and H103C mutant values. In addition, the peaks at high R values (from R + α values between 2.8 and 3.8 Å) for the WT complex, which could be modeled using histidine multiple scattering paths, are less prominent and shifted in both Fra2 mutant complexes. The fits to the first shell for all mutants are quite similar, as one might expect if only one of eight ligands has been modified. All require the presence of a low-Z ligand for the best fit. The difficulty in accurately determining the correct number of nitrogens in the presence of the strongly scattering sulfurs has been previously noted (31). Indeed, the fit minimum is shallow and broad as a function of nitrogen coordination number, varying from 0.5 to 2 depending on the number and type of other scatterers included in the fit. If the nitrogen contribution (10–20%) to the scattering is ignored and the coordination number for both the iron and sulfur shells are allowed to freely refine, the H103C variant refines with the largest S:Fe ratio (Table 1). This minimal fit nicely models the data (supplemental Fig. S4). Finally, small changes in the x-ray absorption near edge spectra can be observed as the His-103 ligand is modified (Fig. 5, inset). Overall, the changes in the x-ray absorption data for the Fra2-His-103 mutants further support the assignment of His-103 as a cluster ligand for the WT complex.
FIGURE 5.
Edges and Fourier transforms of the x-ray absorption data for [2Fe-2S] Fra2-Grx3(H103C) (A), [2Fe-2S] Fra2(H103A)-Grx3 (B), and [2Fe-2S] Fra2-Grx3 (C). The data were transformed over a k range from 1 to 14 Å−1. The vertical line draws attention to small structural perturbations in the Fe-Fe distance resulting in the exchange of the His-103 ligand to cysteine or alanine. The inset shows the edges with the H103C mutant (black line), the H103A mutant (broken line), and the WT (gray line).
TABLE 1.
EXAFS fitting parameters for Grx3-Fra2 H103 mutant heterodimers
Fits B and F allowed the sulfur and iron coordination number to vary; otherwise, only R and σ were varied for each scattering shell. Fits are to unfiltered data over a k range of 1–14 Å−1. Scale factor and ΔE0 were 0.9 and −10 eV, respectively. EXAFS data for fits D, B, H, and F are shown in supplemental Figs. S1–S4. CN, coordination number; R, metal-scatterer distance; σ2, mean square variation in R.
| Fit | Atom | CN | R (Å) | σ2 × 103 | F′ |
|---|---|---|---|---|---|
| Fra2(H103A)-Grx3 | |||||
| A | S | 4 | 2.26 | 7.7 | 0.487 |
| Fe | 1 | 2.69 | 3.9 | ||
| B | S | 3.25 | 2.26 | 6.2 | 0.446 |
| Fe | 1.01 | 2.69 | 4.3 | ||
| C | S | 2 | 2.27 | 4.5 | 0.184 |
| N | 2 | 2.07 | 4.5 | ||
| Fe | 1 | 2.69 | 4.5 | ||
| D | S | 3 | 2.27 | 6.9 | 0.164 |
| N | 1 | 2.09 | 1.7 | ||
| Fe | 1 | 2.69 | 3.1 | ||
| C | 2 | 2.88 | 4.5 | ||
| Fra2(H103C)-Grx3 | |||||
| E | S | 4 | 2.28 | 7.5 | 0.363 |
| Fe | 1 | 2.71 | 4.7 | ||
| F | S | 3.88 | 2.28 | 7.4 | 0.361 |
| Fe | 0.82 | 2.70 | 3.8 | ||
| G | S | 2 | 2.29 | 2.8 | 0.211 |
| N | 2 | 2.07 | 6.1 | ||
| Fe | 1 | 2.70 | 4.9 | ||
| H | S | 3 | 2.28 | 6.1 | 0.181 |
| N | 1 | 2.07 | 2.4 | ||
| Fe | 1 | 2.70 | 4.3 | ||
| C | 2 | 2.91 | 7.7 | ||
His-103 in Fra2 Is Required for in Vivo Iron Signaling
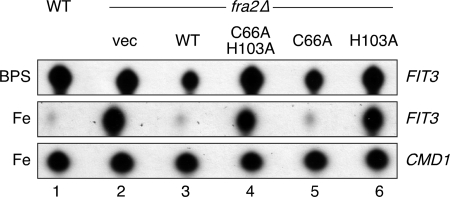
