Abstract
Multispectral widefield optical imaging has the potential to improve early detection of oral cancer. The appropriate selection of illumination and collection conditions is required to maximize diagnostic ability. The goals of this study were to (i) evaluate image contrast between oral cancer∕precancer and non-neoplastic mucosa for a variety of imaging modalities and illumination∕collection conditions, and (ii) use classification algorithms to evaluate and compare the diagnostic utility of these modalities to discriminate cancers and precancers from normal tissue. Narrowband reflectance, autofluorescence, and polarized reflectance images were obtained from 61 patients and 11 normal volunteers. Image contrast was compared to identify modalities and conditions yielding greatest contrast. Image features were extracted and used to train and evaluate classification algorithms to discriminate tissue as non-neoplastic, dysplastic, or cancer; results were compared to histologic diagnosis. Autofluorescence imaging at 405-nm excitation provided the greatest image contrast, and the ratio of red-to-green fluorescence intensity computed from these images provided the best classification of dysplasia∕cancer versus non-neoplastic tissue. A sensitivity of 100% and a specificity of 85% were achieved in the validation set. Multispectral widefield images can accurately distinguish neoplastic and non-neoplastic tissue; however, the ability to separate precancerous lesions from cancers with this technique was limited.
Keywords: autofluorescence, early detection, cancer, computer aided diagnostics
Introduction
Accurate identification and delineation of oral precancerous and cancerous lesions is necessary for successful treatment.1, 2, 3, 4 Multispectral widefield imaging is emerging as an attractive, noninvasive means to inspect at-risk oral mucosa. For example, direct autofluorescence visualization has been shown to reveal biochemical changes associated with oral precancer and cancer.5, 6, 7, 8, 9, 10, 11, 12, 13 Reflectance imaging, including narrowband illumination imaging and polarized imaging, has been shown to aid in visualizing vasculature in the oral cavity,14, 15 which increases during malignant progression.16 To optimize outcomes, it is important to identify the imaging modalities, illumination and collection conditions, and∕or combinations of modalities that provide the most useful diagnostic information. This information is valuable for optimizing multispectral devices based on direct visualization of the tissue, as well as those based on computer-aided analysis of multispectral digital images.
An important goal of multispectral optical imaging is to increase the visual contrast between non-neoplastic and neoplastic (including dysplastic or cancerous) tissue above that available using conventional white-light screening techniques. Several studies have shown that illuminating tissue with blue light and viewing the autofluorescence from the tissue enhances the ability to visually identify neoplastic lesions based on loss of autofluorescence.5, 6, 17, 18 Although these results are encouraging, to date only qualitative observations of image contrast between non-neoplastic and neoplastic oral lesions have been reported; quantitative assessment across multiple modalities is needed to optimize performance.
Our group has previously demonstrated that the normalized ratio of red-to-green autofluorescence intensity at 405-nm excitation is useful to objectively discriminate non-neoplastic oral tissue and dysplastic and cancerous lesions using a simple classification algorithm.19 The biological basis of the observed optical changes likely stems from the degradation of the extracellular matrix by the tumor.13 Additional imaging modalities that target other aspects of malignancy, such as vascularity, may improve the ability to discriminate dysplasia and tumor grades and reduce false positives caused by benign conditions or inflammation.
The goals of this study were twofold. The first was to quantify and compare optical image contrast using autofluorescence, narrowband reflectance, and cross-polarized reflectance imaging modalities for neoplastic lesions in the oral cavity at a variety of illumination and collection wavelengths and conditions. Results of this analysis can help guide the choice of imaging modalities and illumination∕collection wavelengths for visual examination to detect neoplastic oral lesions. The second goal was to develop and evaluate diagnostic algorithms to discriminate neoplastic lesions from analysis of multispectral images of the oral cavity. Results will aid in the determination of the modalities and features that contain the most diagnostic information and will inform the design and construction of new imaging devices for objective diagnosis and delineation of neoplastic oral lesions. Future work is needed to refine and test the ability of these imaging strategies to discriminate neoplastic tissue from benign, potentially confounding lesions, such as inflammation.
Methods
Instrumentation and Data Acquisition
Widefield images were collected in four different modalities from patients and normal volunteers using a multispectral digital microscope (MDM).20 The MDM consists of a commercially available surgical microscope modified to capture digital images in white-light reflectance, autofluorescence, narrowband reflectance, and cross-polarized imaging modalities. The biologic basis for the use of these modalities is described in Roblyer et al.19 Autofluorescence images were acquired at four different excitation wavelengths: 365, 380, 405, and 450 nm, each with ∼50-nm bandwidth. These wavelengths have been previously shown to discriminate oral lesions.21 Narrowband reflectance images were acquired using illumination wavelengths of 430 and 530 nm with 20-nm bandwidth. In a subset of patients, narrowband reflectance images were also acquired at 575-nm illumination. Narrowband illumination wavelengths were chosen to correspond to hemoglobin absorption maxima in order to enhance superficial vascular patterns. White-light reflectance and cross-polarized white-light reflectance images were also captured. White-light reflectance images approximate the clinical appearance of lesions. Cross-polarized or orthogonal polarization reflectance (OPR) imaging was achieved by illuminating the tissue with linearly polarized light and collecting remitted light through a second linear polarizer, oriented orthogonal to the illumination polarization. OPR images detect multiply scattered photons that have typically traveled deeper in tissue. In total, there were nine image types collected by the MDM, accounting for the different excitation or illumination wavelengths in each modality.
