Abstract
In eukaryotic cells the nucleus is separated from the cytoplasm by a double-membraned nuclear envelope (NE). Exchange of molecules between the two compartments is mediated by nuclear pore complexes (NPCs) that are embedded in the NE membranes. The translocation of molecules such as proteins and RNAs through the nuclear membrane is executed by transport shuttling factors (karyopherines). They thereby dock to particular binding sites located all over the NPC, the so-called phenylalanine-glycin nucleoporines (FG Nups). Molecular recognition force spectroscopy (MRFS) allows investigations of the binding at the single-molecule level. Therefore the AFM tip carries a ligand for example, a particular karyopherin whereas the nuclear membrane with its receptors is mounted on a surface. Hence, one of the first requirements to study the nucleocytoplasmatic transport mechanism using MRFS is the development of an optimized membrane preparation that preserves structure and function of the NPCs. In this study we present a stable non-destructive preparation method of Xenopus laevis nuclear envelopes. We use micro-structured polydimethylsiloxane (PDMS) that provides an ideal platform for immobilization and biological integrity due to its elastic, chemical and mechanical properties. It is a solid basis for studying molecular recognition, transport interactions, and translocation processes through the NPC. As a first recognition system we investigate the interaction between an important transport shuttling factor, importin β, and its binding sites on the NPC, the FG-domains.
Keywords: membranes, molecular recognition, molecular recognition force spectroscopy, nuclear pore complexes, scanning probe microscopy
Introduction
Biomembranes attract a great deal of attention in life science since they contain proteins and protein assemblies regulating cellular physiology. One of the most important membranes is the nuclear envelope (NE), a double lipid-bilayer encompassing the nucleus. Nucleocytoplasmatic exchange of molecules is mediated by nuclear pore complexes (NPCs), which are large proteinous assemblies embedded in this NE[1-3]. They consist of about 30 different proteins, the so-called nucleoporines (Nups). These Nups are arranged in several copies and form three essential substructures: i) the cytoplasmatic filaments, ii) the central core, and iii) the nuclear basket. Direct recognition processes of molecules or indirect recognition through coreceptors are responsible for the high selectivity of the particular transport through the NPC. The selective transport mechanism can be pictured by the recognition of the cargo-molecule by karyopherines followed by docking to the NPC. After translocation to the nuclear side the cargo is released and the karyopherin is recycled to bind a new molecule.
Notably, 30% of all NPC-forming proteins are rich in phenylalanine-glycin (FG) repeats. These so-called FG-Nups play a key role in the selective gating since they represent the main binding partners of the karyopherines and are responsible for the highly selective barricade of non karyophilic-macromolecules. Although karyopherin-mediated translocation is already well described, the mechanism of the permeability barrier is still not clear. The determination of molecular properties of the various FG Nups and their interaction to karyopherines will help to understand the permeability barrier mechanism of the NPC.
Atomic force microscopy (AFM)[4] based molecular recognition force spectroscopy (MRFS)[5] has the potential to detect and quantify inter- and intramolecular interaction forces of biomolecules on the single-molecule level under near physiological conditions, thus providing new insights into the dynamics of recognition processes and interaction energy landscapes.[6] In MRFS experiments an AFM tip is equipped with a single ligand molecule using a specially designed coupling procedure.[7,8] This functionalized tip is brought into contact with the sample surface onto which the corresponding receptors are bound for the investigation of ligand-receptor interactions.
MRFS studies of the nucleocytoplasmatic transport require stable immobilization of the nuclear envelope on a solid support, allowing contact of the AFM-tip with the membrane without detaching it from the support. Nuclear membrane preparations on flat surfaces, for example, on glass[9] provide such a stable anchorage of membranes. However, they likely cause deformation or even damage of the proximate face of the membrane. For investigation of transport processes both sides of the nuclear membrane have to be protected against mechanical damage to ensure complete structural and functional integrity since the transport docking sites for molecular translocation are located throughout the NPC.[2,10,11] Hence, an optimal compromise between stable immobilization and preservation of the native NPC is essential to study the nucleocytoplasmatic transport mechanism. In order to overcome NPC degradation, NE preparations on micro-structured surfaces provide near physiological conditions, as they allow the extension of the NPC substructures, for example, the nuclear basket into the cavities.
