Abstract
To prevent photo-oxidative damage to the photosynthetic membrane in strong light, plants dissipate excess absorbed light energy as heat in a mechanism known as non-photochemical quenching (NPQ). NPQ is triggered by the trans-membrane proton gradient (ΔpH), which causes the protonation of the photosystem II light-harvesting antenna (LHCII) and the PsbS protein, as well as the de-epoxidation of the xanthophyll violaxanthin to zeaxanthin. The combination of these factors brings about formation of dissipative pigment interactions that quench the excess energy. The formation of NPQ is associated with certain absorption changes that have been suggested to reflect a conformational change in LHCII brought about by its protonation. The light-minus-dark recovery absorption difference spectrum is characterized by a series of positive and negative bands, the best known of which is ΔA535. Light-minus-dark recovery resonance Raman difference spectra performed at the wavelength of the absorption change of interest allows identification of the pigment responsible from its unique vibrational signature. Using this technique, the origin of ΔA535 was previously shown to be a subpopulation of red-shifted zeaxanthin molecules. In the absence of zeaxanthin (and antheraxanthin), a proportion of NPQ remains, and the ΔA535 change is blue-shifted to 525 nm (ΔA525). Using resonance Raman spectroscopy, it is shown that the ΔA525 absorption change in Arabidopsis leaves lacking zeaxanthin belongs to a red-shifted subpopulation of violaxanthin molecules formed during NPQ. The presence of the same ΔA535 and ΔA525 Raman signatures in vitro in aggregated LHCII, containing zeaxanthin and violaxanthin, respectively, leads to a new proposal for the origin of the xanthophyll red shifts associated with NPQ.
Keywords: Carotenoid, Chloroplast, Fluorescence, Photosynthetic Pigments, Raman Spectroscopy
Introduction
To ensure the efficiency of photosynthesis, even under low light conditions, the photochemically active Chl3 of the photosystem II (PSII) reaction center are served by additional antenna Chl bound to light-harvesting complexes (LHCII) (1). However, under certain environmental conditions, the amount of light absorbed by the LHCs is in excess of that which can be used in photochemistry. Left unchecked, the excess absorbed light energy can lead to the formation of Chl triplet states in the PSII reaction center that sensitize the production of singlet oxygen (2–4). Singlet oxygen damages the PSII reaction center and other components of the photosynthetic membrane, leading to a sustained decrease in photosynthetic efficiency that is known as photoinhibition (2). To mitigate this, plants have evolved a photoprotective mechanism, known as non-photochemical quenching (NPQ), whereby the excess excitation energy is safely dissipated as heat (4, 5). The major component of NPQ is controlled by the level of the trans-membrane proton gradient (ΔpH) formed as a result of photosynthetic electron transport and is known as qE (6). ΔpH formation triggers the protonation of LHCII (7) and of the PsbS protein (8), as well as activating the violaxanthin de-epoxidase enzyme, which converts the LHCII-bound xanthophyll violaxanthin into zeaxanthin (9). The interaction of these three factors brings about formation of dissipative pigment interactions within the PSII antenna, thus shortening the Chl excited-state lifetime (10, 11).
The precise nature of the quenching pigment interactions responsible for qE remain under debate (12–17). Despite this, many of the photophysical and photochemical features of qE are well characterized experimentally. It has been shown that qE is associated with a series of absorption changes in both the Soret and the Qy regions (18–24). These absorption changes have been suggested to reflect an altered environment of bound Chl and xanthophyll molecules, brought about by a protein conformational change within LHCII as a result of ΔpH formation (22). The light-minus-dark recovery absorption difference spectrum in the Soret region contains a series of three negative bands below 500 nm and a positive band at 535 nm (25). The formation and relaxation kinetics of the latter change known as ΔA535 are often monitored as a collective measure of these conformational changes. ΔA535 was originally believed to arise from selective light scattering brought about by a change in the membrane thickness linked to ΔpH formation (26, 27). Later ΔA535 was shown to depend upon the presence of zeaxanthin and was closely correlated with the formation and relaxation of qE (18–20, 22). In the absence of zeaxanthin, qE is greatly reduced and the ΔA535 change has a much smaller amplitude and is blue-shifted to 525 nm (ΔA525) (28). The npq4 mutant of Arabidopsis that lacks the PsbS protein was found to lack rapidly reversible qE and also lacked any ΔA535 providing a further link between these two phenomena (29). Resonance Raman spectroscopy has proved of use in identifying the origin of absorption bands both in isolated light-harvesting complexes and in intact chloroplasts and even leaves (30). Resonance Raman spectroscopy confirmed that ΔA535 belonged to the 0-0 component of the S2 electronic transition of zeaxanthin (31). The size of the zeaxanthin resonance enhancement in the Raman quenched-minus-unquenched difference spectrum at 528 nm excitation was also found to depend upon PsbS (31). In addition, the negative absorption changes in the qE difference spectrum below 500 nm were associated with a loss of xanthophyll resonance confirming that they also arise from true electronic transitions rather than light scattering (31). Native isolated PsbS was found able to bind zeaxanthin, producing a red shift in its absorption spectrum mimicking ΔA535 (32). The putative binding simultaneously affected protein phenylalanine absorption and circular dichroism (CD), leading to the suggestion that ΔA535 may arise from zeaxanthin binding to this hydrophobic protein (32). Indeed, it was suggested that ΔA535 may represent the formation of a zeaxanthin-PsbS quenching complex responsible for qE (8). However, several observations have brought this proposal into question. Firstly, reconstituted PsbS was consistently found unable to bind any xanthophylls (33, 34). Secondly, PsbS was still found to perform its function in the absence of zeaxanthin (35), and thirdly, the npq4 mutant lacking PsbS was found to possess ΔA535 and wild-type levels of NPQ, albeit both forming on a much longer timescale than the wild-type (36).
