SUMMARY
Systematic characterization of cancer genomes has revealed a staggering number of diverse aberrations that differ among individuals, such that the functional importance and physiological impact of most tumor genetic alterations remains poorly defined. We developed a computational framework that integrates chromosomal copy number and gene expression data for detecting aberrations that promote cancer progression. We demonstrate the utility of this framework using a melanoma dataset. Our analysis correctly identified known drivers of melanoma and predicted multiple novel tumor dependencies. Two dependencies, TBC1D16 and RAB27A, confirmed empirically, suggest that abnormal regulation of protein trafficking contributes to proliferation in melanoma. Together, these results demonstrate the ability of integrative Bayesian approaches to identify novel candidate drivers with biological, and possibly therapeutic, importance in cancer.
INTRODUCTION
Large-scale initiatives to map chromosomal aberrations, mutations and gene expression have revealed a highly complex assortment of genetic and transcriptional changes within individual tumors. For example, copy number aberrations (CNAs) occur frequently in cancer due to genomic instability. Genomic data has been collected for thousands of tumors at high resolution using array comparative genomic hybridization (aCGH) (Pinkel et al., 1998), high density single nucleotide polymorphism (SNP) microarrays (Beroukhim et al., 2010; Lin et al., 2008) and massively parallel sequencing (Pleasance et al., 2010). Although multiple new genes have been implicated in cancer through sequencing and CNA analysis (Garraway et al., 2005), these studies have also revealed enormous diversity in genomic aberrations in tumors among individuals. Each tumor is unique and typically harbors a large number of genetic lesions, of which only a few drive proliferation and metastasis. Thus, identifying driving mutations (genetic changes that promote cancer progression) and distinguishing them from passengers (those with no selective advantage) has emerged as a major challenge in the genomic characterization of cancer.
The most widely used approaches are based on the frequency an aberration occurs: if a mutation provides a fitness advantage in a given tumor type, its persistence will be favored and it is likely to be found in multiple tumors. For example, GISTIC identifies regions of the genome that are aberrant more often than would be expected by chance, and has been used to analyze a number of cancers (Beroukhim et al., 2009; Beroukhim et al., 2007; Lin et al., 2008). However, there are limitations to analytical approaches based on CNA data alone: CNA regions are typically large and contain many genes, most of which are passengers that are indistinguishable in copy number from the drivers. CNA data has statistical power to detect only the most frequently recurring drivers above the large number of unrelated chromosomal aberrations that are typical in cancer. Finally, these approaches rarely elucidate the functional importance or physiological impact of the genetic alteration on the tumor. These limitations highlight the need for new approaches that can integrate additional data to identify drivers of cancer. Gene expression is readily available for many tumors, but how best to combine it with information on CNA is not obvious.
We postulate that driving mutations coincide with a “genomic footprint” in the form of a gene expression signature (see Box 1). We developed an algorithm that integrates chromosomal copy number and gene expression data to find these signatures and identify likely driver genes located in regions that are amplified or deleted in tumors. Each potential driver gene is altered in some, but not all tumors and, when altered, is considered likely to play a contributing role in tumorgenesis. Unique to our approach, each driver is associated with a gene module, which is assumed to be altered by the driver. We sometimes gain insight into the likely role of a candidate driver, based on the annotation of the genes in the associated module. We demonstrate the utility of our method using a dataset (Lin et al., 2008) that includes paired measurements of gene expression and copy number from 62 melanoma samples. Our analysis correctly identified known drivers of melanoma and connected them to many of their targets and biological functions. In addition, it predicted novel melanoma tumor dependencies, two of which, TBC1D16 and RAB27A, were confirmed experimentally. Both of these genes are involved in the regulation of vesicular trafficking, which highlights this process as important for proliferation in melanoma.
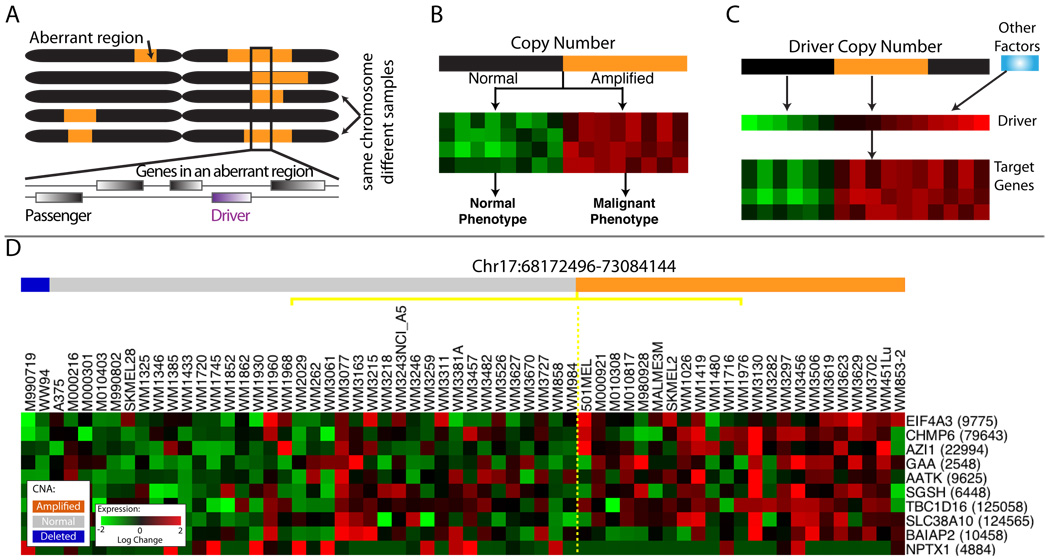
BOX 1: The genomic signature of a driver (combined with Figure 1)
We define a “driving mutation” to be a genetic alteration that provides the tumor cell with a growth advantage during carcinogenesis or tumor progression (Stratton et al., 2009). We reasoned that driving mutations might leave a genomic ‘footprint’ that can assist in distinguishing between driver and passenger mutations based on the following assumptions:
A driving mutation should occur in multiple tumors more often than would be expected by chance (Figure 1A).
A driving mutation may be associated (correlated) with the expression of a group of genes that form a ‘module’ (Figure 1B).
Copy Number Aberrations often influence the expression of genes in the module via changes in expression of the driver (Figure 1C).
RESULTS
Driving mutations are frequently associated with the abnormal regulation of processes such as proliferation, differentiation, motility and invasion. Given that many cancer phenotypes are reflected in coordinated differences in the expression of multiple genes (a module) (Golub et al., 1999; Segal et al., 2004), a driving mutation might be associated with a characteristic gene expression signature or other phenotypic output representing a group of genes whose expression is modulated by the driver. Additionally, CNAs do not typically alter the coding sequence of the driver and so are expected to influence cellular phenotype via changes in the driver’s expression. In consequence, changes in expression of the driver are important and so approaches that measure association between the expression of a candidate driver (as opposed to its copy number) and that of the genes in the corresponding module are likely to promote the identification of drivers.
Gene expression is particularly useful for identifying candidate drivers within large amplified or deleted regions of a chromosome: whereas genes located in a region of genomic copy gain/loss are indistinguishable in copy number, expression permits the ranking of genes based on how well they correspond with the phenotype (Figure 1D). CNA data aids in determining the direction of influence, which cannot be derived based on correlation in gene expression alone. This permits an unbiased approach for identifying candidate drivers from any functional family, beyond transcription factors or signaling proteins.
Figure 1. Modeling assumptions.
