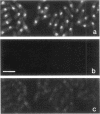
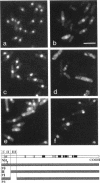
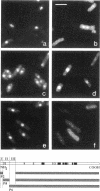
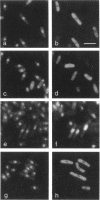
Abstract
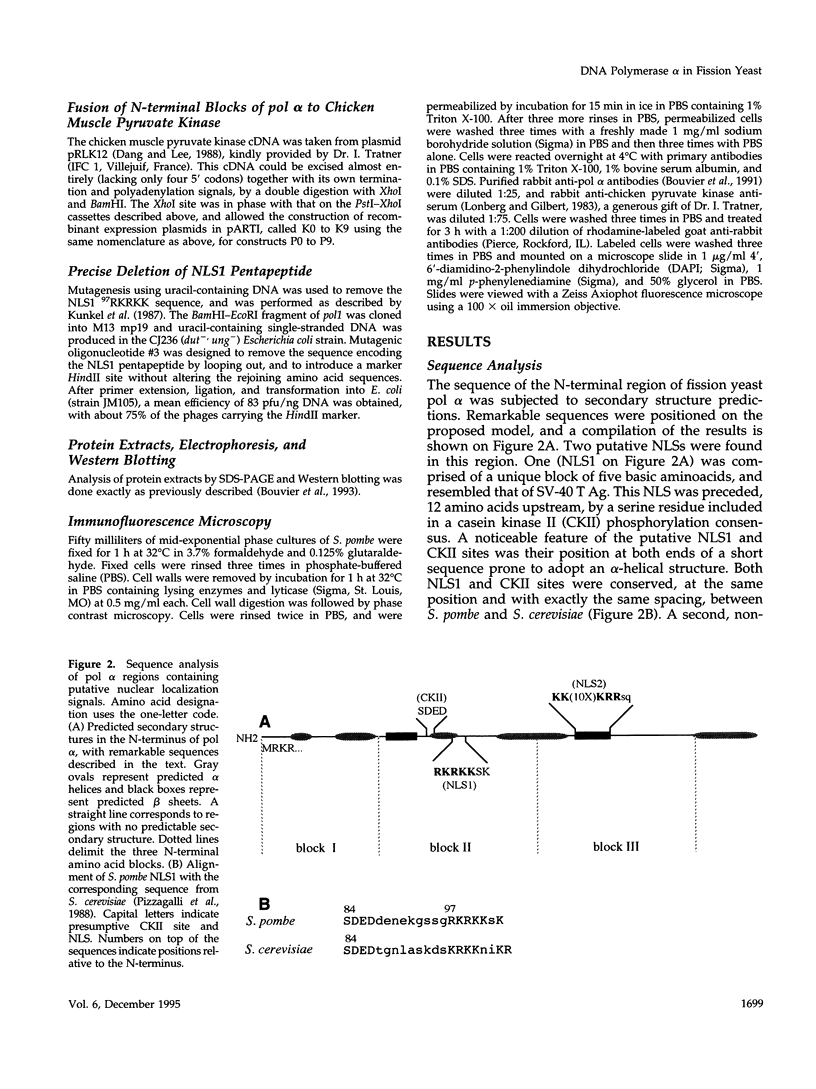
The N-terminal sequence of the catalytic subunit of fission yeast DNA polymerase alpha (pol alpha) contains two putative nuclear localization signals (NLS). To check the functionality of these signals in vivo, the N-terminal sequence was experimentally divided into three amino acid blocks, two of which contain a distinct presumptive NLS. Each block was deleted, either individually or in combination with one of the two others. The deleted gene products were expressed in fission yeast, and assayed by indirect immunofluorescence for their aptitude to localize to the cell nucleus. Block II, which contains the putative NLS pentapeptide 97RKRKK, was both necessary and sufficient to promote nuclear import of pol alpha, as well as of a pyruvate kinase fusion protein. Precise excision of the NLS pentapeptide from block II inhibited the nuclear import of pol alpha, thus confirming the role of this sequence as the functional NLS of the fission yeast enzyme.
