SUMMARY
The repertoire of ~1200 odorant receptors (ORs) is mapped onto the array of ~1800 glomeruli in the mouse olfactory bulb (OB). The spatial organization of this array is influenced by the ORs. Here we show that glomerular mapping to broad domains in the dorsal OB is determined by two types of olfactory sensory neurons (OSNs), which reside in the dorsal olfactory epithelium. The OSN types express either Class I or Class II OR genes. Axons from the two OSN types segregate already within the olfactory nerve and form distinct domains of glomeruli in the OB. These class-specific anatomical domains correlate with known functional odorant response domains. However, axonal segregation and domain formation are not determined by the class of the expressed OR protein. Thus, the two OSN types are determinants of axonal wiring, operate at a higher level than ORs, and contribute to the functional organization of the glomerular array.
INTRODUCTION
OSNs in the main olfactory epithelium (MOE) detect a vast number of odorous chemicals with diverse chemical structures. In the mouse, the repertoire of ORs consists of ~1,200 genes (Buck and Axel, 1991; Mombaerts, 2004a). The MOE is a mosaic of OSNs, each believed to express a single OR gene (Malnic et al., 1999; Mombaerts, 2004b). OSNs that express a given OR are scattered in partially overlapping regions of the MOE (Buck, 1996; Miyamichi et al., 2005). During development, axons of OSNs expressing the same OR coalesce into glomeruli that form at reproducible positions in the OB (Mombaerts, 2006). The spatial arrangement of the ~1800 glomeruli is thought to contribute to odor coding (Mori et al., 2006; Wilson and Mainen, 2006). The mechanisms that determine the organization of the glomerular array and the representation of odorants in the OB remain unclear.
It is now well established that ORs are involved in axon guidance (Mombaerts et al., 1996): mutations that alter the OR amino acid sequence or its expression level affect axonal sorting, often resulting in the appearance of new glomeruli at altered positions in the OB (Mombaerts, 2006). We have proposed that axonal coalescence and glomerular position are determined in part by differential homophilic interactions among OSNs with distinct axon guidance identities, which are defined by the ORs (Feinstein and Mombaerts, 2004). Others have proposed that ORs influence axonal identity by modulating the expression of conventional axon guidance molecules (Imai et al., 2006; Serizawa et al., 2006; Kaneko-Goto et al., 2008).
The dual roles of ORs in odorant transduction and axon guidance would appear to offer a strategy for mapping odorant responsiveness directly onto the surface of the OB. However, the relationship between odorant response and glomerular position is not straightforward. There is a coarse chemotopy in the rodent OB, in which odorants with certain molecular features activate glomerular domains (Mori et al., 2006; Johnson and Leon, 2007). However, the extent to which there is fine-scale mapping remains controversial (Bozza et al., 2004). The molecular and cellular basis for the organization of the observed odorant response domains is unknown.
Mouse ORs are classified into two phylogenetically distinct groups, Class I and Class II, based on homology of deduced amino acid sequences (Zhang et al., 2004; Niimura and Nei, 2005). Class I ORs make up ~10% of intact OR genes in mouse (Niimura and Nei, 2007), and are expressed almost exclusively by OSNs in the dorsal MOE (Zhang et al., 2004; Miyamichi et al., 2005; Tsuboi et al., 2006). In contrast, Class II ORs are expressed throughout the dorsal and ventral MOE. Thus, OSNs in the dorsal MOE (henceforth referred to as dorsal OSNs) express either Class I or II OR genes. Class II OR expression is abolished in mice lacking the LIM-homeodomain protein Lhx2, but Class I OR expression is largely spared (Hirota et al., 2007), implying mechanistic differences in Class I vs II OR gene choice.
It is currently not known if there are systematic functional differences between Class I and II ORs. The hypothesis that mammalian Class I ORs are specialized to detect particular groups of odorants (Freitag et al., 1995) has not been substantiated. Interestingly, OSNs expressing Class I ORs project their axons to glomeruli in a dorsal domain of the OB (Tsuboi et al., 2006; Kobayakawa et al., 2007). The mechanistic basis and functional significance of this domain segregation are unclear.
Here, we describe a set of transgenic and gene-targeted mice that visualize in various ways the mapping of Class I and II ORs to domains in the dorsal OB. We provide genetic evidence that this class-specific mapping results from distinct properties of two types of dorsal OSNs, which are fated to choose either Class I or II OR promoters. Remarkably, the axons of the two OSN types segregate within the olfactory nerve in the lamina propria of the olfactory mucosa, well before they come in contact with the OB. By genetically replacing OR coding regions we show that axonal segregation within the olfactory nerve and axonal coalescence within a domain are independent of the class of the expressed OR protein. We further show that the boundary between the Class I and II anatomical domains correlates with the boundary between functional odorant response domains that have been identified previously. We conclude that the segregation of glomeruli into two domains emanates from intrinsic properties of two OSN types, and is not determined by the class of the expressed ORs.
RESULTS
A Widely Expressed Transgene Reveals Two Domains in the Dorsal OB
We have previously described a series of OR minigenes in which short, candidate OR promoter sequences drive expression of the axonal marker tau-β-galactosidase (βgal) in a pattern that is appropriate for each OR (Vassalli et al., 2002; Rothman et al., 2005). Continuing this approach, we defined a 306 nucleotide (nt) sequence encompassing the transcription start site of the Class II OR gene P3 (MOR263-1, Olfr713) (Lane et al., 2001) that drives expression of βgal in a small subset of OSNs (data not shown). By homology, we further identified a 317 nt sequence that shares 70% nt identity with the P3 promoter, and resides between P3 and the homologous Class II OR gene P4 (MOR263-2, Olfr714) (Figure 1A). We refer to this 317 nt sequence as the ‘P sequence’ (Experimental Procedures). The P sequence has homeodomain and O/E-like binding sites, a feature of Class II OR promoters (Vassalli et al., 2002; Rothman et al., 2005). We thus reasoned that the P sequence may represent a cryptic or displaced OR promoter.
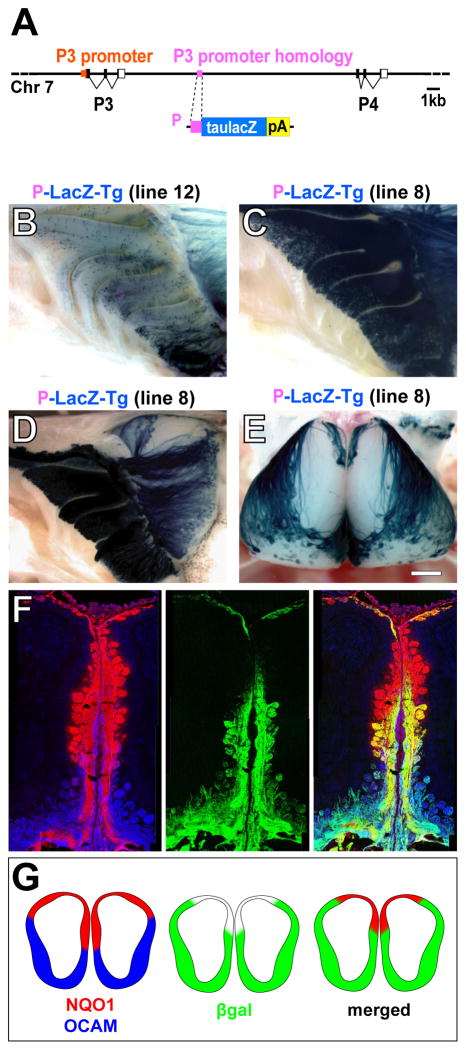
Figure 1. P-LacZ-Tg Reveals Two Domains in the Dorsal Olfactory Bulb.
(A) (Top) Genomic region containing the P3 and P4 OR genes. The P3 promoter is shown in orange, the 317 nt sequence that is homologous to it (P sequence) in purple. (Below) The P-LacZ transgene; the coding sequence for taulacZ (blue box) is preceded by the P sequence (purple box), and followed by a rabbit β-globin polyadenylation sequence (pA; yellow box).
(B) Medial view of X-gal stained wholemount of the MOE of a P-LacZ-Tg mouse of line 12. βgal+ neurons are scattered throughout the dorsal-ventral extent of the MOE.
(C) Medial view of X-gal stained wholemount of the MOE of a P-LacZ-Tg mouse of line 8. This line expresses βgal in ~10% of OSNs throughout the dorsal-ventral extent of the MOE.
(D) Medial view of X-gal stained wholemount of the MOE and OB of a P-LacZ-Tg mouse of line 8. βgal+ axons do not innervate a wedge-shaped region of the dorsal-medial OB.
(E) Dorsal view of X-gal stained wholemount of the OBs of a P-LacZ-Tg mouse of line 8. βgal+ axons do not innervate a dorsal domain, which has a butterfly-like shape in a dorsal view.
(F) Coronal sections from a P-LacZ-Tg mouse (line 8) showing the medial face of the OB stained by IHC with anti-NQO1 (red), anti-βgal (green) and anti-OCAM (blue). NQO1 and OCAM antibodies stain glomeruli in the dorsal and ventral parts of the OB, respectively. βgal+ axons innervate OCAM+ glomeruli and ventral NQO1+ glomeruli. The NQO1+ region can be subdivided into βgal+ and βgal− regions.