Regularly structured nano-porous surfaces such as silicon or modified aluminum have already been used to study membranes and membrane-proteins.[12,13] In case of NE membranes lattice-structured surfaces using modified isoporous filters have been used previously to investigate the translocation-process through the NPC.[14] In this study, the NEs were investigated by optical single transporter recording (OSTR),[15] so as to measure passive transport through the NPCs’ diffusion channels. In another study, Stoffler et al[16] used modified EM grids to avoid sample deformation for the investigation of calcium-mediated structural changes of the NPC. In contrast to the previously mentioned methods we have developed a simple, fast, cheap and reproducible preparation procedure for the generation of microwellstructured surfaces. MRFS experiments can be performed on membrane areas with no mechanical and chemical damage to the NPC, if the measurements are done on areas where the membrane is spanned over a micrometer-sized hole. We found that micro-structured polydimethylsiloxane (PDMS) offers an optimal substrate for this purpose. PDMS, also known as silicone rubber, represents the micromolding materials and it has suitable properties making it attractive for a wide range of applications. The sealing and optical properties of PDMS make it usable for microfluidics,[17,18] and its bio-compatibility is widely used (i.e. for the attachment of cells[19,20]).
In this work we show that chemically modified micro-structured PDMS is an ideal material for nuclear envelope support. The introduced preparation technique offers an optimal system for AFM and MRFS studies of the NPC machinery and it allows further MRFS investigations of molecular interactions accompanying translocation of cargo-importin β complexes through the NPC.
Results and Discussion
Sample Preparation and Imaging
In order to obtain micro-structured surfaces for mounting nuclear membranes, the Si-based polymer polydimethylsiloxane (PDMS) is chosen. Unlike traditional micro-fabrication materials, such as silicon, glass or microtiter plates, PDMS i) allows to generate (sub)microscopic structures, ii) is a low-cost material, iii) can be prepared simply and quickly compared to etching and bonding approaches, and iv) can be easily modified to adhere membranes. PDMS is known to be a rather soft material similar to rubber. By varying the amount of crosslinking agent it is possible to adjust the stiffness. In contrast to the manufacturer’s protocol (3:1, prepolymer : crosslinking agent) we used a ratio of 5:1 and a longer curing time at 70°C, making the PDMS significantly stiffer, due to its higher degree of crosslinking. These parameters obtain micro-structured PDMS with high topographical quality while preserving safe removal of the stamps from the Si-master.
The micro-structural design of PDMS is optimized to meet two important requirements. On one hand the wells are small enough to stably span the NE over it and, on the other hand, they are large enough to enable positioning of the AFM tip via an optical microscope mounted to the AFM. As illustrated in Figure 1 A microchambers, 3 μm in diameter and 0.7 μm in depth were found to be well suited. The PDMS plateaus have the same dimensions as the wells and serve to support the nuclear membrane. Using smaller plateaus and thus bigger wells resulted in membrane folding, collapse and detachment during AFM experiments. Topographical imaging of the bare PDMS surface was used to determine the geometrical parameters and the surface roughness. As illustrated in Figure 2 A the topography revealed a regular arrangement of the microstructures with flat plateaus, which is of key importance for tight attachment of nuclear membranes.
Figure 1.
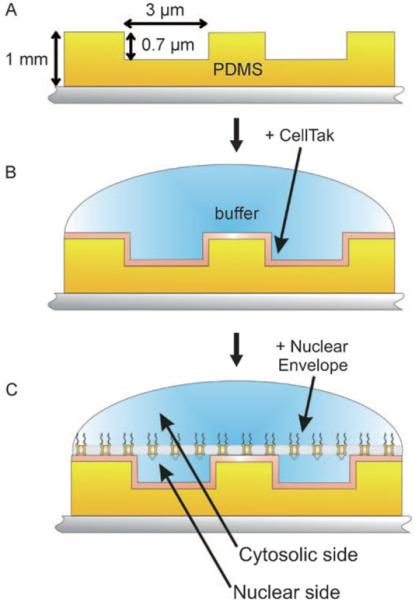
Nuclear envelope preparation procedure. A) Micro-structured PDMS (yellow) is mounted on mica (grey). The thickness of the stamp is about ~1 mm allowing a safe removal from the Si-master. The plateaus as well as the wells are 3 μm in width and 0.7 μm in depth. B) For stable attachment of the NE the micro-structured PDMS was coated with a layer of CellTAK (shown in orange). C) By careful opening of the nucleus and removing the chromatin the NE is spanned over the CellTAK-modified micro-structured PDMS surface, thereby forming microchambers into which the nuclear baskets of the NPCs can extend.