The MDM captures images of tissue with a rectangular field of view of approximately 5×7 cm. The CCD camera utilizes a Bayer mask to collect color images. Images are collected as 1392×1040 pixel, 8-bit per color channel RGB tiff files.
The MDM was used to acquire images from patients with pathologically confirmed squamous oral lesions and normal volunteers with no history of oral lesions, under a protocol reviewed and approved by the Institutional Review Boards at Rice University and the University of Texas MD Anderson Cancer Center. Each measurement sequence consisted of a serial collection of the aforementioned image types. One or more measurements were taken from each study participant, at the lesion site and, when possible, at contralateral or distant clinically normal sites. To ensure that high-quality measurements were obtained, repeat measurements at all sites were usually collected. All images were dark corrected by subtracting a black background image taken with an equivalent exposure time.
For purposes of algorithm development and evaluation, data were divided into a training set and a validation set. The classification algorithms were first developed using data from the training set. The algorithms were then tested using data from the validation set, which was independent from the training set and was collected after the algorithm training. All image processing, statistical calculations, and algorithm development was performed using MATLAB (Natick, Massachusetts).
Preprocessing
Images were collected in vivo, and there was often some subject movement between image capture of different image types. To account for this, image registration was performed for each measurement sequence. An affine transformation was used to translate all of the images taken in a sequence to the white-light base image using up to eight common reference points, chosen manually.
Regions of interest (ROI) were chosen from the white-light images by an expert physician (AG) blinded to the other image types. ROI were considered either “suspicious” or “normal” in appearance using unaided visual inspection. After confirmatory biopsy or tissue resection, these ROI were grouped into one of the following two categories: (i) a histopathologically confirmed lesion or (ii) a non-neoplastic region. Non-neoplastic regions were either histopathologically confirmed, clinically determined by AG to be sufficiently distant to a lesion in a patient or from a normal volunteer. ROI were the approximate size of the corresponding resected tissue specimen obtained from either a circular biopsy or surgical resection. An additional clinically normal ROI, either from the contralateral anatomic site or from a sufficiently distant area (determined by AG) on the same anatomic site, was chosen and used for normalization. In cases where image acquisition was repeated from the same tissue location, feature values extracted from the repeat images were averaged and considered as a single measurement site.
Optical Image Contrast
The optical image contrast between histopathologically determined lesions and non-neoplastic tissue was calculated and compared for each image type. The contrast was computed from ROI chosen from lesions with a pathologic diagnosis of dysplasia or cancer relative to the corresponding clinically normal ROI. Several methods have been proposed to compute image contrast;22, 23 in this study, we explored four different contrast metrics. Each metric used the mean of the gray-scale pixel values inside the ROIs. Simple contrast was calculated as the ratio of abnormal to non-neoplastic. Difference contrast was calculated as the difference between abnormal and non-neoplastic. Weber contrast was calculated as abnormal divided by the sum of abnormal and non-neoplastic. Michaelson contrast was calculated as the difference divided by the sum of abnormal and non-neoplastic. Figure 1 shows several Michaelson contrast values calculated from example images.
Figure 1.
(a), (c), (e), and (g) are white-light, narrowband (NB) 530-nm reflectance, 405-nm excitation autofluorescence, and cross-polarized images of the palate of a normal volunteer. ROI locations are indicated. (b), (d), (f), and (h) are images from the palate of a patient with severe dysplasia. ROI of the biopsy location and a corresponding clinically normal area are shown. Several feature values from these images, computed from the indicated ROI are shown in the table.
The contrast metrics for the autofluorescence, narrowband reflectance, and cross-polarized reflectance image types are reported relative to white-light contrast. This was done to evaluate the contrast of each image type relative to clinical observation. For each image type, the percentage of lesions with a higher optical contrast than white light was computed. One-way analysis of variance (ANOVA) was used to compare the mean contrast metrics for each image type to that for white light.
In order to explore the relationship between optical contrast and pathologic grade, scatter plots of the contrast metrics versus pathologic diagnosis were made. One-way ANOVA was used to compare the statistical difference in mean optical contrast between diagnostic categories.
Classification Algorithms
Figure 2 provides an overview of the algorithm development and evaluation procedure used in this study. After image acquisition, registration, and ROI selection, image features were extracted from each ROI. Image features were designed to quantify commonly observed trends in dysplastic lesions and tumors, including loss of autofluorescence and the presence of high-density small-vessel vascular patterns. Features included statistical measures, texture, and frequency content metrics.
Figure 2.
Flowchart of computer-aided diagnostics procedure. CV denotes cross-validation.
Two different supervised classifiers, a linear classifier and a decision-tree classifier, were used to construct diagnostic algorithms based on features from images in the training set. For each classifier, features were chosen using a feature selection algorithm. The results from the two methods were compared to histopathology, and the algorithm with the highest performance was then used to classify data from the validation set.