For all heat maps, each row represents a gene and each column represents a tumor sample. A. The same chromosome in different tumors, orange represents amplified regions. The box shows regions amplified in multiple tumors. B. An idealized signature in which the target genes are up-regulated (red) when the DNA encoding the driver is amplified (orange). C. A driver may be overexpressed due to amplification of the DNA encoding it, or due to the action of other factors. The target genes correlate with driver gene expression (middle row), rather than driver copy number (top row). D. Data representing amplified region on chromosome 17. Heat maps of expression for 10/24 genes that passed initial expression filtering (Supplementary methods). Samples are ordered according to amplification status of the region (Orange amplified, blue deleted). These genes are identical in their amplification status and while gene expression is correlated with amplification status to some degree, the expression of each gene is unique. It is these differences that facilitate the identification of the driver. See also Supplementary Methods, Figure S1, Table S1.
A Bayesian Network Based Algorithm to Identify Driver Genes
We developed a computational algorithm, COpy Number and EXpression In Cancer (CONEXIC), that integrates matched copy number (amplifications and deletions) and gene expression data from tumor samples to identify driving mutations and the processes they influence. CONEXIC is inspired by Module Networks (Segal et al., 2003), but has been augmented by a number of critical modifications that make it suitable for identifying drivers (see Supplementary Methods). CONEXIC uses a score-guided search to identify the combination of modulators that best explains the behavior of a gene expression module across tumor samples and searches for those with the highest score within the amplified or deleted region (Supplementary methods, Figure S1).
The resulting output is a ranked list of high scoring modulators that both correlate with differences in gene expression modules across samples and are located in amplified or deleted regions in a significant number of these samples. The fact that the modulators are amplified or deleted indicates that they are likely to control the expression of the genes in the corresponding modules (see Figure 3). Since the modulators are amplified or deleted in a significant number of tumors, it is reasonable to assume that expression of the modulator (altered by copy number) contributes a fitness advantage to the tumor. Therefore, the modulators likely include genes whose alteration provides a fitness advantage to the tumor.
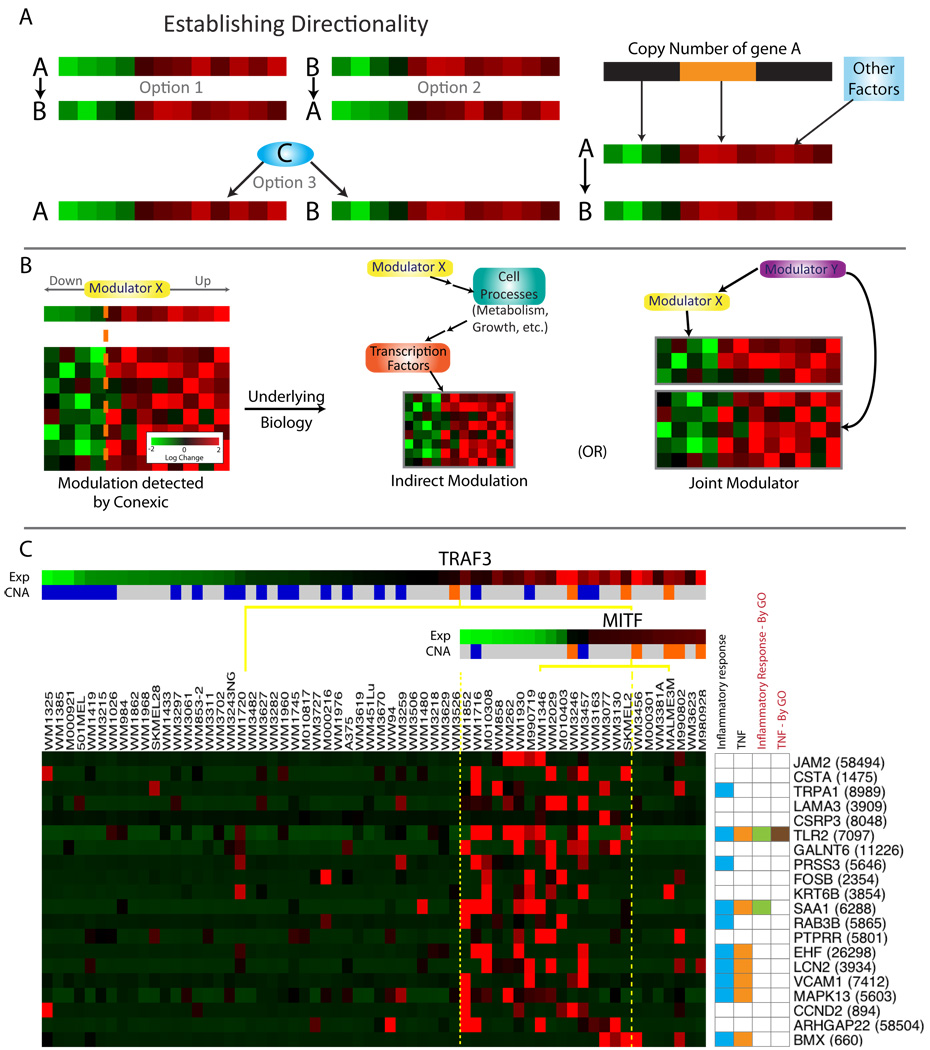
Figure 3. Associating modulators to genes.
A. Three scenarios could explain a correlation between a candidate driver (gene A) and its target (gene B): A could influence B, B influence A, or both could be regulated by a common third mechanism (Pearl, 2000). The availability of both gene expression and chromosomal copy number data allows us to establish the likely direction of influence. If the expression of gene A is correlated with its DNA copy number, and the copy number is altered in a large number of tumors, it is likely that the copy number alteration results in a change in expression of A in these tumors. So the model in which A influences the expression of B and other correlated genes is the most likely. In this way, examination of both copy number and gene expression in a single integrated computational framework facilitates identification of candidate drivers. B. Modulator influence on a module can go beyond direct transcriptional cascades involving transcription factors or signaling proteins and their targets. Genetic alteration of any gene (e.g., a metabolic enzyme) can alter cell physiology, which is sensed by the cell and subsequently leads to a transcriptional response through a cascade of indirect influences and mechanisms. While modules are typically enriched for genes influenced by the modulator, they also contain genes that are coexpressed with the modulator ('joint modulator'). Both types are helpful for annotating the module and determining the functional role of the modulator. C. The TNF module. The modulators include TRAF3 and MITF, where high TRAF3 and low MITF are required for upregulation of the genes in the module. The annotation for each gene is represented in a color-coded matrix. Blue and orange squares represent literature-based annotation (see Table S3); green and brown are from GO. LitVAN associated the genes in this module with TNF and the inflammatory response. See also Figure S3, Table S3.