(G) Schematic drawings of OB sections of a P-LacZ-Tg mouse showing the overlap between βgal, NQO1 and OCAM-labeled domains.
Scale bar in E is 500 μm in E; 420 μm in B and C; 760 μm in D; and 280 μm in F.
To address whether the P sequence has promoter activity, we generated a transgene (P-LacZ-Tg) in which the 317 nt sequence is placed upstream of taulacZ (Figure 1A). Four independent transgenic lines exhibit reproducible and robust expression of βgal in OSNs throughout the dorsal-ventral extent of the MOE. Line 12 shows rates of expression that are typical for an OR (Figure 1B), but lines 8, 11, and 13 show unusually high numbers of labeled OSNs (Figure 1C): as many as ~10% of OSNs throughout the MOE, or two-three orders of magnitude greater than a typical OR promoter. 5′RACE confirms that the βgal-transcript initiates within the 317 nt P sequence. Expression of P-LacZ-Tg is abolished in Lhx2 mutant mice (data not shown), similar to Class II OR genes (Hirota et al., 2007).
Because P-LacZ-Tg lacks an OR coding sequence, OSNs that express this transgene are expected to co-express other OR loci and, as a population, project their axons to multiple glomeruli (Serizawa et al., 2003; Feinstein et al., 2004; Lewcock and Reed, 2004; Shykind et al., 2004). Indeed, βgal+ axons project to a large majority of glomeruli in the OB (Figures 1D and 1E). However, glomeruli in the dorsal-medial and anterior-dorsal OB appear devoid of βgal+ axons. Xgal staining of OB wholemounts thus gives rise to an unlabeled butterfly-shaped pattern in a dorsal view (Figure 1E). This butterfly pattern is observed reproducibly in the four independent transgenic lines (#8, 11, 12, and 13), and at embryonic and neonatal stages (data not shown).
Projections from the MOE can be grossly divided into dorsally-projecting axons, which express NADPH:quinone oxidoreductase (NQO1), and ventrally-projecting axons, which express the olfactory cell adhesion molecule (OCAM) (Gussing and Bohm, 2004; Yoshihara et al., 1997). Immunohistochemistry of OB sections of P-LacZ-Tg mice reveals βgal labeling in OCAM+ glomeruli and in a ventral subset of NQO1+ glomeruli (Figure 1F). Thus, the βgal− domain in P-LacZ-Tg mice is distinct from the domains defined by NQO1 and OCAM (Figure 1G).
P-LacZ-Tg Selectively Labels OSNs That Express Class II OR Genes
The presence of unlabeled glomeruli in P-LacZ-Tg mice strongly suggests that a subpopulation of dorsal OSNs do not express the transgene. We hypothesized that the transgene is expressed differentially in OSNs that express Class I vs II ORs, as the P sequence is 70% homologous to a Class II OR promoter. To examine the co-expression rates of Class I and II OR genes in P-LacZ-Tg+ OSNs, we performed in situ hybridization (ISH) for 52 Class I and 17 Class II probes combined with immunohistochemistry (IHC) for βgal (Figure 2A). We analyzed a total of 29,544 neurons from 10 mice between 3–5 weeks of age.
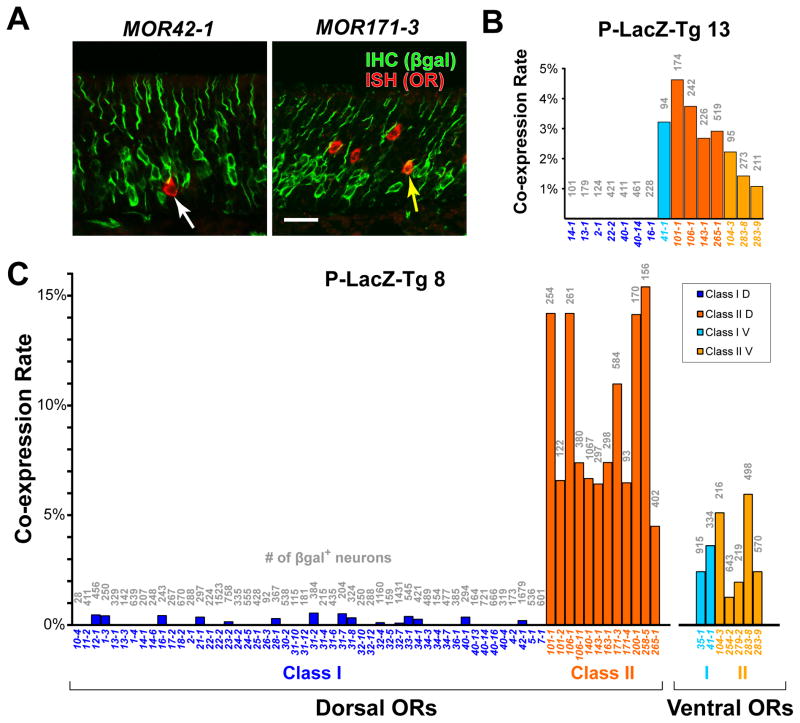
Figure 2. P-LacZ-Tg is a Marker for OSNs that Express Class II ORs.
(A) Confocal image of combined ISH (red) for the Class I OR gene MOR42-1 (=S50) or MOR171-3 (=M72) and IHC for βgal (green) in a coronal section through the MOE of a P-LacZ-Tg mouse of line 8. White arrow in left panel indicates a single-labeled OSN, yellow arrow in right panel shows a double-labeled OSN.
(B and C) Bar graphs showing the percentage of IHC βgal+ cells that are labeled by ISH with OR probes in P-LacZ-Tg mice, of line 13 in (B) and of line 8 in (C). Names of the OR probes are given below the x-axis. Numbers of cells counted for each probe are shown in grey above each bar. Of the 52 Class I probes, 21 were full-length OR coding sequences that should cross-hybridize with at least one other Class I OR due to >85% nucleotide identity. The total number of Class I OR transcripts assayed is estimated at 52–73, which is more than half of the Class I genes. Data are separated into dorsally and ventrally-expressed Class I and Class II OR genes.
Scale bar is 25 μm in A.
We observe a striking dichotomy in co-expression rates for dorsal Class I and dorsal Class II OR genes (Figures 2B and 2C). The average co-expression rate for dorsal Class I genes is 100-fold lower than for dorsal Class II genes in line 8 (Figure 2C): Class I, average 0.08%, with 95% confidence interval (CI) = 0.05–0.14% vs Class II, average 8.9%, CI = 7.1–11.2%. This difference is highly significant (p<0.0001; random effects logistic regression). A similar difference in co-expression rate is observed in line 13 (Figure 2B). Thus, P-LacZ-Tg preferentially labels OSNs that co-express Class II OR genes. This pattern is similar to that of a single transgenic line in which a Class II OR promoter drives Cre recombinase (Kobayakawa et al., 2007).
Two atypical Class I OR genes, MOR35-1 and MOR41-1, are expressed in ventral OSNs (Tsuboi et al., 2006), and are regulated like Class II OR genes (Hoppe et al., 2006; Hirota et al., 2007). Interestingly, the co-expression rate for these ventral Class I genes (2.8%, CI = 1.5–5.3%) is significantly greater than for dorsal Class I genes (p<0.0001), but not significantly different from six ventral Class II genes (2.7%, CI = 1.8–4.2%; p = 0.94) (Figures 2B and 2C).
We interpret the dichotomy in Class I vs II co-expression rates in OSNs that express P-LacZ-Tg as genetic evidence for two OSN types in the dorsal MOE, which are fated to choose Class I vs II OR promoters.
Class-Specific Domains in OR Coding Sequence Deletions
We next asked if the domains revealed by P-LacZ-Tg are also observed with OSNs that choose an endogenous OR promoter that lacks its OR coding sequence. We examined mouse strains in which the coding sequence of a dorsal Class I or II OR gene has been deleted and replaced by a marker via gene targeting. We refer to these mutations as ΔOR, and to the OSNs that express a ΔOR allele as ΔOR OSNs.
To examine the axonal projections of Class I ORs, we studied two genes: S50 (MOR42-1, Olfr545) (Malnic et al., 1999), and MOL2.3 (MOR18-2, Olfr78) (Conzelmann et al., 2000). For comparison, we first generated a mouse strain in which the S50 coding sequence remains intact and is followed by an internal ribosome entry site (IRES) and the sequence for the axonal marker tauGFP (S50-IRES-tauGFP). S50 OSNs typically project to a single dorsal-medial and a single anterior-lateral glomerulus per OB (n = 21 mice, Figure 3A). As expected, the S50 glomeruli reside within the βgal− domain of P-LacZ-Tg mice (Figure 5A). We also examined a strain in which MOR18-2 OSNs are labeled with GFP and taulacZ (Conzelmann et al., 2000). MOR18-2 axons project to multiple medial and lateral glomeruli per OB (n = 8 mice, Figure 3B). The MOR18-2 glomeruli appear to reside just within the βgal− domain of P-LacZ-Tg mice (data not shown). Because these OSNs express both GFP and taulacZ, it is difficult to determine unambiguously whether their axons fall inside or outside the Class I domain.
