Abstract
BACKGROUND
Genome-wide association studies (GWAS) have led to the discovery of multiple SNPs that are associated with prostate cancer (PCa) risk. These SNPs may potentially be used for risk prediction. To date, there is not a stable estimate of their effect on PCa risk and their contribution to the genetic variation both of which are important for future risk prediction.
METHODS
A literature review was conducted to identify SNPs associated with PCa risk with the following criteria: (1) GWAS in the Caucasian population; (2) SNPs with p-value < 1.0×10−6; and (3) one SNP from each independent LD block. A meta-analysis was performed to estimate combined odds ratio (OR) and its 95% confidence interval (CI) for the identified SNPs. The proportion of total genetic variance that is attributable by each of these SNPs was also estimated.
RESULTS
Thirty PCa risk-associated SNPs were identified. These SNPs had OR estimates between 1.12 – 1.47 except for marker rs16901979 (OR = 1.80). Significant heterogeneity in OR estimates was found among different studies for 13 SNPs. The proportion of total genetic variance attributed by each SNP ranged between 0.2% – 0.9%. These 30 SNPs explained ~13 .5% of the total genetic variance of PCa risk in the Caucasian population.
CONCLUSION
This study provides more stable OR estimates for PCa risk-associated SNPs, which is an important baseline for the effect of these SNPs in risk prediction. These SNPs explain a considerable proportion of genetic variance, however, the majority of genetic variance has yet to be explained.
Keywords: PCa, GWAS, meta-analysis, heterogeneity, genetic variation
INTRODUCTION
Prostate cancer (PCa) is the most common solid organ malignancy and the second leading cause of cancer mortality in males in the United States [1]. Using new, high-throughput technologies, GWAS and follow-up fine mapping studies have successfully identified over two dozens of genetic variants or single nucleotide polymorphisms (SNPs) that are associated with PCa risk. The genetic variants detected in this manner have been found to have a moderate effect on PCa risk with ORs between 1.0 –1.5[2]. However, the combinations of these genetic variants can provide a greater cumulative effect on PCa risk and may play an important role in the risk prediction of PCa in addition to established risk factors such as age, ethnicity, and family history.
PCa risk prediction using genetic information relies heavily on the effect size (e.g., OR of each genetic variant), however, some genetic variants have had inconsistent associations among different study populations possibly due to the small magnitude of effect of the SNPs and/or issues with respect to statistical power in these studies. The impact of these inconsistencies can be magnified by the approach employed to examine this cumulative impact, namely, cumulative effect analysis, absolute risk analysis, and proportion of total genetic variance [3–6]. Herein, a meta-analysis was conducted, examining the degree of impact on PCa risk of SNPs identified in GWAS and fine-mapping, taking advantage of the high accuracy of effect size and increased statistical power, which come from these high quality studies performed with large populations [7–9]. Due to increased power and validation, meta-analyses provide more robust risk estimates to distinguish men with high risk of PCa from those with low risk.
Although advances have been made in the discovery of genetic markers, it is important to determine what impact the markers identified to date have on the overall genetic contribution to PCa risk. We estimate the proportion of overall genetic risk that the genetic variants selected in this meta-analysis contribute by using the method of Pharoah et al. [5]
MATERIALS AND METHODS
Selection of Genetic Variants
In order to identify PCa risk SNPs from published GWAS studies, in September of 2009, the national GWAS database maintained by the National Human Genome Research Institute of the NIH, (http://www.genome.gov/gwastudies), was searched for the term "prostate cancer". References in the retrieved articles were also reviewed.
According to the search results of the GWAS database, over two dozens of genetic variants associated with PCa risk have been reported between 2007 and September 2009. The selection of eligible genetic variants was restricted to those meeting the following criteria: (1) GWAS with follow-up fine mapping studies using a case and control study design; (2) studies restricted to individuals of European ancestry; (3) SNPs associated with PCa risk with a p-value cut-off of 1.0×10−6; and (4) a randomly selected single SNP in each independent LD block. We selected the most recently published and/or the largest sample size if the same or overlapping data in multiple research studies were used for one SNP. Using these criteria, we identified 30 SNPs, all of which were from independent linkage LD blocks. We present the main characteristics of the SNPs including locations, related genes, references, p-values of association test, the number of populations studied for each SNP, and the total number of subjects in cases and control in Table 1. The average number of independent populations studied for each SNP was approximately 6. For the eight SNPs in Eeles [10], we included the combined results of 18 European populations from the PRACTICAL Consortium.