To test whether His-103 is important for the in vivo function of Fra2, the H103A mutant was expressed in yeast lacking the chromosomal FRA2 gene and tested for its effect on transcription of the Aft1-regulated gene FIT3, which encodes a cell wall protein involved in siderophore iron uptake (32). RNA levels of FIT3 were measured using the S1 nuclease protein assay (Fig. 6), and expression of the plasmid-encoded Fra2 mutants in yeast was confirmed by Western blotting (supplemental Fig. S5). In WT cells, addition of the iron chelator bathophenanthroline disulfonate leads to iron depletion and activation of the iron regulon as described previously (Fig. 6, lane 1) (5, 6). Under iron-replete conditions, FIT3 mRNA levels are diminished due to Fra2-Grx3/4-mediated inhibition of Aft1 transcriptional activation. In a fra2Δ deletion mutant (Fig. 6, lane 2), iron-dependent inhibition is lost, and the iron regulon is constitutively activated (5). The addition of vector-encoded wild-type Fra2 to the fra2Δ null cells restored iron inhibition of FIT3 expression. In contrast, the Fra2(H103A) mutant protein (Fig. 6, lane 6) failed to support iron inhibition of Aft1 activity, demonstrating that this residue is critical for the in vivo function of Fra2. In contrast, Fra2 Cys-66 is not essential for inhibiting Aft1 activity in iron-replete conditions (Fig. 6, lane 5). These results are consistent with the spectroscopic and mutagenesis evidence for His-103 as a cluster ligand in the complex. Overall, these data strongly suggest that the iron regulon in yeast responds to the cellular iron status by sensing the ability of the Fra2-Grx3/4 complex to assemble a [2Fe-2S] cluster.
FIGURE 6.
His-103 in Fra2 is required for in vivo iron signaling. WT and fra2Δ cells transformed with a low copy plasmid expressing empty vector, WT, or mutant Fra2 were grown in YPD medium in the presence of 0.1 mm bathophenanthroline sulfonate (BPS) or 0.1 mm FeSO4 prior to harvest and mRNA isolation. FIT3 mRNA was quantified by the S1 nuclease protection assay as described under “Experimental Procedures.” Calmodulin (CMD1) is used as an RNA loading control.
Apo-Fra2 Binds Tightly to [2Fe-2S] Grx3 Forming the [2Fe-2S] Fra2-Grx3 Heterodimer
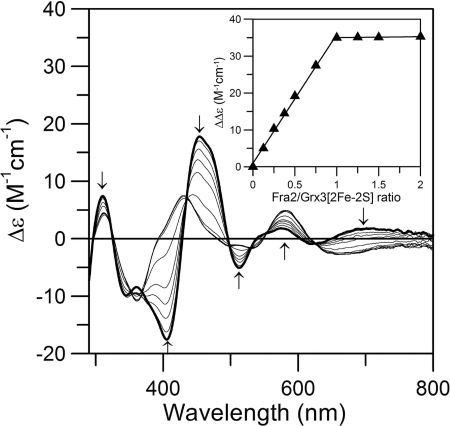
To determine the relative binding affinity and stoichiometry of Fra2 and Grx3 in the presence of the [2Fe-2S] cluster, we monitored changes in the CD spectrum of the [2Fe-2S] Grx3 homodimer upon titration with apo-Fra2 under anaerobic conditions. The homodimeric and heterodimeric [2Fe-2S] complexes have very different CD spectra (9) providing a convenient handle for monitoring changes in cluster ligation upon Fra2 binding. As shown in Fig. 7, increasing concentrations of Fra2 lead to conversion of the [2Fe-2S] Grx3 homodimer CD spectrum to the [2Fe-2S] Fra2-Grx3 heterodimer CD spectrum (compare with the spectrum for as-purified Fra2-Grx3 shown in Fig. 2). The titration experiment clearly demonstrates that ∼1 mol eq of apo-Fra2 binds tightly to the Grx3 homodimer, replacing one Grx3 monomer and forming the [2Fe-2S] Fra2-Grx3 heterodimer in vitro. Formation of the heterodimer and displacement of Grx3 was confirmed by gel filtration analysis (data not shown). Thus, formation of the [2Fe-2S]-bridged heterodimer is thermodynamically and kinetically favored over the [2Fe-2S]-bridged homodimer under these experimental conditions.