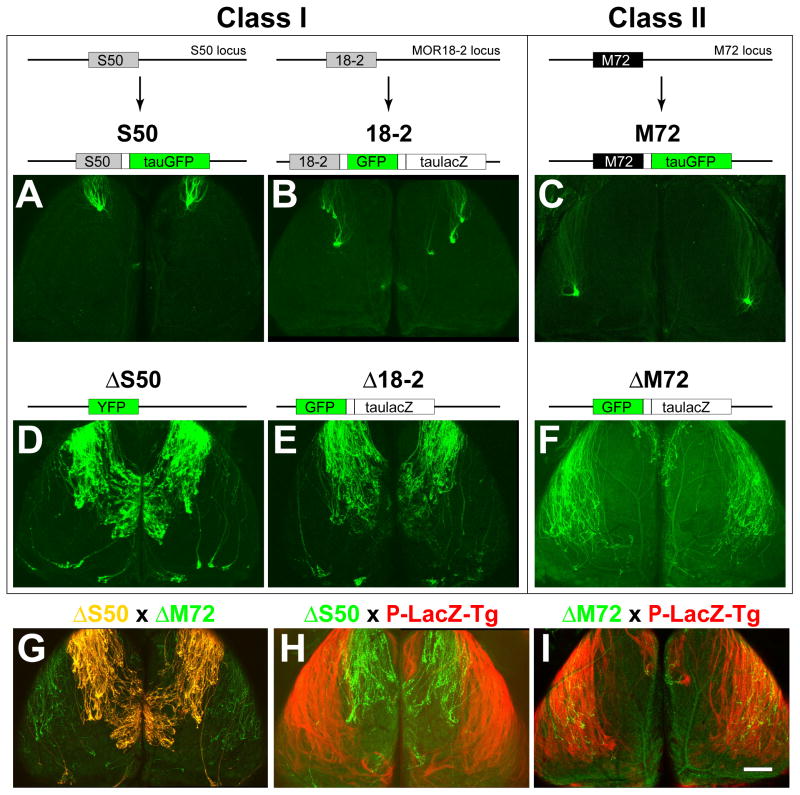
Figure 3. ΔOR Mutations Reveal Complementary Domains in the Dorsal Olfactory Bulb.
Confocal images of wholemounts of left and right OBs, dorsal views, anterior is up. (A) (Top) Structure of the S50 locus and the S50-IRES-tauGFP targeted mutation, in which the S50 coding region (grey box) is followed by an IRES (unlabeled white box) and tauGFP (green box) (Bottom) S50-GFP axons coalesce into glomeruli in the anterior-dorsal OB.
(B) (Top) Structure of the MOR18-2 locus and the MOR18-2-IRES-GFP-IRES-taulacZ targeted mutation (termed MOL2.3-IGITL in Conzelmann et al., 2000), in which the MOR18-2 coding region (grey box) is followed by an IRES, GFP, a second IRES, and taulacZ (unlabeled large white box). (Bottom) 18-2-GFP axons coalesce into 3–4 glomeruli in the dorsal OB.
(C) (Top) Structure of the M72 locus and the M72-IRES-tauGFP targeted mutation (Potter et al., 2001), in which the M72 coding region (black box) is followed by an IRES and tauGFP. (Bottom) M72-GFP axons coalesce into glomeruli in the dorsal-lateral OB.
(D) (Top) Structure of the ΔS50 targeted mutation, in which the S50 coding region is replaced with YFP (green box). ΔS50 axons selectively innervate a circumscribed region of the dorsal-medial OB, and avoid the dorsolateral OB.
(E) (Top) Structure of the Δ18-2 targeted mutation, in which the MOR18-2 coding region is replaced with GFP and followed by IRES and taulacZ. Δ18-2 axons show the same pattern of innervation as ΔS50 axons.
(F) (Top) Structure of the ΔM72 targeted mutation (Feinstein et al., 2004), in which the M72 coding region is replaced with GFP and followed by IRES and taulacZ. ΔM72 axons project to a broad region of the lateral OB, and avoid a discrete domain in the dorsal-medial OB.
(G) OBs of a mouse homozygous both for ΔS50 and ΔM72 alleles. GFP is partially separated from YFP on a spectral basis. GFP is detected in the green channel only, YFP is detected in the red and green channels and appears yellow. Labeled axons project to complementary domains, which cover most of the dorsal OB.
(H) OBs of a P-LacZ-Tg (line 8) mouse that is heterozygous for ΔS50. The βgal+ axons (X-gal/Fast Red Violet staining, red) and ΔS50 axons (intrinsic GFP fluorescence, green) project to complementary domains in the dorsal OB.
(I) OBs of a P-LacZ-Tg (line 8) mouse homozygous for ΔM72. βgal+ axons (red) and ΔM72 axons (green) project to overlapping domains in the dorsal-lateral OB.
Scale bar in I is 470 μm in A–I.
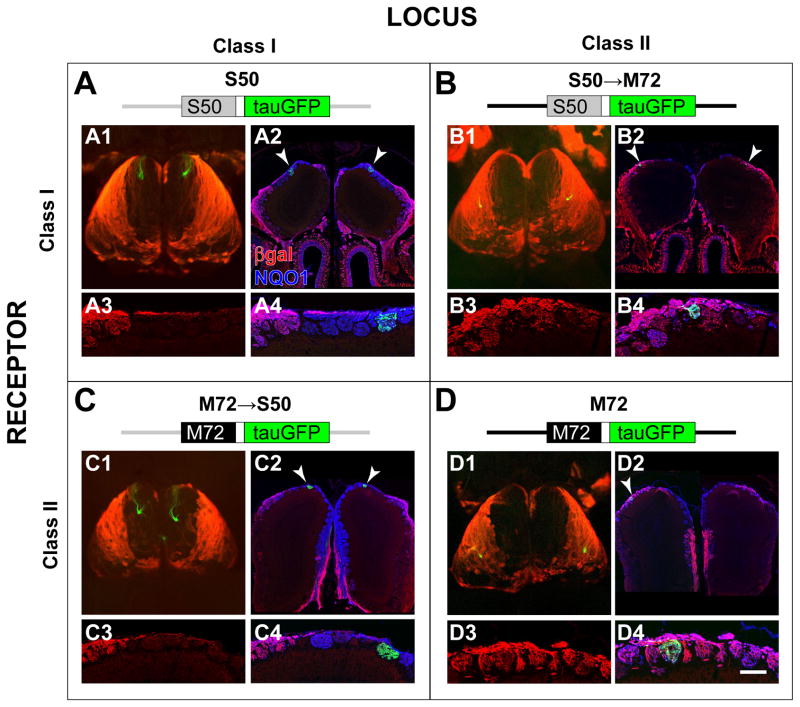
Figure 5. Glomerular Domains are Not Determined by the Class of the Expressed OR Protein.
P-LacZ-Tg (line 8) was crossed to four alleles with an OR-IRES-tauGFP design. A schematic of the alleles is shown at the top of each panel. The S50 (Class I) locus and coding sequence are shown in grey, the M72 (Class II) locus and coding sequence are shown in black. (A1, B1, C1, D1) Wholemounts of the OBs show the relationship between the glomeruli innervated by OR-IRES-tauGFP OSNs (green), and the class domains revealed by X-gal/Fast Red Violet staining (red) for P-LacZ-Tg. (A2, B2, C2, D2) Coronal OB sections labeled with intrinsic GFP fluorescence (green), anti-βgal antibodies (red), and anti-NQO1 antibodies (blue). (A3, B3, C3, D3) High magnification of anti-βgal signals in the glomerular layer. (A4, B4, C4, D4) High magnification of all three markers shows the location of individual glomeruli. Overlay of red and blue is purple. S50 glomeruli (A) and M72→S50 glomeruli (C) are βgal−, reside within the βgal− domain in the dorsal-medial OB, and are surrounded by βgal− glomeruli. In contrast, S50→M72 glomeruli (B) and M72 glomeruli (D) are βgal+, reside within the βgal+ domain in the dorsolateral OB, and are surrounded by βgal+ glomeruli.
Scale bar in D4 is 1000 μm in A1, B1, C1 and D1; 250 μm in A2; 150 μm in A3 and A4; 250 μm in B2; 150 μm in B3 and B4; 580 μm C2; 175 μm in C3 and C4; 700 μm in D2; 100 μm in D3 and D4.
Next, we generated two strains of mice in which the corresponding OR coding sequence is deleted by gene targeting. In the first (YFP→S50, abbreviated ΔS50), the S50 coding sequence is replaced with the sequence encoding the yellow fluorescent protein (YFP) (Figure 3D). In the second (GFP→MOR18-2-IRES-taulacZ, abbreviated Δ18-2), the MOR18-2 coding sequence is replaced with the GFP sequence followed by IRES-taulacZ (Figure 3E). The ΔS50 and Δ18-2 strains exhibit very similar patterns of innervation: a majority of labeled axons project diffusely to a large subset of glomeruli in the dorsal-medial and anterior-medial OB (Figures 3D and 3E). We note that a minor subset of ΔS50 and Δ18-2 axons innervate glomeruli at the caudal margins of the OB (Figures 3D and 3E). The identity of these caudal glomeruli remains unknown; normally, they do not receive axonal input from S50 or MOR18-2 expressing OSNs (Figures 3A and 3B).