Full text
PDF

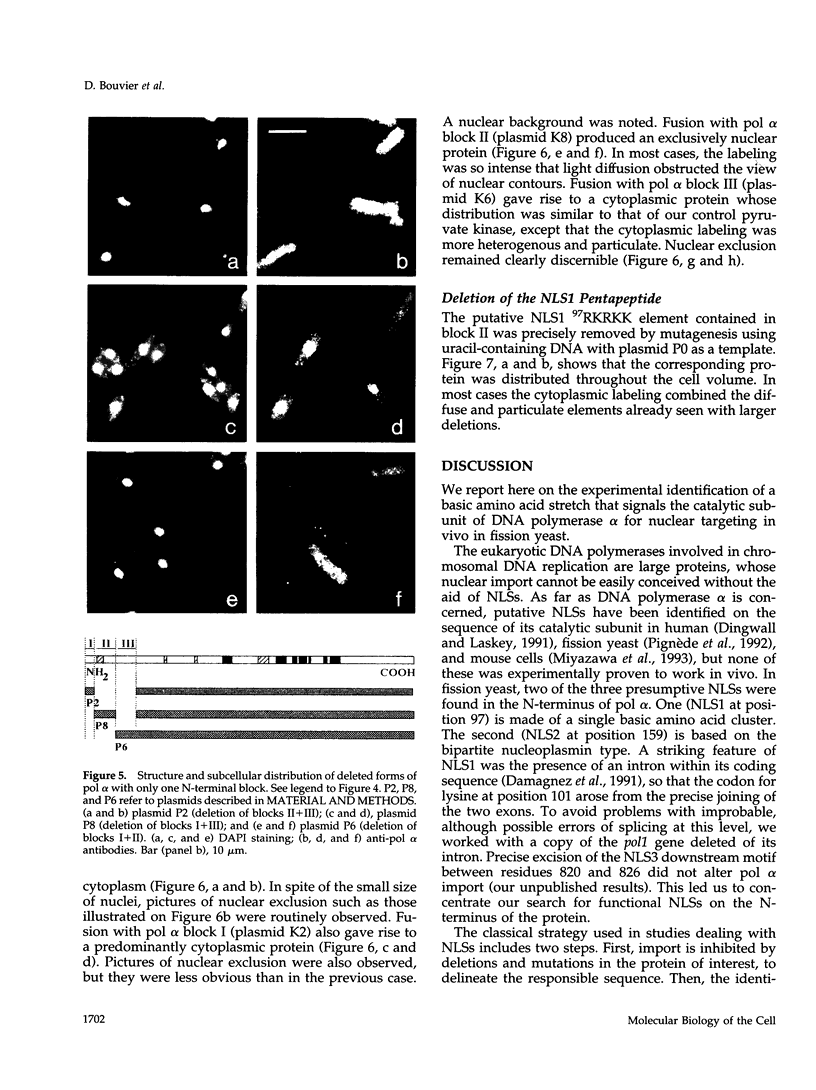
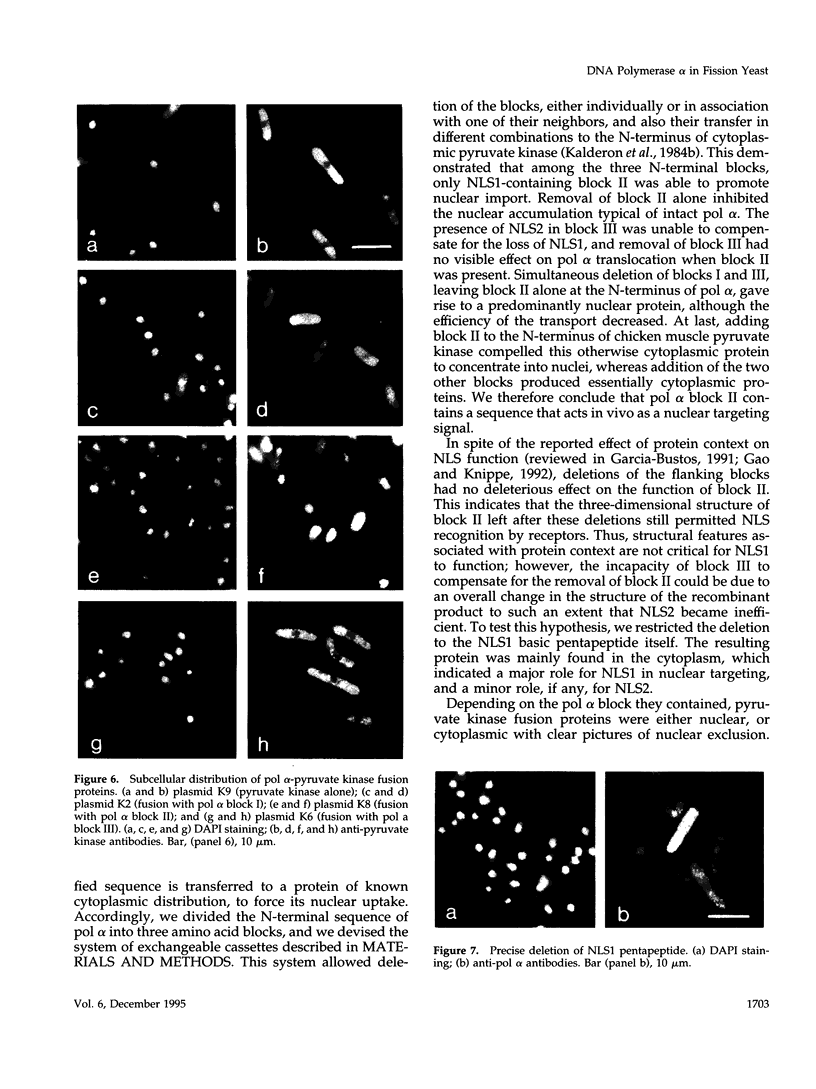


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bouvier D., De Recondo A. M., Baldacci G. In vivo phosphorylation, mitotic behavior, and nuclear binding of the catalytic subunit of DNA polymerase alpha in fission yeast. Exp Cell Res. 1993 Jul;207(1):41–47. doi: 10.1006/excr.1993.1160. [DOI] [PubMed] [Google Scholar]
- Bouvier D., Pignede G., Damagnez V., Tillit J., de Recondo A. M., Baldacci G. DNA polymerase alpha in the fission yeast Schizosaccharomyces pombe: identification and tracing of the catalytic subunit during the cell cycle. Exp Cell Res. 1992 Feb;198(2):183–190. doi: 10.1016/0014-4827(92)90370-n. [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., De Robertis E. M. The nuclear migration signal of Xenopus laevis nucleoplasmin. EMBO J. 1987 Sep;6(9):2617–2625. doi: 10.1002/j.1460-2075.1987.tb02552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damagnez V., Tillit J., de Recondo A. M., Baldacci G. The POL1 gene from the fission yeast, Schizosaccharomyces pombe, shows conserved amino acid blocks specific for eukaryotic DNA polymerases alpha. Mol Gen Genet. 1991 Apr;226(1-2):182–189. doi: 10.1007/BF00273602. [DOI] [PubMed] [Google Scholar]
- Dang C. V., Lee W. M. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988 Oct;8(10):4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen P., Fondrat C., Valencien C., Mugnier C. BISANCE: a French service for access to biomolecular sequence databases. Comput Appl Biosci. 