Identifying candidate driver genes in melanoma
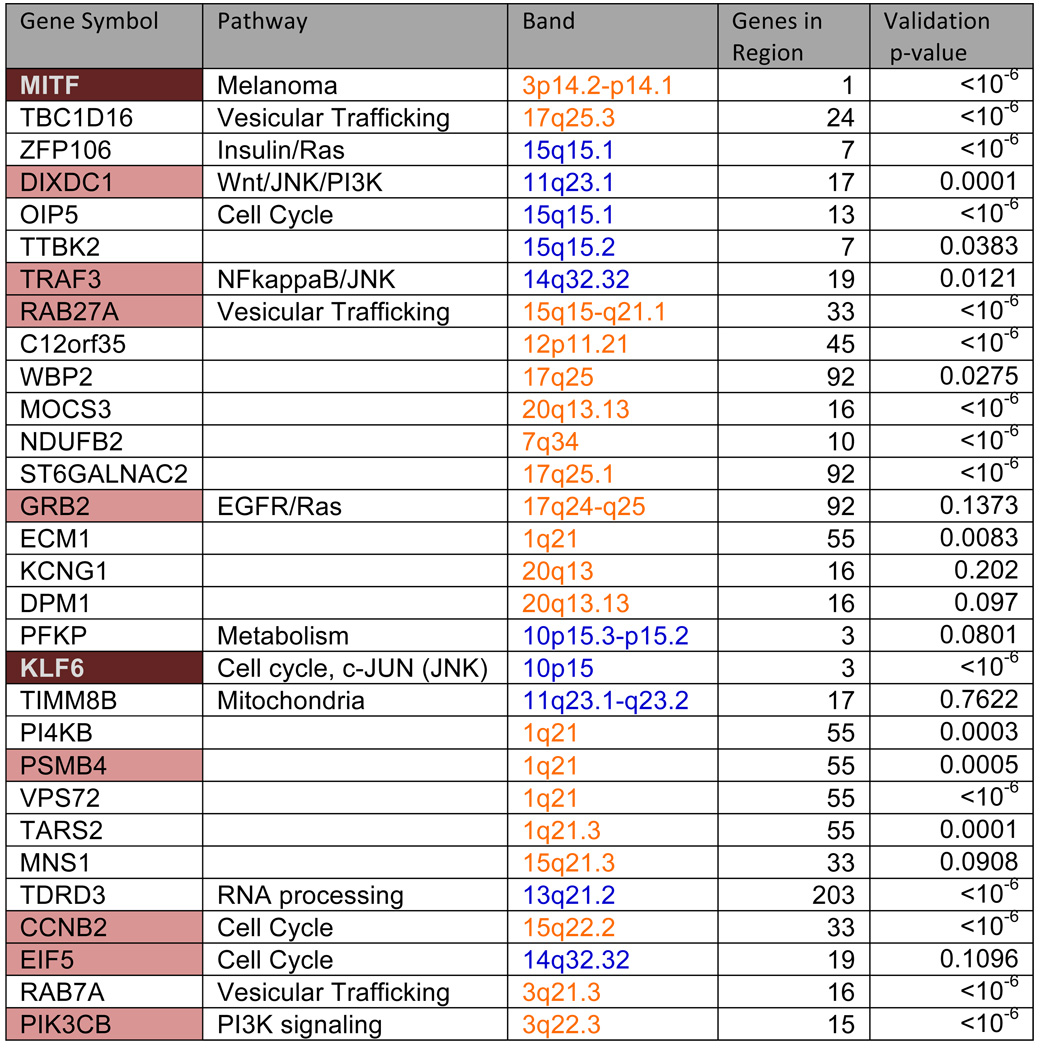
We applied the CONEXIC algorithm to paired gene expression and CNA data from 62 cultured (long and short term) melanomas (Lin et al., 2008). A list of candidate drivers was generated using copy number data available for 101 melanoma samples by applying a modified version (Sanchez-Garcia et al., 2010) of GISTIC (Beroukhim et al., 2007) (see Table S1). Next, we integrated copy number and gene expression data (available for 62 tumors) to identify the most likely drivers (Supplementary methods). Statistical power is gained by integrating all data, and by combining statistical tests on thousands of genes to support the selected modulators. This resulted in the identification of 64 modulators that explain the behavior of 7869 genes. We consider the top 30 scoring modulators, presented in Figure 2, as likely drivers (see Table S2 for the complete list).
Figure 2. The highest scoring modulators identified by CONEXIC.
Gene names are color-coded based on the role of the gene in cancer, 10 genes have been previously identified as oncogenes or tumor suppressors (peach), of these 3 in melanoma (brown). Column 3 represents chromosomal location, where orange represents amplification and blue represents deletion. These genes were identified within regions containing multiple genes, the number of genes in each aberrant region is listed in column 4. Column 5 lists the p-value for modulator validation in independent data (for a full list, see Table S2 and Figure S2C). p-values are shown for the Johansson dataset, unless the modulator was missing from this dataset, and then p-value from the Hoek dataset is shown. See also Supplementary Methods, Table S2, Figure S2.
Many modulators are involved in pathways related to melanoma
The top 30 modulators (likely drivers) include 10 known oncogenes and tumor suppressors (Figure 2). In many cases, CONEXIC chose the cancer related gene out of a large aberrant region containing many genes. For example, DIXDC1, a gene known to be involved in the induction of colon cancer (Wang et al., 2009b), was selected among 17 genes in an aberrant region (Figure S3). CCNB2, a cell cycle regulator, was selected from a large amplified region containing 33 genes. The modulators span diverse functional classes including: signal transducers (TRAF3), transcription factors (KLF6), translation factors (EIF5) and genes involved in vesicular trafficking (RAB27A).
Performing a comprehensive literature search for all genes is tedious and time consuming, so we developed an automated procedure, LitVAn - Literature Vector Analysis, that searches for over-represented terms in papers associated with genes in a gene set. LitVAn uses a manually curated database (NCBI Gene) to connect genes with terms from the complete text of more than 70,000 published scientific articles (Supplementary Methods). LitVAN found a number of over represented terms (Figure S2E) among the top 30 modulators, including ‘PI3K’ and ‘MAPK’, which are known to be activated in melanoma, ’cyclin’, representing proliferation which is common in all cancers and ‘RAB’. Rabs regulate vesicular trafficking, a process not previously implicated in melanoma (Chin et al., 2006).
The association between a modulator and the genes in a module
Beyond generating a list of likely drivers (modulators), the CONEXIC output includes groups of genes that are associated with each modulator (modules). We tested how reproducible the modulators and their associated modules are using gene expression data from two other melanoma cohorts with 45 (Hoek et al., 2006) and 63 (Johansson et al., 2007) samples (see Supplementary methods, Figure S2). We found that 51/64 (80%) of the selected modulators are conserved across datasets in a statistically significant manner. Modules (statistically associated genes) are likely enriched with genes whose expression is biologically affected by the modulator (Figure 3). In consequence, the processes and pathways represented by genes in a module can help us to gain insight into how an aberration in the modulator might alter the cellular physiology and contribute to the malignant phenotype.
Annotation of data-derived sets of genes is typically carried out based on gene set enrichment using Gene Ontology (GO) annotation. Although this approach is useful, there are modules for which GO annotation does not capture the known biology. For example, the 'TNF module' is enriched with the GO terms 'developmental process' and 'cell differentiation' (q-value=0.0014 and 0.004 respectively). We used LitVAn to carry out a systematic literature search and found 11/20 genes in the module related to the TNF pathway, inflammation or both (Figure 3C, Table S3), although only 2 of these genes were annotated for these processes in GO. TRAF3, the modulator chosen by CONEXIC, is known to regulate the NF-kappa-B pathway (Vallabhapurapu et al., 2008), a major downstream target of TNF. Although TRAF3 has not been previously implicated in melanoma, the importance of the NF-kappa-B pathway in melanoma is well supported (Yukiko and Ann, 2006).
A known driver, MITF, is correctly associated with target genes
CONEXIC identified microphthalmia-associated transcription factor (MITF) as the highest scoring modulator. MITF is a master regulator of melanocyte development, function and survival (Levy et al., 2006; Steingrimsson et al., 2004) and the overexpression of MITF is known to have an adverse effect on patient survival (Garraway et al., 2005).