Table 1.
Selected SNPs associated with prostate cancer from GWAS
| CHR | SNPs | BP-build36 | Location | Known genes |
m/M allele |
Risk allele |
References | P-value | # of pops |
Total # of cases |
Total # of controls |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs1465618 | 43,407,453 | 2p21 | THADA | A/G | A | Eeles [10; Gudmundsson [11] | 2×10−8 | 3 | 21733 | 20655 |
| 2 | rs721048 | 62,985,235 | 2p15 | EHBP1 | A/G | A | Gudmundsson [11,12] | 8×10−9 | 8 | 10121 | 42735 |
| 2 | rs12621278 | 173,019,799 | 2q31.1 | ITGA6 | G/A | A | Eeles [12] | 9×10−23 | 3 | 21733 | 20655 |
| 3 | rs2660753 | 87,193,364 | 3p12 | T/C | T | Eeles [13]; Gudmundsson [12] | 3×10−8 | 4 | 6847 | 40622 | |
| 3 | rs10934853 | 129,521,063 | 3q21.3 | A/C | A | Gudmundsson [13] | 3×10−8 | 12 | 13774 | 47614 | |
| 4 | rs17021918 | 95,781,900 | 4q22.3 | PDLIM5 | T/C | C | Eeles [10] | 4×10−15 | 3 | 21733 | 20655 |
| 4 | rs7679673 | 106,280,983 | 4q24 | TET2 | A/C | C | Eeles [10] | 3×10−14 | 3 | 21733 | 20655 |
| 6 | rs9364554 | 160,753,654 | 6q25 | T/C | T | Eeles [13]; Gudmundsson [12] | 6×10−10 | 4 | 6847 | 40659 | |
| 7 | rs6465657 | 97,654,263 | 7q21 | LMTK2 | T/C | C | Eeles [13]; Gudmundsson [12] | 1×10−9 | 4 | 6846 | 40618 |
| 8 | rs2928679 | 23,494,920 | 8p21.2 | NKX3.1 | A/G | A | Eeles [10] | 7×10−8 | 3 | 21733 | 20655 |
| 8 | rs1512268 | 23,582,408 | 8p21.2 | NKX3.1 | T/C | T | Eeles [10] | 3×10−30 | 3 | 21733 | 20655 |
| 8 | rs10086908 | 128,081,119 | 8q24(5) | C/T | T | Olama [14] | 8×10−8 | 8 | 12182 | 11470 | |
| 8 | rs16901979 | 128,194,098 | 8q24(2) | A/C | A | Gudmundsson [12, 15] | 3×10−14 | 4 | 2936 | 37848 | |
| 8 | rs16902094 | 128,389,528 | 8q24.21 | G/A | G | Gudmundsson [12] | 6×10−15 | 11 | 12102 | 16913 | |
| 8 | rs620861 | 128,404,855 | 8q24 (4) | A/G | G | Olama [14]; Yeager [16] | 5×10−8 | 12 | 15760 | 14942 | |
| 8 | rs6983267 | 128,482,487 | 8q24(3) | G/T | G | Yeager [17]; Thomas [18]; Gudmundsson [12] | 9×10−13 | 6 | 6020 | 39666 | |
| 8 | rs1447295 | 128,554,220 | 8q24(l) | A/C | A | Amundadottir[19]; Gudmundsson [12, 15]; Yeager [17] | 2×10−19 | 10 | 8756 | 42989 | |
| 10 | rs10993994 | 51,219,502 | 10q11 | MSMB | T/C | T | Eeles [13]; Thomas [18]; Gudmundsson [12] | 7×10−13 | 9 | 11959 | 45715 |
| 10 | rs4962416 | 126,686,862 | 10q26 | CTBP2 | C/T | C | Thomas [18]; Gudmundsson [12] | 2×10−7 | 6 | 6821 | 40370 |
| 11 | rs7127900 | 2,190,150 | 11p15.5 | IGF2, IGF2AS, INS,TH | G/A | A | Eeles [10] | 3×10−33 | 3 | 21733 | 20655 |
| 11 | rs10896449 | 68,751,243 | 11q13 | A/G | G | Thomas [18]; Gudmundsson [12] | 2×10−9 | 6 | 7053 | 40446 | |
| 17 | rs11649743 | 33,149,092 | 17ql2(2) | A/G | G | Sun [20] | 2×10−9 | 8 | 11193 | 42619 | |
| 17 | rs4430796 | 33,172,153 | 17ql2(l) | TCF2 | A/G | A | Gudmundsson [12, 21]; Thomas [18] | 1×10−11 | 11 | 13049 | 45517 |
| 17 | rs1859962 | 66,620,348 | 17q24.3 | G/T | G | Gudmundsson [12, 21]; Thomas | 2×10−16 | 4 | 3738 | 38178 | |
| 19 | rs8102476 | 43,427,453 | 19q13.2 | T/C | C | Gudmundsson [12] | 2×10−11 | 11 | 13173 | 47198 | |