FIGURE 7.
Titration studies of [2Fe-2S] Grx3 with WT apo-Fra2 monitored by UV-visible CD spectroscopy. [2Fe-2S]2+ cluster-bound Grx3 (50 μm in [2Fe-2S]2+ clusters, thick line) was titrated with a 0.125–10-fold excess of Fra2 (thin lines). The arrows at selected wavelengths indicate the direction of intensity change with increasing Fra2 concentration. Δϵ values are based on the [2Fe-2S]2+ cluster concentrations. The inset shows the maximum difference in CD intensity (at 463 and 403 nm) as a function of the Fra2:Grx3 [2Fe-2S]2+ cluster ratio.
Mutation of Fra2 His-103 Does Not Abolish Fra2 Binding to [2Fe-2S] Grx3
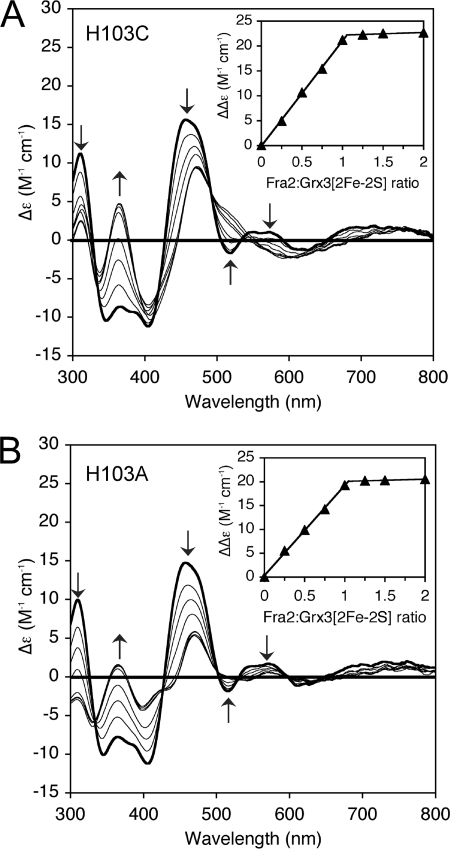
We next tested how mutation of Fra2 His-103 influenced the binding of apo-Fra2 to [2Fe-2S] cluster-bound Grx3 homodimer. Apo-Fra2 H103A and H103C mutants were titrated with [2Fe-2S] Grx3, and cluster coordination changes and binding stoichiometry were monitored by UV-visible CD spectroscopy as described above for WT Fra2 (Fig. 8). Both H103A and H103C Fra2 mutants bound tightly to [2Fe-2S] Grx3 leading to conversion of the homodimer CD spectrum to the heterodimer spectrum for [2Fe-2S] Fra2(H103A)-Grx3 or [2Fe-2S] Fra2(H103C)-Grx3, respectively (compare with as-purified heterodimer spectra shown in Fig. 2). The binding stoichiometry was similar to WT Fra2 because the CD spectral changes reached saturation with the addition of ∼1 mol eq of H103C or H103A Fra2 per [2Fe-2S] cluster (Fig. 8, A and B, inset). Thus, mutation of Fra2 His-103 does not interfere in the tight binding of apo-Fra2 to [2Fe-2S] Grx3. Because mutation of this residue does abolish iron-dependent inhibition of Aft1 in vivo, it is possible that Fra2 His-103 may be critical for a different step in the iron signaling pathway.
FIGURE 8.
Titration studies of [2Fe-2S] Grx3 with H103C and H013A apo-Fra2 monitored by UV-visible CD spectroscopy. [2Fe-2S]2+ cluster-bound Grx3 (200 μm in [2Fe-2S]2+ clusters, thick line) was titrated with a 0.25–2-fold excess of H103C Fra2 (A) or H103A Fra2 (B) (thin lines). The insets show the maximum difference in CD intensity (at 458 and 365 nm) as a function of the Fra2:Grx3 [2Fe-2S]2+ cluster ratio.