Recently, the origin of one of the main negative bands in the qE difference spectrum peaking at 495 nm (ΔA495) was shown to depend upon the presence of lutein (25). In Arabidopsis leaves lacking lutein, the band was shifted to 497 nm (in lut2−, lutein replaced by violaxanthin) and 501 nm (lut2npq2−, lutein replaced by zeaxanthin) (25). Very similar negative absorption changes to those observed in leaves below 500 nm have been observed upon aggregation of isolated LHCII and also in quenched but non-aggregated LHCII, suggesting that both in vivo and in vitro types of quenching may have a common origin (37, 38). However, the characteristic ΔA535 change was not observed upon LHCII aggregation, although this may be due to the absence of zeaxanthin in these samples (37). Thus, the question of the exact origin of all elements of the qE-related absorption changes remains. It is known that the energy of an electronic transition within a molecule is strongly dependent on the refractive index of the solvent environment (39, 40). However, such a strong red shift as seen during the ΔA535 change would require a very dramatic change in the solvent environment, possibly due to the arrival of a static dipole or a point charge in the vicinity of the zeaxanthin (41, 42). Alternatively, this shift in absorption may arise from excitonic interactions between pairs of zeaxanthin molecules (43). These zeaxanthin pairs would occur as a result of aggregation of LHCII trimers within the thylakoid membrane. Because zeaxanthin occupies a relatively peripheral position within the LHCII trimer (44), aggregation could result in close associations between xanthophylls belonging to neighboring trimers. Indeed, it has been shown that the formation of excitonically coupled J-aggregates of zeaxanthin (co-linear or “top-to-tail” chains of molecules) in water-ethanol mixtures gives rise to a red shift in the 0-0 transition to 535 nm (34, 45, 46), whereas lutein, antheraxanthin, and violaxanthin J-type aggregates possess red-shifted bands between 500 and 530 nm (45). In the following study, we specifically address the origin of ΔA525. Firstly, using resonance Raman experiments on intact chloroplasts lacking zeaxanthin, we identify the xanthophyll responsible for ΔA525 as violaxanthin. In addition, we show that the same Raman signature associated with ΔA535 and ΔA525 in vivo is observed upon aggregation of LHCII binding, respectively, zeaxanthin or violaxanthin in the peripheral V1 site.
EXPERIMENTAL PROCEDURES
Fluorescence and Absorption Measurements
Arabidopsis thaliana cv Col0 (wild-type) and two mutants derived from it, npq1 (lacking zeaxanthin) and L17 (PsbS overexpressor) (47), were grown for 8–9 weeks in Sanyo plant growth rooms with an 8-h photoperiod at a light intensity of 100 μmol of photons m−2 s−1 and a day/night temperature of 22/18 °C. The composition of carotenoids was determined by HPLC for leaf disks rapidly frozen in liquid N2 as described previously (48). Leaves were vacuum-infiltrated with 20 mm HEPES buffer (pH 7.0) containing 5 mm dithiothreitol (DTT) to inhibit violaxanthin de-epoxidation, whereas control leaves were vacuum-infiltrated with buffer only. Chl fluorescence kinetic analyses of whole leaves was carried out using a Dual-PAM-100 fluorometer (Walz) using varying actinic intensity together with light saturation pulses (4000 μmol of photons m−2 s−1) as indicated in the figures. The maximum quantum yield of PSII (Fv/Fm) was defined as ((Fm − Fo)/Fm), the quantum yield of PSII (ΦPSII) as ((Fm′ − Fs)/Fm′), qP as ((Fm′ − Fs)/(Fm′ − Fo′)), and NPQ as ((Fm − Fm′)/Fm′). qE-related absorption changes were recorded on whole leaves using an Aminco DW2000 spectrophotometer as described previously (25), and difference spectra were calculated by subtracting the “dark recovery” spectrum (5 min of preillumination at 1000 μmol of photons m−2 s−1 actinic light followed by 5 min of dark relaxation) from the “light” spectrum recorded in the presence of the actinic light following 5 min of preillumination.
LHCII Isolation
Spinach trimeric LHCII binding either violaxanthin or zeaxanthin was isolated from n-dodecyl β-d-maltoside solubilized spinach BBY membranes using sucrose gradient ultracentrifugation as described previously (49). De-epoxidation of violaxanthin to antheraxanthin and zeaxanthin was achieved by incubating the thylakoids (from which BBYs were derived) for 30 min in 0.33 m sorbitol, 1 mm EDTA, 30 mm HEPES, 20 mm MES, 40 mm ascorbate at pH 5.5 at 20 °C. LHCII was desalted to remove sucrose in a PD10 desalting column (GE Healthcare) in a buffer containing 20 mm HEPES (pH 7.8) and 0.03% (w/v) n-dodecyl β-d-maltoside. Quenched LHCII was prepared by removal of detergent by SM-2 bioabsorbent beads (Bio-Rad) allowing for a 10× reduction in fluorescence yield as determined by a PAM-101 fluorometer (Heinz Walz). Chl concentration was determined according to the method of Porra et al. (50).
Resonance Raman Spectroscopy
Low temperature (77 K) resonance Raman spectra were measured on leaves immediately frozen following 5 min of preillumination at 1000 μmol of photons m−2 s−1 (light) or alternatively preilluminated and then frozen following an additional period of 5 min of dark relaxation (dark recovery) and on isolated LHCII prepared in the trimeric and aggregated states as described above. Purified xanthophylls prepared as described previously (51, 52) were dissolved in pyridine for 77 K measurements. Samples were measured in a helium flow cryostat (Air Liquide, France) using a Jobin-Yvon U1000 Raman spectrophotometer equipped with a liquid nitrogen-cooled CCD detector (Spectrum One, Jobin-Yvon, France) as described (51). Excitation was provided by a Coherent Argon (Innova 100) laser (488.0 and 528.7 nm).