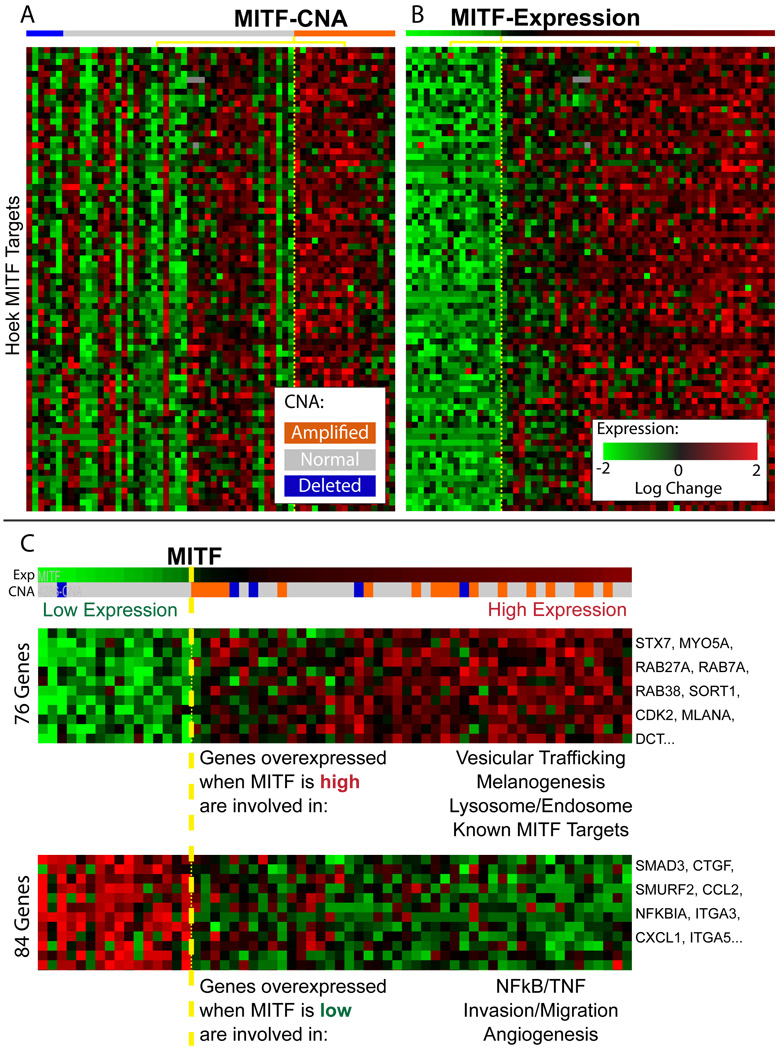
To test the association between modulator and module, we obtained an experimentally derived list of MITF targets (Hoek et al., 2008b) and asked whether the modules identified by CONEXIC associate MITF with its known targets. The MITF associated modules contained 45/80 previously identified targets (p-value < 1.5e−45) supporting a match between the transcription factor (TF) and its known targets. However, a few targets (TBC1D16, ZFP106 and RAB27A) are both associated with MITF and are themselves modulators of additional modules. CONEXIC limits each gene to a single module, so association with an MITF target would preclude association with MITF. If we permit indirect association to MITF through the modules of these additional modulators, CONEXIC correctly identifies 76 of the 80 targets identified by Hoek et. al. (p-value < 1.5e−78). Similar target sets are not available for any other modulator, precluding a more rigorous evaluation of our other predictions.
MITF expression correlates with targets better than copy number
Expression of MITF correlates with the expression of its targets better than MITF copy number, though both correlations are statistically significant (p-value of 0.0001 versus 0.04, Figures 4A and 4B). This relationship is unidirectional: MITF is significantly overexpressed when its DNA is amplified (p-value 0.0004), but over-expressed MITF does not always correspond with MITF amplification. We find that MITF is less correlated with its copy number (rank 294th) than most other genes in aberrant regions (see Table S1C) and more than half of the tumors that over-express MITF do not have a CNA that spans the MITF gene. Comparison of MITF target expression between samples with and without MITF amplification did not show an effect of DNA amplification on expression of the targets (Supplementary methods).
Figure 4. MITF expression correlates with expression of the genes in the associated module.
A. Each row represents the gene expression of one of 78 MITF targets identified by Hoek (Hoek et al., 2008b); the tumor samples are split into two groups based on the copy number of MITF (Welch t-test p-value=0.04) B. The rows represent the same genes, in the same order as in A, but here the tumor samples are split into a group of samples that express MITF at high (n=46) or low levels (n=16) Welch t-test p-value=0.0001). C. Two modules associated with MITF, showing a selected subset of genes. LitVAN annotation for the genes in each module is shown below the heat map. The complete modules with all genes are available in Figure S4.
MITF correctly annotated with its known role in melanoma
We used LitVAN to identify the biological processes and pathways represented in each module associated with MITF. The module containing the genes most significantly upregulated by MITF (Figure 4B, Figure S4A) is significantly enriched for the terms 'melanosome' and 'pigment granule' (q-value= 4.86e−6 for each). It includes targets involved in proliferation such as CDK2, consistent with the observation that MITF can promote proliferation via lineage specific regulation of CDK2 (Du et al., 2004). The module containing genes most strongly inhibited by MITF (Figure 4B, Figure S4B) has a metastatic signature strongly associated with invasion, angiogenesis, the extracellular matrix and NF-kappa-B signaling. These modules and their annotation suggest that MITF serves as a developmental switch between two types of melanoma, where high MITF expression promotes proliferation and low MITF expression promotes invasion. Thus our automated, computationally derived findings dissect a complex response and accurately recapitulate the known literature, including the experimental characterization of MITF (Hoek et al., 2008a).
LitVAN annotated additional modulators with their known role (e.g., CCNB2 with cell cycle and mitosis, data not shown). The detailed match between the CONEXIC output and empirically derived knowledge of the role of known modulators in melanoma provides confidence in CONEXIC’s predictions for modulators that are not well characterized.
Identification of TBC1D16 as a tumor dependency in melanoma
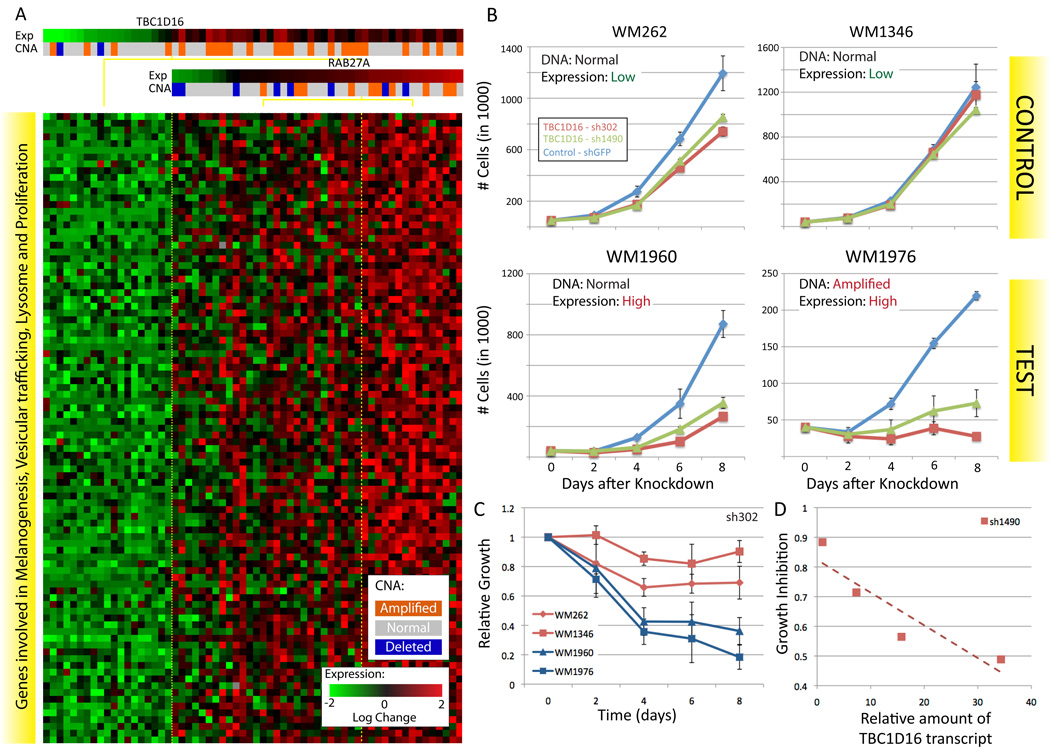
The second highest scoring modulator identified by CONEXIC is TBC1D16, a Rab GTPase-activating protein of unknown biological function. Rabs are small monomeric GTPases, involved in membrane transport and trafficking. TBC1D16 is well conserved and although its targets are not known, a close paralog, TBC1D15, regulates RAB7A (also selected as a modulator, Figure 2) (Itoh et al., 2006). We used a module associated with TBC1D16 to infer its potential role in melanoma (Figure 5A), and discovered that diverse biological processes are represented by genes in the module and that more than half are annotated for processes such as melanogenesis, vesicular trafficking and survival/proliferation (Table S4A). This suggests that TBC1D16 plays a role in cell survival and proliferation.