DISCUSSION
The cytosolic proteins Fra2 and Grx3/4 relay cellular iron status to the transcriptional activators Aft1 and Aft2 via an unknown mechanism (5–7). We sought to characterize the physical and functional interactions between Fra2 and Grx3/4 to ultimately uncover this mechanism for sensing and regulating intracellular iron levels in yeast. Our previous results demonstrated that these proteins form a novel [2Fe-2S]-bridged heterodimeric complex with the Fe-S cluster ligated at the heterodimer interface by a conserved Cys in the CGFS active site motif of Grx3/4 and a Cys from GSH. The identities of the other ligands were unclear; however, the spectroscopic evidence indicated that the Fe-S cluster is ligated by one histidine, presumably provided by Fra2 (9). Using site-directed mutagenesis coupled with biochemical and spectroscopic characterization, we confirm here that His-103 in Fra2 is an Fe-S cluster ligand in the Fra2-Grx3 complex. Mutation of this residue causes a change in the Fe-S cluster coordination that results in destabilization of the Fe-S cluster. Furthermore, in vivo studies demonstrate that replacement of His-103 in Fra2 disrupts iron-dependent inhibition of Aft1 activity, suggesting that the histidine coordination and the resultant stability of the Fe-S cluster in the Fra2-Grx3/4 complex are critical for its in vivo role in iron regulation.
Despite identification of the two Cys ligands provided by GSH and Grx3, and the His ligand from Fra2, the nature of the fourth ligand in the [2Fe-2S] Fra2-Grx3/4 heterodimer is still unclear. Our previously published results ruled out an additional His ligand, a second GSH molecule, and an additional Cys residue provided by Grx3/4 (9). Cys-66 in Fra2 was a likely candidate tested in this study, yet mutation of this residue has no effect on Fra2-Grx3 heterodimer formation or the in vivo function of Fra2. Because Grx3/4 interacts with both Aft1 and the Fra1-Fra2 complex in vivo (5, 6), it is possible that Aft1 or Fra1 provides the fourth cluster ligand under physiological conditions. In the absence of these interacting proteins, water or another residue from Grx3/4 or Fra2 may occupy this ligation site. Additional biochemical and structural studies will be required to resolve this issue.
Given the data presented in this study, why is His-103 important for the regulatory function of [2Fe-2S] Fra2-Grx3/4? Interestingly, our results demonstrate that replacement of the conserved His ligand in Fra2 does not prohibit Fe-S cluster binding to Fra2-Grx3 or prevent tight binding of apo-Fra2 with [2Fe-2S] Grx3 homodimers. However, unlike the WT heterodimer, the [2Fe-2S] in Grx3-Fra2(H103A/C) heterodimers is rapidly degraded upon reduction with dithionite indicating that the Fe-S cluster stability is compromised in these mutants complexes. Furthermore, the EXAFS analysis presented in this work suggests that cluster destabilization results from expansion of the Fe-S cluster dimensions for Grx3-Fra2(H103A/C) mutants. These results thus suggest that a stable and/or redox-active cluster may be important for the in vivo function of Fra2-Grx3/4. These results also lead to the question of how [2Fe-2S] Fra2-Grx3/4 influences the transcriptional activity of Aft1/2. As mentioned earlier, the Fra-Grx signaling pathway is proposed to induce oligomerization of Aft1 (and presumably Aft2) under iron-replete conditions (5, 33). This conformational change, in turn, may promote export of Aft1/2 from the nucleus or prevent interaction of Aft1/2 with the target DNA. Although genetic and biochemical evidence demonstrates that the interactions between Fra2-Grx3/4 and Fra1-Fra2 are not strictly iron-dependent (5, 9), yeast two-hybrid and coimmunoprecipitation studies indicate that the physical interactions between Grx3/4-Aft1 and Aft1-Aft1 are dependent on the Fe-S binding Cys of Grx3/4 and the conserved Cys-Xaa-Cys motif of Aft1 (6, 33). Taken together, these observations strongly suggest a role for redox chemistry or metal binding in control of Aft1 function. For example, [2Fe-2S] Fra2-Grx3/4 may catalyze redox modification of the Cys-Xaa-Cys motif or transfer iron or an Fe-S cluster to Aft1/2. If the Fra2-Grx3/4 heterodimer has redox activity, replacement of the His ligand is likely to significantly alter the redox potential and pH dependence of the Fe-S cluster as demonstrated for other His-ligated [2Fe-2S] cluster proteins (30, 34). Alternatively, the histidine-ligated [2Fe-2S] cluster may allow the Fra2-Grx3/4 complex to adopt a conformation that facilitates stable complex formation with Aft1/2 or other components of the iron-sensing pathway (e.g. Fra1), which ultimately inhibits Aft1/2 transcriptional activity. Unraveling the nature of the molecular interactions between Fra2, Grx3/4, Fra1, and Aft1/2 and their effect on the structure, subcellular localization, and DNA binding activity of Aft1/2 will be key in understanding the mechanism of iron regulation in yeast.