Figure 2.
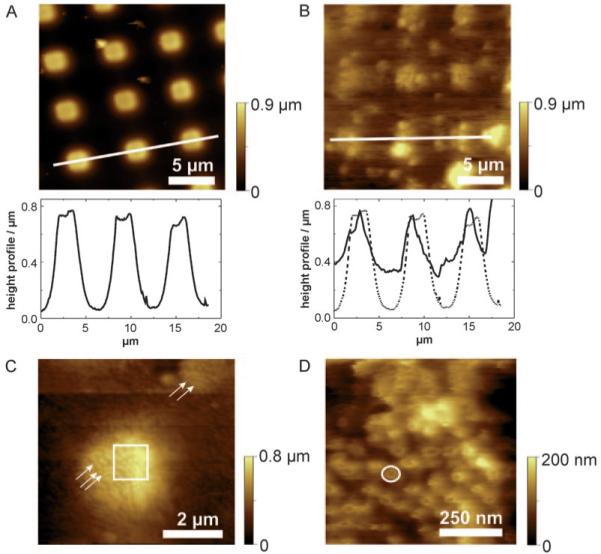
NE attached on CellTAK modified PDMS microstructures. A) Overview scan of bare PDMS in contact mode in dry state. The image reveals flat and regular arrangements of the plateaus. Below the cross-section along the white line. B) Overview scan image of the membrane preparation performed in contact mode. The less height differences and the cloudy appearance indicate a patch of NE. Below the cross-section along the white line (solid line). The dashed line denotes the profile of bare PDMS. The comparison of these two section analysis indicates the stable immobilization of the NE membrane on micro-structured PDMS. C) The small spots appearing in the image can be identified as NPCs (highlighted with arrows). The image demonstrates that the NE is stably spanned over the wells. The square indicates the scan area of Figure 2 D. D) A dense layer of NPCs embedded in the membrane is observed. A single NPC is framed with a white circle.
After generation of micro-structured PDMS (Figure 1 A) an additional step for the membrane attachment is required, since the NE does not adhere stably on bare PDMS. For tight attachment of the NE we used CellTAK, a non-specific cell adhesive protein derived from shells, as a “bio-glue” (Figure 1 B). In comparison to the protocol in Kramer et al. 2007[21] CellTAK was 100 times more diluted, thereby only coating the surface with a thin layer. In contrast to the CellTAK preparation on glass mentioned before, this variation just slightly increases the roughness of the surface and it additionally prevents the wells from being obstructed with CellTAK protein. Finally the nuclei from freshly harvested Xenopus leavis oocytes were placed onto the chemically modified micro-structured PDMS surface and their membranes were subsequently spanned over the surface as flat as possible (Figure 1 C).
AFM contact mode imaging in buffer resolved the NPC structures and revealed that the nuclear membrane is stably spanned across the PDMS cavities. In Figure 2 B an overview scan of a membrane suspended over the micro-structured PDMS is shown. In comparison to bare PDMS (Figure 2 A) this image shows less height differences and has a more “cloudy” appearance. The corresponding height profiles (cross-sections of Figures 2 A and B) emphasize that the NE membrane just slightly sagged into the wells and withstood the force applied by the scanning AFM tip. By reducing the scanrange, (Figure 2 C) details on the attached nuclear membrane become more and more visible and single NPCs can already be identified (marked with arrows) on the plateau as well as in the non-supported areas. This image shows that the membrane is stably fixed on the upper plateaus of PDMS and nicely spans over the wells. After further zooming into a 2.8×2.8 μm (Figure 2 D) image, single NPCs can clearly be observed with an outer diameter of ~120 nm.[1] These AFM images prove that this newly developed NE preparation procedure provides a solid basis for studying recognition, transport interactions, and translocation processes through the nuclear pore.
