Abstract
Recent improvements in methods of single-particle fluorescence tracking have permitted detailed studies of molecular motion on the nanometer scale. In a quest to introduce these tools to the burgeoning field of DNA nanotechnology, we have exploited fluorescence imaging with one-nanometer accuracy (FIONA) and single-molecule high-resolution colocalization (SHREC) to monitor the diffusive behavior of synthetic molecular walkers, dubbed “spiders”, at the single-molecule level. Here we discuss the imaging methods used, results from tracking individual spiders on pseudo-one-dimensional surfaces, and some of the unique experimental challenges presented by the low velocities (~3 nm/min) of these nanowalkers. These experiments demonstrate the promise of fluorescent particle tracking as a tool for the detailed characterization of synthetic molecular nanosystems at the single-molecule level.
1. INTRODUCTION
During the last few decades, there has been remarkable growth in the use of fluorescence spectroscopy in biophysical studies. Fluorescence-based tools are now being employed to understand the properties and dynamics of proteins (Min et al., 2005; Giepmans et al., 2006; Schuler and Eaton, 2008; Shi et al., 2008) and nucleic acids (Ditzler et al., 2007; Joo et al., 2008; Pljevaljcic and Millar, 2008; Zhao and Rueda, 2009). They are implemented in cutting-edge applications in medical and clinical chemistry for high-throughput screening and detection (Gribbon et al., 2004; Hintersteiner and Auer, 2008), as well as in cellular imaging for characterizing the localization and movement of intracellular components (Lippincott-Schwartz et al., 2003; Michalet et al., 2005; Moerner, 2007; Walter et al., 2008). Continued advances in fluorescence techniques and instrumentation have fueled applications of fluorescence spectroscopy to more detailed characterization of biomolecules. For example, there has been rapid expansion in the use of nucleotide analogs as fluorescent probes (Rist and Marino, 2002) to detect and characterize single nucleic acid molecules in real-time. Fluorescence resonance energy transfer (FRET) has emerged as a particularly powerful tool to probe distances (Stryer and Haugland, 1967; Ha et al., 1996; Deniz et al., 1999), conformational changes (Weiss, 2000; Truong and Ikura, 2001; Kim et al., 2002), and dynamics (Zhuang et al., 2002; Al-Hashimi and Walter, 2008) of macromolecules on the order of 1–10 nm. To measure larger distances of typically 10 nm or more, single-particle fluorescence tracking has proven useful (Barak and Webb, 1982; Yildiz et al., 2003; Gordon et al., 2004; Churchman et al., 2005).
The accuracy of particle tracking techniques is determined in part by the finite resolution of light microscopy. When light passes through a lens with a circular aperture as it does in a single molecule fluorescence microscope, the focused light emitted from a pointlike source forms a diffraction pattern known as an Airy pattern. The radius of the bright central region, called the Airy disk, can be approximated as λ/(2×N.A.) where λ is the wavelength of the light source and N.A. is the numerical aperture of the lens. Within this radius, according to Rayleigh’s criterion, no features may be resolved (Hecht, 2002). When imaging in the visible spectrum with a typical N.A. of 1.2, the radius of the Airy disk, and therefore the optical resolution limit, is ~250 nm. However, so-termed super-resolution methods have been developed in the last few years that overcome this optical resolution barrier and bring the localization accuracy of a single particle down to the low nanometer range (Betzig and Chichester, 1993; Heintzmann et al., 2002; Qu et al., 2004; Hofmann et al., 2005; Hess et al., 2006; Rust et al., 2006; Moerner, 2007; Huang et al., 2009). It should be noted that we are using the term “super-resolution” here in its broader sense for all techniques that localize (and track over time) one or more single molecule emitters at higher than the diffraction limit of accuracy (Moerner, 2007), while the more narrow sense of the term refers only to imaging techniques that resolve many closely spaced emitters by observing only few of them at a time over a time series of images (Huang et al., 2009).
Low-nanometer localization accuracy can allow for tracking slowly moving molecular devices in real-time. Several years ago, we suggested applying modern single-molecule fluorescence microscopy tools to nanotechnology as a way of monitoring and ultimately enabling control of the assembly and function of the desired structures and devices (Rueda and Walter, 2005). Here, we present in detail the implementation of nanometer-scale tracking of a novel type of autonomously walking DNA nano-assembly termed a “spider” (Lund et al., 2010).
1.1. History of fluorescent single-particle tracking
Fluorescent single-particle tracking entails using the fluorescence emission from a pointlike source to accurately determine its location, typically over a span of time ranging from milliseconds to minutes that depends on the speed of particle motion. One of the first examples of fluorescent particle tracking was accomplished by Barak and Webb in their study of the diffusion of intensely fluorescent (~45 fluorophores) low density lipoprotein (LDL)-receptor complexes along human fibroblasts in which they were able to observe the movement of as few as 1–3 “molecules” in a given region (Barak and Webb, 1982). By using low temperatures, single chromophores were first optically detected in solids by Moerner (Moerner and Kador, 1989). Advancements in single-molecule fluorescence techniques allowed for single-molecule detection in more biologically relevant conditions with ever-improving localization accuracy. For example, near-field optical microscopy (NSOM) brought the tracking error down to ~14 nm in solution at room temperature (Betzig and Chichester, 1993). The high temporal as well as spatial resolution often required for single-particle tracking was accomplished by Schindler and coworkers who successfully combined a low localization error of ~30 nm with a high time resolution of 40 ms, enabling them to study the diffusion of single phospholipids in a phospholipid membrane (Schmidt et al., 1996).
Experimental fluorescent single particle tracking called for the development of its theoretical counterpart. Using a maximum likelihood estimation analysis, Bobroff developed a quantitative method for analyzing the statistical error in position measurements made with light and particle signals, taking into consideration the measurement signal, noise distribution, and instrument resolution. This was done particularly for a Gaussian signal (Bobroff, 1986). Based on Bobroff’s least-squares fitting approach, Webb and coworkers derived a simple equation for calculating the standard error of the mean of the position measurements (σμ) that depends on the instrumentation parameters and features of the Gaussian fit:
| (1) |
where si is the standard deviation of the Gaussian distribution of the ith index that indicates either the x- or y-direction, a is the pixel size, and b is the standard deviation of the background. The first term (si2/N) arises from photon noise, the second term represents the effect of the finite pixel size of the camera, and the third term arises from the background signal of the sample (Thompson et al., 2002; Yildiz et al., 2003). Webb and coworkers were able to determine, according to this equation, the position of stationary beads with ~2 nm localization accuracy (Thompson et al., 2002).
These advances in single-molecule fluorescence imaging and analysis laid a firm foundation for the development of fluorescence imaging with one-nanometer accuracy (FIONA) and related techniques. Developed by Selvin and coworkers, FIONA enabled the localization of singly fluorophore-labeled myosin V motor proteins along microtubules with <1.5 nm error and 0.5 s temporal resolution, using a total internal reflection fluorescence (TIRF) microscope at room temperature. This accomplishment was achieved by maximizing the number of photons collected (to ~5,000–10,000 photons) while optimizing the camera pixel size (to 86 nm) and minimizing the background noise (to a standard deviation of ~11 photons) (Yildiz et al., 2003). Complementing FIONA, Spudich and coworkers developed a technique, termed single-molecule high-resolution colocalization (SHREC), which utilizes two fluorophores of differing emission spectra mapped onto the same space and measures interfluorophore distances with 1-nm precision (based on 482 molecules). To demonstrate this technique, they verified the expected ~36 nm distance between the two heads of myosin V (Churchman et al., 2005).
We adapted FIONA and SHREC, both of which have previously been used to study ATP-fueled biological motor proteins (Yildiz et al., 2003; Churchman et al., 2005), to study synthetic DNA-fueled nanowalkers termed spiders (Lund et al., 2010).