| 19 | rs887391 | 46,677,464 | 19q13(1) | 10MbtoKLK3 | C/T | T | Hsu [22] | 3×10−7 | 7 | 9516 | 7252 |
| 19 | rs2735839 | 56,056,435 | 19ql3(2) | KLK3 | A/G | G | Eeles [13]; Gudmundsson [12] | 2×10−18 | 4 | 6848 | 40636 |
| 22 | rs9623117 | 38,782,065 | 22q13 | C/T | C | Sun [20]; Gudmundsson [12] | 5×10−7 | 8 | 10689 | 42589 | |
| 22 | rs5759167 | 41,830,156 | 22q13.2 | TTLL1, BIK, MCAT, PACSIN2 | T/G | G | Eeles [10] | 6×10−29 | 3 | 21733 | 20655 |
| 23 | rs5945619 | 51,258,412 | Xp11 | NUDT10, NUDT11, LOC340602 | C/T | C | Eeles [13]; Gudmundsson [12] | 2×10−9 | 4 | 7021 | 40644 |
P-value was reported in the GWAS database(www.genome.gov/gwastudies); # of pops denotes the number of populations used in each SNP study.
Statistical Analysis
A meta-analysis was performed to obtain the pooled estimate of OR and its CI for the 30 SNPs identified by GWAS as being associated with PCa risk. The OR was used to evaluate the extent of this association between study populations. If the raw data, e.g., allele counts of case and control, were available, we used this information for calculating the OR and its standard error for each study population. Otherwise, we calculated these estimates using the reported OR and 95% CI. The results from both approaches are statistically comparable [23].
We assessed heterogeneity among study populations with the p-value of the Q-statistic for test of heterogeneity and I2 statistic which measures the proportion of total variance in estimated ORs due to heterogeneity. The I2 statistic provides the degree of heterogeneity while the Q-statistic only provides the presence or absence of heterogeneity. A value of the I2 statistic greater than 50% indicates the high degree of heterogeneity in estimated ORs across study populations [24].
In this meta-analysis, the fixed effects method was used to calculate the OR and 95% CI estimates by weighing each study with the inverse of variance of logarithm of OR while the random effects method additionally incorporated between variance in that weight. Although the random effects method is preferred for meta-analyses because this method can capture the heterogeneity among research studies, the random effects method for GWAS is likely to suffer from the winner’s curse problem in the sense that the random effects method may be too conservative due to additional variability compared to the fixed effect [8]. Nonetheless, if there was a high degree of heterogeneity in OR estimates, then we considered the results of the random effects method. We obtained the pooled estimates of OR and its 95% CI for each SNP using both fixed and random effects methods. In order to evaluate the statistical significance of the pooled estimates of OR, we used the two-sided Z-test with the corresponding p-value of 2(1-Φ(|Z|)) where Φ(·) is the standard normal cumulative distribution. All meta-analyses and forest plots in this paper were performed using R software where forest plots provide the OR and 95% CI of each study population and the pooled estimate of OR and its 95% CI for each SNP.