Molecular Recognition Force Spectroscopy
For molecular recognition force spectroscopy (MRFS) studies on the immobilized NE membrane the interaction between importin β and FG domains of the NPC is investigated. The FG domains are natively unfolded motifs present in 30% of all nucleoporines. They are located all over the NPC and represent the main binding partners for karyopherines. In order to study the importin β–FG motif binding on single molecule basis the AFM tip is modified following a typical four step procedure: Firstly, amine groups are introduced to the tip surface. Then an adaptor molecule is used to couple the distensable PDP –PEG –NTA crosslinker to the tip and finally the His6-tagged importin β is bound to the outer end of the tether by forming an NTA–Ni–His6 complex.[22] The corresponding receptor was part of the immobilized native nuclear membrane (Figure 3 A). The force of the importin β–FG domain bond was measured in so-called force–distance cycles in which the functionalized AFM tip is continously approached and withdrawn from the membrane while recording the cantilevers force as a function of the tip-surface distance. A typical force–distance cycle showing a specific unbinding event of the importin β–FG domain interaction is presented in Figure 3 B. The inset in Figure 3 B shows a force–distance cycle where no interaction between importin β and the FG domains of the NPC takes place or if the complex formation is hindered.
Figure 3.
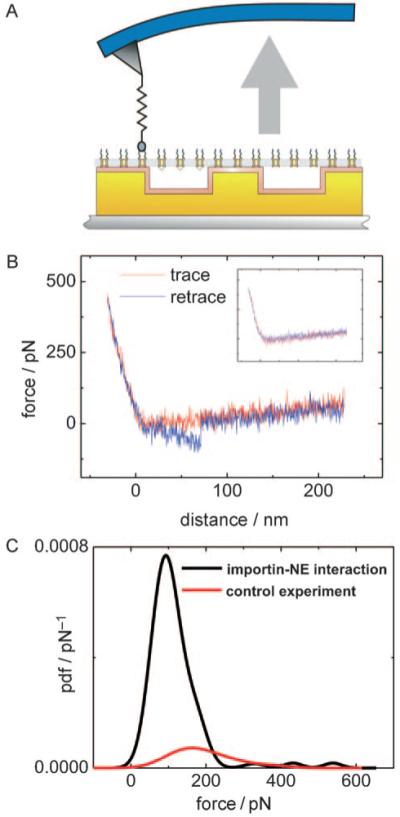
Molecular recognition force spectroscopy (MRFS). A) Scheme of tip-bound importin β and surface attached NE. By approaching and retracting the functionalized tip, importin β–FG complexes are repeatedly formed and dissociated while measuring the interaction force. B) Typical force–distance cycle containing a single molecular recognition event in the retraction period (blue line). Inset: force–distance cycle without importin β–FG domain interaction. C) Probability density function (pdf) of the unbinding force between tip-bound importin β and the NE binding sites (black line). The red line shows the control experiment in which importin β was removed from the tip by imidazol.
For statistical evaluation of the MRFS measurements between tip-bound importin β and the FG motifs of the NPC, 1000 force–distance cycles per data set were recorded, resulting in an average binding probability of 12.3±6.2% (mean ± standard deviation) and a most probable unbinding force of 110.7±29.3 pN. Figure 3 C represents a typical unbinding force distribution [probability density function (pdf), normalized to an area of 1.0] with specific unbinding events in 8.9% of the force–distance cycles and a maximum at 92.4 pN. This value was clearly different from the number of “apparent unbinding events” after cleaving off importin β from the tip by replacing the nickel with imidazol (1.5%), thus showing the specificity of the interaction. The unbinding force distributions (pdf) measured with blocked tips were re-normalized such that their smaller area reflects the much lower unbinding probability of these blocked tips. Overall, the results of the MRFS experiments suggest specific interactions between importin β and the FG repeats of the proteins forming the cytoplasmic filaments.