To examine the projections for a Class II OR, we studied the dorsally expressed Class II OR gene M72 (MOR171-3, Olfr160). In mice with an intact M72 coding sequence (M72-IRES-tauGFP, Potter et al., 2001), labeled axons project to glomeruli within the caudal-medial and dorsal-lateral OB (Figure 3C). As expected, the M72 glomeruli are located within the βgal+ domain of P-LacZ-Tg mice (Figure 5D). In the M72 deletion (abbreviated ΔM72, Feinstein et al., 2004), a majority of labeled axons project diffusely to the caudal-medial and dorsal-lateral OB (Figure 3F).
Crossing ΔS50 and ΔM72 strains reveals that ΔS50 OSNs and ΔM72 OSNs project their axons to complementary domains with minimal overlap (Figure 3G). Crossing these strains separately with P-LacZ-Tg mice shows that ΔS50 axons fill in the βgal− domain (Figure 3H), and that, by contrast, ΔM72 axons avoid this βgal− domain but project to the βgal+ domain (Figure 3I).
Thus, axons of OSNs that choose an endogenous OR promoter that lacks its OR coding sequence reveal the same domains as OSNs that express P-LacZ-Tg. Moreover, the domains that we observe appear consistent with the proposed Class I and II domains (Kobayakawa et al., 2007).
OR Co-expression in OR Coding Sequence Deletions
OSNs that express a ΔOR targeted mutation or a ΔOR transgene are known to co-express other OR genes (Serizawa et al., 2003; Feinstein et al., 2004; Lewcock and Reed, 2004; Shykind et al., 2004). We next asked if ΔOR OSNs co-express OR genes selectively from the corresponding class, as we observed with P-LacZ-Tg and Class II OR genes.
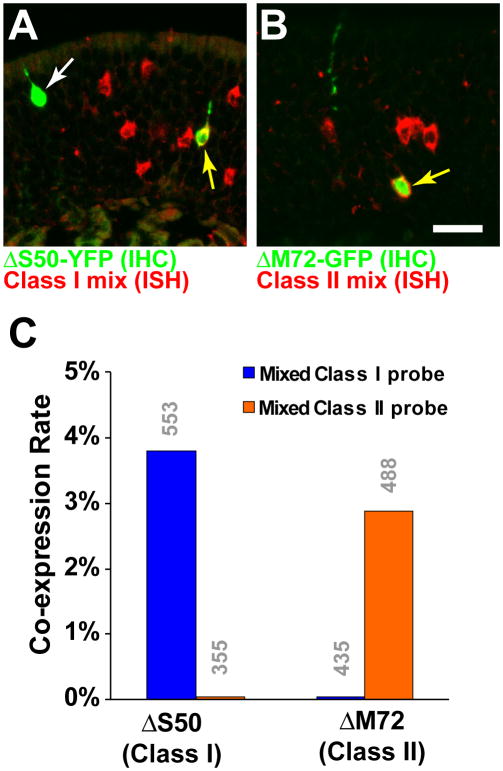
We determined the co-expression rates of Class I and II OR genes in ΔS50 and in ΔM72 OSNs, using a combination of IHC for YFP/GFP and ISH with probes for OR coding sequences (Figures 4A and 4B). Co-expression rates for a given OR are low (Feinstein et al., 2004; Serizawa et al., 2003; Shykind et al., 2004) due to the size of the repertoire. To increase the probability of observing double-labeled OSNs, we used a mix of five probes for dorsal Class I ORs (MOR22-2, MOR31-2, MOR31-6, MOR34-7, and MOR40-14) and a mix of five probes for dorsal Class II ORs (MOR101-1, MOR106-1, MOR140-1, MOR143-1, and MOR 265-1). We observe that 3.8% of ΔS50 OSNs are labeled with the Class I mix, but none with the Class II mix (Fisher Exact Test, p<0.001.) By contrast, no ΔM72 OSNs are labeled with the Class I mix, but 2.9% with the Class II mix (Fisher Exact Test, p<0.001).
Figure 4. ΔOR OSNs Are Skewed to Co-express OR Genes of the Same Class.
(A) Combined ISH and IHC in the MOE of a ΔS50 homozygous mouse, using a mixed Class I OR probe (red). ΔS50 cells are labeled green. White arrow indicates a ΔS50 neuron that is not labeled with the mixed probe, yellow arrow indicates a ΔS50 OSN that co-expresses one (or more) Class I OR genes.
(B) Combined ISH and IHC in the MOE of a ΔM72 homozygous mouse, using a mixed Class II OR probe (red). Yellow arrow indicates a ΔM72 OSN that co-expresses one (or more) Class II OR genes.
(C) Percentage of ΔS50 or ΔM72 neurons that are double-labeled with the mixed Class I and Class II OR probes. OR co-expression in ΔOR OSNs is strongly skewed by OR class. Numbers of OSNs counted are indicated in grey above the bars.
Scale bar in B is 20 μm in A, and 18 μm in B.
Thus, OSNs that express a ΔOR targeted mutation selectively co-express ORs of the same class, as we observed with the P-LacZ-Tg and Class II OR genes. These observations support our model of two dorsal OSN types that are fated to choose either Class I or II promoters.
Domain Organization is Not Determined by the Class of the Expressed OR Protein
How do axons of two dorsal OSN types sort out into two domains? Given the close involvement of ORs in axon guidance, one intuitive hypothesis is that the class of the expressed OR protein, in and of itself, determines the domain to which the axons project. An alternative hypothesis is that domain segregation reflects intrinsic properties of OSN types, regardless of the class of the expressed OR protein. To distinguish between these two hypotheses (class of expressed OR vs OSN type), we asked if replacing the coding sequence of a Class I OR with a Class II OR, or vice versa, directs axons to form a glomerulus in the domain corresponding to the expressed OR protein. We performed these OR coding sequence replacements with S50 and M72.
As noted above, the S50 glomeruli reside well within the βgal− domain (Figure 5A); their βgal− phenotype is consistent with the low co-expression rate between S50 and P-LacZ-Tg (0.2%, MOR42-1 in Figure 2). In contrast, both the medial (data not shown) and lateral M72 glomeruli reside within the βgal+ domain (Figure 5D); their βgal+ phenotype is consistent with the high co-expression rate between M72 and P-LacZ-Tg (11%, MOR171-3 in Figure 2).
We generated two strains of mice: one in which the S50 coding sequence replaces that of M72 (S50→M72-IRES-tauGFP, abbreviated S50→M72), and vice versa (M72→S50-IRES-tauGFP, abbreviated M72→S50). These alleles were crossed individually with P-LacZ-Tg. We observe that S50→M72 glomeruli fall within the βgal+ domain, and receive extensive input from βgal+ axons (Figure 5B), and conversely, that M72→S50 glomeruli are situated well within the βgal− domain, and receive little or no input from βgal+ axons (Figure 5C).
Thus, for both OR coding sequence replacements, the domain to which axons project is determined, not by the class of the expressed OR protein, but by the OSN type. These observations support a model in which two dorsal OSN types exhibit intrinsic differences in axon guidance identity that cannot be overridden by the class of the expressed OR protein. We refer to these OSN types tentatively as DI-OSN and DII-OSN (consistent with the nomenclature of Kobayakawa et al., 2007), reflecting their dorsal location within the MOE and the preferred class of OR promoter choice.
Axons of Dorsal OSN Types Segregate Within the Olfactory Nerve
OSN axons cross the basal lamina of the MOE, and form axon bundles that course via the lamina propria through the cribriform plate to glomeruli. NQO1+ and OCAM+ OSNs are segregated within the MOE; their axons maintain this segregation in the olfactory nerve, and project to different parts of the OB (Yoshihara et al., 1997; Gussing and Bohm, 2004). In contrast, DI- and DII-OSNs project their axons to distinct domains of the OB, although they are intermingled diffusely in the same region of the dorsal MOE, without obvious compartmentalization or patterning (Figure 6A, and data not shown). We therefore asked if DI and DII axons also segregate in the olfactory nerve.
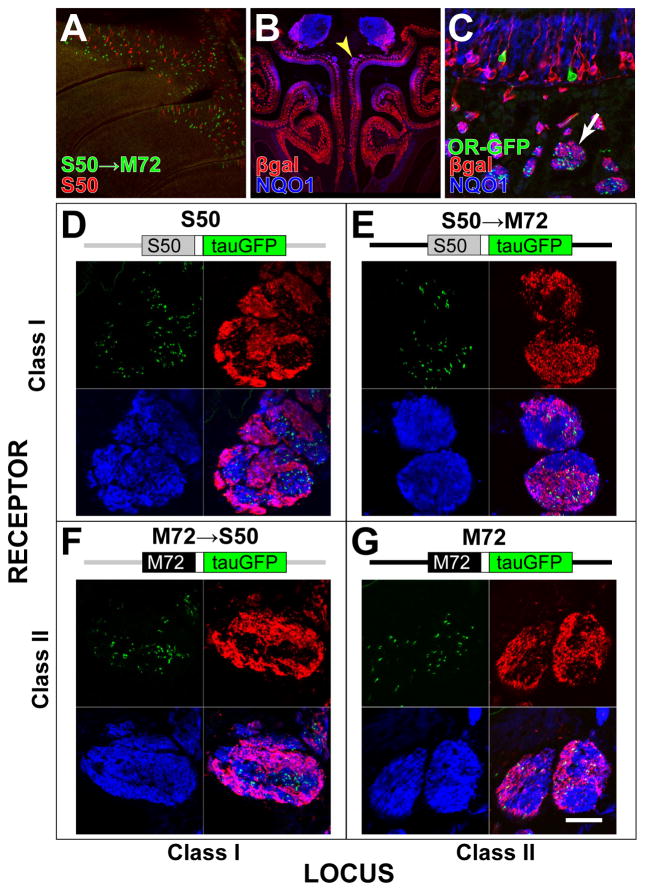
Figure 6. Class I and Class II Axons Segregate Within the Olfactory Nerve.