Figure 5. TBC1D16 is necessary for melanoma growth.
A. A module associated with TBC1D16 and RAB27A, the genes in the module are involved in melanogenesis, survival/proliferation, lysosome and protein trafficking (see Table S4A for details). B. Representative growth curves for each of the 4 STCs infected with TBC1D16 shRNA, each curve represents 3 technical replicates. RT-PCR was used to confirm that the reduction in the amount of the TBC1D16 transcript was similar for all of the STCs (Figure S5). C. Change in growth over time, relative to the number of cells plated, averaged over all replicates (Supplementary methods). Mean over 3 biological replicates X 3 technical replicates for each STC, see Figure S5 and Table S4B for additional replicates and hairpins. D. Growth inhibition at 8 days is directly proportional to the amount of the TBC1D16 transcript and is independent of the TBC1D16 copy number.
TBC1D16 is an uncharacterized gene located in an amplified region that contains 23 other genes, including CBX4, which is known to play a role in cancer (Satijn et al., 1997). Expression of TBC1D16 is not highly correlated with TBC1D16 copy number, compared to other genes in the region (ranked 7th out of 24) or to all candidate drivers (252th out of 428). Nevertheless, TBC1D16 is the top scoring gene in the region and the 2nd highest scoring modulator, so it was selected for experimental verification
The module exhibits a dose-response relationship between TBC1D16 expression and the expression of genes in the module such that higher expression of TBC1D16 is correlated with higher expression of genes in the module (correlation coefficient 0.76). We carried out western blotting and RT-PCR on some of the short term cultures (STCs) used to generate the Lin dataset and asked whether the TBC1D16 transcript correlates with protein levels. The results confirmed that the expression of TBC1D16 corresponds well with the amount of the 45kD form of TBC1D16 (data not shown). These results suggest that knockdown of TBC1D16 expression in tumors that have high levels of TBC1D16 will lead to a reduction in proliferation.
TBC1D16 is required for proliferation
To test whether TBC1D16 is required for proliferation of melanoma cultures we carried out a knockdown experiment. We selected two STCs with high levels of TBC1D16, WM1960 (16-fold greater expression than WM1346, DNA not amplified) and WM1976 (34-fold greater expression, amplified DNA) and control STCs, WM262 and WM1346 that express TBC1D16 at a lower level. We used two shRNAs to knock down TBC1D16 expression in each of the four STCs and measured growth over 8 days (Supplementary methods). RT-PCR was used to confirm that the reduction in the amount of the TBC1D16 transcript was similar for all of the STCs (Figure S6). Knockdown of TBC1D16 expression reduced cell growth in WM1960 and WM1976 to 16% and 40%, respectively, relative to controls infected with GFP shRNA in the same STCs (Figures 5B, C and D). This result is specific for cultures with high levels of TBC1D16, as the controls, WM262 and WM1346, grow at similar rates to cultures infected with shGFP (75%–90%). As predicted, growth inhibition at day 8 is proportional to the amount of the TBC1D16 transcript and is independent of TBC1D16 copy number (Figures 5C and D). Taken together, these results support CONEXIC’s prediction that TBC1D16 is required for proliferation in melanomas that over express the gene.
RAB27A identified and experimentally confirmed as a tumor dependency
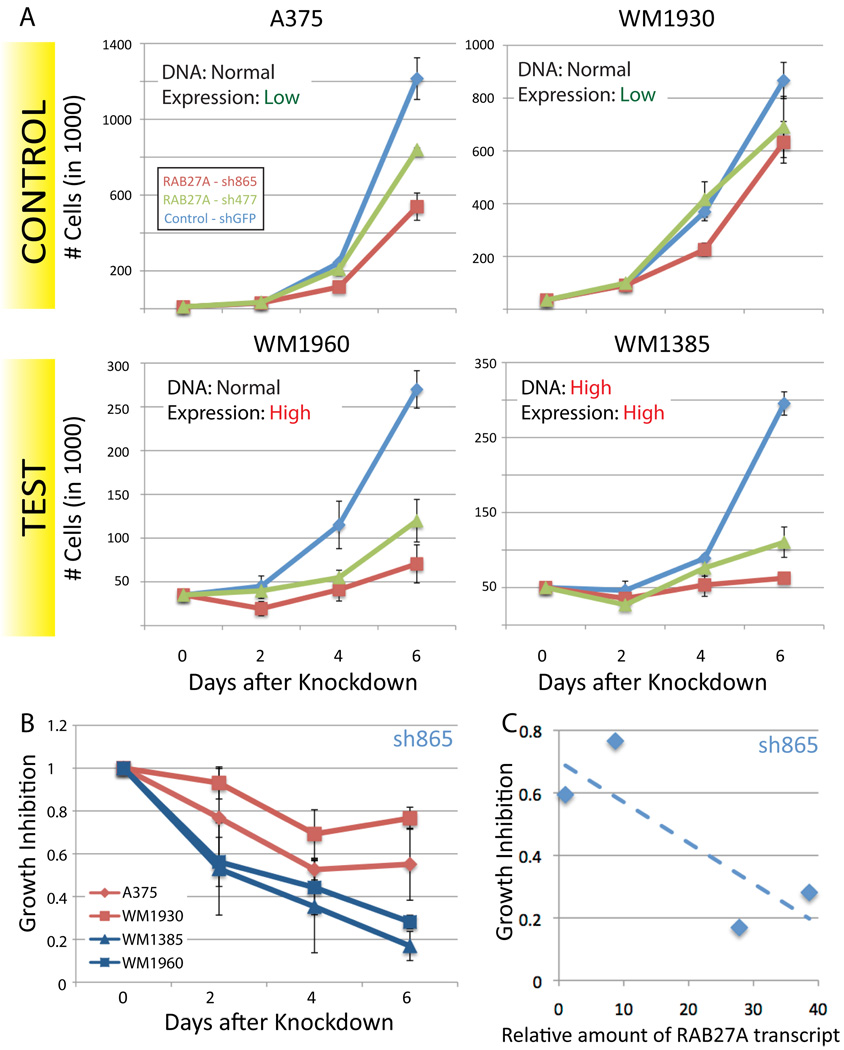
The TBC1D16 module contains a second modulator, RAB27A, also known to be involved in vesicular trafficking (Figure 5A). RAB27A functions, with RAB7A, to control melanosome transport and secretion. RAB7A localises to early melanosomes, while RAB27A is found in mature melanosomes (Jordens et al., 2006). CONEXIC selected both RAB27A and RAB7A as modulators.