RESULTS
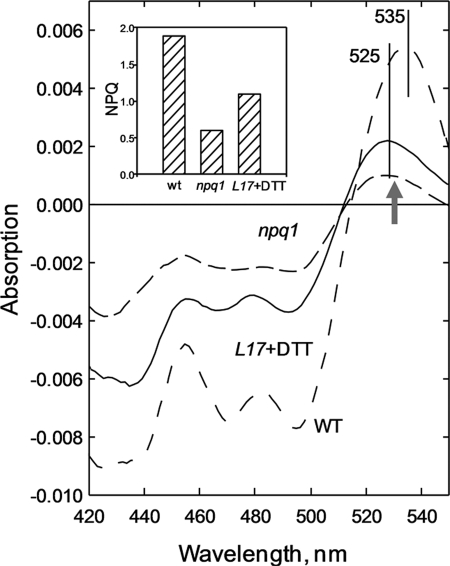
The well known light-minus-dark recovery absorption difference spectrum in Arabidopsis wild-type leaves is presented in Fig. 1, exhibiting the usual positive maximum at 535 nm consistent with the presence of zeaxanthin (Table 1). In the npq1 difference spectrum, the positions of the negative bands below 500 nm are virtually identical, whereas the 535 nm maximum is blue-shifted to 525 nm, consistent with the absence of zeaxanthin (and antheraxanthin) (Fig. 1, Table 1). The amplitude of all bands in the difference spectrum is also smaller in npq1 than in the wild type, consistent with the lower NPQ in the former sample (Fig. 1). Using resonance Raman difference spectroscopy, the origin of the 525 nm band in the npq1 absorption difference spectrum was probed. Two Raman spectra were recorded using 528.7 nm excitation, one for leaves frozen under illumination (5 min of preillumination at 1000 μmol of photons m−2 s−1) and the second one frozen after a further 5 min of dark recovery. A light-minus-dark recovery Raman difference spectrum should then reveal a selective gain of resonance for the xanthophyll species responsible for the absorption difference (30). However, although a resonance gain was detected in the npq1 leaves, the signal-to-noise ratio was such that it was impossible to identify accurately the xanthophyll species responsible (data not shown). Thus, to identify the origin of the ΔA525 band, we used the L17 mutant that overexpresses PsbS and thus has a much higher level of NPQ when compared with the wild type (47). It has been shown previously that PsbS overexpression in this mutant also enhances qE in the absence of zeaxanthin (35). Using the same principle, leaves were vacuum-infiltrated with DTT, an inhibitor of xanthophyll cycle activity, such that no zeaxanthin (or antheraxanthin) was formed during illumination (Table 1). In these leaves, the level of ΔA525 was enhanced by around a factor of 2 when compared with npq1, reflecting the larger NPQ, although there was no difference in the de-epoxidation state between the two samples (Fig. 1). Consistent with this enhanced ΔA525, the light-minus-dark recovery resonance Raman difference spectrum for L17 leaves treated with DTT had a significantly improved signal-to-noise ratio. Thus, a meaningful comparison with individual pigment spectra was possible, allowing us to attribute the band(s) (see below).
FIGURE 1.
Light-treated-minus-dark recovery absorption difference spectra in Arabidopsis leaves. Wild-type npq1 and L17+DTT leaves were measured immediately after 5 min of illumination at 1000 μmol of photons m−2 s−1 (light-treated) or following a further 5 min of dark relaxation (dark recovery). Inset, amplitude of NPQ for each sample, ± S.E.
TABLE 1.
Pigment composition of npq1 and L17 + DTT leaves
Samples were taken from leaves, either dark-adapted for 120 min or after 5 min of illumination at 1000 μmol of photons m−2 s−1. Data are normalized to 100 Chl a + b molecules and are means ± S.E. from four replicates. Neo, Lut, Vio, Ant, Zea, DEPs and Chl a/b: neoxanthin, lutein, violaxanthin, antheraxanthin, zeaxanthin, de-epoxidation state (Z + 0.5A)/(V + A + Z), and Chl a/b ratio, respectively; ND, none detected.
| Plant | Neo | Lut | Vio | Ant | Zea | DEPs | Chl a/b |
|---|---|---|---|---|---|---|---|
| % | |||||||
| WT dark | 5.2 ± 0.5 | 17 ± 1 | 4.4 ± 0.2 | 0.2 ± 0.1 | 0 | 4 ± 0.9 | 3.1 |
| WT light | 5.1 ± 0.9 | 16 ± 1 | 2.0 ± 0.4 | 0.8 ± 0.3 | 1.7 ± 0.4 | 46 ± 1.2 | 3.1 |
| npq1 | 5.3 ± 0.9 | 19 ± 2 | 4.4 ± 0.8 | ND | ND | 0 | 3.1 |
| L17 + DTT dark | 5.2 ± 0.6 | 16 ± 1 | 4.4 ± 0.2 | ND | ND | 0 | 3.1 |
| L17+ DTT light | 5.2 ± 0.5 | 17 ± 2 | 4.6 ± 0.4 | ND | ND | 0 | 3.1 |
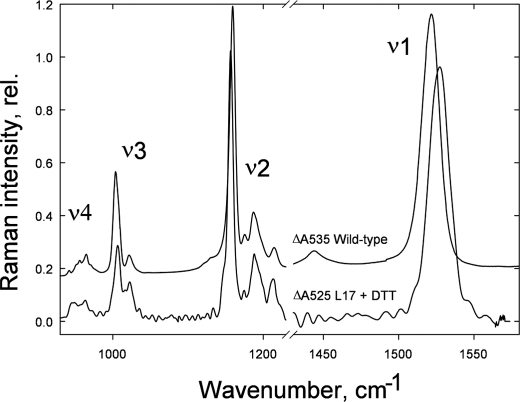
Fig. 2 presents the total light-minus-dark recovery resonance Raman difference spectrum using 528.7 nm excitation of L17 leaves treated with DTT. The four characteristic frequency regions of carotenoids have been assigned as follows: ν1, C=C stretching mode; ν2, C–C stretches coupled with C–H in-plane bending or C–CH3 stretching; ν3, CH3 in-plane rocking vibrations; ν4, C–H out-of-plane bending modes (30). The light-minus-dark recovery resonance Raman difference spectrum at 528.7 nm excitation of wild-type leaves containing zeaxanthin had a ν1 position at 1522 cm−1 (Fig. 2), consistent with the position of isolated zeaxanthin in pyridine as observed previously (31). However, the ν1 position in spectra of L17+DTT leaves was significantly different, peaking at 1528 cm−1. Because the ν1 position is highly dependent upon the conjugation length of the C=C chain (conjugated double bonds) of a carotenoid, lutein, violaxanthin, and neoxanthin have respectively higher ν1 positions (51). In the case of neoxanthin, the presence of a 9-cis configuration results in a further up-shift in ν1 when compared with violaxanthin, despite them both possessing nine conjugated double bonds. The ν1 position is thus a unique “molecular fingerprint” of the xanthophyll species involved in a particular resonance enhancement in the Raman difference spectrum resulting from an absorption change (30). Therefore, the up-shifted ν1 position in L17+DTT leaves when compared with the wild-type confirms that a xanthophyll other than zeaxanthin is involved in the ΔA525 change (Fig. 2).