(A) Wholemount confocal image of the turbinates showing the distribution of OSNs expressing S50→M72-IRES-tauGFP (green) and S50-IRES-tauRFP (red). OSNs are intermingled in the dorsal MOE, without obvious compartmentalization or patterning. (B) Coronal section through the olfactory mucosa of a P-LacZ-Tg hemizygous mouse of line 8, labeled by IHC with anti-βgal antibodies (red) and NQO1 antibodies (blue). Yellow arrowhead indicates the location of large segregated axon bundles.
(C) Coronal section through the dorsal olfactory mucosa of a P-LacZ-Tg hemizygous (line 8), S50-IRES-tauGFP homozygous mouse labeled with intrinsic GFP fluorescence (green), and IHC with anti-βgal antibodies (red) and NQO1 antibodies (blue). The distribution of GFP, βgal and NQO1 signals can be seen in cross-sections through axon bundles (white arrow) in the lamina propria underneath the MOE.
(D–G) A diagram of the targeted mutation is above each panel. Confocal images of cross-sections through axon bundles show the distribution of OR-defined axons (green) with respect to βgal+ axons (red) and NQO1+ axons (blue). βgal preferentially labels sub-compartments of the axon bundles. (D) Axons of OSNs expressing S50 from the endogenous S50 locus are associated preferentially in βgal− axonal compartments. (E) Axons of OSNs expressing S50 from the M72 locus are found preferentially in βgal+ compartments. (F) Axons of OSNs expressing M72 from the S50 locus are found preferentially in βgal− compartments. (G) Axons of OSNs expressing M72 from the endogenous M72 locus are found preferentially in βgal+ compartments.
Scale bar in G is 400 μm in A; 700 μm in B; 35 μm in C; 25 μm in D–G.
We examined the spatial distribution of DI-OSN (βgal−) and DII-OSN (βgal+) axons in dorsally-projecting (NQO1+) axon bundles in the olfactory mucosa by IHC (Figure 6B). The spatial distribution of axons in the lamina propria was analyzed in cross-sections of axon bundles using confocal microscopy. We find that βgal+ (DII) axons are distributed non-homogeneously within NQO1+ (dorsal) axon bundles, and are often segregated into discrete compartments. This segregation is most obvious in large (>100 μm diameter) axon bundles as they approach the cribriform plate. However, smaller axon bundles also show evidence of axonal segregation (Figure 6C).
To analyze this axonal segregation in detail, we used endogenous GFP fluorescence to visualize specifically the axons from OSNs that express tagged Class I and II OR alleles (Figures 6C–G). As expected, S50 axons are preferentially found in NQO1+, βgal− compartments (Figure 6D), and M72 axons tend to occupy βgal+ compartments (Figure 6G). We quantified axonal segregation by counting the numbers of S50 and M72 axons that fall within the βgal+ and βgal− compartments of NQO1+ axon bundles. For S50, 18% of axons (98/533, n = 9 axon bundles) reside within βgal+ compartments. In contrast, for M72, 84% of axons (359/429, n = 14 axon bundles) reside within βgal+ compartments. This difference is statistically significant (Fisher’s Exact test, p < 0.001). We confirmed this observation for another dorsal Class II OR gene, S1 (MOR106-1, Olfr749): in S1-IRES-tauGFP mice, 91% of axons (320/350, n = 22 bundles) reside within βgal+ compartments (data not shown).
As was the case with glomerular position in either domain, we find that axonal segregation into compartments is not determined by the class of expressed OR protein. The proportion of S50→M72 axons in βgal+ compartments (1430/1684 axons, 85%; n = 18 bundles) is similar as for M72 axons (84%). M72→S50 axons show a similarly low proportion of axons in βgal+ compartments (257/1473 axons, 17%; n = 10 bundles) as S50 axons (18%). There is no correlation between the class of the expressed OR and the proportion of axons in βgal+ compartments (M72 vs. S50→M72, p = 0.45; S50 vs. M72→S50, p = 0.64; Fisher’s Exact test). In contrast, there is a significant correlation between the class of the chosen OR promoter (DI- or DII-OSN type, in our model) and the proportion of axons in βgal+ compartments (M72 vs. M72→S50, p < 0.001; S50 vs. S50→M72, p <0.001).
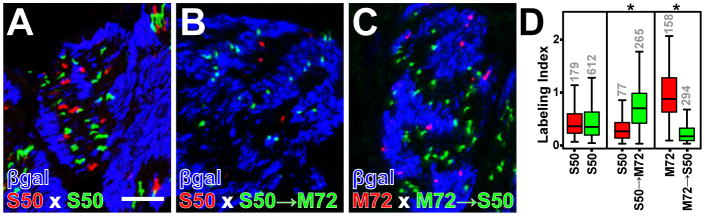
We confirmed these observations by simultaneously examining two alternately labeled populations of OR-defined axons within the same axon bundles. We generated a series of crosses in which S50-expressing OSNs and M72-expressing OSNs are labeled with GFP and RFP respectively, on a P-LacZ-Tg backgound. Cross-sections of axon bundles were stained with antibodies against βgal and analyzed using three-color confocal microscopy (Figure 7). The position of each axon with respect to the βgal+ compartment was quantified by calculating a Labeling Index (LI) (Figure 7D, see Experimental Procedures for details). A greater value of the mean LI indicates a higher degree of overlap of the axonal population with the βgal+ compartment. In a control experiment, there is no significant difference in mean LI between S50-RFP axons (0.48 ± 0.028, n=179) and S50-GFP axons (0.47 ± 0.015, n = 612; t(283)= 0.27, p = 0.79; unequal variance t-test) (Figures 7A and 7D), ruling out an effect of the fluorescent marker used. In contrast, there is a significant difference in mean LI between S50-RFP axons (0.39 ± 0.04, mean ± SEM, n = 77) and S50→M72-GFP axons (0.75 ± 0.026, n = 265; t(142)= −7.5, p<0.001) (Figures 7B and 7D). Similarly, there is a significant difference in the reciprocal experiment (Figures 7C and 7D), between M72-RFP axons (0.94 ± 0.037, n = 158) and M72→S50-GFP axons (0.27 ± 0.15, n = 294); t(211) = 16.9, p<0.001).
Figure 7. Quantification of Axonal Segregation by OR Locus.
(A–C) Confocal images of axon bundles in which axons from OSNs expressing tagged OR genes are labeled with RFP (red) or GFP (green). Axons expressing P-LacZ-Tg are labeled by IHC for βgal (blue). A Labeling Index (LI) is calculated for each OR-defined axon, ranging from 0 for axons located in a βgal− compartment and ≥1 for axons located in a βgal+ compartment. (D) Distribution of the LI values observed for red or green axons in three pairwise crosses. Number of axons in each group is indicated in grey above each bar. Asterisks signify p-values <0.001 from unequal variance t-tests comparing the two means from each pairwise experiment.
Scale bar in A is 12 μm for A–C.
Thus, despite the intermingling of DI- and DII-OSNs within the MOE, their axons segregate at the level of the olfactory nerve, and this segregation is independent of the class of the expressed OR protein.
Segregation of DI and DII Axons Underlies Odorant Response Domains
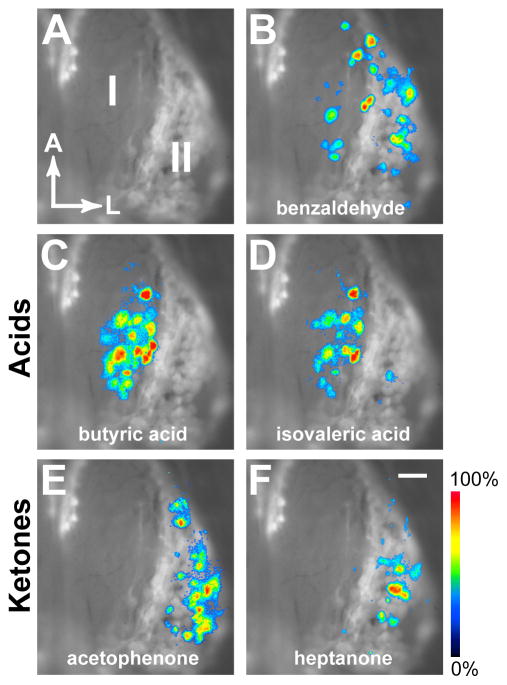
Finally, we asked if the glomerular domains formed by axons of DI- and DII-OSNs correlate with odorant-evoked activity patterns in the OB. We crossed P-LacZ-Tg mice with mice in which all mature OSNs express synaptopHluorin, a genetically-encoded indicator for synaptic transmission (Bozza et al., 2004). We imaged odorant-evoked activity in the dorsal OBs of freely breathing, anesthetized mice (Figure 8). The DI and DII domains were revealed by staining the OBs with the fluorescent βgal substrate Fast Red Violet (Feinstein and Mombaerts, 2004) immediately after imaging (Figure 8A). Many odorants, such as benzaldehyde (Figure 8B) and methylbenzoate (data not shown), activated spatially distributed glomeruli in both domains. Previous studies in rat and mouse have shown a distinct boundary between groups of glomeruli that respond to organic acids and ketones (Uchida et al., 2000; Wachowiak and Cohen, 2001; Mori et al., 2006; Johnson and Leon, 2007). We therefore tested acids and ketones that have been shown to activate these response domains in rat (Takahashi et al, 2004). We find that butyric acid and isovaleric acid activate preferentially (but not exclusively) a subset of glomeruli in the βgal− domain (DI), even at high stimulus concentrations (Figures 8C and 8D). In contrast, acetophenone and heptanone elicit activity preferentially in a subset of glomeruli in the βgal+ domain (Figures 8E and 8F). The lateral boundary of the group of acids-responsive glomeruli correlates closely with the boundary between the DI and DII domains.