Conclusions
The investigation of native nuclear membranes with single-molecule techniques like MRFS requires stable and non-destructive attachment of the nuclear envelope. Micro-structured PDMS turned out to be perfectly suited as a support for AFM investigations of the transport mechanism. The process of PDMS fabrication as well as the modification with CellTAK is fast, simple and efficient. It was shown that the NE can be stably spanned over the wells allowing the extension of the nuclear substructures into the cavities. Hence, this preparation method preserves the structure of the entire NPC on the cytosolic (filaments) as well as on the nucleoplasmic side (nuclear baskets), resulting in integrity of its biological function. Furthermore, first MRFS experiments have shown that tip-bound importin β interacts specifically with the NPCs FG domains. As a future perspective it appears to be attractive to investigate cargo-translocation processes through the NPC and to employ force mapping[23] and TREC imaging[24-26] for the localization of adhesion and affinity sites.
Experimental Section
Preparing Micro-Structured PDMS Surfaces: The PDMS prepolymer and the crosslinking agent [Ge Bayer Silicones (Leverkusen, Germany)] were mixed in the ration 5:1. To ensure the complete mixing between the two components the solution was vortexed for about 5 min. In order to receive thin PDMS, the bottom of a 35 mm petri dish was cut off and positioned on the favoured microstructure on a Si wafer. The master was fabricated using conventional photolithography. The PDMS solution was poured into the petri dish, so that it covers the whole base. For curing the PDMS silicon wafers with PDMS were kept in the oven at approximately 70°C for 15 min. After removing the PDMS an additional curing time of 3 h at 70°C was applied.
CellTAK Modification: The modification with CellTAK [BD Biosciences (Bedford, USA)] was adapted form Kramer et al.[21] The micro-structured PDMS was cut and mounted in a commercial AFM-fluid cell. To mark the microstructured surface (which cannot be identified in the wet state with the preparation microscope), the bottom side of the PDMS surface was circled with a marker pen. To adhere the nuclear membrane on the PDMS CellTAK solution (30 μL of 21 μg mL−1 CellTAK protein in 100 mm NaHCO3, pH 8.08) were pipetted on the microstructured surface and incubated for at least 15 min. Then the sample was washed 15 times with NIM (nuclear isolation medium) buffer (90 mm KCl, 10 mm NaCl, 2 mm MgCl2, 10 mm HEPES, pH 7.4) to remove excess CellTAK, and NIM was added to the sample (~200 μL).
Nuclear Envelope Preparation: Freshly harvested Xenopus leavis oocytes (4 days old at the most) were intermediately stored in ND96+ buffer (2 mm KCl, 96 mm NaCl, 1 mm MgCl2, 5 mm HEPES, 1.8 mm CaCl2, pH 7.65). A single oocyte was then transferred into a BSA coated petri dish filled with NIM buffer. This oocyte was opened with two small tweezers to bare the nucleus. With the help of the “preparation cone” (e.g. a modified Eppendorf pipette) the nucleus was transferred to another NIM buffer solution where the excessive cytoplasm was removed from the nuclear membrane. The clean and swollen nucleus was then transferred to the CellTAK modified PDMS surface. There the nucleus was opened with two preparation needles (e.g. minutiae needles from entomology) to remove the chromatin and to subsequently span the membrane over the surface, in a way that the cytoplasmic face of the membrane is facing up and as flat as possible. To relocate the membrane with the optical device of the AFM, the membrane was circled with cuts in the PDMS surface done by a scalpel. Prior to AFM experiments the buffer was exchanged by NIM buffer containing 8% PVP, which flattens the membrane. Alternatively the sample was washed carefully with water, before the fluid droplet was removed with paper tissue. The sample was then stored in ambient environment for 2 h and incubated with MI buffer (NIM buffer including 0.75 mm CaCl2, and 1.1 mm EGTA, pH 7.4).
AFM Experiments
All measurements were done using an AFM 5500 (Agilent Technologies, USA) in which an optical microscope is included. The optical device was required for the localization of the membrane on the PDMS surface and for AFM-tip positioning. Imaging was performed with a bare cantilever with a nominal spring constant of 20 pNnm−1 [Si3N4 AFM measuring cantilevers were purchased from Veeco (Santa Barbara, USA) and Olympus (Japan)].