2. DNA-BASED NANOWALKERS
DNA nanotechnology, the study of constructing programmable nanometer-scale structures and devices based on the Watson-Crick base-pairing rules of DNA, has continuously accelerated in pace since its conception in the early 1980’s (Rothemund, 2006; Seeman, 2007; Douglas et al., 2009). DNA nanotechnology has recently yielded synthetic molecular machines that mimic naturally occurring bipedal nanowalkers (Sherman and Seeman, 2004; Shin and Pierce, 2004; Yin et al., 2004; Green et al., 2008; Bath et al., 2009; Omabegho et al., 2009). These DNA-based nanowalkers, consisting of two single-stranded DNA (ssDNA) “legs,” traverse tracks composed of ssDNA strands complementary to the legs in an experimentally controlled direction using thermodynamically favored strand displacement (or exchange) by an ssDNA fuel strand as an energy source (Fig 1a).
Figure 1.
Example mechanisms for processive movement of DNA-based nanowalkers. (A) Biped nanowalker from Green et al. (2008) that utilizes fuel consisting of two complementary DNA hairpins. Colors represent complementary sequences. (i) The competition between identical feet to bind to the track permits the exposure of a toehold region in the left foot (ii). A hairpin hybridizes to the toehold region (iii), displacing the left foot from the track. A second, complementary hairpin hybridizes to the first hairpin (iv) to form a waste product, allowing the foot to rebind to the track with equal probability to the left or right. (B) Single-stranded deoxyribozyme-based nanowalker from Tian et al. (2005). (i) The 10–23 deoxyribozyme (red with orange active site) is able to cleave its substrate (green) at a specific site (purple) in the presence of Mg2+. The shorter end dissociates from its product (ii) and hybridizes to a neighboring strand (iii). Via displacement of the cleavage product by neighboring substrates, the deoxyribozyme progresses along the track (iv). (C) Spider moving along a three-substrate-wide origami track (Lund et al., 2010). The spider is composed of a streptavidin body, a capture leg, spacers, all shown in blue, and three 8–17 deoxyribozyme legs (red binding arms with orange active site). (i) The deoxyribozyme spider legs hybridize to substrates (green) that are attached to the origami scaffold via hybridization to staple overhangs (black). In the presence of Zn2+, each leg of the spider cleaves its substrate, dissociates from its products (ii) and hybridizes to a neighboring strand (iii). The greater affinity of the leg for the substrate than the cleavage products makes it energetically favorable for the strand to bind to the full substrate, generating a biased-random walk from the cleaved towards the uncleaved substrate (iv).
2.1. The molecular “spider”
Due to slow kinetics, strand displacement is not an ideal source of energy for molecular walkers. Biological motor proteins have average velocities of ~20–80 nm/s in vitro (Yildiz et al., 2003; Yildiz et al., 2004; Reck-Peterson et al., 2006), while these synthetic nanowalkers are limited by the kinetics of unwinding one DNA duplex and forming another, both ~15–50 base pairs in length (Sherman and Seeman, 2004; Shin and Pierce, 2004; Yin et al., 2004; Green et al., 2008; Bath et al., 2009; Omabegho et al., 2009), leading to velocities of ~10 nm/hr (Shin and Pierce, 2004). In addition, while it has been predicted that they have the ability to traverse longer tracks, as yet they have only been shown to accomplish a few successive steps along short tracks typically on the lower end of tens of nanometers (Green et al., 2008; Omabegho et al., 2009).
The catalytic power of deoxyribozymes, or DNAzymes, offers an attractive alternative to strand displacement for driving locomotion. DNAzymes are DNA sequences with the ability to site-specifically cleave chimeric DNA-RNA substrates in the presence of an appropriate divalent metal ion cofactor. Tian et al. (2005) were the first to incorporate DNAzymes into their nanowalker in the form of the 10–23 DNAzyme (Santoro and Joyce, 1997). As it cleaves an oligonucleotide on its track, the DNAzyme dissociates from the shorter cleavage product and is able to progress along the track by displacement of the still bound longer product portion by an adjacent substrate strand (Fig. 1b) (Tian et al., 2005). The speed of movement may still be limited by the kinetics of strand displacement despite the shorter cleavage product, and the processivity limited by the risk of complete dissociation of the single leg from its track.
To overcome these limitations, Stojanovic and coworkers recently developed a polypedal DNAzyme-based nanowalker dubbed a “spider”(Santoro and Joyce, 1997; Pei et al., 2006). Spiders consist of a streptavidin “body” bound to multiple biotinylated 8–17-based DNAzyme (Santoro and Joyce, 1997; Li et al., 2000) “legs”. The spider’s multivalent binding allows it to remain securely bound to the surface even as individual DNAzymes cleave their substrate and detach, and therefore allows a large number of substrate sites to be visited and cleaved by a single spider before it dissociates from the surface (Pei et al., 2006). Once a leg cleaves its bound substrate, it can more rapidly dissociate from the 10-nucleotide long products and bind another 18 nucleotide-long substrate in the vicinity. Mathematical modeling of this system suggests that these properties will result in spiders undergoing biased movement on a substrate-field, avoiding sites they have previously visited (Fig. 1c) (Antal and Krapivsky, 2007).
2.2. Behavior-based molecular robots: programming tasks into the walker’s environment
Due to their ability to sense and respond to stimuli (for instance, leg substrates), nano-assemblies such as spiders can be considered behavior-based molecular robots (Brooks, 1991). Although they cannot themselves store complex programs or instructions, one can influence their behavior by exposing them to controlled environmental cues. It is possible to, for example, direct a spider to complete simple tasks such as “start,” “stop,” and “turn” by precisely controlling the position and sequence of the substrates to which the spider has access (Lund et al., 2010). Hence, tracks of substrate can define a program of movement consisting of local environmental cues (e.g., bends in the track, tight binding sites acting as “fly paper”) which are executed by the spider, and represent an initial step towards a world of useful molecular robots.
To assess the capability of spiders to execute such programs, tracks with a feature resolution of 6 nm were engineered using DNA origami technology (Rothemund, 2006). A rectangular origami scaffold was constructed using the 7-kilobase single-stranded M13mp18 genomic DNA, which was shaped and held in place with the aid of 202 oligodeoxynucleotide staple strands that hybridize to complementary regions of the M13mp18 DNA. Specific staple strands in the array were extended on their 5′ end with specific sequences to position three types of surface features on the scaffold. First, a single staple near one corner of the origami tile was extended to contain the START sequence that is partially complementary to the non-catalytic, ssDNA “capture” leg of the spider, which positions the spider at the start of the track. In assembling the spider-origami complex, the spider was first allowed to bind to the START position before any other substrates were added in order to ensure specific binding at the START site. Second, a number of staples were extended by a single specific sequence to permit the site-specific hybridization of cleavable substrate strands, creating a track for the spider to graze. These substrates contain a single ribonucleotide (rA) upstream of G at the cleavage site to permit cleavage by the three DNAzyme legs (Fig. 1c). Six staples at the end of the track were extended with a unique sequence to similarly attach non-cleavable all-DNA substrate analogs (GOAL) intended to trap the spider once it reaches the end of the track (“fly paper”). The GOAL strands and spiders were labeled with the cyanine fluorophores Cy5 and Cy3, respectively, to allow the spider to be tracked by super-resolution fluorescence microscopy relative to the GOAL position. For purposes of immobilizing the origami-spider complexes on avidin-coated quartz slides for TIRF microscopy, four staples near the corners of the origami tile were biotinylated.