As a utilization of the result of this meta-analysis, we estimated the proportion of a single SNP of the total genetic variance (V) and the proportion of the combination of selected SNPs. An advantage of using the results of the meta-analysis is that the pooled estimate of OR obtained from the meta-analysis provides more robust results in the calculation of genetic variance than the OR of a single study population. Assuming the selected SNPs were independent of each other, the proportion of variance explained by the combination of selected SNPs was calculated by adding the proportion of each SNP. In order to estimate the proportion of the total genetic variance explained by each SNP, we first estimated the variance of risk allele for each SNP (Vi) using the risk allele frequency in the HapMap CEU population and the pooled estimate of OR from the meta-analysis [Table 2]. If the p-value of the Q-statistic was greater than 0.05, then the results of fixed effect were used. Otherwise, the results of random effect were used to consider the heterogeneity in the estimates. The total genetic variance for all the genetic factors was calculated as 2.5 for PCa risk [25]. The proportion of each SNP to the total genetic variation was calculated as the value of Vi divided by V.
Table 2.
Result of meta-analysis for selected SNPs
| Chr | SNPs | Position | m/M alleles |
Risk allele |
Fixed effect |
Random effect |
Heterogeneity |
Genetic variation |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | P-valofZ | P-valofQ | I2 | Freq | Proportion | |||||
| 2 | rs1465618 | 43,407,453 | A/G | A | 1.11 | 1.07 – 1.16 | 1.15 | 1.04 – 1.26 | 4.92E-03 | 8.66E-03 | 79% | 0.212 | 0.003 |
| 2 | rs721048 | 62,985,235 | A/G | A | 1.16 | 1.11 – 1.22 | 1.18 | 1.10 – 1.28 | 1.22E-05 | 0.084 | 44% | 0.137 | 0.002 |
| 2 | rs12621278 | 173,019,799 | A/G | A | 1.35 | 1.27 – 1.44 | 1.35 | 1.27 – 1.44 | 3.36E-19 | 0.576 | 0% | 0.960 | 0.003 |
| 3 | rs2660753 | 87,193,364 | T/C | T | 1.22 | 1.14 – 1.31 | 1.24 | 1.04 – 1.48 | 0.015 | 3.87E-04 | 84% | 0.102 | 0.003 |
| 3 | rs10934853 | 129,521,063 | A/C | A | 1.12 | 1.08 – 1.16 | 1.12 | 1.06 – 1.18 | 7.43E-05 | 0.016 | 53% | 0.239 | 0.002 |
| 4 | rs17021918 | 95,781,900 | T/C | C | 1.14 | 1.10 – 1.18 | 1.14 | 1.10 – 1.18 | 1.54E-15 | 0.473 | 0% | 0.646 | 0.003 |
| 4 | rs7679673 | 106,280,983 | A/C | C | 1.13 | 1.10 – 1.17 | 1.14 | 1.09 – 1.20 | 1.29E-07 | 0.153 | 47% | 0.624 | 0.003 |
| 6 | rs9364554 | 160,753,654 | T/C | T | 1.16 | 1.10 – 1.21 | 1.17 | 1.06 – 1.29 | 1.91E-03 | 5.96E-03 | 76% | 0.274 | 0.004 |
| 7 | rs6465657 | 97,654,263 | T/C | C | 1.13 | 1.08 – 1.18 | 1.14 | 1.05 – 1.23 | 1.42 E-03 | 0.031 | 66% | 0.509 | 0.003 |