In order to determine the image types and modalities or combination of modalities most capable of discriminating neoplastic lesions, algorithms were developed and evaluated using features extracted from each of the four modalities alone, as well as in combination. The following feature subsets were used: (i) Features obtained from white-light reflectance images; (ii) features obtained from narrowband reflectance images at 430 and 530 nm illumination; (iii) features obtained from cross-polarized reflectance images; (iv) features obtained from autofluorescence images at 365, 380, 405, and 450 nm excitation; and (v) features obtained from all of the modalities and image types.
ROI were first classified into one of two diagnostic categories: non-neoplastic and neoplastic. The neoplastic class included lesions diagnosed histopathologically as mild, moderate, or severe dysplasia; carcinoma in situ; or invasive carcinoma. The non-neoplastic class included clinically non-neoplastic sites in patients and normal volunteers. The non-neoplastic sites in patients could include inflammation, hyperplasia, and∕or hyperkeratosis. No known benign lesions were measured in normal volunteers. We then attempted the more difficult problem of classifying the ROI into one of three diagnostic categories: non-neoplastic, dysplasia, and cancer.
Feature extraction
For each ROI site, 98 image features were computed for each image type, as described later in detail. The Michaelson and Weber contrasts were computed from the gray-scale images and from each color channel of the RGB images, resulting in eight features for each image type. Eighteen first-order statistical features were calculated using the gray-scale pixel values from the ROI, including the mean, standard deviation, entropy (defined as −∑i Pi ln Pi where P is a vector in which each element contains the number of pixels in the gray-scale image belonging to one of 256 evenly spaced bins), variance, skewness, and kurtosis. These features were used to quantify intensity differences between neoplastic and normal tissue, which might be due to hemoglobin absorption (narrowband reflectance images) or changes in extracellular matrix (autofluorescence). These features were calculated for each ROI and as normalized by corresponding clinically normal ROI. Normalization was performed in two ways; the first was calculated as the difference between the ROI (difference-normalization), and the second was the ratio of the ROI (ratio-normalization).
Eighteen features were obtained using the color channels of measurements. The mean values of the red, green, and blue channels of each ROI were used as features. The ratio of the mean red-to-green, red-to-blue, and green-to-blue pixel values were also utilized. Both normalized and non-normalized feature values were calculated.
Features for texture and frequency content were also utilized. These features were explored because vascular patterns were commonly observed to be different on lesions compared to normal tissue. Texture and frequency metrics are designed to quantify intensity patterns in images.
Features representing texture in the images were obtained by using gray-scale-level co-occurrence matrices (GLCMs). GLCMs are useful for quantifying how pixel intensities vary spatially. A pixel separation, d, and angle, θ, are specified for a particular GLCM. The size of the GLCM is determined by the number of discrete intensity values contained in the gray-scale image. Each entry (i, j) in the GLCM is a count of the number of times a pixel of intensity i occurred at the specified pixel separation d and angle θ away from a pixel with intensity j. Statistical measures including contrast, correlation, energy, and homogeneity were computed from the GLCMs. More detail is provided in Argenti et al.24
Twenty-four features were created based on these statistical measures from GLCMs where d varied from 1 to 6. The features were averaged at angles θ = 0, 45, 90, and 135 deg to account for the fact that these multispectral images do not have a specific spatial orientation.
A 2-D discrete Fourier transform (DFT) was performed on a rectangular region whose center corresponded to the approximate center location of the selected ROI. This 2-D DFT was converted into a 1-D plot of frequency content by integrating the pixel intensities at discrete radii from the origin. The 1-D plot was then partitioned into 10 frequency ranges, and the frequency content was integrated inside each range. The contribution of each partition was calculated by dividing by the total integrated 1-D plot so that the sum for all 10 partitions added to unity. Thirty features were computed using the relative frequency content for the partitions. Normalized and non-normalized feature values were included. Variations of this method have been used by Gossage et al.25 and Srivastava et al.26
Linear classifier
We implemented a linear classifier (LC) based on empirical Bayesian parameter estimation. This method assumes multivariate normal densities and equal covariance for each class. The LC is trained on a data set which is used to estimate the mean μi for each class and a pooled covariance matrix Σ for all classes. A priori probabilities are determined from the relative proportion of each class in the training set; posterior probabilities are output for each measurement and used by a linear discriminant function to separate the measurements into classes.
Decision tree classifier
We utilized a decision-tree classification method based on the widely used Classification and Regression Tree induction technique. This method has the attractive attribute of classifying data without assumptions of the underlying statistical distributions of the observations.27 We used Gini impurity to determine splits.27, 28
To help avoid overtraining the decision tree in the training set, it was pruned to find the smallest tree at which adding further nodes does not statistically decrease the cost of the tree. The cost of the tree is defined in the zero-one sense, where the cost of misclassifying an observation is 1 and the cost of correctly classifying an observation is 0.
Feature dimension and selection
A forward sequential search (FSS) algorithm was used on the training set to determine the optimal feature dimension: the minimum number of features needed to maximize a chosen classifier performance criterion without overtraining.28, 29 Starting with one feature, classification of the training set was performed with fivefold cross-validation using the FSS algorithm to find the single feature that maximized the criterion value. The area under the curve (AUC) of the receiver-operating characteristic (ROC) was used as the criteria for the FSS algorithm for the LC. For the decision tree, the sum of the sensitivity and specificity was used. This was repeated with an additional feature added at each iteration until classifier performance did not increase. This entire procedure was repeated 25 times, with a random membership selection to each of the five folds at each iteration, to provide statistically significant results. One-way ANOVA, with multiple comparison tests, was used to determine the optimal feature dimension. The final feature sets for each classifier was determined by the most commonly chosen features by the FSS algorithm in the 25 iterations. This entire procedure was repeated for each of the five imaging modality-based feature subsets.