To initiate motion of the spider by displacement from the START, a “TRIGGER” DNA strand that is fully complementary to the START sequence was added (“start” command). Zinc (II) ions were then added to promote cleavage of bound substrates by the DNAzyme legs and thus walking (“walk” command). Spiders were predicted to graze the surface until they reach and become trapped at the GOAL position (“stop” command). Scaffolded tracks were designed with either a linear shape or incorporating one left- or right-handed turn, resulting in a program of motion for the spider. All of the tracks were three substrates wide and at least 14 substrates long, thus actualizing a pseudo-one-dimensional path or program of motion for the spiders to follow.
2.3. Methods for characterizing nanowalker behavior
Surface plasmon resonance (SPR) was previously employed to detect movement of spiders with 2–6 legs in a polymer matrix containing a high density of DNAzyme substrate, yielding ensemble estimates of cleavage rate and processivity for spiders (Pei et al., 2006). Native polyacrylamide gel electrophoresis (PAGE) is commonly used to detect procession of DNA-based nanowalkers along tracks, since the site at which the walker is bound to the track will influence the overall topology of the complex and, hence, its electrophoretic mobility (Omabegho et al., 2009). While useful, these ensemble-averaging techniques do not directly report on movement and provide limited information concerning the distribution of possible behaviors for individual nano-assemblies.
One technique Lund et al. (2010) have used to study individual molecular spiders is atomic force microscopy (AFM), which yields detailed, high-resolution “snapshots” of trajectories followed by the nano-assemblies. While AFM yields insight into the behavior of individual spiders, real-time observation of the spider walk is not readily achieved. This is thought to be due to the inhibition of walking by the mica surface necessary for sample immobilization and the potential for mechanical disruption of the sample associated with repeated scanning by the AFM probe.
A complementary single-molecule technique is TIRF microscopy, which permits the visualization of individual fluorescent molecules on a microscope slide by limiting the excitation volume to a thin (~100–200 nm) sheet near the surface of the slide, thereby suppressing background noise. By fluorescently labeling the spider and origami track, we have used this technique to monitor the motion of spiders along prescriptive DNA origami tracks.
3. CONSIDERATIONS FOR FLUORESCENCE IMAGING OF SLOWLY MOVING PARTICLES
While spiders have experimentally been shown to be faster and more processive than previous DNA-based nanowalkers, traversing a 100 nm track with speeds on the order of several nm/min, they are still significantly slower than Myosin V, resulting in heightened challenges for fluorescence imaging due to fluorophore photobleaching and stage drift.
3.1. Prolonging fluorophore lifetime
Fluorophores are subject to permanent photobleaching: After a limited number of excitation events (~106) (Willets et al., 2003) they will remain in a permanent dark state, often induced by reaction of the excited state with molecular oxygen. Optimizing the fluorophore lifetime in order to track a specific fluorescently labeled particle for an extended period of time may be accomplished in multiple ways. The first intuitive way is by illuminating the sample only periodically, rather than continuously. However, this method has the often undesired consequence of reducing the experimental time resolution. The goal is, therefore, to strike a balance between the time resolution commensurate with the velocity of the moving particle, the camera exposure (or photon integration) time necessary to obtain super-resolution position accuracy, and fluorophore longevity. For our spider origami applications, the photon integration time is typically 2.5 s with a 12.5-s dark period between acquisitions of successive images, resulting in a time resolution of 15 s. With velocities of ~3 nm/min, the spider moves ~1 nm per frame, which constitutes sufficient time resolution considering the net travel distance of 100 nm.
A complementary means of extending fluorophore lifetime is to introduce an oxygen scavenging system (OSS), which reduces the rate of oxygen-dependent photobleaching by sequestering molecular oxygen from solution. A widely used OSS is a coupled enzyme system consisting of glucose oxidase and catalase that converts glucose and molecular oxygen into gluconic acid and water (Benesch and Benesch, 1953). However, Aitken et al. (2008) found an improved OSS that consists of 25 nM protocatechuate dioxygenase (PCD), 2.5 mM protocatechuate (PCA), and 1 mM Trolox. The PCD employs a nonheme iron center that catalyzes the conversion of PCA and molecular oxygen into β-carboxy-cis,cis-muconic acid (Aitken et al., 2008), while the antioxidant Trolox suppresses slow blinking and photobleaching of cyanine dyes (Rasnik et al., 2006). We use as much as five-fold the concentrations of the components specified above to further prolong fluorophore lifetime.
While sufficient fluorophore longevity is the primary concern for lengthy single molecule fluorescence tracking experiments, other fluorophore properties should also be considered. The fluorophores should be photostable to reduce excessive blinking (access to reversible dark states) that will interrupt tracking. Also, it is preferable to choose fluorescent probes with high brightness (molar extinction coefficient times quantum yield) to maximize the number of photons emitted and, hence, position determination accuracy. The fluorescent signal may also be increased by multiply labeling a tracked particle, although it has been noted that Cy5 can self-quench (Gruber et al., 2000). We label the spider with 2–3 Cy3 molecules and each of the six GOAL strands at the end of the DNA origami track with one Cy5 molecule. We do not have a detrimental self-quenching problem with these redundant probes.
3.2. Control for stage and focal drift
When imaging for long periods of time (several minutes to hours), stage and focal drift due to thermal fluctuations and mechanical instability become problematic. A practical approach to correct for such drift is to use a fiduciary marker(s) and determine the relative motion between the moving particle and stationary marker by subtracting the trajectory of the marker from that of the particle. We use the cluster of 6 Cy5 fluorescent probes on the GOAL strands as a fiduciary marker for spider motion on the corresponding origami track. Controls should be performed in which the essential divalent metal ion cofactor is omitted to verify that the spider trajectory is stationary in its absence. In addition, since determination of spider movement is based on only its corresponding fiduciary marker, it is important to control for possible aberrant movement of the marker by comparing its trajectory with those of neighboring markers to ensure that the trajectories exhibit the same drift pattern.
4. SINGLE-MOLECULE FLUORESCENCE TRACKING OF NANOWALKERS
The following super-resolution fluorescence imaging protocol permits real-time tracking of fluorescently labeled, slowly-moving nanowalkers along DNA origami paths.
4.1. Slide preparation
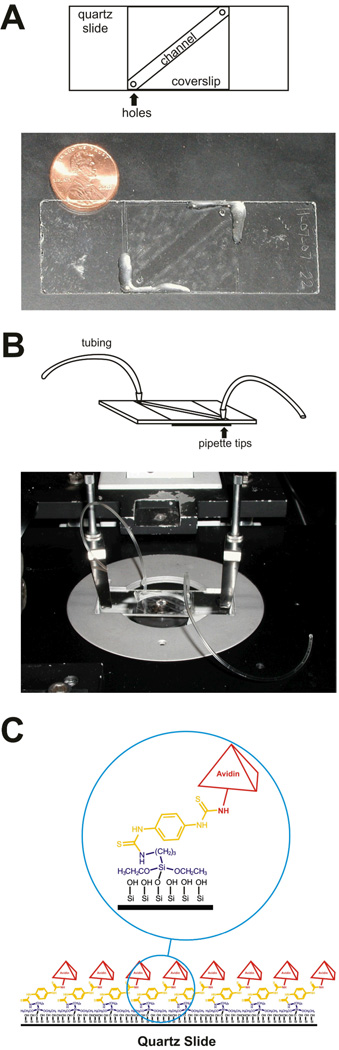
First, the origami tiles need to be immobilized so as to lie flat on the microscope slide’s surface. This is accomplished via a strong non-covalent bond between the avidin-coated slide and four biotinylated staples located near the four corners of the underside of each origami tile. To maximize the probability that all biotins bind securely to the surface, the avidin coating on the slide surface must be dense, which we accomplish here by aminosilanization followed by reaction with a bifunctional diisothiocyanate to covalently anchor avidin (Fig. 2a).