| 8 | rs2928679 | 23,494,920 | A/G | A | 1.09 | 1.06 – 1.12 | 1.13 | 1.02 – 1.25 | 0.017 | 3.89E-04 | 87% | 0.456 | 0.003 |
| 8 | rs1512268 | 23,582,408 | T/C | T | 1.17 | 1.14 – 1.21 | 1.17 | 1.12 – 1.23 | 1.56E-12 | 0.205 | 37% | 0.420 | 0.005 |
| 8 | rs10086908 | 128,081,119 | C/T | T | 1.13 | 1.09 – 1.18 | 1.13 | 1.08 – 1.19 | 6.25E-07 | 0.243 | 23% | 0.625 | 0.003 |
| 8 | rs16901979 | 128,194,098 | A/C | A | 1.80 | 1.57 – 2.06 | 1.82 | 1.44 – 2.30 | 6.32E-07 | 0.163 | 41% | 0.031 | 0.009 |
| 8 | rs16902094 | 128,389,528 | G/A | G | 1.21 | 1.16 – 1.27 | 1.20 | 1.12 – 1.30 | 1.12E-06 | 0.015 | 54% | 0.271 | 0.005 |
| 8 | rs620861 | 128,404,855 | A/G | G | 1.16 | 1.11 – 1.20 | 1.16 | 1.11 – 1.20 | 1.18E-13 | 0.618 | 0% | 0.619 | 0.004 |
| 8 | rs6983267 | 128,482,487 | G/T | G | 1.20 | 1.14 – 1.26 | 1.20 | 1.14 – 1.26 | 8.13E-12 | 0.758 | 0% | 0.487 | 0.006 |
| 8 | rs1447295 | 128,554,220 | A/C | A | 1.49 | 1.40 – 1.57 | 1.47 | 1.33 – 1.62 | 3.82E-14 | 0.013 | 57% | 0.071 | 0.008 |
| 10 | rs10993994 | 51,219,502 | T/C | T | 1.26 | 1.22 – 1.31 | 1.25 | 1.12 – 1.40 | 6.80E-05 | 1.22E-15 | 91% | 0.341 | 0.009 |
| 10 | rs4962416 | 126,686,862 | C/T | C | 1.14 | 1.09 – 1.20 | 1.15 | 1.04 – 1.27 | 7.85 E-03 | 1.79E-04 | 80% | 0.257 | 0.003 |
| 11 | rs7127900 | 2,190,150 | G/A | A | 1.25 | 1.20 – 1.30 | 1.25 | 1.20 – 1.30 | <1.00E-30 | 0.713 | 0% | 0.235 | 0.007 |
| 11 | rs10896449 | 68,751,243 | A/G | G | 1.16 | 1.11 – 1.22 | 1.16 | 1.11 – 1.22 | 8.67E-10 | 0.898 | 0% | 0.532 | 0.004 |
| 17 | rs11649743 | 33,149,092 | A/G | G | 1.16 | 1.11 – 1.22 | 1.16 | 1.11 – 1.22 | 1.19E-09 | 0.922 | 0% | 0.757 | 0.003 |
| 17 | rs4430796 | 33,172,153 | A/G | A | 1.22 | 1.17 – 1.26 | 1.22 | 1.17 – 1.26 | 1.13E-25 | 0.645 | 0% | 0.491 | 0.008 |
| 17 | rs1859962 | 66,620,348 | G/T | G | 1.20 | 1.13 – 1.27 | 1.21 | 1.12 – 1.30 | 7.94E-07 | 0.214 | 33% | 0.527 | 0.007 |
| 19 | rs8102476 | 43,427,453 | T/C | C | 1.12 | 1.08 – 1.15 | 1.12 | 1.08 – 1.15 | 2.27E-12 | 0.729 | 0% | 0.504 | 0.002 |
| 19 | rs887391 | 46,677,464 | C/T | T | 1.14 | 1.08 – 1.20 | 1.14 | 1.08 – 1.20 | 6.86E-07 | 0.826 | 0% | 0.757 | 0.003 |
| 19 | rs2735839 | 56,056,435 | A/G | G | 1.30 | 1.22 – 1.39 | 1.30 | 1.11 – 1.51 | 7.47E-04 | 6.37E-04 | 83% | 0.863 | 0.006 |
| 22 | rs9623117 | 38,782,065 | C/T | C | 1.12 | 1.07 – 1.17 | 1.13 | 1.05 – 1.22 | 1.22E-03 | 0.019 | 58% | 0.221 | 0.002 |
| 22 | rs5759167 | 41,830,156 | T/G | G | 1.18 | 1.14 – 1.21 | 1.18 | 1.14 – 1.21 | 1.44E-27 | 0.682 | 0% | 0.549 | 0.005 |
| 23 | rs5945619 | 51,258,412 | C/T | C | 1.26 | 1.18 – 1.34 | 1.27 | 1.12 – 1.43 | 2.05E-04 | 0.012 | 73% | 0.385 | 0.005 |
m = minor allele, M = major allele; OR and 95% CI were calculated in terms of risk allele; and Freq is the risk allele freq in HapMap.