Classification performance
The classifier performance for the training set was determined using fivefold cross-validation. For two-class classification, we utilized sensitivity and specificity as the performance metrics [or figures-of-merit (FOM)] to evaluate and compare the performance of the classifiers on the training set for each of the feature subsets. Sensitivity and specificity were determined at the Q point on the ROC curve, the location on the curve with the shortest Euclidean distance to the upper left-hand corner of the plot. For three-class classification, we utilized the FOM of total correct classification rates and correct classification rates for each class. On the basis of these results, the best performing classifier for the two- and three-class problems was retrained on the entire training set and then used to evaluate the validation set.
Results
Multispectral Images and ROIs
In total, images were acquired from 72 subjects, including 61 patients with pathologically confirmed oral lesions and 11 normal volunteers. From these images, we defined 175 ROI measurements sites. The following anatomic sites were included: 67 ROI from tongue, 31 ROI from buccal mucosa, 26 ROI from floor of mouth, 9 ROI from gingiva, 18 ROI from lip, and 24 ROI from palate. There were 93 non-neoplastic ROI and 82 neoplastic ROI. The neoplastic ROI consisted of 22 ROI with mild dysplasia, 13 ROI with moderate dysplasia, 16 ROI with severe dysplasia or carcinoma in situ, and 31 ROI with invasive cancer.
The ROI were divided into training and validation sets. There were 102 ROI from 46 subjects in the training set (∼ 2∕3 of the subjects) and 73 ROI from 26 subjects in the validation set (∼ 1∕3 of the subjects).
Figure 1 shows representative multispectral images from the palate of a normal volunteer and the palate of a patient with severe dysplasia. White-light [Figs. 1a, 1b], 530-nm narrowband reflectance [Fig. 1c, 1d], 405-nm autofluorescence [Figs. 1e, 1f], and white-light cross-polarized reflectance images [Figs. 1g, 1h] are shown. ROI locations are indicated by circles in the white-light images. A chart of example features and contrast metric values, which will be described next, are shown. In this example, all contrast and feature values are increased in the patient with severe dysplasia.
Optical Contrast
The results from the different contrast definitions were very similar (data not shown); thus, we report only results from Michaelson contrast. Figure 3a shows a box plot of the Michaelson contrast metric by image type; contrast for each image type is reported relative to the contrast achieved using white-light illumination. For each image type, Fig. 3a also indicates the percentage of lesions where contrast was greater than in white light. On average, the contrast for each of the image types was greater than that available in white-light mode. Autofluorescence imaging at 405 nm showed the greatest average increase in contrast and the greatest percentage of abnormal lesions with increased contrast relative to white-light imaging. The mean contrast value for autofluorescence at 365-, 380-, 405-, and 450-nm excitation was statistically different from that of white-light imaging, using one-way ANOVA with a 95% confidence interval.
Figure 3.
(a) A boxplot of the increase in Michaelson contrast compared to white light when using narrowband (NB) reflectance imaging, autofluorescence (FL) imaging, and cross-polarized (CPOL) reflectance imaging. The imaging type is indicated on the x-axis. For each modality, the percentage of lesions where the contrast was increased over white light is indicated near the top of the plots. The three horizontal lines on each of the boxes represent the lower quartile, median, and upper quartile of the data from bottom to top. The whiskers extending from the box indicate the remaining data except for outliers, which are indicated by the dots. (b) The Michaelson gray-scale contrast computed from the 405-nm autofluorescence images. The contrast values are displayed by graded diagnostic category. The shaded squares in each diagnostic cluster indicate the location of the mean contrast value. There is a statistically significant difference in contrast between non-neoplastic and the other diagnostic categories, but not between the grades of dysplasia or carcinoma.
A subset of 26 patients was imaged with narrowband illumination at 575 nm. From this subset there were 41 lesion sites. The 575-nm narrowband imaging provided an increase in Michaelson contrast over white light in 80% of these lesions. The median increase was 1.15, which was similar to the other narrowband illuminations and was significantly less than that achieved from the autofluorescence modality.
Autofluorescence images at 405-nm excitation show the greatest increase in optical contrast compared to white-light images when discriminating non-neoplastic from neoplastic tissue. In order to explore whether optical contrast in this image type increased with increasing grade of dysplasia or cancer, we plotted the contrast of each lesion by diagnostic category ]Fig. 3b]. The means for the diagnostic categories are shown in the shaded boxes. The mean contrast of non-neoplastic tissue was significantly lower than that of each of the grades of dysplasia and cancer, when calculated using one-way ANOVA at 5% type 1 error rate. However, the mean contrast values for each of the dysplastic categories and cancer were not statistically different from each other.
Classification
Two-class classification: Non-neoplastic versus neoplastic
Table 1 lists the number of features selected and summarizes the classifier performance for both the LC and decision tree methods using fivefold cross-validation for the training set. For both classifiers, the best performance was obtained using the autofluorescence feature subset. Furthermore, the only feature chosen by the FSS in the combined feature subset was a single feature from the autofluorescence modality subset.