Because the Grx3/4 and Fra2 residues required for iron-dependent control of Aft1 activity are specifically required for binding the Fe-S cluster in the Fra2-Grx3 heterodimer, these results provide strong evidence that the iron regulon in yeast is controlled by the ability of the Fra2-Grx3/4 complex to assemble a stable histidine-ligated [2Fe-2S] cluster. How is the [2Fe-2S] Fra2-Grx3/4 complex formed in vivo? Previous genetic studies demonstrate that control of the yeast iron regulon is dependent on mitochondrial Fe-S cluster biogenesis rather than cytosolic iron levels (8). The Fe-S-dependent signal emanating from the mitochondria is proposed to be an unknown sulfur-containing species exported by the mitochondrial ABC transporter Atm1. Whether or not this species is involved in assembly of a [2Fe-2S] cluster on Fra2-Grx3/4 heterodimers or Grx3/4 homodimers is unclear. However, the available evidence demonstrates that the cytosolic Fe-S cluster assembly (CIA) machinery is not required for iron signaling (8), suggesting that a parallel pathway exists for assembly of Fe-S clusters on Fra2-Grx3/4 heterodimers and Grx3/4 homodimers.
It is interesting to note that the phenotypes for grx3Δgrx4Δ mutants are significantly different from fra2Δ mutants. Although both mutant strains exhibit misregulation of the iron regulon, grx3Δgrx4 mutants are sensitive to peroxides, accumulate high levels of intracellular iron, and grow poorly on synthetic medium (6, 7), whereas fra2Δ mutants display no detectable growth defect (5). These results suggest that the cytosolic CGFS-type Grxs play an additional role beyond iron sensing and regulation. Chloroplast and mitochondrial monothiol Grxs (Grx5 in yeast) are proposed to be involved in assembly, storage, and/or delivery of [2Fe-2S] clusters (10, 35–37). Although this function has not been established for cytosolic Grxs, yeast Grx3 and Grx4 can partially rescue the Fe-S cluster biogenesis and growth defects of yeast grx5Δ mutants when artificially targeted to the mitochondria, suggesting a conservation of function among proteins in the CGFS-type Grx family (38). If yeast Grx3 and Grx4 do play a role in cytosolic [2Fe-2S] cluster biogenesis or trafficking, the levels of [2Fe-2S] cluster-bound Grx3/4 can be assessed by interaction with Fra2 without perturbing Grx3/4 function provided that Fra2 can replace one of the Grx molecules in the homodimer with high binding affinity and the cellular concentration of Fra2 is much less than that of Grx3 or Grx4. In fact, the absolute protein abundances of Grx3 (11,000 molecules/cell) and Grx4 (7800 molecules/cell) determined in a proteome-wide study are significantly higher than Fra2 (2050 molecules/cell) (39). Hence, the tight and stoichiometric binding of Fra2 to holo-Grx3 to form the Fra2-Grx3 heterodimeric complex that we present here provides a plausible hypothesis for an initial step in the iron or Fe-S cluster sensing mechanism that controls the yeast iron regulon. Changes in the protein levels of Grx3/4 and Fra2 under iron-deplete versus iron-replete conditions may also provide an additional level of control; however, previous results suggest that Fra2, Grx3, and Grx4 protein levels show little variation in response to cytosolic iron levels (5). Although the control of the iron regulon via Aft1/2 is unique to yeast, similar types of regulation based on iron or Fe-S cluster sensing are likely to be found in other organisms based on the identification of BolA-type proteins as physiological partners of monothiol Grxs in other organisms (40–42).
Supplementary Material
Acknowledgments
We thank F. Wayne Outten (University of South Carolina) for helpful discussions and Jerry Kaplan (University of Utah) for Fra2 antibodies. The Stanford Synchrotron Radiation Laboratory is a national user facility supported by the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by Department of Energy and the National Institutes of Health-NCRR Biomedical Technology Program.