RESULTS
Meta-analysis
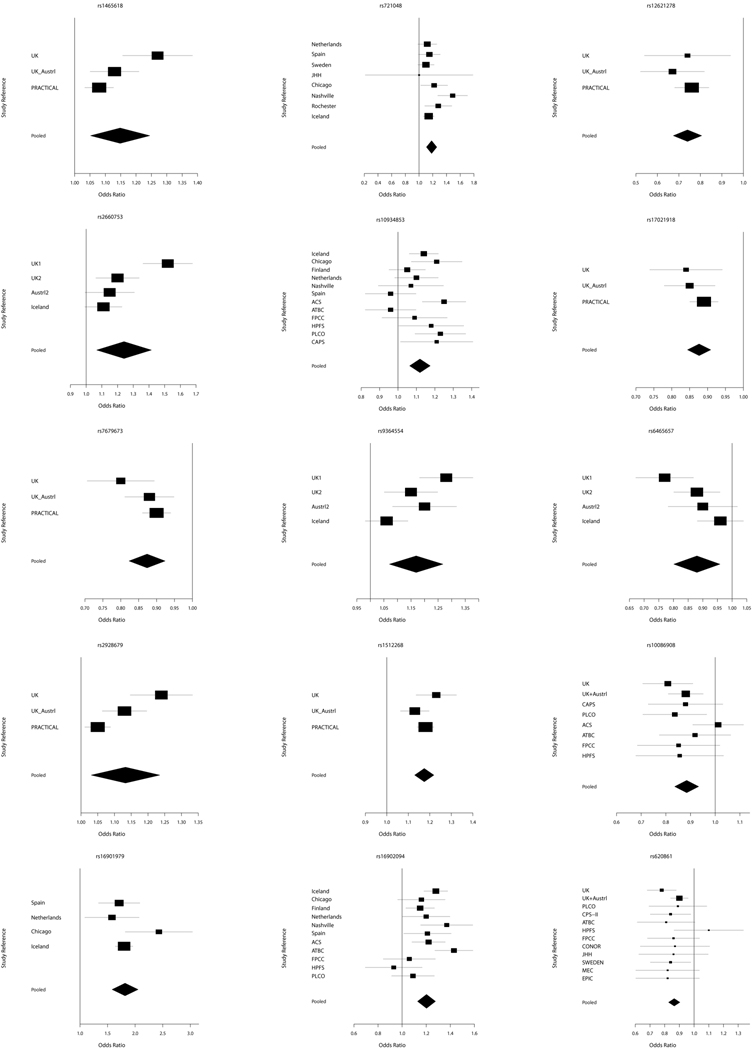
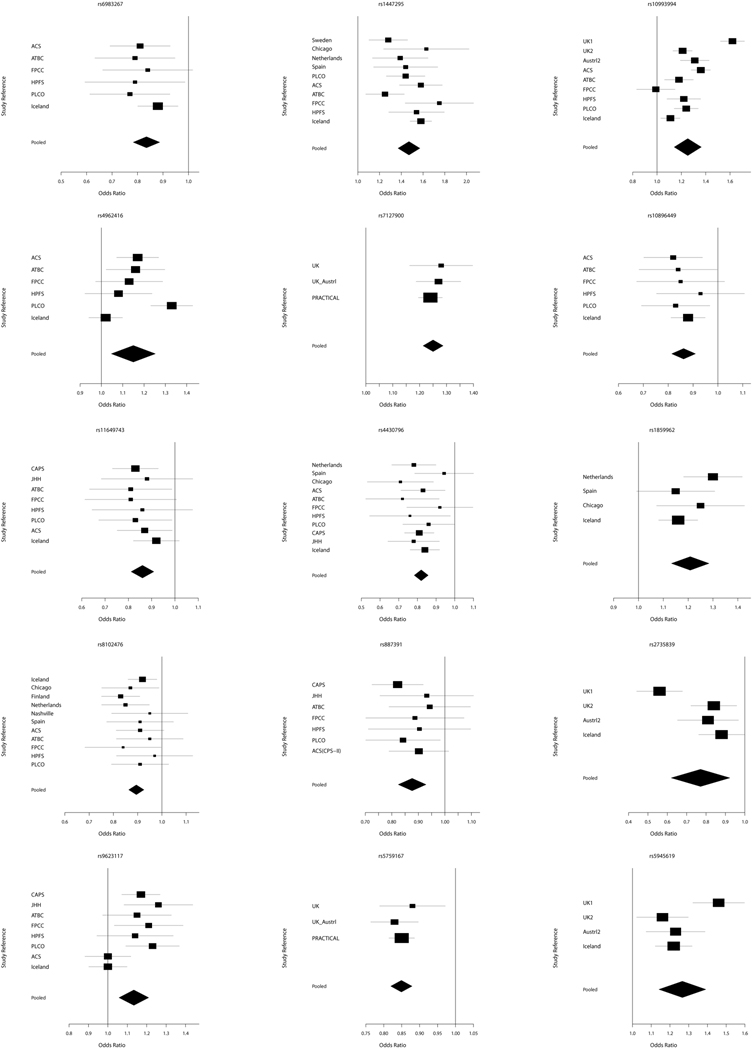
The fixed and random effects meta-analyses for the 30 SNPs identified by searching the National Human Genome Research Institute database as being associated with PCa risk can be seen in Table 2. The pooled estimates of OR and its CI were calculated in terms of risk allele. All meta-analysis results were statistically significant by both the fixed and random effect methods. There was no significant difference in pooled estimates of OR between the two methods. We thus reported the p-values for the random effect in Table 2. The marker rs7127900 on 11p15.5, which was selected with the smallest p-value of 3.0×10−33, achieved the lowest p-value from the random effects meta-analysis. The markers rs2928679 on 8p21 and rs2660753 on 3p12 were marginally significant at the 5% significance level compared to other markers. These markers were identified using populations with relatively small number of study subjects for this meta-analysis and thus have a substantially high degree of heterogeneity which exceeds 80%.
There were 13 SNPs which have the p-value of the Q-statistic less than 0.05 and the I2 statistic greater than 50%. For these SNPs, we considered only the results of the random effects method although the OR estimates of both fixed and random effects were significant. The heterogeneity pattern of these SNPs is illustrated in Figure 1a–b. This result implies that the Q-statistic and I2 statistic are compatible for