Drill two 1-mm holes in each slide (Finkenbeiner, fused silica, 1”×3” ×1mm) to be used for buffer exchange. It is helpful to have a template to follow when drilling the holes to ensure that the coverslip will fit over both holes (Fig. 2b).
Fluorescent and other impurities must be removed from the slide surface prior to imaging. Submerge the slides in a boiling “piranha” solution (5% (v/v) ammonium hydroxide (Aldrich, 231-704-8) and 14% (v/v) hydrogen peroxide (Acros, 202465000)) for 20 min. Rinse well with deionized water, flame sides with a propane torch for 1 min each. Sonicate slides in 1 M potassium hydroxide (KOH; Aldrich, 215813) for 30 min. Rinse again thoroughly with deionized water, then rinse with acetone (Fisher, A949-4) and sonicate for ~15–20 min in acetone.
To aminosilanize the slides, immerse clean slides in a 5% (v/v) 3-aminopropyltriethoxysilane (APTES; Sigma-Aldrich, A3648-100ML) acetone solution for 1 h. Rinse aminosilanized slides thoroughly with acetone. Dry at 80 °C for 1 h.
Para-diisothiocyanatobenzene (PDTIC; Acros, 417510050) covalently couples to the aminosilanized surface based on the approach developed by Guo et al. (Guo et al., 1994). Incubate the slides in 0.2% (w/v) PDTIC and 10% (v/v) pyridine (Fisher, P368-500) in dimethylformamide (Acros, spectroscopic grade, 40832-5000) for 2 h. Rinse thoroughly with acetone and methanol (Fisher, A452-4), and allow to dry.
Coat the slides with avidin. Pipette 70 µL of 0.5 mg/mL avidin (Sigma-Aldrich, A9275-2MG) on the center of each slide. Cover each slide with a glass coverslip (VWR, 48404-466) and incubate at room temperature for 2 h in a closed container above a water bath to maintain a humid environment. Remove the coverslips. Thoroughly rinse slides with deionized water. To quench the PDTIC not bound to avidin, apply 1 M NaCl in 40 mM NaOH to each slide for a few min. Thoroughly rinse the slides with deionized water. Dry with nitrogen.
Construct a flow channel between the drilled holes on each slide using two pieces of double-sided tape (Scotch, permanent, ½ inch) spaced 2–3 mm apart (Fig. 2c). Securely place a coverslip over the tape to cover the channel. Use epoxy glue (Hardman Adhesives 04001) along the edges of the coverslip to seal the channel. These avidin-coated slides may be stored at 4 °C in an evacuated desiccator for up to four weeks.
Figure 2.
Microscope slide preparation. (A) A fluidic channel is created between the two 1-mm holes in the quartz slide by sandwiching two pieces of double-sided sticky tape between the slide and the coverslip. (B) Tubing for fluid introduction and disposal is attached to the holes in microscope slide by using pipette tips. Epoxy resin is added to generate a tight seal between the tubing and pipette tips as well as between the pipette tips and holes. Note that, when placed on the microscope stage, the coverslip is face-down, allowing the tubing on the microscope slide to be face-up. (C) The slide is densely coated with avidin (red) covalently bound via a PDTIC linker (yellow) to an aminosilanized (blue) quartz slide (black) surface.
4.2. Experimental procedure
When choosing an appropriate imaging buffer, it is important to perform control experiments to ensure that the nano-assemblies are active under the buffer conditions. If a divalent transition metal cation such as Zn2+ is required to catalyze the reaction, be aware of possible chelation by buffering agents such as citrate in buffers such as saline-sodium citrate (SSC; 150 mM NaCl, 15 mM sodium citrate, pH 7.0). The imaging buffer used in our protocol is HEPES buffered saline (HBS) (10 mM HEPES, 150 mM NaCl, pH 7.4). This protocol assumes that the origami-spider complex, with a Cy3-labeled spider and Cy5-labeled GOAL position on the origami, has already been assembled as described in Lund et al. (2010).
Let 1 µL of 10 nM origami-spider complex incubate with 1 µL of 1–100 µM TRIGGER strand for at least 30 min to allow adequate time for the TRIGGER to hybridize to the START strand.
For a volume of 200 µL (the recommended minimum volume for any solution that will be flushed through the fluidic channel to ensure that the solution makes it all the way through the ~10 cm of tubing and to minimize the formation of air pockets), 1× OSS is created by combining 5 µL 100mM protocatechuate acid (PCA; Sigma, P5630-10G), 5 µL 400 nM protocatechuate-3,4-dioxygenase (PCD; Sigma, P8279-25UN), and 2 µL 100 mM Trolox (Acros, 218940050). We use concentrations as high as 5× OSS.
Add 1×–5× OSS and 1× HBS to the sample to result in a final concentration of 100 pM spider-origami complex. This solution will henceforth be referred to as the sample.
Prepare the OSS buffer (1× HBS supplemented with OSS) as well as the reaction buffer (1× HBS supplemented with OSS and 1 mM ZnSO4). If the zinc precipitates (the solution turns opaque), lower the concentration of ZnSO4 or buffer pH accordingly.
Attach tubing (Tygon, 0.02’ ID, 63018-044) on both sides of the fluidic channel using pipette tips and epoxy. Plastic tubing should be just wide enough for a syringe needle (Fig. 2c).
Using a syringe with needle, flow 200 µL imaging buffer in the absence of OSS through the channel. Using excitation from the 532-nm and 638-nm lasers, image the slide with a TIRFM microscope (described in detail in section 5.3) prior to adding the sample. Inspect the slide to see if there is any fluorescent debris that may interfere with the signal emitted by the fluorophores. If so, photobleach these spots by exciting the location at a high laser intensity for several min until the background fluorescence has stabilized. If there are still many contaminants, use a new slide.
Flow the sample onto the slide in the dark and incubate for 2–10 min. To optimize the incubation time for an optimal density of nano-assemblies on the surface, excite the sample every few minutes with the lasers to see how many nano-assemblies have bound to the surface. Ideally, the nano-assemblies are dense enough to have a large number of PSFs to analyze, but sparse enough that Gaussian functions may reliably be fit to the individual PSFs. We normally work with densities of ~0.03 molecules/µm2.
Once the optimal density is achieved, flush out the extra unbound sample using the OSS buffer. Image the origami at room temperature using a 2.5-s camera integration time and 12.5-s delay during which a shutter is used to protect the fluorophores from excessive photobleaching.
Add the reaction buffer after imaging for 20 min. Omitting the divalent metal cofactor for the first several min provides an important control for movement in the absence of catalysis. Continue imaging for at least 60 min to allow ample time for the spider to reach the GOAL position.