RAB27A is in an amplified region that did not pass the standard GISTIC q-value threshold for significance and expression of the gene is not highly correlated with RAB27A copy number, compared to other candidate drivers (323th out of 428). Nevertheless, CONEXIC identified it as the top-scoring modulator out of the 33 genes in this region, and ranked it 8th out of 64 modulators and it was therefore selected for empirical assessment.
To test the prediction that RAB27A is important for proliferation in tumors with high levels of RAB27A, we tested the effect of shRNA knockdown of the RAB27A transcript on proliferation. We chose two STCs in which the gene is highly expressed WM1385 (28-fold greater expression compared with A375, DNA amplified) and WM1960 (38-fold greater expression, DNA not amplified) and two controls that express RAB27A at a lower level (A375 and WM1930). Western blots show that expression of RAB27A correlates with expression of the cognate gene in these cultures (data not shown).
Knockdown of RAB27A expression using shRNA was similar for all cultures (Figure S6), but only reduced cell growth significantly in the STCs that overexpress RAB27A (18% or 35% in WM1385 or WM1960 relative to the same cultures infected with GFP shRNA). RAB27A shRNA had less impact (growth rates of 65–80%) in the control STCs that have low RAB27A (Figures 6A and B). Growth inhibition at 6 days is correlated with the amount of the RAB27A transcript and is independent of RAB27A copy number (Figures 6B and C). Taken together, these results support CONEXIC’s prediction that RAB27A is a tumor dependency in melanomas that overexpress RAB27A.
Figure 6. RAB27A is necessary for melanoma growth.
A. Representative growth curves for each of the 4 STCs infected with RAB27A shRNA, each curve represents 3 technical replicates. RT-PCR was used to confirm that the reduction in the amount of the RAB27A transcript was similar in all of the STCs (Figure S6). B. Change in growth over time, relative to the number of cells plated, averaged over all replicates. Knockdown of RAB27A expression in cells that express this gene at high levels reduces proliferation. Data averaged over all replicates for each STC, see Figure S6 and Table S5 for all data. C. Growth inhibition at 6 days is dependent on the amount of the RAB27A transcript and is independent of RAB27A copy number.
RAB27A affects the expression of genes in associated modules
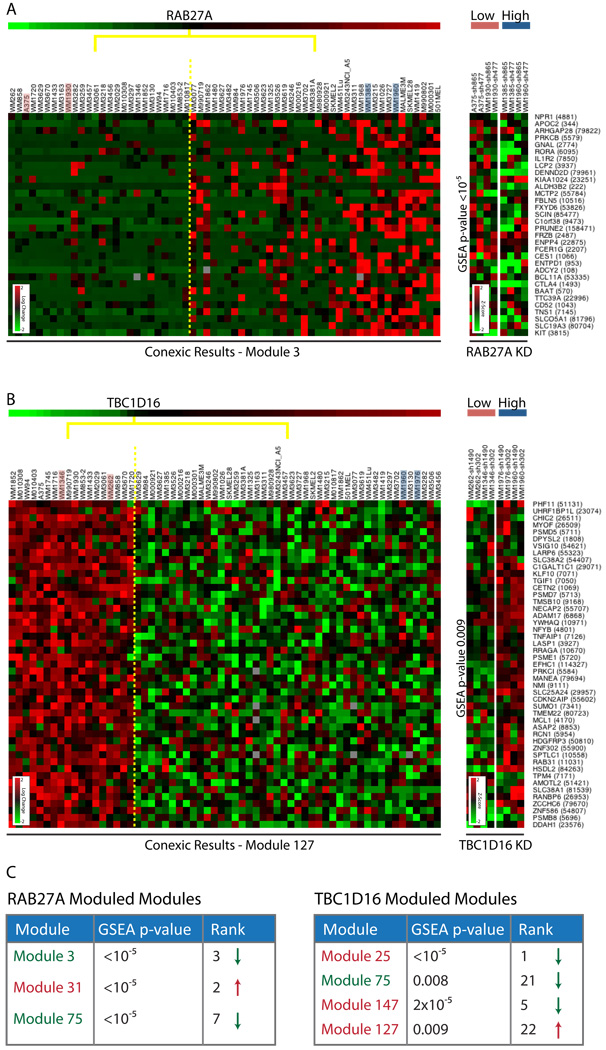
To test whether RAB27A affects the expression of genes in associated modules, as predicted by CONEXIC, we carried out microarray profiling after knockdown of RAB27A in the test STCs (WM1385, WM1960). We compared the expression profile after RAB27A knockdown to a control profile generated by infecting the same STC with GFP shRNA. We used Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) to test whether each of the 3 modules associated with RAB27A are enriched with genes that are differentially expressed (DEG) after knockdown (see Supplementary Methods). We found that all 3 RAB27A associated modules are significantly enriched for genes affected by RAB27A (p-values < 10−5 for all 3 modules, see Figure 7C), and that these modules responded in the direction predicted by CONEXIC.
Figure 7. Results of knockdown microarrays for RAB27A and TBC1D16.
A. To the left is one of the modules associated with RAB27A and to the right data generated following knockdown (KD) of RAB27A for the same genes in the STCs indicated (pink and blue). The expression of genes in the module goes down relative to shGFP as predicted, KD expression heatmap shows Z-scores (see Supplementary Materials) showing that these are some of the most differentially expressed genes (DEGs) in the genome. B. To the left is one of the modules associated with TBC1D16 and to the right data generated following KD of TBC1D16 in the STCs indicated. The expression of genes in the module goes up relative to shGFP, as predicted. The test STCs (blue) and control STCs (pink) respond differently demonstrating the importance of context (TBC1D16 over-expression status) in determining the response. C. GSEA p-value and ranking (relative to 177 CONEXIC modules) for RAB27A and TBC1D16 associated modules (see Figure S7 for data). GSEA was calculated using the median of 4 profiles (2 cell lines X 2 hairpins) on the test STCs. Significant p-values indicate that knockdown of RAB27A and TBC1D16 each affect the subset of genes predicted by CONEXIC (note that 10−5 is the smallest p-value possible given that 100,000 permutations are used). The color of the module name represents the predicted direction of response to knockdown (red and green represent up and down regulated, respectively). The arrow represents the observed response to knockdown. The direction of response was correctly predicted for 2/4 TBC1D16 modules and for all RAB27A modules. See also Figure S7, Table S6.
These results support our computational prediction that the expression of RAB27A affects the expression of the genes in the associated modules. We note that RAB27A functions as vesicular trafficking protein, suggesting that it influences gene expression through an unknown, and likely indirect, mechanism. We used LitVAN to identify the biological processes and pathways represented among the DEGs. Cell cycle related terms are significant among the down-regulated genes, which might be expected given the reduced growth after RAB27A knockdown. In addition, we found that genes annotated for the Erk pathway are up-regulated (including MYC, FOSL1 and DUSP6). We used GSEA to measure enrichment of an experimentally derived set of genes that respond to MEK inhibition in melanoma (Pratilas et al., 2009). The resulting p-value < 4.7e−5 suggests that ERK signaling is altered after RAB27A knockdown in these STCs.
TBC1D16 influences the expression of genes in associated modules
We carried out microarray profiling after knockdown of TBC1D16 to evaluate whether expression of TBC1D16 affects the expression of genes in the 4 modules associated with it. We used two shRNAs to knock down TBC1D16 in the test STCs (WM1960, WM1976) and compared the gene expression to controls infected with GFP shRNA (in the same STCs). GSEA analysis established that all 4 modules are significantly enriched for genes affected by differences in TBC1D16 expression (p-values < 10−5, 0.0002, 0.008 and 0.009 respectively, see Figure 7). Two modules responded to TBC1D16 knockdown in the direction predicted by CONEXIC. In addition, GSEA analysis ranked genes in the TBC1D16 module (Module25) highest out of 177 (based on the GSEA p-value), demonstrating that the genes in this module are the most highly differentially expressed genes in the data set.