For MRFS experiments, the AFM-tip has to be functionalized with His6-tagged importin β. This was done in a four step procedure: At first, the cantilevers were functionalized with active amino-groups via ethanolamine or APTES (3-aminoprobyl triethoxysilane) silanization like previously described in.[27] Secondly, thiol-groups were introduced on the tip via SATP (N-succinimidyl-3-(S-acetylthio) propionate). Three mg SATP was dissolved in 1 mL chloroform and the aminofunctionalized cantilevers were placed in the solution. To start the reaction triethylamine (5μl) was added. The cantilevers were incubated in the solution for two hours at room temperature. After incubation the tips were washed three times with chloroform and dried under a stream of nitrogen. Afterwards the cantilevers were incubated for one hour in 100 mm hydroxylamine solution and washed three times in buffer A (100 mm NaCl, 50 mm NaH2PO4, 1 mm EDTA.Na2, pH 7.5). In the third step, the heterobifunctional PDP-PEG-NTA-linker[28] was coupled via its PDP-group to the AFM-tip via a disulfide bond. Therefore NTA-PEG-PDP, was dissolved in buffer A (1 mg mL−1). The cantilevers were added and incubated for one hour. The tips were then washed in 50 mm Tris, 5mm NiCl2, pH 7.5 and incubated in the solution for 5 min. The presence of nickel makes the NTA group reactive and allows the coupling of the protein to the distal end of the crosslinker by forming a His6-Ni-NTA complex. His-tagged human importin β was expressed and purified as described in.[29] For the protein binding the AFM tips were dried with a piece of paper tissue and placed on a parafilm. A droplet of 30 μL of importin β (0.75 mg mL−1) was pipetted on the cantilevers, so that the tips were completely immersed. After incubating for about three hours the cantilevers were washed and stored in 50 mm Tris, 150 mm NaCl, pH 7.5 up to three days for further use.
For MRFS experiments the modified cantilever with His6 tagged importin β was positioned above the same location which was imaged before. Force–distance cycles were recorded at 1 Hz vertical sweeping frequency and 300 nm z-range. During one data set of 1000 force distance curves, the tip position was changed for a few 100 nm for several times to ensure that the binding events were statistically reasonable. After performing force distance cycles a specificity proof of the binding events was performed. For this the cantilever chip was removed and immersed in an imidizol solution (200 mm in PBS, pH 7.5) for about 5 min, so that the nuclear membrane did not get damaged. Then the tips were again placed above the right position of the membrane and FDC were performed. Data evaluation and analysis was performed according to ref. [30].
Acknowledgements
Many thanks to Viktoriia Kolotovska and Thomas Frühwirth for providing the micro-structured Si-masters. This work was supported by the Austrian Science Fond (FWF W1201 N13), the Human Science Frontier Program of the European Union (HFSP project RGP0053), and the EU project Single Molecule Workstation (NMP4-SE-2008-213717).
Footnotes
PDMS: polydimethylsiloxane
References
- [1].Weis K. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- [2].Suntharalingam M, Wente SR. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- [3].Tran EJ, Bolger TA, Wente SR. Cell. 2007;131:420. doi: 10.1016/j.cell.2007.10.015. [DOI] [PubMed] [Google Scholar]
- [4].Binnig G, Quate CF, Gerber C. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- [5].Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Proc. Nat. Acad. Sci. (U SA) 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hinterdorfer P, Reich Z. In: Molecular Recognition Force Microscopy: From Simple Bonds to Complex Energy Landscapes. Bushan B, editor. Springer-Verlag; Berlin–Heidelberg: 2007. pp. 767–787. Chapter 7. [Google Scholar]