4.3. Instrumentation for imaging
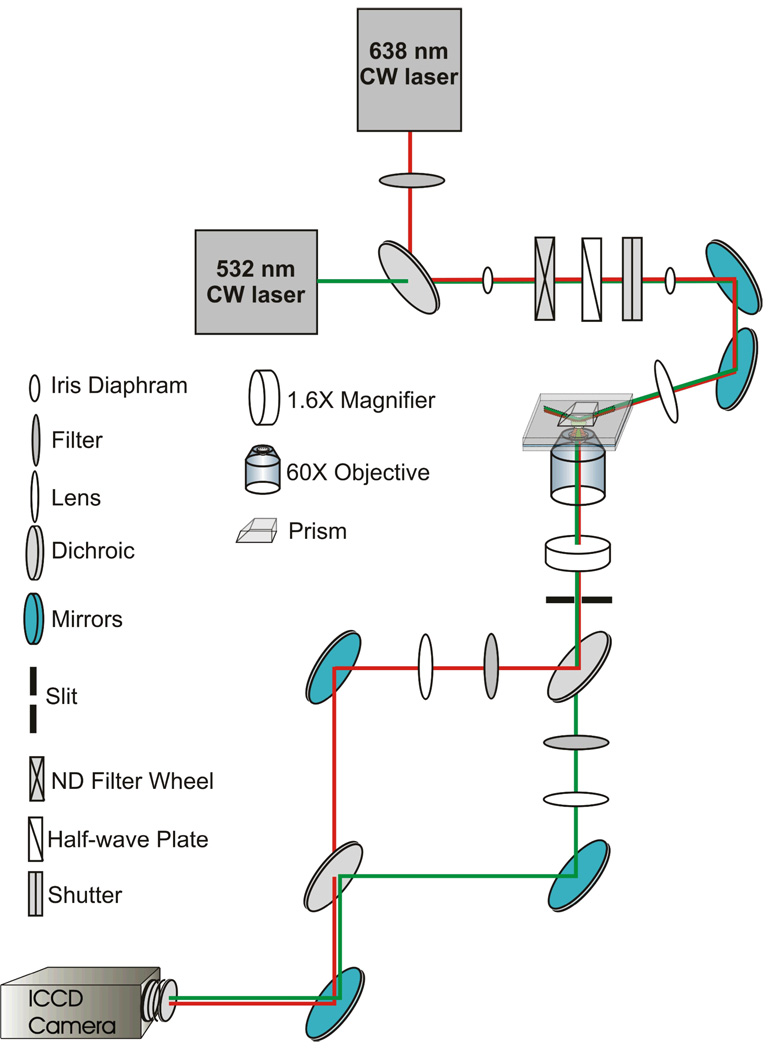
We use a Newport ST-UT2 vibration isolation table to minimize disturbances from vibrations in the microscope’s external environment and place it into a temperature controlled room. The table holds a home-built prism-based TIRF microscope equipped with a 1.2 NA 60× water objective (Olympus Uplanapo) for imaging (Fig. 3). A 532-nm ultra-compact diode-pumped Nd:YAG laser (GCL-025-S, CrystaLaser) is used to excite Cy3, while a 638-nm red diode laser (Coherent CUBE 635-25C, Coherent Inc.) is used to excite Cy5.
Figure 3.
Single-molecule prism-based TIRFM setup. Linearly polarized light from the 630-nm red diode laser passes through a 638 ± 10 nm clean-up filter and is reflected off a 610?-nm cutoff dichroic. 532-nm light from the Nd:YAG passes through same dichroic to join the same beam path. Light from both lasers then passes through the following components, in order: an iris diaphragm, a neutral density filter, a λ/2-wave plate for 532-nm light, a shutter, an iris diaphragm, a series of mirrors (two are shown for ease of representation; three were used during our experiments), a focusing lens, the TIR prism, the microscope slide containing the sample, the 60X objective, a 1.6X magnifier (along with additional filters, mirrors, and image transferring lenses contained within the microscope), a slit, and a 610-nm cutoff dichroic mirror. The emission from Cy3 passes through a band-pass filter, while that of Cy5 passes through a long pass filter. These separated images are projected side-by-side onto an intensified charge-coupled device (ICCD) camera.
The linearly polarized light from the red diode laser passes through a 638 ± 10 nm clean-up filter (Chroma, z640/20) before reflecting off a dichroic that the 532 nm light from the Nd:YAG laser passes through. The light from both lasers passes through an iris diaphragm that blocks excess stray light, followed by a neutral density filter to help regulate the laser power exciting the sample (Fig. 3). It continues through a λ/2-wave plate for 532-nm light and a shutter that is used to limit exposure time. As an extra measure for avoiding unwanted scattered light, a second iris diaphragm is used followed by a series of mirrors to redirect the light to the microscope where it passes through a focusing lens before entering the TIR prism and exciting the fluorescently-labeled sample. The emitted light enters the 60× objective and passes through a 1.6× magnifier along with additional filters, mirrors, and image transferring lenses, resulting in an effective pixel size of 133 nm. The emission from the Cy3 and Cy5 fluorophores are then separated with a dichroic mirror with 610-nm cutoff (Chroma, 610DCXR). The Cy3 emission additionally passes through a band-pass filter (HQ580/60m, Chroma), while the Cy5 emission passes through a long pass filter (HQ655LP, Chroma). These spectrally separated images are projected side-by-side onto an intensified charge-coupled device (ICCD) camera (IPentamax HQ Gen III, Roper Scientific, Inc.). An aperture constructed of two razor blades and positioned at the microscope sideport image is used to adjust the size of the spectrally separated images on each half of the CCD chip. WinView32 software (Roper Scientific, Inc) or suitable home-programmed software is used to immediately visualize and store the signal received from the camera. Camera gain values, or conversion values for converting the camera signals from arbitrary digital numbers (DN) to collected photoelectrons, are determined using the photon transfer method (Janesick, 1997).
5. EXTRACTING SUPER-RESOLUTION POSITION INFORMATION
After collection, the raw particle tracking data must be subjected to a series of analysis steps to yield and interpret super-resolution tracking data. A number of software routines custom-written in our laboratory implement these steps (Lund et al., 2010) and are available upon request.
5.1. Mapping the two emission channels onto one another
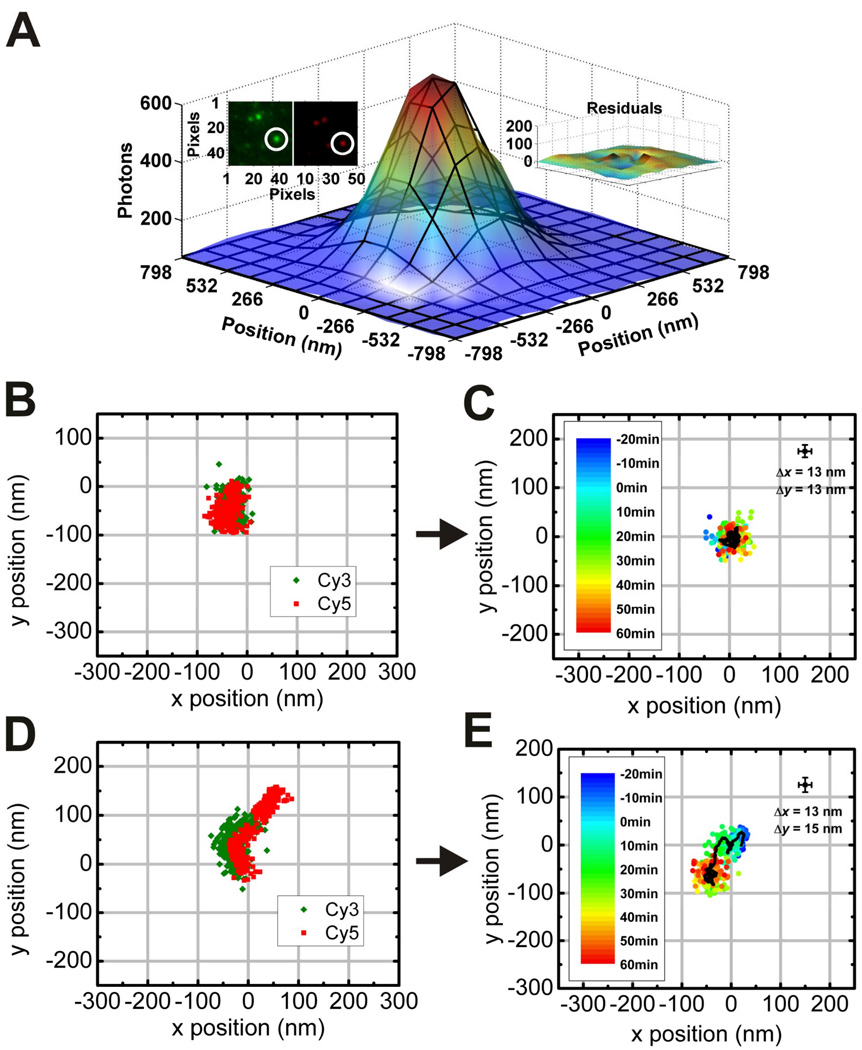
Since images of the Cy3 and Cy5 PSFs are projected onto separate halves of the ICCD camera and passed through different sets of optics, careful mapping of one channel onto the other is needed to determine which spider resides on a given origami. For this purpose we use a SHREC approach (Churchman et al., 2005) wherein fluorescent beads (FluoSpheres, F8810) visible in both channels are used as fiduciary markers to establish a locally weighted mean transformation between the channels in MATLAB (Mathworks, Natick, MA). Unlike global mapping routines, which attempt to establish a valid single transformation over the entire field, a locally weighted mean transformation accounts for local variations and distortions in the image due to, for example, aberrations in the optical path. Mapping the Cy5 channel onto the Cy3 channel enables us to pair each Cy3-labeled spider with its (most likely) Cy5-labeled GOAL. Once this is performed, the motion of each spider relative to its GOAL is determined by first applying a Gaussian fitting routine (Yildiz et al., 2003; Rust et al., 2006) to the individual Cy3 and Cy5 PSFs in MATLAB, followed by subtracting the Cy5 trajectory from that of the Cy3 (Fig. 4).