1990 Oct;6(4):355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991 Dec;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Fleck O., Michael H., Heim L. The swi4+ gene of Schizosaccharomyces pombe encodes a homologue of mismatch repair enzymes. Nucleic Acids Res. 1992 May 11;20(9):2271–2278. doi: 10.1093/nar/20.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S., Copeland W. C., Wang T. S. In vivo species specificity of DNA polymerase alpha. Mol Gen Genet. 1993 Nov;241(3-4):457–466. doi: 10.1007/BF00284700. [DOI] [PubMed] [Google Scholar]
- Gao M., Knipe D. M. Distal protein sequences can affect the function of a nuclear localization signal. Mol Cell Biol. 1992 Mar;12(3):1330–1339. doi: 10.1128/mcb.12.3.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M. N. Nuclear protein localization. Biochim Biophys Acta. 1991 Mar 7;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lonberg N., Gilbert W. Primary structure of chicken muscle pyruvate kinase mRNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3661–3665. doi: 10.1073/pnas.80.12.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod M., Stein M., Beach D. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987 Mar;6(3):729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
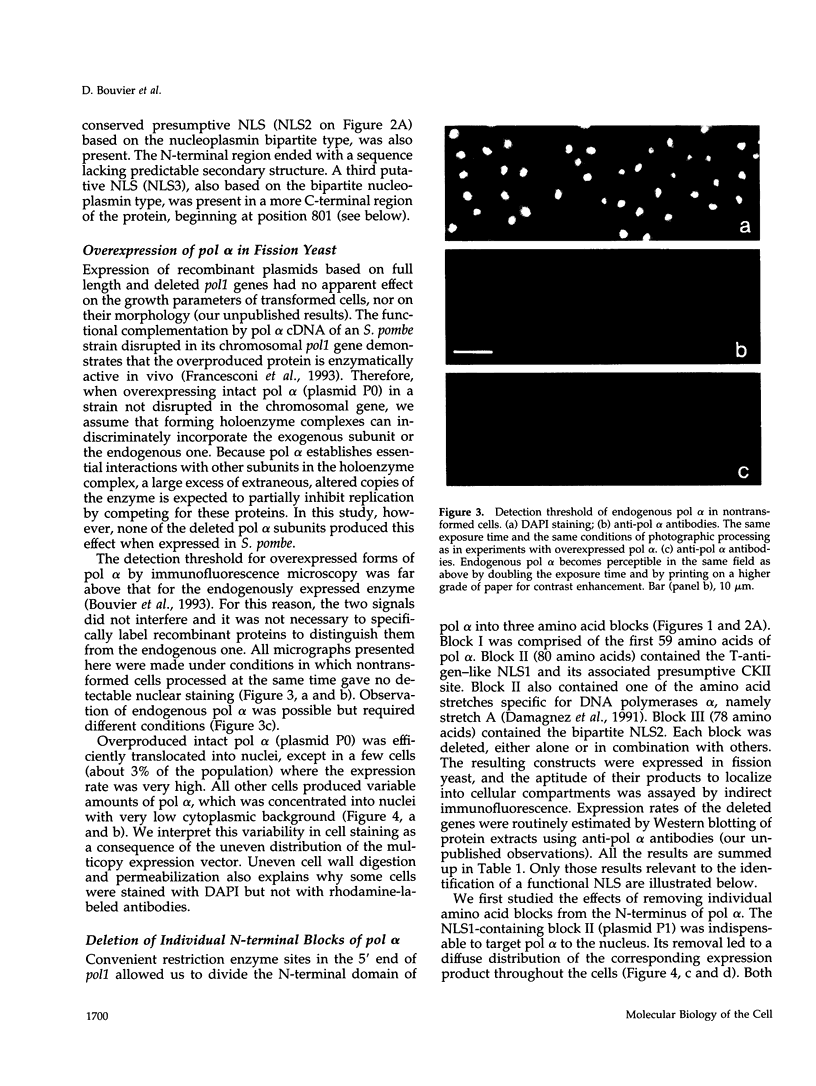
- Melchior F., Gerace L. Mechanisms of nuclear protein import. Curr Opin Cell Biol. 1995 Jun;7(3):310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- Melov S., Vaughan H., Cotterill S. Molecular characterisation of the gene for the 180 kDa subunit of the DNA polymerase-primase of Drosophila melanogaster. J Cell Sci. 1992 Aug;102(Pt 4):847–856. doi: 10.1242/jcs.102.4.847. [DOI] [PubMed] [Google Scholar]