- [7].Hinterdorfer P, Dufrene YF. Nat. Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- [8].Ebner A, Wildling L, Zhu R, Rankl C, Haselgrübler T, Hinterdorfer P, Gruber HG. In: STM and AFM Studies on (Bio) molecular Systems: Unravelling the Nanoworld. Samori P, editor. Vol. 285. Springer-Verlag; Berlin–Heidelberg: 2008. pp. 29–76. (Top Curr. Chem). [DOI] [PubMed] [Google Scholar]
- [9].Kramer A, Ludwig Y, Shahin V, Oberleithner H. J Biol Chem. 2007;282:31437–31443. doi: 10.1074/jbc.M703720200. [DOI] [PubMed] [Google Scholar]
- [10].Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Patel SS, Belmont BJ, Sante JM, Rexach MF. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- [12].Gonçalves R, Agnus G, Sens P, Houssin C, Bartenlian B, Scheuring S. Nat. Methods. 2006;3:1007. doi: 10.1038/nmeth965. [DOI] [PubMed] [Google Scholar]
- [13].Hennesthal C, Drexler J, Steinem C. ChemPhysChem. 2002;3 doi: 10.1002/1439-7641(20021018)3:10<885::AID-CPHC885>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- [14].Keminer O, Peters R. Biophys J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tschodrich-Rotter M, Peters R. J. Microsc. 1998;192:114–125. doi: 10.1046/j.1365-2818.1998.00411.x. [DOI] [PubMed] [Google Scholar]
- [16].Stoffler D, Goldie KN, Feja B, Aebi U. J Mol Biol. 1999;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- [17].Whitesides G, Stroock A. Phys. Today. 2001;54:42. [Google Scholar]
- [18].Renaud P, van Lintel H, Heuschkel M, Gu rin L. In: Photo-polymer Microchannel Technologies and Applications. Harrison DJ, van den Berg A, editors. Kluwer Academic Publishers; Twente: 1998. pp. 17–22. [Google Scholar]
- [19].Leclerc E, Sakai Y, Fujii T. Biomed. Microdevices. 2003;5:109–114. [Google Scholar]
- [20].Bhatia S, Balis U, Yarmush M, Toner M. Biotechnol. Prog. 1998;14 doi: 10.1021/bp980036j. [DOI] [PubMed] [Google Scholar]
- [21].Kramer A, Liashkovich I, Ludwig Y, Shahin V. Pflugers Arch. 2008;456:155–162. doi: 10.1007/s00424-007-0396-y. [DOI] [PubMed] [Google Scholar]
- [22].Kienberger F, Kada G, Gruber HJ, Pastushenko V, Riener C, Trieb M, Knaus H-G, Schindler H, Hinterdorfer P. Single Mol. 2000;1:59–65. [Google Scholar]
- [23].Shahin V, Ludwig Y, Schafer C, Nikova D, Oberleithner H. J. Cell Sci. 2005;118:2881–2889. doi: 10.1242/jcs.02429. [DOI] [PubMed] [Google Scholar]
- [24].Stroh CM, Ebner A, Geretschlager M, Freudenthaler G, Kienberger F, Kamruzzahan ASM, Smith-Gil SJ, Gruber HJ, Hinterdorfer P. Biophys. J. 2004;87:1981–1990. doi: 10.1529/biophysj.104.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ebner A, Kienberger F, Kada G, Stroh CM, Geretschlager M, Kam-ruzzahan AS, Wildling L, Johnson WT, Ashcroft B, Nelson J, Lindsay SM, Gruber HJ, Hinterdorfer P. ChemPhysChem. 2005;6:897–900. doi: 10.1002/cphc.200400545. [DOI] [PubMed] [Google Scholar]
- [26].Ebner A, Nikova D, Lange T, Häberle J, Falk S, Dübbers A, Bruns R, Hinterdorfer P, Oberleithner H, Schillers H. Nanotechnology. 2008;19:384017. doi: 10.1088/0957-4484/19/38/384017. [DOI] [PubMed] [Google Scholar]
- [27].Ebner A, Hinterdorfer P, Gruber HJ. Ultramicroscopy. 2007;107:922–927. doi: 10.1016/j.ultramic.2007.02.035. [DOI] [PubMed] [Google Scholar]
- [28].Riener CK, Kienberger F, Hahn CD, Buchinger GM, Egwim IOC, Haselgrubler T, Ebner A, Romanin C, Klampfl C, Lackner B, Gruber HJ. Anal. Chim. Acta. 2003;497:101–114. [Google Scholar]
- [29].Nevo R, Stroh C, Kienberger F, Kaftan D, Brumfeld V, Elbaum M, Reich Z, Hinterdorfer P. Nat. Struct. Mol. Biol. 2003;10:553–557. doi: 10.1038/nsb940. [DOI] [PubMed] [Google Scholar]
- [30].Baumgartner W, Hinterdorfer P, Schindler H. Ultramicroscopy. 2000;82:85–95. doi: 10.1016/s0304-3991(99)00154-0. [DOI] [PubMed] [Google Scholar]