Figure 4.
Extraction of trajectories from a single nanowalker on a linear origami track imaged by TIRF microscopy (Lund et al., 2010). (A) Point spread functions (PSFs) of Cy3-labeled spiders and Cy5-labeled origami are colocalized (inset, upper left) and fit separately to two-dimensional Gaussian functions in each movie frame to determine their coordinates over time. The fit has low residuals (inset, upper right). (B)–(E) Centroids from Gaussian fitting are plotted as a function of time to yield nanowalker trajectories. Even in the absence of the divalent metal ion cofactor, Cy3 and Cy5 coordinates show considerable drift (B), but when the trajectory of Cy5 is subtracted from that of Cy3 it becomes clear that the nanowalker is stationary on its track (C). In contrast, in the presence of 5 mM ZnSO4, subtraction of the raw Cy5 and Cy3 trajectories (D) yields net movement of about 90 nm. In (C) and (E), HBS buffer containing either 0 or 5 mM ZnSO4 was added to the sample at time t = 0 min.
5.2. Quality control
In single molecule microscopy, it is generally necessary to identify and reject traces that, due to poor tracking accuracy, fluorescent contamination, or failed assembly of the complexes, do not accurately represent the molecular behavior of interest. Suitable selection criteria are, however, challenging to implement without introducing some form of bias. To record spider motion along origami tracks, we employ a minimal yet essential set of selection criteria to analyze the data based on the following parameters (Lund et al., 2010):
Intensity. The signal-to-noise ratio must be sufficient to obtain high-resolution results from the Gaussian fitting routine (Thompson et al., 2002). In our current experiments, the minimum number of photons required for the peak intensity of a PSF is 1,000.
Ellipticity. The ellipticity is calculated using the equation ε = 1-σmin/σmax where σmin and σmax are the standard deviations along the major and minor axes of the ellipse (Gordon et al., 2004) in a fixed plane. We demand that the maximum ellipticity be 0.3 for the included data points. Note that this cutoff may be dependent on the fluorophore distribution on the surface and may be modified to ensure that reliable data are not unintentionally removed. This filters out immobile dyes whose fixed polarization orientations result in elliptical PSFs and provides reassurance that we are tracking single molecules and not aggregates that rarely accumulate to form a perfectly circular PSF.
Displacement. The origami tracks are 100 nm in length. Position measurements that far exceed this net displacement do not likely represent a single spider moving along an intact origami track. A typical displacement cutoff is less than three standard deviations from the mean of all position measurements. However, to reduce the influence of extreme outliers, such as those caused by aberrant Gaussian fitting of transiently binding fluorescent contaminants, the median is used instead of the mean. To further reduce the influence of aberrant fitting, we also discard any position measurements greater than 500 nm from the spider’s initial position.
Colocalization. SHREC analysis must confirm that each spider is within a reasonable distance of its GOAL partner. Considering the track length of ~100 nm, we discard any Cy3 PSF that is more than 150 nm away from the nearest Cy5 PSF at any point in its trajectory.
5.3. Presentation
Single-molecule fluorescence experiments uniquely allow for a comparison of individual molecular behaviors, while averaging information from individual (diverse) traces is imperative to characterize the ensemble behavior and ensure statistical significance of individual trajectories (Lund et al., 2010).
5.3.1. Trajectory plots
Trajectory plots consist of the spider position values (x,y) as a function of time, which is color-coded to ease visualization (Fig. 5a). We have also found it useful to incorporate an origami track representation drawn to scale into the background of these plots. Such plots provide intuitive insight concerning the range of behaviors exhibited by individual walkers; features such as start time, velocity, end time, total distance, and path traversed may be approximated across the samples at a glance. However, quantitative comparisons require additional representation tools.
Figure 5.
Characterization of nanowalker movement (Lund et al., 2010). (A) The two-dimensional trajectory of an individual spider is plotted as a function of time and compares reasonably well to a scale-drawn schematic of the origami track design (gray rectangle). HBS buffer containing 5 mM ZnSO4 was added at time t = 0 min. (B) Displacement of the spider in panel (A) measured with respect to its position at the time of adding 5 mM ZnSO4. Displacement plots calculated from both the raw trajectory (green dots) and a 4-min rolling average of the raw trajectory (black line) are shown. (C) Ensemble mean-square displacement (MSD) calculated from 16 spiders observed in HBS buffer containing 5 mM (green squares) or 0 mM (red diamonds) ZnSO4 added at time t = 0 min. The former plot is fit with a power law function (green solid line), and the latter with a straight line (red line). To clearly illustrate the concave-up shape of the MSD in 5 mM ZnSO4, a straight line is also fit to the first 15 min of this plot (green dotted line).
5.3.2. Displacement plots
The displacement plot is generated by calculating the distance between the initial position and each subsequent position and plotting these displacements as a function of time (Fig. 5b). When determining the displacement of a slowly moving object in the presence of significant noise, it is important to carefully consider how to define the initial position since it will significantly affect the overall displacement calculated. A Monte Carlo simulation that compares the displacement calculated with and without the noise level of a typical experiment may be helpful in making this decision. Currently, we define the initial position as the arithmetic mean of the first 16 data points. Because noise artificially increases these displacements, the data are smoothed using a 16-point rolling average; this method is based on previous observations about measuring distances only slightly above the noise level (Churchman et al., 2005). A bin size of 16 (4 min at 4 frames per min) is appropriate because, with a typical velocity of 3 nm/min, a spider will move only 12 nm, which is generally within the error of our position measurements. The final or net displacement can be determined by heuristic or direct quantitative methods, or by inspection, and will depend on the system under study. Because many spiders on a 100-nm origami track will reach and become trapped on the GOAL, we define the net displacement as the first local maximum in the rolling average to reach within 20 nm (a typical error value for displacement, calculated as described in section 5.4) of the global maximum of the rolling average, and define the accompanying time coordinate as the stopping time. The average velocity of the spider, which provides a quantitative means for directly comparing individual molecules, is estimated by dividing the final net displacement by the stopping time.