FIGURE 2.
Comparison of the qE-associated Raman spectrum (difference spectrum of light-treated-minus-dark recovery) induced by 528.7 nm excitation in Arabidopsis leaves from wild type and L17 treated with DTT. rel., relative.
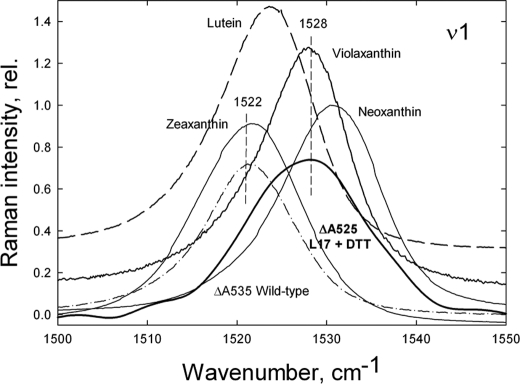
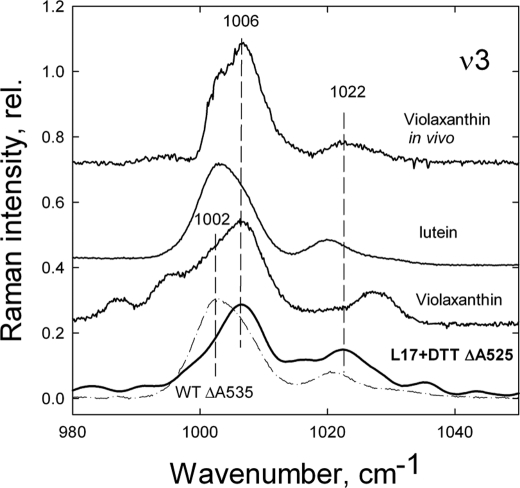
Several features of the Raman difference spectrum using 528.7 nm excitation in L17+DTT leaves give clues as to the identity of the xanthophyll species responsible for the ΔA525 change. Firstly, the ν1 position at 1528 cm−1 is identical to that of isolated violaxanthin dissolved in pyridine (Fig. 3), whereas ν1 for isolated lutein is at 1524 cm−1 and ν1 for neoxanthin is at 1533 cm−1 (Fig. 3). Examination of the ν3 region of the spectrum provides further evidence of the involvement of violaxanthin in the ΔA525 change (Fig. 4). The ν3 peak position is at 1006 cm−1 in the L17+DTT Raman difference spectrum (Fig. 4), again consistent with the position of isolated violaxanthin (Fig. 4, trace 2), whereas for both isolated lutein and isolated zeaxanthin, ν3 is at 1003 cm−1 (Fig. 4). It should be noted, however, that the satellite of the main ν3 band in the L17+DTT difference spectrum is at ∼1022 cm−1, deviating slightly from the corresponding band for isolated violaxanthin (1030 cm−1), whereas it is still higher than that of isolated lutein at 1020 cm−1 (Fig. 4). To account for this deviation, the L17+DTT spectrum was compared with the previously reported in vivo violaxanthin Raman spectrum (dark adapted-minus-dark recovery spectrum of wild-type leaves, where a preillumination period has caused violaxanthin de-epoxidation). The resulting Raman difference spectrum at 488.0 nm excitation reveals a selective loss of resonance from violaxanthin, thus allowing the in vivo features of this pigment to be observed. Comparison of the in vivo violaxanthin spectrum with that of the L17+DTT leaves presented here reveals that all features of the ν3 region are identical, including the position of this satellite band (Fig. 4).
FIGURE 3.
Comparison of the ν1 region of the qE-associated Raman spectrum (difference spectrum of light-treated-minus-dark recovery) induced by 528.7 nm excitation in Arabidopsis leaves from wild type and L17 treated with DTT. The ν1 region of the Raman spectra induced by 528.7 nm excitation of isolated zeaxanthin, violaxanthin, lutein, and neoxanthin dissolved in pyridine are also shown for comparison. rel., relative.
FIGURE 4.
Comparison of the ν3 region of the qE-associated Raman spectrum (difference spectrum of light-treated-minus-dark recovery) induced by 528.7 nm excitation in Arabidopsis leaves from wild type and L17 treated with DTT. The ν3 region of the Raman spectra induced by 528.7 nm excitation of isolated violaxanthin and lutein dissolved in pyridine and the in vivo violaxanthin spectrum induced by 488.0 nm excitation (difference spectrum of wild-type dark adapted-minus-dark recovery leaves) are shown for comparison. rel., relative.