- Miyazawa H., Izumi M., Tada S., Takada R., Masutani M., Ui M., Hanaoka F. Molecular cloning of the cDNAs for the four subunits of mouse DNA polymerase alpha-primase complex and their gene expression during cell proliferation and the cell cycle. J Biol Chem. 1993 Apr 15;268(11):8111–8122. [PubMed] [Google Scholar]
- Moreland R. B., Langevin G. L., Singer R. H., Garcea R. L., Hereford L. M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987 Nov;7(11):4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
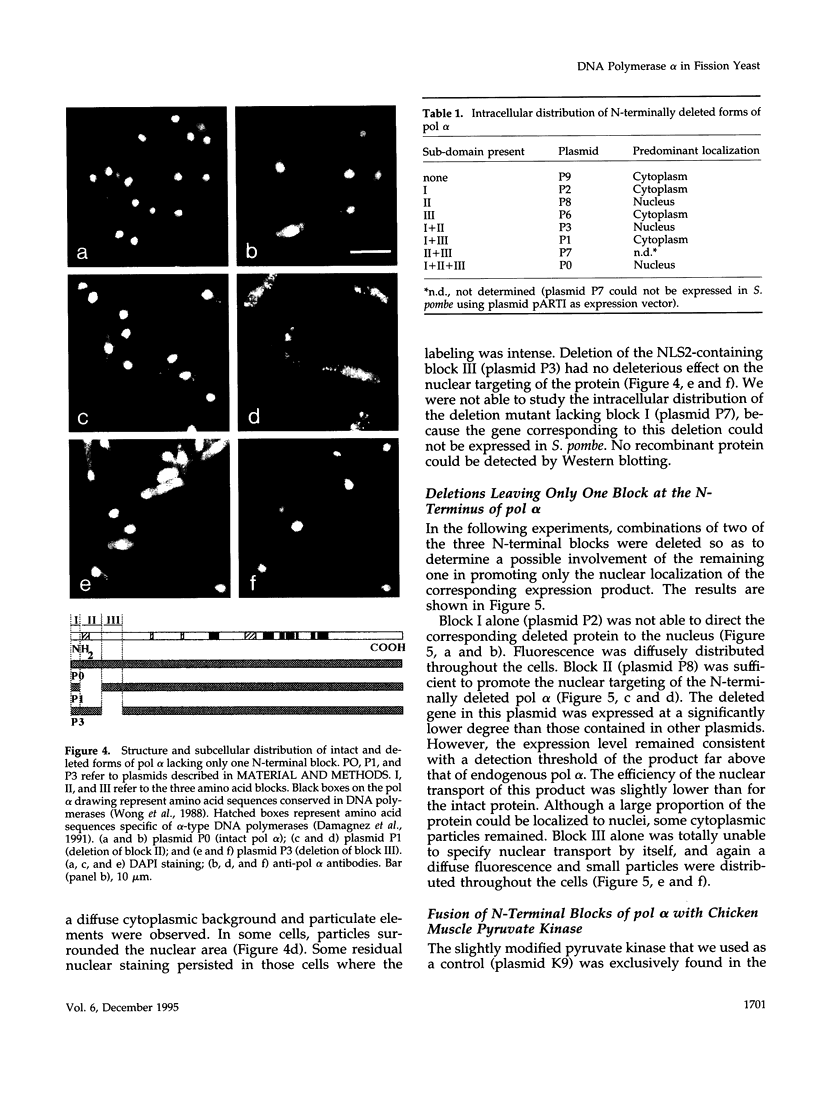
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moussy G., De Recondo A. M., Baldacci G. Inter-species DNA polymerase delta chimeras are functional in Saccharomyces cerevisiae. Eur J Biochem. 1995 Jul 1;231(1):45–49. [PubMed] [Google Scholar]
- Paine P. L., Moore L. C., Horowitz S. B. Nuclear envelope permeability. Nature. 1975 Mar 13;254(5496):109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- Pignède G., Moussy G., Bouvier D., Tillit J., de Recondo A. M., Baldacci G. Expression of the catalytic subunits of pol alpha and pol delta from fission yeast Schizosaccharomyces pombe. Chromosoma. 1992;102(1 Suppl):S128–S132. doi: 10.1007/BF02451796. [DOI] [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W. D., Roberts B. L., Smith A. E. Nuclear location signals in polyoma virus large-T. Cell. 1986 Jan 17;44(1):77–85. doi: 10.1016/0092-8674(86)90486-1. [DOI] [PubMed] [Google Scholar]
- Rihs H. P., Jans D. A., Fan H., Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 1991 Mar;10(3):633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihs H. P., Peters R. Nuclear transport kinetics depend on phosphorylation-site-containing sequences flanking the karyophilic signal of the Simian virus 40 T-antigen. EMBO J. 1989 May;8(5):1479–1484. doi: 10.1002/j.1460-2075.1989.tb03531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991 Feb 8;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V., Molinete M., Boeuf H., de Murcia G., Ménissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992 Sep;11(9):3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K., Yanagida M. Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J Cell Biol. 1992 Dec;119(5):1023–1036. doi: 10.1083/jcb.119.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A. G., Downey K. M. Eukaryotic DNA replication. Crit Rev Biochem Mol Biol. 1992;27(1-2):129–155. doi: 10.3109/10409239209082561. [DOI] [PubMed] [Google Scholar]
- Sugino A. Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci. 1995 Aug;20(8):319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- Underwood M. R., Fried H. M. Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J. 1990 Jan;9(1):91–99. doi: 10.1002/j.1460-2075.1990.tb08084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S., Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994 May 19;369(6477):207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Wang T. S. Eukaryotic DNA polymerases. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]