The function of TBC1D16 is unknown, but it is predicted to be involved in vesicular trafficking. In our knockdown analysis LitVAN annotated the up-regulated genes with terms related to vesicular trafficking. These include RAB3C, RAB7A, CHMP1B, RAB18, SNX16, COPB1 and CAV1 (see Table S6A). However, it is not clear how TBC1D16 affects gene expression or how changes in expression impact vesicular trafficking.
DISCUSSION
We have demonstrated that combining tumor gene expression and copy number data into a single framework increases our ability to identify likely drivers in cancer and the processes affected by them. Gene expression allows us to distinguish between multiple genes in an amplified or deleted region (many of which are indistinguishable based on copy number) and to identify those that are likely to be drivers. The combination of data types allows us to identify regions that would be overlooked using methods based on DNA copy number alone.
Expression of a driver, not its copy number, drives phenotype
The novelty of our method and the key to its success is our modeling paradigm: the expression of a driver should correspond with the expression of genes in an associated module. Examination of MITF and its targets supports our assumptions. Expression of MITF best correlates with the expression of its targets, but MITF overexpression does not always correspond with MITF amplification. A change in DNA copy number is only one of many ways that gene expression can be altered. For example, MITF expression can be upregulated via signaling from the Ras/Raf (oncogenic BRAF occurs frequently in melanoma) (Wellbrock et al., 2008) and Frizzled/Wnt pathways (Chin et al., 2006).
Most methods for identifying drivers within aberrant regions focus on genes whose expression is well correlated with the copy number of the cognate DNA (Lin et al., 2008; Turner et al., 2010). The expression of many of the predicted drivers we identify is poorly correlated with their copy number, relative to other genes in the region and to all other candidate drivers MITF (294th), TBC1D16 (252th) and RAB27A (323th) (see Table S1C). We believe the discrepancies between CNA and expression arise because there are multiple ways to up or down-regulate a gene. For example, TBC1D16 and RAB27A were both identified as transcriptional targets of MITF (Chiaverini et al., 2008; Hoek et al., 2008b), and are therefore up-regulated when MITF is over-expressed. Moreover, we postulate that many drivers are less correlated with their copy number than passengers due to selective pressure; if there is a fitness advantage to up or down regulate expression, the tumor will find a mechanism to do so.
TBC1D16 and RAB27A are required for proliferation
We tested two drivers predicted by CONEXIC with knockdown experiments, and showed that tumors that express either TBC1D16 or RAB27A at high levels are dependent on the corresponding gene for growth. Our results demonstrate that these dependencies are determined by expression of the gene (in both cases), rather than DNA amplification status, further supporting the assumptions underlying our approach. Thus, we not only identify tumor dependencies, but also the tumors in which these genes are crucial for proliferation. Identifying dependencies that are critical for tumor survival is needed for drug targeted therapies. For example, FLT3 inhibitors in AML, which have had successful phase II trials (Fischer et al., 2010). Our approach is unbiased with respect to protein function and does not incorporate prior knowledge, thus enabling the identification of dependencies in genes involved with vesicular trafficking. TBC1D16 and RAB27A validate the ability of our approach to correctly identify tumor dependencies and the genes that they affect.
Association between modulator and module
A key feature of our approach is that CONEXIC goes beyond identifying drivers. By associating candidate drivers with gene modules and annotating them using information from the literature, CONEXIC provides insight into the physiological roles of drivers and associated genes. We used LitVAn to find biological processes and pathways overrepresented in each module and to associate drivers with functions, accurately identifying targets of MITF and annotating the functions of known drivers (MITF, CCBN2 and TRAF3).
The results of microarray profiling following knockdown further support the association between modulator and module and confirm our ability to identify genes affected by TBC1D16 and RAB27A. We successfully connected genes involved in vesicular trafficking to their effects on gene expression, likely through a cascade of indirect influences. In addition to profiling the STCs that highly express each of these genes (test STCs), we also profiled two lower expressing STCs (control STCs), in which the effect of knockdown is less detrimental to growth. For TBC1D16, there is substantial overlap in the DEGs in the test STCs (p-value < 6.6e−22), but not in the DEGs between control and test STCs (p-value > 0.76). This reflects the complexity of the transformed state and demonstrates that genetic context has a fundamental impact on the effect of a perturbation.
Genes involved in trafficking are important in melanoma
Of the top 30 drivers selected by CONEXIC, three genes (TBC1D16, RAB27A and RAB7A) are known to be involved in vesicular trafficking (Itoh et al., 2006; Jordens et al., 2006). All of these genes are amplified (DNA) and highly expressed (RNA) in multiple melanomas. There is increasing evidence that genes controlling trafficking play a role in melanoma. Germline variation in GOLgi PHosphoprotein 3 (GOLPH3), a gene involved in vesicular trafficking, is associated with multiple cancers (Scott et al., 2009). Our data identifies two novel dependencies that are encoded in somatic CNAs, demonstrates the dependency of melanoma on TBC1D16 and RAB27A expression for proliferation and highlights the potential role of vesicular trafficking in this malignancy.
The role of vesicular trafficking in melanoma has yet to be characterized. Vesicular trafficking regulates many receptor tyrosine kinases (RTKs) both spatially and temporally and thus determines both the duration and intensity of signaling (Ying et al., 2010). For example, RAB7A is involved in the regulation of ERK signaling (Taub et al., 2007), and ERK is known to play an important role in melanoma (Chin et al., 2006). Tight control of ERK expression could potentially be important in melanocytes because of its influence on MITF: ERK is required for the activation of MITF, but high levels of ERK lead to MITF degradation (Wellbrock et al., 2008). It is possible that recurrent aberrations in vesicular trafficking genes might involve control of ERK signaling intensity. This is further supported by the up-regulation of an ERK signature (Pratilas et al., 2009) following RAB27A knockdown in our data (p-value < 4.7e−5).
CONEXIC and other approaches
CONEXIC differs from other methods in a number of ways. First, it uses the gene expression of a candidate driver, rather than its copy number as a proxy to report on the status of the gene, e.g., two tumors that overexpress a driver are treated equivalently, even if there is amplification in the DNA of only one of them. Second, it associates a candidate driver with a module of genes whose expression corresponds with that of the predicted driver, which was critical for identification of TBC1D16 as a modulator. Third, combining copy number and gene expression provides greater sensitivity for identifying significantly aberrant regions that would not be selected based on DNA alone, this was critical for the identification of RAB27A.
Methods based on copy number data are limited to detecting large regions containing multiple genes, such that the driver cannot be readily identified among them. Recent efforts have focused on integrating additional sources of information into the analysis. Some methods use prior information, such as the role of a gene in other cancers (Beroukhim et al., 2010). Others, like CONEXIC, integrate gene expression data (Adler et al., 2006), but the results of these methods fall short of CONEXIC's. We systematically compared CONEXIC to other methods using the same data and found that they did not identify MITF, or any other known driver in melanoma (see Supplementary Methods).
Statistical dependencies in gene expression have been used to connect a regulator to its target (Friedman et al., 2000; Lee et al., 2006; Segal et al., 2003) and for uncovering important regulators in cancer (Adler et al., 2006; Carro et al., 2010; Wang et al., 2009a). These approaches typically only detect transcription factors and signaling molecules and do not connect the altered regulatory networks to upstream genetic aberrations.