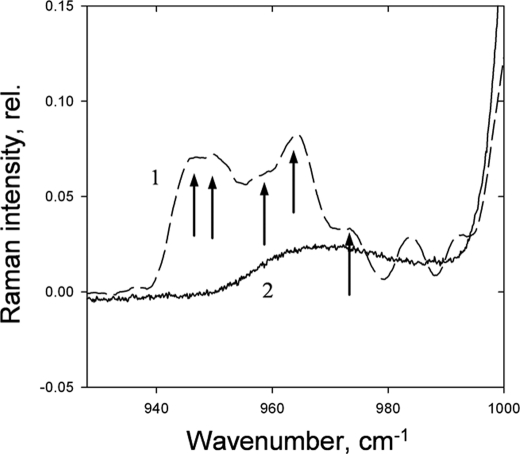
The ν4 region of the Raman spectrum has been shown to provide information on the molecular distortion of xanthophylls upon binding to proteins because the C–H wagging modes are formally resonance-forbidden in planar carotenoids (30). In L17+DTT leaves, the ν4 region of the spectrum shows the two sharp modes at 949 and 962 cm−1, characteristic for the in vivo violaxanthin Raman spectrum previously reported (51) and in contrast to the structureless ν4 region of violaxanthin dissolved in lipid micelles (Fig. 5). In addition, the ν4 region in L17+DTT leaves is slightly broadened with additional modes appearing (Fig. 5, arrows). The data therefore indicate that the red-shifted violaxanthin responsible for ΔA525 is selectively distorted through protein binding.
FIGURE 5.
ν4 region of the qE-associated Raman spectrum (difference spectrum of light-treated-minus-dark recovery) induced by 528.7 nm excitation in Arabidopsis leaves from L17 plants. The ν4 region of the Raman spectra induced by 528.7 nm excitation (difference spectrum of wild-type dark adapted-minus-dark recovered leaves) (trace 1) is presented in comparison with the spectrum of isolated violaxanthin dissolved in lipid micelles (trace 2). rel., relative.
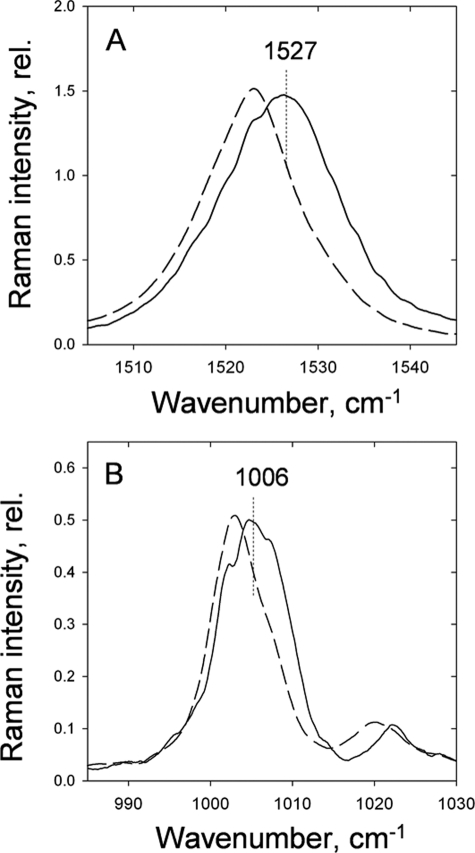
Around 75% of PSII-associated xanthophyll cycle carotenoids have been shown by numerous biochemical studies to be bound to the major trimeric LHCII complex (49). To investigate whether this complex could represent the source of the ΔA525 and ΔA535 change, we compared the Raman spectra of aggregated and trimeric LHCII binding violaxanthin and zeaxanthin in the V1 sites (Table 2). Remarkably, the same selective resonance enhancements in the aggregated-minus-trimeric LHCII Raman difference spectrum were observed as those associated with ΔA525 and ΔA535 in vivo for both the ν1 and the ν3 regions, providing a strong link between the two phenomena (Fig. 6, A and B).
TABLE 2.
Pigment composition of isolated LHCII antenna proteins
LHCII trimers enriched in either violaxanthin or zeaxanthin were obtained from sucrose gradients following solubilization of BBY membranes in n-dodecyl β-d-maltoside (see “Experimental Procedures”). De-epoxidation of violaxanthin to antheraxanthin and zeaxanthin was achieved by incubating the thylakoids (from which BBYs were derived) for 30 minutes in 0.33 m sorbitol, 1 mm EDTA, 30 mm HEPES, 20 mm MES, 40 mm ascorbate at pH 5.5 at 20 °C (de-epoxidation state of thylakoids following this procedure was ∼80%). Neo, Vio, Ant, Lut, Zea, DEPs, and Chl a/b: neoxanthin, violaxanthin, antheraxanthin, lutein, zeaxanthin, de-epoxidation state (Z + 0.5A)/(V + A + Z), and Chl a/b ratio. Carotenoid data are presented normalized to 14 molecules of Chl a + b ± S.E. from four replicates, DEPs (Zea + 0.5Ant)/ total Vio + Zea + Ant)% and Chl a/b presented as molar ratio; ND, none detected.
| Complex | Neo | Vio | Ant | Lut | Zea | DEPs | Chl a/b |
|---|---|---|---|---|---|---|---|
| LHCII V | 1.04 ± 0.1 | 0.69 ± 0.1 | ND | 2.1 ± 0.1 | ND | 0 | 1.33 ± 0.1 |
| LHCII Z | 1.05 ± 0.1 | 0.08 ± 0.03 | 0.08 ± 0.04 | 2.1 ± 0.1 | 0.58 ± 0.1 | 84% | 1.33 ± 0.1 |
FIGURE 6.