5.3.3. Ensemble-averaged mean square displacement plots
The characteristics of single particle motion can be determined from the sequence of positions and corresponding times made accessible by single particle tracking methods. The ability to observe the motions of individual particles allows one to sort trajectories into various modes of motion, and the stochastic nature of the particles call for statistical methods to be used for analysis (Qian et al., 1991; Saxton and Jacobson, 1997) by finding distributions of quantities characterizing the motion, such as diffusion coefficients, distances, velocities, anomalous diffusion exponents, corral sizes, etc. (Qian et al., 1991; Kusumi et al., 1993; Saxton and Jacobson, 1997). The mean square displacement (MSD) is a particularly useful quantity for sorting and characterizing diffusive particle trajectories (Qian et al., 1991; Kusumi et al., 1993; Saxton and Jacobson, 1997), and can help uncover the spider walking mechanism. As the name would suggest, the MSD is produced by squaring the displacements between increasing time steps, averaging them over single trajectories for time-averaged MSD (TA MSD) or averaging over all the trajectories in a set (ensemble MSD), and plotting them as a function of time (Fig. 5c). For a trajectory in two dimensions, the MSD curve can be fit with the equation MSD = <r2> = 4Dtα, where the diffusion coefficient D and the α parameter help characterize the motion of the particle(s). A molecule exhibiting subdiffusive behavior, which is exemplified, for example, by confined particles, corresponds to the condition 0<α <1. Brownian diffusion corresponds to α=1. Superdiffusion corresponds to the case for α>1, where the MSD grows faster than it does in normal (Brownian) diffusion. Such processes are characterized by broad distributions of both waiting time and step size (with no meaningful mean), as opposed to subdiffusion, which is characterized by a broad distribution of only the waiting time but a narrow (Gaussian) step size distribution (Klafter and Sokolov, 2005). Diffusive processes with α≠1 are generally referred to as anomalous diffusion and are commonplace in Nature (Klafter and Sokolov, 2005). Finally, there are various modifications that the MSD fitting equation can have depending on the system under study, such as an additive velocity term for particles exposed to a continuous drift (directed transport) (Saxton and Jacobson, 1997).
Simple Brownian motion is an example of a stationary system (which implies that it is also ergodic, i.e., the time average of a single particle equals the ensemble average) and either TA MSD or ensemble MSD methods can be applied (Qian et al., 1991). By contrast, if applied to non-stationary systems (which implies non-ergodicity), the TA MSD may yield misleading/incorrect results (Gross, 1988; He et al., 2008; Lubelski et al., 2008). The modification (cleavage) of a pseudo-one-dimensional origami track featuring START and GOAL positions upon interaction with a spider allows the system to have a memory of previously visited locations and a bias to move toward new locations, which may be expected to result in the spider MSD increasing faster than that of a simple Brownian diffusion process (Antal and Krapivsky, 2007).
Complicated diffusion processes and noisy data can affect the MSD curves by further adding to the complexity of the underlying MSD model in various ways. For example, transitions between diffusive and non-diffusive segments of a trajectory (Saxton and Jacobson, 1997), crossover between anomalous dynamics on various time scales (Dieterich et al., 2008), and either noise inherent to the system (e.g., “biological noise”) (Dieterich et al., 2008) or measurement noise can affect MSD curves at short time scales (Qian et al., 1991; Martin et al., 2002). Even for “perfect data” with no errors in the position measurements, the MSD and MSD-derived parameters will still have expected statistical variances due to the stochastic nature of a random walk; an estimate of these statistical variances is important to the analysis and should be assessed from the reproducibility of a series of corresponding measurements (Qian et al., 1991; Saxton and Jacobson, 1997). In addition, computer (e.g., Monte Carlo) simulations can generate random walks and provide a powerful tool to help assess the expected MSD curve and the processes and properties underlying a random walker’s behavior (Qian et al., 1991; Saxton and Jacobson, 1997; Ritchie et al., 2005; Dix and Verkman, 2008). We use Monte Carlo simulations in MATLAB for this purpose (Lund et al., 2010). Because the proposed biased-walk mechanism is dependent on the difference in affinity of the spider’s legs for the cleaved versus the intact substrate, a track composed completely of cleaved substrates should not exhibit this bias and would be predicted to undergo Brownian diffusion (Antal et al., 2007). To test for differences between the walking mechanism on cleaved and non-cleaved substrate surfaces, a control origami track can be designed that contains cleaved substrate; the walking mechanism on the cleaved substrate is expected to be distinct from that on the intact substrate (Lund et al., 2010).
5.4. Error analysis
The error in tracking single particles is well defined by equation (1), which can be used to determine the accuracy of individual position determinations. For measurements that represent an average of multiple determinations for a stationary object over a span of time, a simple standard error of the mean would suffice. However, for an object such as a spider with a poorly defined velocity on short time scales, an alternative approach should be used. For the displacement plots, since a 16-point rolling average is used in smoothing the data, we use the following approach (Lund et al., 2010):
Define x- and y-errors, Δxi and Δyi, as the standard deviations in the x- and y- dimension of all raw position measurements going into each averaged measurement; the resulting values for Δxi and Δyi can then be averaged (arithmetic mean) across all 16-point bins within the trajectory to get overall errors Δx and Δy as shown in Fig. 4, panels C and E.
- Propagate these errors to the displacement measurement according to the equation:
where di, xi and yi are the displacement and coordinate measurements for an individual point in the rolling average, and x0, y0, Δx0 and Δy0 are coordinate measurements and errors for the initial point defined as having d = 0.(2) Calculate the arithmetic mean of all individual displacement errors Δdi to obtain an overall displacement error Δd for the entire trajectory. The result is a measure of the spread of individual displacement measurements around the 16-frame rolling average in a trajectory. An alternative approach is to sum the pixel intensities from individual images of a PSF in rolling 16-frame bins, fit these to 2-D Gaussian functions, and then calculate the error using equation (1). This may, however, underestimate the error as it assumes that all position determinations in each bin report on a single stationary object; that is, it ignores “biological noise”.
6. CONCLUDING REMARKS
While autonomous deoxyribozyme-based walkers demonstrate the promise of molecular robotics, it is even more exciting to imagine applications such robots may see in the future. It has previously been shown how deoxyribozymes may be utilized for programming purposes (Stojanovic and Stefanovic, 2003). We now seek to expand the applications of the spider world while incorporating other single molecule fluorescence techniques such as Stochastic Optical Reconstruction Microscopy (STORM) to monitor and characterize differing types of behavior with high spatial accuracy.
ACKNOWLEDGEMENTS
This work was funded by the National Science Foundation (NSF) Collaborative Research: Chemical Bonding Center, award 0533019; and NSF Collaborative Research: EMT/MISC, award CCF-0829579. We thank Dr. Steven Taylor from Dr. Milan Stojanovic’s lab at Columbia University for providing the spiders, Dr. Kyle Lund and Jeanette Nangreave from Dr. Hao Yan’s lab at Arizona State University for preparing the origami tracks, and Dr. David Rueda for assembling the TIRF microscope.