Figure 8. Class Domains Correlate With Odorant Response Modules.
(A) Class I/II domains revealed by X-gal/Fast Red Violet staining for βgal enzymatic activity in vivo, after removal of the skull and dura. Right OB is shown. Anterior is up, lateral is to the right.
(B) Overlay of (A) with an activity pattern measured as the change in fluorescence of synaptopHluorin, following a 5 sec application of 5% benzaldehyde. Responses are seen in glomeruli in both domains.
(C) Butyric acid at 5% and (D) isovaleric acid at 5% prefentially activate glomeruli in the Class I domain.
(E) Acetophenone at 2% and (F) heptanone at 5% selectively activate glomeruli in the Class II domain.
Response images were thresholded at 30% maximal response, pseudocolored and overlayed. Scale (bottom right) shows the synaptopHluorin signal intensity from blue (low) to red (high).
Scale bar in F is 200 μm in A–F.
Thus, the mapping of the acids to a cluster or module in the OB is mediated by the axon guidance properties of the DI- and DII-OSN types.
DISCUSSION
OSN Types Underlie OR Choice and Axon Guidance
Previous genetic experiments support the idea that Class I and II OR genes are differentially regulated. In mutant mice that lack the homeodomain protein Lhx2, Class II OR expression is abolished while Class I OR expression is largely spared (Hirota et al., 2007). The number of Class I expressing OSNs is not increased however, suggesting that Class I and II OSNs derive from distinct lineages.
Here, we provide genetic evidence for two dorsal OSN types, which we refer to as DI and DII. We propose a model in which DI- and DII-OSNs exhibit class-specific OR gene choice, differential dependence on Lhx2 for maturation and/or OR gene choice, and distinct axon guidance identities (Figure 9). In our model, a given dorsal OSN cannot choose indiscriminately between dorsal Class I and dorsal II OR genes, but is restricted, by lineage and by type, to the choice of a Class I or II promoter. Further, axonal segregation in the olfactory nerve and domain segregation are not determined by the class of the expressed OR protein, but by other properties of the type of OSN, which remain to be defined. We have previously suggested that various OSN types (referred to as positional cell types) may affect axonal wiring of OSNs, regardless of the ORs they express (Feinstein and Mombaerts, 2004).
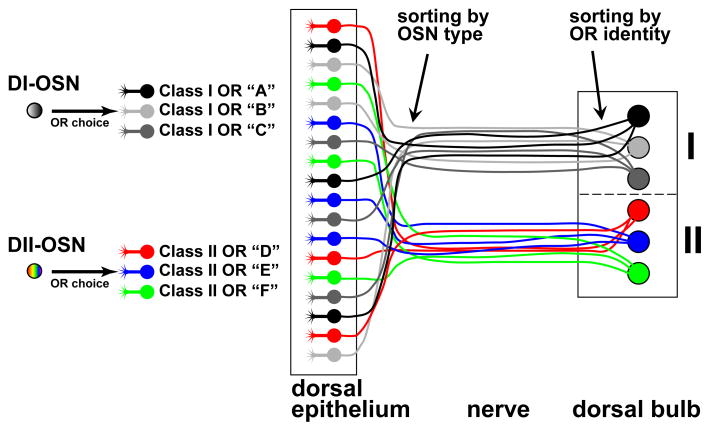
Figure 9. Model for Mapping of OSN Types to Glomerular Domains.
The dorsal MOE contains two OSN types, which we refer to as DI and DII to reflect their dorsal position and choice of OR promoter. DI-OSNs choose promoters of Class I OR genes (grayscale), and DII-OSNs promoters of Class II OR genes (color). The cell bodies of DI- and DII-OSNs are intermingled in the dorsal MOE, and their axons segregate within the olfactory nerve via a mechanism that does not depend on the class of the expressed OR protein. DI-axons form glomeruli in the dorsal-medial OB (I), and DII-axons form glomeruli in the dorsal-lateral OB (II). Within each domain, axons sort into glomeruli via a mechanism that depends on the identity of the expressed OR protein.
Class-specific OR Gene Expression in OSN Types
OSNs that express an endogous OR gene or transgene in which the OR coding sequence is deleted or otherwise crippled, can co-express other OR genes (Serizawa et al., 2003; Feinstein et al., 2004; Lewcock and Reed, 2004; Shykind et al., 2004). We find that co-expression is heavily skewed by OR class in our P-LacZ-Tg mice. This phenomenon is most readily observed and quantified in these mice because the transgene exhibits an unusually high rate of expression. A similar class-specific co-expression likely underlies the Class II OSN-specific genetic manipulation that was reported for a mouse strain with a Class II OR transgene that lacks an OR coding sequence (Kobayakawa et al., 2007). Our ΔOR gene-targeted mutations further show that class-specific OR gene choice also occurs with endogenous OR promoters that lack their coding sequence.
As is often the case in biological systems, the dichotomy in Class I and II OR expression among dorsal OSNs is not absolute. We detect a very low rate of co-expression of P-LacZ-Tg with typical Class I OR probes in dorsal OSNs (8/10,000 cells), consistent with rare βgal+ axons projecting to individual glomeruli in the DI-OSN domain. Our interpretation is that DI-OSNs can occasionally activate a Class II promoter. We note that co-labeling between an OR probe and the βgal reporter is not necessarily the result of concurrent transcription of an OR gene and the transgene (Shykind et al., 2004).
Two ventral Class I OR genes show a high co-expression rate with P-LacZ-Tg, providing further evidence that they are regulated like Class II genes (Hoppe et al., 2006; Hirota et al., 2007). Our interpretation is that they encode ORs that are phylogenetically Class I, but have Class II-like promoters that are chosen by a ventral OSN type. We speculate that, conversely, there may be a few OR genes that are driven by Class I-like promoters but that encode OR proteins that are phylogenetically Class II.
OSN Types and Axonal Identity
It is now well established that ORs are intimately involved in axonal coalescence into glomeruli (Mombaerts et al., 1996; Mombaerts, 2006). Axons from OSNs expressing the same OR fasciculate after traversing from the outer to the inner nerve layer of the OB (Treloar et al., 2002). Axonal sorting culminates within the glomeruli, where it is believed to be nearly absolute (Treloar et al., 2002). The sorting and coalescence of OR-defined axons may occur via differential, homophilic interactions among OR-containing complexes (Feinstein and Mombaerts, 2004) or among axon guidance molecules that are themselves modulated by the expressed OR proteins (Serizawa et al., 2006). Models for OR-dependent convergence of OSN axons have been proposed that involve OR signaling through G-proteins and the cAMP pathway, with the level of signaling modulating the expression of conventional axon guidance molecules (Imai et al., 2006; Serizawa et al., 2006; Chesler et al., 2007; Kaneko-Goto et al., 2008).
We hypothesize that DI and DII axons sort by axon-axon interactions, attractive and/or repulsive. This kind of axonal sorting, however, appears to involve different molecular mechanisms than OR-mediated axonal sorting into glomeruli, for three reasons. First, this sorting likely involves just two (or a few) populations of axons. Second, axonal segregation according to OSN type occurs well before axons enter the inner nerve layer of the OB, where OR-specific sorting is thought to occur (Treloar et al., 2002). Third, OR coding sequence replacements argue that axonal segregation and axonal coalescence in a domain are independent of the identity of the expressed OR protein.
Sorting within the olfactory nerve has been reported between ventrally and dorsally projecting OSN axons in the mouse (Gussing et al., 2004), and between axons of presumptive Class I and Class II OR-expressing OSNs in Xenopus laevis (Gaudin and Gascuel, 2005), suggesting that it is a conserved feature of OSN axon guidance.
Self-Organization and Axonal Pre-sorting
OSN have a remarkable capacity for self-organization: axonal sorting and glomerular formation can occur in the absence of second-order neurons (Bulfone et al., 1998), even in the absence of the OB (St John et al., 2003; Chehrehasa et al., 2006). A similar phenomenon of pre-target sorting has been reported in the mouse retinotectal projection (Plas et al., 2005), but its functional significance is unknown. The gross sorting of OSN axons within the olfactory nerve could partition or channel axons to discrete domains in the OB. Once axons arrive within a domain, they would sort according to OR identity into glomeruli. Pre-target sorting could promote axonal coalescence by concentrating axons of a defined OSN population within the same region of the OB, thereby increasing the likelihood of axonal interactions.