Table 1.
Two-class classification results of the training data for the LC and the decision-tree classifiers. The AUC of the receiver operating characteristic curve, sensitivity, and specificity are shown for the LC. Sensitivity and specificity are indicated at the Q point on the ROC curve. The sensitivity and specificity are shown for the decision tree classifier. The number of features chosen is indicated for each feature subset.
| Linear Classifier |
Decision Tree |
||||||
|---|---|---|---|---|---|---|---|
| Feature Subset | No. Features | AUC | Se (%) | Sp (%) | No. Features | Se (%) | Sp (%) |
| White Light | 5 | .931 | 87.8 | 88.7 | 3 | 87.8 | 84.9 |
| Narrowband | 4 | .916 | 87.8 | 83.0 | 4 | 83.7 | 83.0 |
| Cross Polarized | 6 | .898 | 83.7 | 75.5 | 3 | 85.7 | 77.4 |
| Autofluorescence | 1 | .981 | 93.9 | 98.1 | 1 | 95.9 | 92.5 |
| Combined | 1 | .981 | 93.9 | 98.1 | 1 | 95.9 | 92.5 |
Features extracted from white-light images provided the second best performance after autofluorescence for both of the classifiers on the training set; classification required five features for the LC and three features for the decision tree. Narrowband reflectance provided the third best classification with four features. Cross-polarized provided the worst classification with six features for the LC and three features for the decision tree.
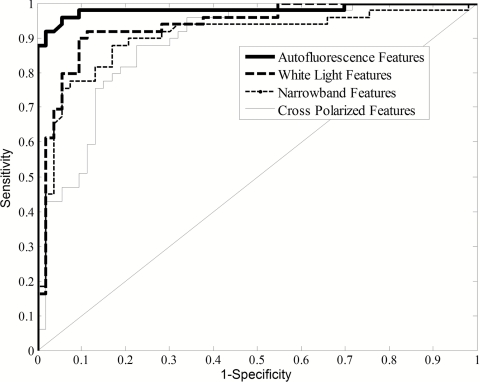
Figure 4 shows ROC curves produced from the LC on the training set. The autofluorescence feature subset produced the highest AUC followed by white-light, narrowband, and cross-polarized features.
Figure 4.
ROC curves from the two-class LC for different feature subsets on the training set. When considering the AUC of the ROC plots, the autofluorescence features performed the best, followed by white light, NB, and cross polarized. Note that these ROC plots are from a single fivefold cross-validation run on the training set, and therefore, the parameters may not match exactly to Table 1.
The autofluorescence feature chosen for the LC was the ratio of red-to-green intensity at 405-nm excitation (difference normalized). For the training set, this single feature provided an AUC of 0.981, a sensitivity of 93.9%, and a specificity of 98.1%. The feature chosen from the decision tree was very similar, the ratio of ratio of red-to-green intensity at 405-nm excitation (ratio normalized). For the training set, this single feature provided a sensitivity of 95.9% and a specificity of 92.5%.
The LC was used on the validation set because it provided a slightly higher sum of sensitivity and specificity than the decision tree. Using these same features, the algorithm was retrained on the entire training set and applied to the validation set yielding an AUC of 0.949, a sensitivity of 100%, a specificity of 85.0%, a positive predictive value of 84.6%, and a negative predictive value of 100%.
Three-class classification: Non-neoplastic versus dysplasia versus cancer
Table 2 lists the number of features chosen for the three-class LC and the decision tree classifier using fivefold cross-validation. Algorithm performance is also summarized in Table 2, as the percent of sites correctly classified for all sites and for each of the three diagnostic categories.
Table 2.
Three-class classification results of training set for LC and decision tree classifier. All results are computed using fivefold cross-validation.
| Linear Classifier |
Decision Tree |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Feature Subset | No. Features | Total Correct (%) | Non-neo. Correct (%) | Dys. Correct (%) | Cancer Correct (%) | No. Features | Total Correct (%) | Non-neo. Correct (%) | Dys. Correct (%) | Cancer Correct (%) |
| White Light | 4 | 81.4 | 92.5 | 65.5 | 75.0 | 2 | 72.6 | 81.1 | 75.9 | 45.0 |
| Narrowband | 5 | 65.6 | 88.7 | 37.9 | 45.0 | 2 | 72.6 | 79.3 | 69.0 | 60.0 |
| Cross Polarized | 5 | 61.8 | 90.6 | 34.5 | 25.0 | 3 | 69.6 | 84.9 | 58.6 | 45.0 |
| Autofluorescence | 4 | 84.3 | 98.1 | 79.3 | 55.0 | 2 | 86.3 | 94.3 | 75.9 | 80.0 |
| Combined | 4 | 77.5 | 94.3 | 69.0 | 45.0 | 2 | 83.3 | 96.2 | 79.3 | 55.0 |
The best performance from both classifiers was from the autofluorescence feature subset using the decision tree, which provided correct classification of 94.3% of the non-neoplastic ROI, 75.9% of the dysplasia ROI, and 80% of the cancer ROI. The first feature chosen was the ratio of red-to-green intensity (ratio normalized) from the 405-nm autofluorescence image; this was the same feature chosen for the two-class problem by the decision tree. The second feature was the GLCM homogeneity with d = 6 calculated from the 380-nm autofluorescence image. For both classifiers in the training set, autofluorescence features and white-light features were chosen in the combined feature subset, but this combination did not improve performance, based on overall correct classification rate, compared to the autofluorescence only feature subset.