heterogeneity of study populations in meta-analysis. In addition, as the I2 statistic approaches to zero, the results of the random effect become similar to the results of fixed effect. In case that the I2 statistic is zero, the results of the two methods are the same [Table 2].
Figure 1.
a Forest plots of selected PCa risk associated SNPs. Pooled is the result of meta-analysis.
b Forest plots of selected PCa risk associated SNPs. Pooled is the result of meta-analysis.
There was a moderate effect on PCa risk, with the identified SNPs in this study with ORs between 1.0 and 1.5. However, marker rs16901979 exhibited the highest pooled OR (95% CI) of 1.80 (1.57 – 2.06). Overall, the CI of estimated ORs became narrower than that of individual study populations [Figure 1a–b], which indicated that the meta-analysis achieved a more robust result. The marker rs16901979 has a low minor allele frequency in the HapMap CEU sample of 0.03 [Table 2] and a small number of subjects in the case compared to the control, 2936 vs. 37848 [Table 1], and as a result, the estimated OR and CI is relatively higher and wider than other SNPs, respectively. The second highest estimate of OR (95% CI) is 1.47 (1.33 – 1.1.62) in random effect for the marker rs1447295. These two highest estimates of OR are from loci 8q24, a region known for its association with PCa susceptibility [14 and references therein]. The functional relevance of SNPs within this region is not well understood, however, the MYC oncogene is located >250 kilobases from this region.