Pre-target sorting may work in conjuction with target-derived cues, which may lay out a broad, permissive framework for self-organization. We cannot exclude the possibility that retrograde cues from the OB cause the appearance of sorting via effects on OSN survival in embryonic or early postnatal stages. It will be interesting to determine if axonal segregation in the olfactory nerve occurs early in development, before the axons have contacted the OB, and also in mice that do not develop OBs.
Conclusion
The preferential mapping of acids responses to a domain in the dorsal OB (Mori et al., 2006) appears to be accomplished by mapping the axons of an OSN type to this domain. Our findings are generally consistent with Kobayakawa et al., 2007. Odorant mapping to this domain is OR-independent in the sense that it is governed not by the identity or specificity of the ORs, but by the OSN type that expresses them.
We speculate that additional OSN types (dorsal or ventral) choose among various subsets of the OR repertoire, exhibit intrinsic axonal identities, and consequently project to distinct anatomical domains. These other hypothetical OSN types may be revealed through comparative analysis of ΔOR strains: by determining the subset of OR genes that are co-expressed, and the subset of glomeruli that are innervated. Such OSN types would also be determinants of axonal wiring at a higher level than ORs. These OSN types would then strongly influence the functional organization of the glomerular array, independently of OR-determined odorant response properties.
EXPERIMENTAL PROCEDURES
Transgenic Mice
In P-LacZ-Tg, the 317 nt P sequence precedes the taulacZ coding sequence, and is followed by a 556 nt rabbit β-globin polyadenylation sequence. The P sequence is: TCCCTGTGCAGCAGAGTAGCTCATTAACAACTTTTTAATGAAGTCACAGATAGTGAC TGCTCATTAATAAATTTATTAGTCTGCAGCCCCAGAGATTAAAGAAACTGAAAACAG AAGTCAGTGACAACAGTGTTTAGCCAGAGTAGCTGCATCAGAGTGAAAAGAGGGAA CTTCTCTTGGCAGACTCTGAAACTTTTACTGTTGGACCTCTAGGGTCCTTGCAGGTCC CAGAAACTGTAATTCTGAAAATAGAAACCTGGGGGGACTAAGAACTTGATGGGATG AAGGAAAAACAGAGCCAAATGATCCCAGGCTTG. Transgenic mice were produced by microinjection into B6DF2 pronuclei.
Gene-targeted Mice
Strains OMP-tau-lacZ, M72-IRES-tauGFP, M72-IRES-tauRFP and GFP→M72-IRES-tauLacZ (ΔM72) mice have been described (Mombaerts et al., 1996; Potter et al., 2001; Feinstein et al., 2004). The IRES-taulacZ sequence of ΔM72 has undergone an inadvertent mutation, which is not present in the targeting vector, and most likely a partial deletion within taulacZ. Cells that express the targeted mutation do not exhibit βgal enzymatic activity. This inadvertent mutation does not affect the interpretation of Feinstein et al., 2004.
S50-IRES-tauGFP
A 8.2 kb BamHI/BglII genomic fragment containing the S50 coding sequence was isolated from a 129/SvJ BAC clone (Genome Systems). An AscI restriction site was introduced three nt downstream of the stop codon using recombinant PCR. A cassette with an IRES, the sequence for tauGFP, and a self-excising neo gene (ACNf) was inserted into the AscI site (Bozza et al., 2002; Bunting et al., 1999).
S50-IRES-tauRFP2
An IRES-RFP-ACNf cassette encoding a tau-DsRed-DsRed fusion protein was inserted into the AscI site.
ΔS50
The coding sequence for venus-YFP (Nagai et al., 2002) was amplified by PCR as a BsaHI/AscI fragment, and inserted into the S50 targeting vector. This fragment replaces the S50 coding sequence exactly from start to stop codon, leaving the native S50 Kozak consensus sequence. An ACNf cassette was inserted into the AscI site.
ΔMOR18-2
A BspEI/StuI genomic fragment containing the MOR18-2 (also known as MOL2.3) locus was subcloned into pBluescript-SK(−). A PstI shuttle was subcloned, and the MOR18-2 coding sequence was replaced with the GFP coding sequence, simultaneously introducing an AscI restriction site three nt downstream of the GFP coding sequence. The modified shuttle was ligated to the BspEI/StuI fragment. An IRES-taulacZ-ACNf cassette was cloned into the AscI site.
M72→S50-IRES-tauGFP
The M72 coding sequence was amplified from a 129/SvJ targeting vector, with a BsaHI site appended just 5′ of the coding sequence and an AscI site three nt downstream of the stop codon. The amplified sequence was then inserted into the S50 targeting vector as a BsaHI/AscI fragment such that it replaced the S50 coding sequence exactly from start to stop codon, leaving the native S50 Kozak consensus sequence. An IRES-tauGFP-ACNf cassette was inserted into the Asc I site.
S50→M72-IRES-tauGFP
A BglII-PacI fragment containing the S50 coding sequence fused to 184 bp 5′ of the M72 coding sequence was generated by PCR. The fragment was inserted into the M72 targeting vector (Feinstein et al., 2004) with BglII/PacI, replacing the M72 coding region exactly from start to stop codon, leaving the M72 native Kozak consensus sequence. An IRES-tauGFP-ACNf cassette was inserted into the Pac I site located three nt downstream of the stop codon.
Gene targeting was performed with E14 embryonic stem cells as described (Mombaerts et al., 1996). Mice are in a mixed 129/B6 background.
The strains are publicly available from The Jackson Laboratory:
S50-IRES-tauGFP, clone S50G-37, JAX #6712, B6;129P2-Olfr545tm1Mom/MomJ
S50-IRES-tauRFP2, clone S50R-8, JAX #6713, B6;129P2-Olfr545tm2Mom/MomJ
venusYFP→S50, clone ΔS50-101, JAX #6716, B6;129P2-Olfr545tm4Mom/MomJ
M72→S50-IRES-tauGFP, clone 7250-75, JAX #6715, B6;129P2-Olfr545tm3(Olfr160)Mom/MomJ
S50→M72-IRES-tauGFP, clone 5072-15, JAX #6714, B6;129P2-Olfr160tm11(Olfr545)Mom/MomJ
GFP→MOR18-2-IRES-taulacZ, clone D18-268, JAX #6722, B6;129P2-Olfr78tm1Mom/MomJ
P-LacZ-Tg, line 8, JAX #6742, B6;CBA-Tg(P-taulacZ)8Mom/MomJ
P-LacZ-Tg, line 11, JAX #6743, B6;CBA-Tg(P-taulacZ)11Mom/MomJ
P-LacZ-Tg, line 13, JAX #6793, B6;CBA-Tg(P-taulacZ)13Mom/MomJ.
ISH and IHC
OR coding sequence probes were amplified by PCR from olfactory cDNA or genomic DNA, subcloned and sequenced. ISH combined with IHC was performed as described (Ishii et al., 2004). Three to five week-old P-LacZ-Tg mice were anesthetized and perfused transcardially with cold 4% paraformaldehyde. Olfactory epithelia were dissected, postfixed overnight at 4°C, decalcified, cryoprotected, and sectioned at 12 μm. Each OR probe was tested on representative sections from the anterior-posterior extent of the MOE.
For combined ISH and IHC, proteinase K treatment was reduced to 4 min at 37°C. Hybridization at 62°C and washing were performed as described (Ishii et al., 2004). For ISH detection, sections were incubated with an anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche) at 1:1000, along with a rabbit anti-βgal antibody (Cappel) at 1:500 to detect βgal in P-LacZ-Tg mice, or a rabbit anti-GFP antibody (Molecular Probes) at 1:100 to detect GFP/YFP in ΔOR mice. After staining with HNPP/Fast Red (Roche), tissue was incubated with a goat-anti-rabbit AlexaFluor 488 or an HRP-goat-anti-rabbit antibody (Molecular Probes) at 1:500 for 1 hr at room temperature. The GFP/YFP signal was amplified using the Tyramide Signal Amplification kit (Perkin Elmer), and detected using an AlexaFluor 488-streptavidin conjugate (Molecular Probes) at 1:300 for 30 min at room temperature. Imaging was performed using a Zeiss LSM 510 or LSM5 Pascal confocal microscope and Zeiss AIM software. Candidate double-labeled neurons were examined via high-resolution, confocal z-stacks to confirm co-localization of ISH and IHC signals.
OR probes were classified into four groups based on OR Class and dorsal-ventral expression. The co-expression rate for each group of OR genes was estimated using a random effects logistic regression model, in which the observed co-expression rate for each OR is treated as an independent estimate of the true rate for a given group. The model allows us to test for group differences while accounting for variability among the probes. Data were analyzed using the R statistical software package (www.r-project.org).