REFERENCES
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal T, Krapivsky PL. Molecular spiders with memory. Phys. Rev. E. 2007;76:021121. doi: 10.1103/PhysRevE.76.021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal T, Krapivsky PL, Mallick K. Molecular spiders in one dimension. J. Stat. Mech.- Theory Exp. 2007:P08027. doi: 10.1088/1742-5468/2007/08/P08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak LS, Webb WW. Diffusion of Low-Density Lipoprotein-Receptor Complex on Human-Fibroblasts. J. Cell Biol. 1982;95:846–852. doi: 10.1083/jcb.95.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Green SJ, Allen KE, Turberfield AJ. Mechanism for a directional, processive, and reversible DNA motor. Small. 2009;5:1513–1516. doi: 10.1002/smll.200900078. [DOI] [PubMed] [Google Scholar]
- Benesch RE, Benesch R. Enzymatic removal of oxygen for polarography and related methods. Science. 1953;118:447–448. doi: 10.1126/science.118.3068.447. [DOI] [PubMed] [Google Scholar]
- Betzig E, Chichester RJ. Single Molecules Observed by near-Field Scanning Optical Microscopy. Science. 1993;262:1422–1425. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- Bobroff N. Position measurement with a resolution and noise-limited instrument. Rev. Sci. Instrum. 1986;57:1152–1157. [Google Scholar]
- Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz AA, Dahan M, Grunwell JR, Ha TJ, Faulhaber AE, Chemla DS, Weiss S, Schultz PG. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Forster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 1999;96:3670–3675. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich P, Klages R, Preuss R, Schwab A. Anomalous dynamics of cell migration. Proc. Natl. Acad. Sci. USA. 2008;105:459–463. doi: 10.1073/pnas.0707603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Aleman EA, Rueda D, Walter NG. Focus on function: single molecule RNA enzymology. Biopolymers. 2007;87:302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- Dix JA, Verkman AS. Crowding effects on diffusion in solutions and cells. Annu. Rev. Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes (vol 459, pg 414, 2009) Nature. 2009;459:1154–1154. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Review - The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SJ, Bath J, Turberfield AJ. Coordinated Chemomechanical Cycles: A Mechanism for Autonomous Molecular Motion. Phys. Rev. Lett. 2008;101:238101. doi: 10.1103/PhysRevLett.101.238101. [DOI] [PubMed] [Google Scholar]
- Gribbon P, Schaertl S, Wickenden M, Williams G, Grimley R, Stuhmeier F, Preckel H, Eggeling C, Kraemer J, Everett J, Keighley WW, Sewing A. Experiences in implementing uHTS--cutting edge technology meets the real world. Curr. Drug Discov. Technol. 2004;1:27–35. doi: 10.2174/1570163043484798. [DOI] [PubMed] [Google Scholar]
- Gross DJ, Webb WW. Cell surface clustering and mobility of the liganded LDL receptor measured by digital video fluorescence microscopy. In: Loew LM, editor. Spectroscopic membrane probes. Boca Raton, Fla.: CRC Press; 1988. pp. 19–45. [Google Scholar]
- Gruber HJ, Hahn CD, Kada G, Riener CK, Harms GS, Ahrer W, Dax TG, Knaus HG. Anomalous fluorescence enhancement of Cy3 and cy3.5 versus anomalous fluorescence loss of Cy5 and Cy7 upon covalent linking to IgG and noncovalent binding to avidin. Bioconjug. Chem. 2000;11:696–704. doi: 10.1021/bc000015m. [DOI] [PubMed] [Google Scholar]
- Guo Z, Guilfoyle RA, Thiel AJ, Wang RF, Smith LM. Direct Fluorescence Analysis of Genetic Polymorphisms by Hybridization with Oligonucleotide Arrays on Glass Supports. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Burov S, Metzler R, Barkai E. Random time-scale invariant diffusion and transport coefficients. Physical Review Letters. 2008;101:058101. doi: 10.1103/PhysRevLett.101.058101. [DOI] [PubMed] [Google Scholar]
- Hecht E. Optics. Reading, Mass: Addison-Wesley; 2002. [Google Scholar]
- Heintzmann R, Jovin TM, Cremer C. Saturated patterned excitation microscopy - a concept for optical resolution improvement. J. Opt. Soc. Am. A. 2002;19:1599–1609. doi: 10.1364/josaa.19.001599. [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintersteiner M, Auer M. Single-bead, single-molecule, single-cell fluorescence: technologies for drug screening and target validation. Ann. N. Y. Acad. Sci. 2008;1130:1–11. doi: 10.1196/annals.1430.055. [DOI] [PubMed] [Google Scholar]
- Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl. Acad. Sci. USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick J. CCD transfer method - Standard for absolute performance of CCDs and digital CCD camera systems. Proc. SPIE. 1997;3019:70–102. [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klafter J, Sokolov IM. Anomalous diffusion spreads its wings. Physics World. 2005;18:29–32. [Google Scholar]
- Kusumi A, Sako Y, Yamamoto M. Confined Lateral Diffusion of Membrane-Receptors as Studied by Single-Particle Tracking (Nanovid Microscopy) - Effects of Calcium-Induced Differentiation in Cultured Epithelial-Cells. Biophys. J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2003:S7–S14. [PubMed] [Google Scholar]
- Lubelski A, Sokolov IM, Klafter J. Nonergodicity mimics inhomogeneity in single particle tracking. Phys. Rev. Lett. 2008;100:250602. doi: 10.1103/PhysRevLett.100.250602. [DOI] [PubMed] [Google Scholar]
- Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic MN, Walter NG, Winfree E, Yan H. Molecular Robots Guided by Prescriptive Landscapes. Nature. 2010 doi: 10.1038/nature09012. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DS, Forstner MB, Kas JA. Apparent subdiffusion inherent to single particle tracking. Biophys. J. 2002;83:2109–2117. doi: 10.1016/S0006-3495(02)73971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W, English BP, Luo G, Cherayil BJ, Kou SC, Xie XS. Fluctuating enzymes: lessons from single-molecule studies. Acc. Chem. Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- Moerner WE. New directions in single-molecule imaging and analysis. Proc. Natl. Acad. Sci. USA. 2007;104:12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE, Kador L. Optical-Detection and Spectroscopy of Single Molecules in a Solid. Phys. Rev. Lett. 1989;62:2535–2538. doi: 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- Omabegho T, Sha R, Seeman NC. A bipedal DNA Brownian motor with coordinated legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R, Taylor SK, Stefanovic D, Rudchenko S, Mitchell TE, Stojanovic MN. Behavior of polycatalytic assemblies in a substrate-displaying matrix. J. Am. Chem. Soc. 2006;128:12693–12699. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- Pljevaljcic G, Millar DP. Single-molecule fluorescence methods for the analysis of RNA folding and ribonucleoprotein assembly. Methods Enzymol. 2008;450:233–252. doi: 10.1016/S0076-6879(08)03411-3. [DOI] [PubMed] [Google Scholar]
- Qian H, Sheetz MP, Elson EL. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XH, Wu D, Mets L, Scherer NF. Nanometer-localized multiple single-molecule fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rist MJ, Marino JP. Fluorescent nucleotide base analogs as probes of nucleic acid structure, dynamics and interactions. Curr. Org. Chem. 2002;6:775–793. [Google Scholar]
- Ritchie K, Shan XY, Kondo J, Iwasawa K, Fujiwara T, Kusumi A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 2005;88:2266–2277. doi: 10.1529/biophysj.104.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Rueda D, Walter NG. Single molecule fluorescence control for nanotechnology. J. Nanosci. Nanotechnol. 2005;5:1990–2000. doi: 10.1166/jnn.2005.505. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schutz GJ, Baumgartner W, Gruber HJ, Schindler H. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA. 1996;93:2926–2929. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC. An overview of structural DNA Nanotechnology. Mol. Biotechnol. 2007;37:246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman WB, Seeman NC. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203–1207. [Google Scholar]
- Shi J, Dertouzos J, Gafni A, Steel D. Application of single-molecule spectroscopy in studying enzyme kinetics and mechanism. Methods Enzymol. 2008;450:129–157. doi: 10.1016/S0076-6879(08)03407-1. [DOI] [PubMed] [Google Scholar]
- Shin JS, Pierce NA. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, He Y, Chen Y, Yin P, Mao CD. Molecular devices - A DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Ed. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- Truong K, Ikura M. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr. Opin. Struct. Biol. 2001;11:573–578. doi: 10.1016/s0959-440x(00)00249-9. [DOI] [PubMed] [Google Scholar]
- Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- Willets KA, Ostroverkhova O, He M, Twieg RJ, Moerner WE. Novel fluorophores for single-molecule imaging. J. Am. Chem. Soc. 2003;125:1174–1175. doi: 10.1021/ja029100q. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- Zhao R, Rueda D. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods. 2009;49:112–117. doi: 10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]