The decision-tree classifier was then retrained on the entire training set and applied to the validation set. Results are summarized in Table 3. In the training set, this method misclassifies 5.7% (3 of 53) non-neoplastic ROI as dysplasia, 17.2% (5 of 29) dysplasia ROI as cancer, 3.4% (1 of 29) dysplasia ROI as non-neoplastic, and 20% (4 of 20) cancer ROI as dysplasia. No cancerous ROI were misclassified as non-neoplastic, and no non-neoplastic ROI were misclassified as cancer.
Table 3.
Confusion matrices of three-class classification results from the decision tree using two autofluorescence features. Numeric values indicate the number of classified measurement sites. Results from both the training set and validation set are shown without cross-validation. The decision tree was trained on the training set and then applied to the validation set.
| Decision Tree Results | Training SetTruth |
Validation SetTruth |
|||||
|---|---|---|---|---|---|---|---|
| Normal | Dysplasia | Cancer | Normal | Dysplasia | Cancer | ||
| Normal | 50 | 1 | 0 | 32 | 0 | 0 | |
| Predicted | Dysplasia | 3 | 23 | 4 | 8 | 22 | 9 |
| Cancer | 0 | 5 | 16 | 0 | 0 | 2 | |
In the validation set, 20% (8 of 40) non-neoplastic ROI were misclassified as dysplasia and 81.8% (9 of 11) cancerous ROI were misclassified at dysplasia. 100% (22 of 22) dysplastic ROI were correctly classified. No cancerous ROI were misclassified as non-neoplastic and no non-neoplastic ROI were misclassified as cancer.
Discussion
Oral cancer is a major global health problem, ranking as one the most common malignancies in developing nations. Optical imaging is being explored to improve early detection and diagnosis of oral cancer for improved patient outcomes. To meet these goals, optical imaging devices need to be optimized to provide high discriminant ability between early cancer and non-neoplastic tissue. In this study, multispectral wide-field imaging data from 72 study subjects were analyzed to determine the imaging modalities and specific image types that provide the greatest optical image contrast between oral dysplastic and cancerous lesions and surrounding non-neoplastic tissue, and to determine if objective classification algorithms could be used to diagnostically classify oral lesions.
Quantitative optical contrast calculated from grayscale autofluorescence images at 405-nm excitation was greater for 78% of the lesions compared to white light. These lesions included all grades of dysplasia and carcinoma. When the contrast was analyzed by diagnostic category, contrast was significantly greater for all pathologies compared to non-neoplastic areas but contrast was not significantly different between pathologic grades. This suggests that the contrast observed using autofluorescence may be due to changes that occur early in malignant transformation. Narrowband images and cross-polarized images showed an increase in contrast over white light in the majority of images but not to the extent of the autofluorescence images.
Two-class classification performance using both the LC and decision trees was excellent in the training set, providing sensitivities and specificities of >90% for both classifiers. It is clear from the analysis that color-based features extracted from autofluorescence images at 405 nm were highly dominant, demonstrated by the fact that a single feature of this type was chosen using both classifiers, even with access to the entire feature set. The combination of features from multiple subsets did not improve two-class classifier performance above the autofluorescence feature subset alone. The LC was used on the validation set, and a sensitivity of 100% and a specificity of 85.0% were achieved, helping to confirm the diagnostic capability of the 405-nm autofluorescence image type.
For the three-class problem, the decision tree modestly outperformed the LC in the training set. Two features based on autofluorescence were selected after pruning in the two-step decision tree. The first feature, the ratio of red-to-green intensity (ratio normalized) at 405-nm excitation, was used to classify non-neoplastic sites from dysplastic and carcinoma sites, just as in the two-class problem. The second feature was then used to separate the dysplastic lesions from the carcinoma lesions. This second feature was produced by texture analysis and was from the 380-nm excitation autofluorescence image. In the validation set, 0% (0 of 22) of the dysplastic lesions were misclassified but 81.8% (9 of 11) of the carcinoma lesions were misclassified as dysplasia. This suggests that the second feature may not generalize well to new data to discriminate dysplasia from cancer.
Results from both the contrast analysis and classification portions of this study indicate that dysplastic and cancerous oral lesions can be discriminated from non-neoplastic tissue using the autofluorescence imaging modality at 405-nm excitation, confirming the results of our previous analysis.19 It was shown that the reflectance white-light, narrowband, and cross-polarized images included in this analysis did not improve the diagnostic ability available from the autofluorescence images. This is somewhat surprising given that the signal from autofluorescence and reflectance modalities are likely dominated by two different biological phenomenon during malignant progression: the loss of collagen fluorescence in the stroma13 and the increase in vascularity in and around lesions,16 respectively.