For visualization of GFP fluorescence and IHC, mice were perfused with cold PLP (paraformaldehyde/lysine/periodate), postfixed for 1 h at 4°C, and processed as above. Sections were incubated overnight at 4°C with goat-anti NQO1 (AbCAM) at 1:250, and rabbit-anti-βgal (Cappel) at 1:1000. Tissue was stained with donkey anti-rabbit AlexaFluor 568 or donkey anti-goat AlexaFluor 633 (Molecular Probes) as secondary antibodies for 45 min at room temperature. Co-localization of labeling in axon bundles was performed using images from single confocal optical sections.
For three-color IHC imaging with ORs tagged red and green, images from the red, green, and blue channels were imported into Matlab, and a 3×3 pixel median filter was applied to reduce pixel noise. A mask was applied to each channel and individual axons were identified using an object selection algorithm. For each axon, we calculated a Labeling Index as the mean pixel value of the unmasked blue channel (βgal) surrounding the axon, normalized by the average pixel intensity over the entire axon bundle. Thus, axons within a βgal+ compartment have LI values towards 1 or greater, and those within a βgal− compartment have LI values approaching 0.
For three-color IHC imaging with an OR tagged green together with βgal (red) and NQO1 (blue), a mask was applied to the green and red channels, and axons were identified using an object selection algorithm. The proportion of axons falling either inside or outside the red mask (βgal+ area) was determined. An axon was considered within the domain if ≥ 50% of the axons pixels axon fell within the βgal masked area.
Wholemount Staining and Imaging
Visualization of endogenous GFP fluorescence and of βgal enzymatic activity with X-gal/Fast Red Violet in wholemounts has been described previously (Feinstein and Mombaerts, 2004). Images of OB wholemounts were collected as z-stacks and projected into a single image for display.
Imaging of SynaptopHluorin Activity
In vivo imaging was performed essentially as described (Bozza et al., 2004) using 5–12 week old P-LacZ-Tg mice that are homozygous for the OMP-synaptopHluorin allele. OBs were imaged using a custom Nikon epifluorescence microscope, a 4× (0.2 NA) objective, and a 150W Xenon arc lamp (Opti-Quip) attenuated with ND filters. Fluorescent filters were HQ470/40 (exciter), Q505LP (dichroic), HQ525/50 (emitter). Optical signals were recorded using a cooled CCD camera (NeuroCCD, SM-256) at 256×256 pixel resolution and a frame rate of 7 Hz, controlled by Neuroplex software (RedShirtImaging). Odor stimuli were delivered as 5 sec pulses at ≥60 sec interstimulus interval, using a computer-controlled, flow-dilution olfactometer. Odorants (95–99% pure) are from Sigma or Fluka, and were diluted from saturated vapor in cleaned, desiccated air using mass flow controllers (Aalborg). Concentrations are expressed as percent dilutions of saturated vapor. Response maps were constructed by subtracting the temporal average of a 1.7 sec time window preceding stimulus onset from a 1.7 sec temporal average beginning just after odorant offset. The difference images (ΔF) were pseudocolored, thresholded, and overlayed onto resting fluorescence images. Immediately after recording, head-fixed mice were overdosed with pentobarbital, the dura was removed, and additional images of the resting synaptopHluorin fluorescence were taken. Then the OBs were stained for 5–10 min with X-gal (0.2 mg/ml) and Fast Red Violet LB (1 mg/ml) to label P-LacZ-Tg axons. Odorant response maps and Fast Red Violet images were aligned manually using glomeruli as fiduciary marks. Data were acquired, processed, and displayed using Neuroplex software, Adobe Photoshop, and custom software written in MATLAB (Mathworks).
Acknowledgments
This research was supported by grants from NIDCD to T.B, A.V., S.F, P.F. and P.M. The authors thank Jessica Wu, Jinsong Li, and Wei Tang for performing blastocyst injections; Stephen Fischboek and Katherine Wofsey for help with ISH and IHC; Masayo Omura for ISH probes; Atsushi Miyawaki for providing Venus YFP; Matt Wachowiak for providing software and advice for odor delivery and optical imaging; and Hyung Woo Kim and the Northwestern University Biostatistics Collaboration Center for help with statistical analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression dfines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, McGann JP, Mombaerts P, Wachowiak M. In vivo imaging of neronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 2004;42:9–21. doi: 10.1016/s0896-6273(04)00144-8. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Buck LB. Information coding in the vertebrate olfactory system. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Wang F, Hevner R, Anderson S, Cutforth T, Chen S, Meneses J, Pedersen R, Axel R, Rubenstein JL. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21:1273–1282. doi: 10.1016/s0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- Chehrehasa F, St John JA, Key B. Implantation of a scaffold following bulbectomy induces laminar organization of regenerating olfactory axons. Brain Res. 2006;1119:58–64. doi: 10.1016/j.brainres.2006.08.060. [DOI] [PubMed] [Google Scholar]
- Chesler AT, Zou DJ, Le Pichon CE, Peterlin ZA, Matthews GA, Pei X, Miller MC, Firestein S. A G protein/cAMP signal cascade is required for axonal convergence into olfactory glomeruli. Proc Natl Acad Sci USA. 2007;104:1039–1044. doi: 10.1073/pnas.0609215104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann S, Levai O, Bode B, Eisel U, Raming K, Breer H, Strotmann J. A novel brain receptor is expressed in a distinct population of olfactory sensory neurons. Eur J Neurosci. 2000;12:3926–3934. doi: 10.1046/j.1460-9568.2000.00286.x. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Freitag J, Krieger J, Strotmann J, Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Gaudin A, Gascuel J. 3D atlas describing the ontogenic evolution of the primary olfactory projections in the olfactory bulb of Xenopus laevis. J Comp Neurol. 2005;489:403–424. doi: 10.1002/cne.20655. [DOI] [PubMed] [Google Scholar]
- Gussing F, Bohm S. NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci. 2004;19:2511–2518. doi: 10.1111/j.0953-816X.2004.03331.x. [DOI] [PubMed] [Google Scholar]
- Hirota J, Omura M, Mombaerts P. Differential impact of Lhx2 deficiency on expression of class I and class II odorant receptor genes in mouse. Mol Cell Neurosci. 2007;34:679–688. doi: 10.1016/j.mcn.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Hoppe R, Breer H, Strotmann J. Promoter motifs of olfactory receptor genes expressed in distinct topographic patterns. Genomics. 2006;87:711–723. doi: 10.1016/j.ygeno.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Ishii T, Omura M, Mombaerts P. Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocyt. 2004;33:657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Leon M. Chemotopic odorant coding in a mammalian olfactory system. J Comp Neurol. 2007;503:1–34. doi: 10.1002/cne.21396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57:834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Lane RP, Cutforth T, Young J, Athanasiou M, Friedman C, Rowen L, Evans G, Axel R, Hood L, Trask BJ. Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc Natl Acad Sci USA. 2001;98:7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci USA. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004a;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004b;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Mori K, Takahashi YK, Igarashi KM, Yamaguchi M. Maps of odorant molecular features in the mammalian olfactory bulb. Physiol Rev. 2006;86:409–433. doi: 10.1152/physrev.00021.2005. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DT, Lopez JE, Crair MC. Pretarget sorting of retinocollicular axons in the mouse. J Comp Neurol. 2005;491:305–319. doi: 10.1002/cne.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SM, Zheng C, Koos DS, Feinstein P, Fraser SE, Mombaerts P. Structure and emergence of specific olfactory glomeruli in the mouse. J Neurosci. 2001;21:9713–9723. doi: 10.1523/JNEUROSCI.21-24-09713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- St John JA, Clarris HJ, McKeown S, Royal S, Key B. Sorting and convergence of primary olfactory axons are independent of the olfactory bulb. J Comp Neurol. 2003;464:131–140. doi: 10.1002/cne.10777. [DOI] [PubMed] [Google Scholar]
- Takahashi YK, Kurosaki M, Hirono S, Mori K. Topographic representation of odorant molecular features in the rat olfactory bulb. J Neurophysiol. 2004;92:2413–2427. doi: 10.1152/jn.00236.2004. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Feinstein P, Mombaerts P, Greer CA. Specificity of glomerular targeting by olfactory sensory axons. J Neurosci. 2002;22:2469–2477. doi: 10.1523/JNEUROSCI.22-07-02469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A, Miyazaki T, Imai T, Sakano H. Olfactory sensory neurons expressing class I odorant receptors converge their axons on an antero-dorsal domain of the olfactory bulb in the mouse. Eur J Neurosci. 2006;23:1436–1444. doi: 10.1111/j.1460-9568.2006.04675.x. [DOI] [PubMed] [Google Scholar]
- Uchida N, Takahashi YK, Tanifuji M, Mori K. Odor maps in the mammalian olfactory bulb: domain organization and odorant structural features. Nat Neurosci. 2000;3:1035–1043. doi: 10.1038/79857. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Wachowiak M, Cohen LB. Representation of odorants by receptor neuron input in the mouse olfactory bulb. Neuron. 2001;32:723–735. doi: 10.1016/s0896-6273(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Wilson RI, Mainen ZF. Early events in olfactory processing. Annu Rev Neurosci. 2006;29:163–201. doi: 10.1146/annurev.neuro.29.051605.112950. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K. OCAM: A new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci. 1997;17:5830–5842. doi: 10.1523/JNEUROSCI.17-15-05830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zhang X, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci USA. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]