This work was supported, in whole or in part, by National Institutes of Health Grants ES13780 (to C. E. O.), GM62524 (to M. K. J.), ES03817 (to D. R. W.), and P20 RR016461 (P. J. R.-G.). This work was also supported by The Camille and Henry Dreyfus Foundation Scholar Award (to P. J. R.-G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- Grx
- glutaredoxin
- EXAFS
- extended x-ray absorption fine structure.
REFERENCES
- 1. Yamaguchi-Iwai Y., Dancis A., Klausner R. D. (1995) EMBO J. 14, 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rutherford J. C., Jaron S., Ray E., Brown P. O., Winge D. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14322–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blaiseau P. L., Lesuisse E., Camadro J. M. (2001) J. Biol. Chem. 276, 34221–34226 [DOI] [PubMed] [Google Scholar]
- 4. Yamaguchi-Iwai Y., Ueta R., Fukunaka A., Sasaki R. (2002) J. Biol. Chem. 277, 18914–18918 [DOI] [PubMed] [Google Scholar]
- 5. Kumánovics A., Chen O. S., Li L., Bagley D., Adkins E. M., Lin H., Dingra N. N., Outten C. E., Keller G., Winge D., Ward D. M., Kaplan J. (2008) J. Biol. Chem. 283, 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ojeda L., Keller G., Muhlenhoff U., Rutherford J. C., Lill R., Winge D. R. (2006) J. Biol. Chem. 281, 17661–17669 [DOI] [PubMed] [Google Scholar]
- 7. Pujol-Carrion N., Belli G., Herrero E., Nogues A., de la Torre-Ruiz M. A. (2006) J. Cell Sci. 119, 4554–4564 [DOI] [PubMed] [Google Scholar]
- 8. Rutherford J. C., Ojeda L., Balk J., Mühlenhoff U., Lill R., Winge D. R. (2005) J. Biol. Chem. 280, 10135–10140 [DOI] [PubMed] [Google Scholar]
- 9. Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., Hoffman B. M., Riggs-Gelasco P. J., Huynh B. H., Johnson M. K., Outten C. E. (2009) Biochemistry 48, 9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bandyopadhyay S., Gama F., Molina-Navarro M. M., Gualberto J. M., Claxton R., Naik S. G., Huynh B. H., Herrero E., Jacquot J. P., Johnson M. K., Rouhier N. (2008) EMBO J. 27, 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Picciocchi A., Saguez C., Boussac A., Cassier-Chauvat C., Chauvat F. (2007) Biochemistry 46, 15018–15026 [DOI] [PubMed] [Google Scholar]
- 12. Iwema T., Picciocchi A., Traore D. A., Ferrer J. L., Chauvat F., Jacquamet L. (2009) Biochemistry 48, 6041–6043 [DOI] [PubMed] [Google Scholar]
- 13. Gibson L. M., Dingra N. N., Outten C. E., Lebioda L. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riemer J., Hoepken H. H., Czerwinska H., Robinson S. R., Dringen R. (2004) Anal. Biochem. 331, 370–375 [DOI] [PubMed] [Google Scholar]
- 15. Beinert H. (1983) Anal. Biochem. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 16. Broderick J. B., Henshaw T. F., Cheek J., Wojtuszewski K., Smith S. R., Trojan M. R., McGhan R. M., Kopf A., Kibbey M., Broderick W. E. (2000) Biochem. Biophys. Res. Commun. 269, 451–456 [DOI] [PubMed] [Google Scholar]
- 17. Outten C. E., Culotta V. C. (2004) J. Biol. Chem. 279, 7785–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cosper M. M., Jameson G. N., Hernández H. L., Krebs C., Huynh B. H., Johnson M. K. (2004) Biochemistry 43, 2007–2021 [DOI] [PubMed] [Google Scholar]
- 19. Dohrmann P. R., Butler G., Tamai K., Dorland S., Greene J. R., Thiele D. J., Stillman D. J. (1992) Genes Dev. 6, 93–104 [DOI] [PubMed] [Google Scholar]
- 20. Chin K. H., Lin F. Y., Hu Y. C., Sze K. H., Lyu P. C., Chou S. H. (2005) J. Biomol. NMR 31, 167–172 [DOI] [PubMed] [Google Scholar]