LHCII aggregation-associated Raman spectra (difference spectrum of aggregated-minus-trimeric) for LHCII binding zeaxanthin (dashed line) and violaxanthin (solid line) in the peripheral V1 binding site in the ν1 (A) and the ν3 (B) regions. Excitation is at 528.7 nm. rel., relative.
DISCUSSION
In this study, the origin of the ΔA525 absorption change that is characteristic for qE in the absence of zeaxanthin was studied by resonance Raman difference spectroscopy using 528.7 nm excitation. Using the same technique, the ΔA535 change was previously identified as belonging to a subpool of red-absorbing zeaxanthin molecules (31). Here, evidence was provided that the ΔA525 change is associated with a similar red-shifted pool of violaxanthin that is formed under qE conditions. A resonance enhancement of a xanthophyll species with ν1 and ν3 maxima consistent with that of violaxanthin and significantly different from that of lutein and the other xanthophylls was observed. The similarities in the absorption difference spectra below 500 nm and above 600 nm between qE in vivo and that of LHCII aggregation in vitro are well documented (11, 37). However, previously, the origin of the ΔA525 and ΔA535 bands has remained a mystery because they were absent in LHCII aggregates (37). However, the LHCII aggregates previously studied, unlike the ones used here, lacked peripheral xanthophyll cycle carotenoids. In this study, the same Raman signature associated with ΔA525 in leaves, and indeed that previously associated with ΔA535, was observed in LHCII aggregates binding xanthophyll cycle carotenoids at the peripheral V1 site, providing further evidence of a common origin for quenching in both systems.
It is known that xanthophyll absorption is sensitive to the solvent environment. The 0-0 transition for violaxanthin in vivo peaks at ∼490 nm (52), suggesting an ∼35-nm red shift upon ΔA525 formation, similar to that associated with ΔA535 attributable to zeaxanthin (45). To produce such large red shifts, a dramatic change in xanthophyll environment would be required, such as the appearance of a static dipole or point charge near the pigment responsible (41, 42).
An alternative explanation for a large red shift would be an excitonic splitting of the S2 excited state of the xanthophyll (46). This so-called Davydov splitting is a result of strong Coulombic coupling between the excited states of closely associated molecules. Such Coulomb coupling causes a mixing of the excited states of the individual molecules to form new excited states (excitonic states) that are delocalized across the whole molecular ensemble (53). The result of this quantum mechanical mixing is that the original single-molecule excited states move apart in energy (split).
A characteristic feature of excitonic splitting is that the new states have greatly different oscillator strengths when compared with the original single-molecule states, arising from different superpositions of the single-molecule transition dipole moments. If we consider the simple case of excitonic interactions between two identical xanthophyll models, then the excitonic splitting will be dominated by the highly dipole-allowed S2 states. The lower lying S1 state does not play as significant a role because it has no dipole connection with the ground state, meaning that the Coulomb coupling between S1 states will be much weaker than that between S2 states. The mixing of the two S2 states will result in the appearance of two new states, one strongly dipole-allowed, or “bright,” and the other only weakly dipole-allowed, or “dark”. Both of these states are split symmetrically about the original S2 state, and the energy ordering depends entirely on the relative geometry of the two molecules. An H-type (parallel or “card-packed”) geometry yields a dark state lower in energy than the bright state, whereas a J-type (collinear or top-to-tail) geometry results in the bright state being the higher in energy. It is clear that the formation of an H-aggregate is associated with an overall blue shift of the S2 absorption peak because the bright state lies above the original S2 state. By the same logic, the formation of a J-aggregate is associated with a corresponding red shift. We therefore propose that both ΔA535 and ΔA525 are the result of the formation of xanthophyll J-aggregates (zeaxanthin in the case of ΔA535 and violaxanthin for ΔA525) within the membrane.
Indeed, it has previously been shown that isolated zeaxanthin can mimic ΔA535 upon formation of J-type aggregates in vitro (45). A casual formation of J-type aggregates without protein interaction can be excluded for both ΔA525 and ΔA535 in vivo because the structure of the ν4 region of the Raman difference spectrum indicates that that the pigments remain bound to protein. The distinct lack of structure of the ν4 region in J-type aggregates formed between zeaxanthin molecules isolated in vitro when compared with in vivo supports this view (32, 34). The data are rather more consistent with the hypothesis made recently, that upon LHCII aggregation, pairs of either zeaxanthin or violaxanthin, depending upon the de-epoxidation state, may come into contact in such a way that a J-type aggregate is formed at the trimer-trimer interface (43).
In vitro, zeaxanthin enhances LHCII aggregation and violaxanthin restricts it, consistent with an enhancement of the qE-related structural changes by the former (22, 54). A notable feature regarding both ΔA525 and ΔA535 is that neither depends upon the presence of a specific LHCII, minor antenna complex, or PsbS; rather they are features of the ensemble of LHC(-like) proteins in the membrane, just like NPQ (25, 36). This would be anticipated given the high level of homology between LHCII and the minor antenna proteins (55). Indeed, CP26 and possibly CP29 are predicted to have V1 sites occupied by xanthophyll cycle carotenoids, according to biochemical studies (49). Thus, involvement of the V1 site in both ΔA525 and ΔA535 formation could explain why a signal is observed at ΔA530 in leaves of the Arabidopsis szl1 mutant, which lacks zeaxanthin but compensates by greatly increasing levels of lutein at the expense of violaxanthin (56). In this mutant, we predict that the extra lutein would replace violaxanthin in the V1 site, providing an explanation of the slight red shift of ΔA525 to ΔA530 in these plants (56).