Unfortunately, the data suggest that it may not be feasible for wide-field multispectral optical imaging to discriminate between dysplasia and cancer using the specific modalities and wavelengths tested here. On the basis of results from Pavlova et al.13 and Schwarz et al.,30 it may be necessary to selectively probe the epithelium in order to discriminate oral precancer and cancer using optical techniques. It is also possible that the patient sample size may have been too small or too homogeneous to benefit from these additional modalities. An additional limitation of this study is that confounding factors, such as inflammation or bacterial infection, were not separately considered or classified. It has previously been shown that inflammation may cause a decrease in autofluorescence signal and may lead to false positives.13 A combination of imaging modalities may be able to help discriminate true precancerous and cancerous lesions from confounding benign lesions. Rahman et al.31 demonstrated the importance of combining data from white-light and autofluorescence images to avoid misclassification of confounding lesions such as oral mucosal pigmentation.
Several groups have previously demonstrated that increased contrast of lesions observed using autofluorescence imaging with blue excitation light (400–460 nm) is an effective diagnostic means for high-grade oral lesions.6, 7, 17 Poh et al.18 reported that an observed visual decrease in autofluorescence signal often extended beyond clinical tumor margins up to 25 mm. Lane et al.5 showed a sensitivity of 98% and a specificity of 100% for the discrimination of severe dysplasia and carcinoma from direct autofluorescence visualization. This work has achieved comparable diagnostic performance using an objective discrimination technique while including low-grade as well as high-grade lesions.
Multispectral wide-field optical imaging has the potential to improve on current oral cancer screening and delineation methods. The use of 405-nm excitation autofluorescence imaging of the oral mucosa can increase the optical image contrast of lesions above that observed using white light and can provide objective classification of neoplastic lesions with high accuracy.
Acknowledgments
This work was supported in part by the National Institutes of Health under Grants No. R21DE16485, No. R01CA124319, and No. P50CA097007.
References
- Institute N. C., SEER Program: Oral Cancer, N. C. Institute, pp. 96–99, Bethesda, MD: (2003). [Google Scholar]
- Lippman S. M. and Hong W. K., “Second malignant tumors in head and neck squamous cell carcinoma: the overshadowing threat for patients with early-stage disease,” Int. J. Radiat. Oncol. Biol. Phys. 17(3), 691–694 (1989). [DOI] [PubMed] [Google Scholar]
- Day G. L. and Blot W. J., “Second primary tumors in patients with oral cancer,” Cancer 70(1), 14–19 (1992). [DOI] [PubMed] [Google Scholar]
- Slaughter D. P., Southwick H. W., and Smejkal W., “Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin,” Cancer 6(5), 963–968 (1953). [DOI] [PubMed] [Google Scholar]
- Lane P. M., Gilhuly T., Whitehead P., Zeng H., Poh C. F., Ng S., Williams P. M., Zhang L., Rosin M. P., and MacAulay C. E., “Simple device for the direct visualization of oral-cavity tissue fluorescence,” J. Biomed. Opt. 11(2), 024006 (2006). 10.1117/1.2193157 [DOI] [PubMed] [Google Scholar]
- Svistun E., Alizadeh-Naderi R., El-Naggar A., Jacob R., Gillenwater A., and Richards-Kortum R., “Vision enhancement system for detection of oral cavity neoplasia based on autofluorescence,” Head Neck 26(3), 205–215 (2004). 10.1002/hed.10381 [DOI] [PubMed] [Google Scholar]
- Utzinger U., Bueeler M., Oh S., Heintzelman D. L., Svistun E. S., Abd-El-Barr M., Gillenwater A., and Richards-Kortum R., “Optimal visual perception and detection of oral cavity neoplasia,” IEEE Trans. Biomed. Eng. 50(3), 396–399 (2003). 10.1109/TBME.2003.808832 [DOI] [PubMed] [Google Scholar]
- Rosin M. P., Poh C. F., Guillard M., Williams P. M., Zhang L., and MacaUlay C., “Visualization and other emerging technologies as change makers for oral cancer prevention,” Ann. N Y Acad. Sci. 1098, 167–183 (2007). 10.1196/annals.1384.039 [DOI] [PubMed] [Google Scholar]
- Andersson-Engels S., “Medical diagnostic system based on simultaneous multispectral fluorescence imaging,” Appl. Opt. 33(34), pp. 8022–8029 (1994). 10.1364/AO.33.008022 [DOI] [PubMed] [Google Scholar]
- Kulapaditharom B. and Boonkitticharoen V., “Laser-induced fluorescence imaging in localization of head and neck cancers,” Ann. Otol. Rhinol. Laryngol. 107(3), 241–246 (1998). [DOI] [PubMed] [Google Scholar]
- Onizawa K., Saginoya H., Furuya Y., and Yoshida H., “Fluorescence photography as a diagnostic method for oral cancer,” Cancer Lett. 108(1), 61–66 (1996). 10.1016/S0304-3835(96)04388-1 [DOI] [PubMed] [Google Scholar]