Incorporating information on amplification or deletion status allows us to consider any functional class of genes and thus permits detection of vesicular trafficking genes that would not be identified using other methods. It also allows us to relate the malignant phenotype to genetic aberrations from which it is likely to have originated.
We tuned our method towards reducing the selection of modulators that are not drivers. To gain this specificity, we do not detect all genes and pathways that drive tumors. First, some drivers in amplified and deleted regions do not pass the stringent statistical tests employed in our method. Second, CONEXIC only identifies candidate drivers that are encoded in amplified or deleted regions. In consequence, it would not detect drivers of melanoma such as BRAF and NRAS that are typically associated with point mutations. Third, CONEXIC detects drivers based on the assumptions delineated in Box 1; while these hold for many drivers, it is likely that they are not appropriate for all drivers.
To meet the challenge of finding all driving alterations in cancer, a number of complementary approaches are needed. Experimental approaches such as screening using pooled short hairpin RNAs (shRNAs) (Bric et al., 2009; Zender et al., 2008) are likely to detect a set of drivers different from those detected by CONEXIC. These screens are dependent on the genetic background and are limited to drivers that influence processes that can be readily measured, such as proliferation, whereas CONEXIC scans all the genetic data together and can potentially identify drivers of any function across different genetic backgrounds. In the future we envision that CONEXIC will be used to guide in vivo screening initiatives and to assist in the choice of regions, functional assays and genetic backgrounds probed.
Beyond Melanoma
The challenge of finding candidate drivers is considerable: tumors are heterogeneous, the data are noisy and highly correlated and there are a large number of possible combinations of drivers and genes in modules. Our approach is successful because it couples simple modeling assumptions with powerful computational search techniques and rigorous statistical evaluation of the results at each step.
Both the principles underlying CONEXIC and the software can be applied to any tumor cohort containing matched data for copy number aberrations and gene expression. The principle of associating any type of mutation (e.g., epigenetic alterations, coding sequence) with gene expression signatures or other phenotypic outputs that differ among samples will be of increasing importance as sequence and epigenetic data accumulates. Not only does this help to distinguish between driving and passenger mutations, but the genes in the associated module can also provide insight into the role of the driver. This approach can be used to identify the genetic aberrations responsible for tumorigenesis and to find those that relate to any other measurable phenotype, such as the resistance of tumors to drugs. We anticipate our approach will make an important contribution towards a basic mechanistic understanding of cancer and in revealing associations of clinical significance. Cancer is a heterogeneous disease in which we are only just beginning to appreciate the importance of genetic background and the myriad ways in which the cellular machinery can be redirected towards the transformed state. Methods that begin to dissect this complexity move us another step closer to a world where personalized therapies are routine.
EXPERIMENTAL PROCEDURES
Statistical Methods
A detailed description of the statistical methods and computational algorithms used can be found in the Supplementary Materials. The CONEXIC and LitVAN algorithms were developed for this research, the software is available at: www.c2b2.columbia.edu/danapeerlab/html/software.html
Experimental Methods
Cells were grown using standard culture conditions and knockdown was carried out by infection with lentivirus using RNAi sequences designed by the RNAi Consortium. shRNA lentivirus were prepared according to TRC protocols (http://www.broadinstitute.org/rnai/trc), with minor modifications. Cell proliferation assays, RT-PCR, microarrays and immunoblotting were carried out using standard techniques. Primer sequences and detailed methods can be found in Supplementary Experimental Procedures.
All primary data are available at the Gene Expression Omnibus (GSE23884).
Supplementary Material
Acknowledgements
The authors will like to thank Nir Hacohen, Antonio Iavarone, Daphne Koller, Liz Miller, Itsik Pe’er, Suzanne Pfeffer, Neal Rosen and Olga Troyanskaya for valuable comments. This research was supported by the National Institutes of Health Roadmap Initiative, NIH Director's New Innovator Award Program through grant number 1-DP2-OD002414-01 and National Centers for Biomedical Computing Grant 1U54CA121852-01A1. DP holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and Packard Fellowship for Science and Engineering.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Brunet J-P, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. Patterns of Gene Expression and Copy-Number Alterations in von-Hippel Lindau Disease-Associated and Sporadic Clear Cell Carcinoma of the Kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al. Functional Identification of Tumor-Suppressor Genes through an In Vivo RNA Interference Screen in a Mouse Lymphoma Model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaverini C, Beuret L, Flori E, Busca R, Abbe P, Bille K, Bahadoran P, Ortonne J-P, Bertolotto C, Ballotti R. Microphthalmia-associated Transcription Factor Regulates RAB27A Gene Expression and Controls Melanosome Transport. J Biol Chem. 2008;283:12635–12642. doi: 10.1074/jbc.M800130200. [DOI] [PubMed] [Google Scholar]
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes & Development. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Fischer T, Stone RM, Deangelo DJ, Galinsky I, Estey E, Lanza C, Fox E, Ehninger G, Feldman EJ, Schiller GJ, et al. Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J Clin Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo Switching of Human Melanoma Cells between Proliferative and Invasive States. Cancer Res. 2008a;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008b;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, Satoh M, Kanno E, Fukuda M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes to Cells. 2006;11:1023–1037. doi: 10.1111/j.1365-2443.2006.00997.x. [DOI] [PubMed] [Google Scholar]
- Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20:216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- Jordens I, Westbroek W, Marsman M, Rocha N, Mommaas M, Huizing M, Lambert J, Naeyaert JM, Neefjes J. Rab7 and Rab27a control two motor protein activities involved in melanosomal transport. Pigment Cell Research. 2006;19:412–423. doi: 10.1111/j.1600-0749.2006.00329.x. [DOI] [PubMed] [Google Scholar]
- Lee S-I, Pe'er D, Dudley AeM, Church GM, Koller D. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc Natl Acad Sci USA. 2006;103:14062–14067. doi: 10.1073/pnas.0601852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in Molecular Medicine. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl J. Causality : models, reasoning, and inference. Cambridge, U.K. ; New York: Cambridge University Press; 2000. [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W-L, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia F, Akavia UD, Mozes E, Pe'er D. JISTIC: Identification of Significant Targets in Cancer. BMC Bioinformatics. 2010;11:189. doi: 10.1186/1471-2105-11-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Olson DJ, van der Vlag J, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang M-C, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub N, Teis D, Ebner HL, Hess MW, Huber LA. Late Endosomal Traffic of the Epidermal Growth Factor Receptor Ensures Spatial and Temporal Fidelity of Mitogen-activated Protein Kinase Signaling. Mol Biol Cell. 2007;18:4698–4710. doi: 10.1091/mbc.E07-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010 doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng P-H, Keats JJ, Wang H, Vignali DAA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-[kappa]B signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, Rajbhandari P, Shen Q, Nemenman I, Basso K, Margolin AA, et al. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009a;27:829–839. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cao XX, Chen Q, Zhu TF, Zhu HG, Zheng L. DIXDC1 targets p21 and cyclin D1 via PI3K pathway activation to promote colon cancer cell proliferation. Cancer Sci. 2009b;100:1801–1808. doi: 10.1111/j.1349-7006.2009.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF Regulates Melanoma Proliferation through the Lineage Specific Factor MITF. PLoS ONE. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, Huang J, Dhakal S, Ivanova E, Xiao Y, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci USA. 2010;107:6912–6917. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukiko U, Ann R. NF-kappaB activation in melanoma. Pigment Cell Research. 2006;19:112–124. doi: 10.1111/j.1600-0749.2006.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An Oncogenomics-Based In Vivo RNAi Screen Identifies Tumor Suppressors in Liver Cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.