The correlation between ΔA525/ΔA535 and qE has led to the suggestion that this change may monitor directly the formation of the quencher (5). An excitonic interaction between xanthophylls that lowers the energy of the S2 excited state may also affect the energy of the S1 state. Because the dissipation of energy during qE in vivo has been linked to the population of a carotenoid S1 state (12, 13, 16), a red-shifted xanthophyll species could indeed be involved. However, several lines of evidence indicate that ΔA525 and ΔA535 are not quenchers. Firstly, the xanthophylls responsible for these changes do not appear in the qE-related fluorescence excitation difference spectrum, suggesting that the S2 (and by inference S1) states of these xanthophylls are not strongly energetically coupled to Chl (11), as concluded previously for the bulk xanthophyll cycle pool (57) and consistent with their peripheral location and proposed role in the allosteric structural regulation of qE (58). Secondly, quenching still occurs in LHCII aggregates in vitro even in the absence of xanthophyll cycle carotenoids (37). Thirdly, the lut2npq2 mutant of Arabidopsis, which possesses zeaxanthin as the only xanthophyll, has a similar level of ΔA535 to the wild-type but significantly less qE (25). Finally, the S1 energy level of all carotenoids reconstituted within the LHCII complex has already been demonstrated experimentally to be lower than that of the lowest Chl Qy transition (59). Thus, it is likely that other factors, such as distance, are more important should carotenoids be quenchers. Rather, we suggest that the ΔA525 and ΔA535 monitors the tighter interaction between LHCII units formed upon aggregation both in vivo and in vitro. The fact that all four LHCII xanthophylls undergo some change in their absorption, excitation, and Raman spectra upon formation of quenching (12, 25, 31) indicates that the associated structural changes are felt across the protein complex in each xanthophyll-binding domain.
Acknowledgment
We thank Krishna Niyogi (Berkeley, CA) for the kind gift of L17 plant seeds.
This work was supported by research and equipment grants from the United Kingdom Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council, The Netherlands Organization for Scientific Research via the Foundation of Earth and Life Sciences (to C. I. and R. v. G.), the European Union (EU) FP7 Marie Curie Reintegration Grant (ERG 224796) (to C. I.), and an EU FP7 Marie Curie HARVEST Network grant.
- Chl
- chlorophyll(s)
- PSII
- photosystem II
- LHCII
- light-harvesting antenna complex of photosystem II
- NPQ
- nonphotochemical Chl fluorescence quenching
- qE
- rapidly reversible component of NPQ
- ΔpH
- proton gradient across the thylakoid membrane
- Zea
- zeaxanthin
- Fm
- maximum Chl fluorescence in the dark
- Fm′
- maximum Chl fluorescence after illumination.
REFERENCES
- 1. Dekker J. P., Boekema E. J. (2005) Biochim. Biophys. Acta 1706, 12–39 [DOI] [PubMed] [Google Scholar]
- 2. Ledford H. K., Niyogi K. K. (2005) Plant Cell Environ. 28, 1037–1045 [Google Scholar]
- 3. Müller P., Li X. P., Niyogi K. K. (2001) Plant Physiol. 125, 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horton P., Ruban A. V., Walters R. G. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655–684 [DOI] [PubMed] [Google Scholar]
- 5. Holt N. E., Fleming G. R., Niyogi K. K. (2004) Biochemistry 43, 8281–8289 [DOI] [PubMed] [Google Scholar]
- 6. Briantais J. M., Vernotte C., Picaud M., Krause G. H. (1979) Biochim. Biophys. Acta 548, 128–138 [DOI] [PubMed] [Google Scholar]
- 7. Walters R. G., Ruban A. V., Horton P. (1994) Eur. J. Biochem. 226, 1063–1069 [DOI] [PubMed] [Google Scholar]
- 8. Li X. P., Gilmore A. M., Caffarri S., Bassi R., Golan T., Kramer D., Niyogi K. K. (2004) J. Biol. Chem. 279, 22866–22874 [DOI] [PubMed] [Google Scholar]
- 9. Jahns P., Latowski D., Strzalka K. (2009) Biochim. Biophys. Acta 1787, 3–14 [DOI] [PubMed] [Google Scholar]
- 10. Gilmore A. M., Hazlett T. L., Govindjee (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2273–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson M. P., Ruban A. V. (2009) J. Biol. Chem. 284, 23592–23601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruban A. V., Berera R., Ilioaia C., van Stokkum I. H., Kennis J. T., Pascal A. A., van Amerongen H., Robert B., Horton P., van Grondelle R. (2007) Nature 450, 575–578 [DOI] [PubMed] [Google Scholar]
- 13. Ma Y. Z., Holt N. E., Li X. P., Niyogi K. K., Fleming G. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4377–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holt N. E., Zigmantas D., Valkunas L., Li X. P., Niyogi K. K., Fleming G. R. (2005) Science 307, 433–436 [DOI] [PubMed] [Google Scholar]
- 15. Ahn T. K., Avenson T. J., Ballottari M., Cheng. Y. C., Niyogi K. K., Bassi R., Fleming G. R. (2008) Science 320, 794–797 [DOI] [PubMed] [Google Scholar]
- 16. Bode S., Quentmeier C. C., Liao P. N., Hafi N., Barros T., Wilk L., Bittner F., Walla P. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12311–12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller M. G., Lambrev P., Reus M., Wientjes E., Croce R., Holzwarth A. R. (2010) Chem. Phys. Chem. 11, 1289–1296 [DOI] [PubMed] [Google Scholar]