- Betz C. S., Mehlmann M., Rick K., Stepp H., Grevers G., Baumgartner R., and Leunig A., “Autofluorescence imaging and spectroscopy of normal and malignant mucosa in patients with head and neck cancer,” Lasers Surg. Med. 25(4), 323–334 (1999). [DOI] [PubMed] [Google Scholar]
- Pavlova I., Williams M., El-Naggar A., Richards-Kortum R., and Gillenwater A., “Understanding the biological basis of autofluorescence imaging for oral cancer detection: high-resolution fluorescence microscopy in viable tissue,” Clin. Cancer Res. 14(8), 2396–2404 (2008). 10.1158/1078-0432.CCR-07-1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhash N., Mallia J. R., Thomas S. S., Mathews A., Sebastian P., and Madhavan J., “Oral cancer detection using diffuse reflectance spectral ratio R540∕R575 of oxygenated hemoglobin bands,” J. Biomed. Opt. 11(1), 014018 (2006). 10.1117/1.2165184 [DOI] [PubMed] [Google Scholar]
- Lindeboom J. A., Mathura K. R., and Ince C., “Orthogonal polarization spectral (OPS) imaging and topographical characteristics of oral squamous cell carcinoma,” Oral. Oncol. 42(6), 581–585 (2006). 10.1016/j.oraloncology.2005.10.014 [DOI] [PubMed] [Google Scholar]
- Pazouki S., Chisholm D. M., Adi M. M., Carmichael G., Farquharson M., Ogden G. R., Schor S. L., and Schor A. M., “The association between tumour progression and vascularity in the oral mucosa,” J. Pathol. 183(1), 39–43 (1997). [DOI] [PubMed] [Google Scholar]
- de Veld D. C., Witjes M. J., Sterenborg H. J., and Roodenburg J. L., “The status of in vivo autofluorescence spectroscopy and imaging for oral oncology,” Oral. Oncol. 41(2), 117–131 (2005). 10.1016/j.oraloncology.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Poh C. F., Zhang L., Anderson D. W., Durham J. S., Williams P. M., Priddy R. W., Berean K. W., Ng S., Tseng O. L., MacAulay C., and Rosin M. P., “Fluorescence visualization detection of field alterations in tumor margins of oral cancer patients,” Clin. Cancer Res. 12(22), 6716–6722 (2006). 10.1158/1078-0432.CCR-06-1317 [DOI] [PubMed] [Google Scholar]
- Roblyer D., Kurachi C., Stepanek V., Williams M. D., El-Naggar A. K., Lee J. J., Gillenwater A. M., and Richards-Kortum R., “Objective detection and delineation of oral neoplasia using autofluorescence imaging,” Cancer Prev. Res. 2(5), pp. 423–431 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roblyer D., Richards-Kortum R., Sokolov K., El-Naggar A. K., Williams M. D., Kurachi C., and Gillenwater A. M., “Multispectral optical imaging device for in vivo detection of oral neoplasia,” J. Biomed. Opt. 13(2), 024019 (2008). 10.1117/1.2904658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzelman D. L., Utzinger U., Fuchs H., Zuluaga A., Gossage K., Gillenwater A. M., Jacob R., Kemp B., and Richards-Kortum R. R., “Optimal excitation wavelengths for in vivo detection of oral neoplasia using fluorescence spectroscopy,” Photochem. Photobiol. 72(1), 103–113 (2000). [DOI] [PubMed] [Google Scholar]
- Peli E., “Contrast in complex images,” J. Opt. Soc. Am. A 7(10), 2032–2040 (1990). 10.1364/JOSAA.7.002032 [DOI] [PubMed] [Google Scholar]
- Bex P. J. and Makous W., “Spatial frequency, phase, and the contrast of natural images,” J. Opt. Soc. Am. A 19(6), 1096–1106 (2002). 10.1364/JOSAA.19.001096 [DOI] [PubMed] [Google Scholar]
- Argenti F., Alparone L., and Benelli G., “Fast Algorithms for Texture Analysis Using cooccurrence matrices,” IEE Proc. F 137(6), p. 443–448 (1990). [Google Scholar]
- Gossage K. W., Tkaczyk T. S., Rodriguez J. J., and Barton J. K., “Texture analysis of optical coherence tomography images: feasibility for tissue classification,” J. Biomed. Opt. 8(3), 570–575 (2003). 10.1117/1.1577575 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Rodriguez J. J., Rouse A. R., Brewer M. A., and Gmitro A. F., “Computer-aided identification of ovarian cancer in confocal microendoscope images,” J. Biomed. Opt. 13(2), 024021 (2008). 10.1117/1.2907167 [DOI] [PubMed] [Google Scholar]
- Breiman L., Friedman J., Olshen R., and Stone C., Classification and Regression Trees. CRC Press, Boca Raton, FL: (1984). [Google Scholar]
- Duda R. O., Hart P. E., and Stork D. G., Pattern Classification, 2nd ed., Wiley, Hoboken, NJ: (2001). [Google Scholar]
- Albert A. and Harris E. K., Multivariate Interpretation of Clinical Laboratory Data, Marcel Dekker, New York: (1987). [Google Scholar]
- Schwarz R. A., Arifler D., Chang S. K., Pavlova I., Hussain I. A., Mack V., Knight B., Richards-Kortum R., and Gillenwater A. M., “Ball lens coupled fiber-optic probe for depth-resolved spectroscopy of epithelial tissue,” Opt. Lett. 30(10), 1159–1161 (2005). 10.1364/OL.30.001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. S., Ingole N., Roblyer D., Stepanek V., Richards-Kortum R., Gillenwater A., Shastri S., and Chaturvedi P., “Evaluation of a low-cost, portable imaging system for early detection of oral cancer,” Head Neck Oncol. 2, p. 10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]