- 21. Kasai T., Inoue M., Koshiba S., Yabuki T., Aoki M., Nunokawa E., Seki E., Matsuda T., Matsuda N., Tomo Y., Shirouzu M., Terada T., Obayashi N., Hamana H., Shinya N., Tatsuguchi A., Yasuda S., Yoshida M., Hirota H., Matsuo Y., Tani K., Suzuki H., Arakawa T., Carninci P., Kawai J., Hayashizaki Y., Kigawa T., Yokoyama S. (2004) Protein Sci. 13, 545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer J., Fujinaga J., Gaillard J., Lutz M. (1994) Biochemistry 33, 13642–13650 [DOI] [PubMed] [Google Scholar]
- 23. Bertrand P., Gayda J. P. (1979) Biochim. Biophys. Acta 579, 107–121 [DOI] [PubMed] [Google Scholar]
- 24. Bertrand P., Gayda J. P. (1980) Biochim. Biophys. Acta 625, 337–342 [DOI] [PubMed] [Google Scholar]
- 25. Werth M. T., Cecchini G., Manodori A., Ackrell B. A., Schröder I., Gunsalus R. P., Johnson M. K. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8965–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Werth M. T., Sices H., Cecchini G., Schröder I., Lasage S., Gunsalus R. P., Johnson M. K. (1992) FEBS Lett. 299, 1–4 [DOI] [PubMed] [Google Scholar]
- 27. Link T. A. (1999) Adv. Inorg. Chem. 47, 83–157 [Google Scholar]
- 28. Cosper N. J., Eby D. M., Kounosu A., Kurosawa N., Neidle E. L., Kurtz D. M., Jr., Iwasaki T., Scott R. A. (2002) Protein Sci. 11, 2969–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsang H. T., Batie C. J., Ballou D. P., Penner-Hahn J. E. (1989) Biochemistry 28, 7233–7240 [DOI] [PubMed] [Google Scholar]
- 30. Kounosu A., Li Z., Cosper N. J., Shokes J. E., Scott R. A., Imai T., Urushiyama A., Iwasaki T. (2004) J. Biol. Chem. 279, 12519–12528 [DOI] [PubMed] [Google Scholar]
- 31. Clark-Baldwin K., Tierney D. L., Govindaswamy N., Gruff E. S., Kim C., Berg J., Koch S. A., Penner-Hahn J. E. (1998) J. Am. Chem. Soc. 120, 8401–8409 [Google Scholar]
- 32. Protchenko O., Ferea T., Rashford J., Tiedeman J., Brown P. O., Botstein D., Philpott C. C. (2001) J. Biol. Chem. 276, 49244–49250 [DOI] [PubMed] [Google Scholar]
- 33. Ueta R., Fujiwara N., Iwai K., Yamaguchi-Iwai Y. (2007) Mol. Biol. Cell 18, 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bak D. W., Zuris J. A., Paddock M. L., Jennings P. A., Elliott S. J. (2009) Biochemistry 48, 10193–10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mühlenhoff U., Gerber J., Richhardt N., Lill R. (2003) EMBO J. 22, 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim K. D., Chung W. H., Kim H. J., Lee K. C., Roe J. H. (2010) Biochem. Biophys. Res. Commun. 392, 467–472 [DOI] [PubMed] [Google Scholar]
- 37. Ye H., Jeong S. Y., Ghosh M. C., Kovtunovych G., Silvestri L., Ortillo D., Uchida N., Tisdale J., Camaschella C., Rouault T. A. (2010) J. Clin. Invest. 120, 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molina M. M., Bellí G., de la Torre M. A., Rodríguez-Manzaneque M. T., Herrero E. (2004) J. Biol. Chem. 279, 51923–51930 [DOI] [PubMed] [Google Scholar]
- 39. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 40. Couturier J., Jacquot J. P., Rouhier N. (2009) Cell. Mol. Life Sci. 66, 2539–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huynen M. A., Spronk C. A., Gabaldón T., Snel B. (2005) FEBS Lett. 579, 591–596 [DOI] [PubMed] [Google Scholar]
- 42. Rouhier N., Couturier J., Johnson M. K., Jacquot J. P. (2010) Trends Biochem. Sci. 35, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lambert C., Léonard N., De Bolle X., Depiereux E. (2002) Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.