- 18. Bilger W., Bjorkman O. (1990) Photosynth. Res. 25, 173–185 [DOI] [PubMed] [Google Scholar]
- 19. Krause G. H. (1973) Biochim. Biophys. Acta 292, 715–728 [DOI] [PubMed] [Google Scholar]
- 20. Bilger W., Björkman O., Thayer S. S. (1989) Plant Physiol. 91, 542–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bilger W., Bjorkman O. (1994) Planta 193, 238–246 [Google Scholar]
- 22. Horton P., Ruban A. V., Rees D., Pascal A. A., Noctor G., Young A. J. (1991) FEBS Lett. 292, 1–4 [DOI] [PubMed] [Google Scholar]
- 23. Ruban A. V., Horton P. (1992) Biochim. Biophys. Acta 1102, 30–38 [Google Scholar]
- 24. Ruban A. V., Young A. J., Horton P. (1993) Plant Physiol. 102, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson M. P., Pérez-Bueno M. L., Zia A., Horton P., Ruban A. V. (2009) Plant Physiol. 149, 1061–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heber U. (1969) Biochim. Biophys. Acta 180, 302–319 [DOI] [PubMed] [Google Scholar]
- 27. Murakami S., Packer L. (1970) J. Cell Biol. 47, 332–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noctor G., Ruban A. V., Horton P. (1993) Biochim. Biophys. Acta 1183, 339–344 [Google Scholar]
- 29. Li X. P., Björkman O., Shih C., Grossman A. R., Rosenquist M., Jansson S., Niyogi K. K. (2000) Nature 403, 391–395 [DOI] [PubMed] [Google Scholar]
- 30. Robert B., Horton P., Pascal A. A., Ruban A. V. (2004) Trends Plant Sci. 9, 385–390 [DOI] [PubMed] [Google Scholar]
- 31. Ruban A. V., Pascal A. A., Robert B., Horton P. (2002) J. Biol. Chem. 277, 7785–7789 [DOI] [PubMed] [Google Scholar]
- 32. Aspinall-O'Dea M., Wentworth M., Pascal A., Robert B., Ruban A., Horton P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16331–16335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dominici P., Caffarri S., Armenante F., Ceoldo S., Crimi M., Bassi R. (2002) J. Biol. Chem. 277, 22750–22758 [DOI] [PubMed] [Google Scholar]
- 34. Bonente G., Caffarri S., Finazzi G., Niyogi K., Bassi R. (2007) Photosynth. Res. 91, 292–292 [Google Scholar]
- 35. Crouchman S., Ruban A., Horton P. (2006) FEBS Lett. 580, 2053–2058 [DOI] [PubMed] [Google Scholar]
- 36. Johnson M. P., Ruban A. V. (2010) Plant J. 61, 283–289 [DOI] [PubMed] [Google Scholar]
- 37. Ruban A. V., Rees D., Pascal A. A., Horton P. (1992) Biochim. Biophys. Acta 1102, 39–44 [Google Scholar]
- 38. Ilioaia C., Johnson M. P., Horton P., Ruban A. V. (2008) J. Biol. Chem. 283, 29505–29512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le Rosen A. L., Reid C. E. (1952) J. Chem. Phys. 20, 233–236 [Google Scholar]
- 40. Bayliss N. S. (1950) J. Chem. Phys. 18, 292–296 [Google Scholar]
- 41. Gottfried D. S., Steffen M. A., Boxer S. G. (1991) Biochim. Biophys. Acta 1059, 76–90 [DOI] [PubMed] [Google Scholar]
- 42. Reich R., Sewe K. U. (1977) Photochem. Photobiol. 26, 11–17 [Google Scholar]
- 43. Ruban A. V. (2009) in Carotenoids: Physical, Chemical and Biological Functions and Properties (Landrum J. T. ed), CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 44. Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004) Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 45. Ruban A. V., Horton P., Young A. J. (1993) J. Photochem. Photobiol. B 21, 229–234 [Google Scholar]
- 46. Billsten H. H., Sundström V., Polívka T. (2005) J. Phys. Chem. A 109, 1521–1529 [DOI] [PubMed] [Google Scholar]
- 47. Li X. P., Muller-Moule P., Gilmore A. M., Niyogi K. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Johnson M. P., Havaux M., Triantaphylidès C., Ksas B., Pascal A. A., Robert B., Davison P. A., Ruban A. V., Horton P. (2007) J. Biol. Chem. 282, 22605–22618 [DOI] [PubMed] [Google Scholar]
- 49. Ruban A. V., Lee P. J., Wentworth M., Young A. J., Horton P. (1999) J. Biol. Chem. 274, 10458–10465 [DOI] [PubMed] [Google Scholar]
- 50. Porra R. J., Thompson W. A., Kriedemann P. E. (1989) Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 51. Ruban A. V., Pascal A. A., Robert B., Horton P. (2001) J. Biol. Chem. 276, 24862–24870 [DOI] [PubMed] [Google Scholar]
- 52. Ruban A. V., Pascal A. A., Robert B. (2000) FEBS Lett. 477, 181–185 [DOI] [PubMed] [Google Scholar]
- 53. Davydov A. S. (1962) Theory of Molecular Excitons, McGraw-Hill Inc., New York [Google Scholar]
- 54. Ruban A. V., Phillip D., Young A. J., Horton P. (1997) Biochemistry 36, 7855–7859 [DOI] [PubMed] [Google Scholar]
- 55. Jansson S. (1999) Trends Plant Sci. 4, 236–240 [DOI] [PubMed] [Google Scholar]
- 56. Li Z., Ahn T. K., Avenson T. J., Ballottari M., Cruz J. A., Kramer D. M., Bassi R., Fleming G. R., Keasling J. D., Niyogi K. K. (2009) Plant Cell 21, 1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Caffarri S., Croce R., Breton J., Bassi R. (2001) J. Biol. Chem. 276, 35924–35933 [DOI] [PubMed] [Google Scholar]
- 58. Horton P., Ruban A. V., Wentworth M. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Polívka T., Zigmantas D., Sundström V., Formaggio E., Cinque G., Bassi R. (2002) Biochemistry 41, 439–450 [DOI] [PubMed] [Google Scholar]