Proportion of Genetic Variation
The results of the meta-analysis for the 30 identified SNPs were applied to the estimation of the proportion of total genetic variance. The proportion of each SNP to the total genetic effect on PCa risk is reported in Table 2. The proportion of total genetic variance attributed by each SNP ranges between 0.2% – 0.9%. Markers rs16901979 and rs10993994 have the highest proportion of 0.9%. The proportion of the total variance depends on both OR and risk allele frequency. The combination of the 30 selected SNPs in this meta-analysis explains 13.5% of the total genetic variance in the European ancestry population associated with PCa risk. The first 5 SNPs and 10 SNPs with the highest proportion explain 4.1% and 7.1% of the total genetic variance, respectively.
DISCUSSION AND CONCLUSION
Herein we identify 30 genetic variants in independent loci consistently associated with PCa risk in multiple Caucasian populations by exploiting the results of published studies discovered by searching the National Human Genome Research Institute database. We calculated the pooled estimate of OR for each SNP which was statistically significant by both fixed and random effect methods. These pooled estimates can be utilized in future risk prediction models to predict PCa risk in individuals of Caucasian population. These estimates will be more accurate due to the more robust OR estimates of this meta-analysis compared to those from an individual study.
There are two main approaches to incorporate the results of multiple studies: meta-analysis and mega-analysis. The former uses the result of each study, and the latter uses the raw data of each study for calculating an effect size. Meta-analysis has been proven as valid as mega-analysis in the sense that these two approaches have approximately the same variance [23]. For example, the results of markers, rs10934853, rs16902094, and rs8102476 in Table 2 are almost the same as the results in Gudmundsson [12], which used the mega-analysis approach. Thus, meta-analysis is useful especially when it is not possible to access the raw data set of an individual study.
The meta-analysis in this paper has a few limitations. Some SNPs have been identified using study populations with relatively small numbers of study subjects. The numbers of case and control subjects are asymmetric for some SNPs. These factors may lead to bias and inefficiency in this meta-analysis. Since we only used the reported SNPs in the GWAS database, we might have missed some SNPs that have not been reported or that were not captured in the database. In addition, publication bias, which is an intrinsic problem of meta-analysis, could not be addressed because the reported ORs of the selected SNPs showed similar effects, e.g., either greater than or less than 1 [1a–b].
The contributions of single and multiple SNPs to the total genetic variance of PCa risk were not very high in this analysis. The assessment of genetic variation incorporating additional SNPs discovered from ongoing GWAS and fine-mapping studies may provide beneficial information for future PCa risk prediction studies.
These results allow for many opportunities to capitalize on the stable estimates of OR for the PCa associated SNPs. First, this meta-analysis can be extended to the reported SNPs that have p-values greater than 1.0×10−6 because they could be potentially associated with PCa risk. The best combination of potential SNPs is crucial to risk prediction analyses using multiple genetic variants. Thus, more research is needed to construct the best set of SNPs associated with PCa risk. Second, this meta-analysis can be analyzed using the OR and CI after adjusting with other covariates such as age, family history, sub-ethnicity type, geographical factor, etc. These two approaches combined will allow researchers to conduct a more precise meta-analysis and an improved risk prediction analysis.
Acknowledgments
Grants: National Cancer Institute (CA129684, CA140262, CA148463 to J.X.) and Department of Defense (W81XWH-09-1-0488 to J.S.)
REFERENCES
- 1.Gurel B, Iwata T, Koh CM, Yegnasubramanian S, Nelson WG, De Marzo AM. Molecular alterations in prostate cancer as diagnostic, prognostic, and therapeutic targets. Adv Anat Pathol. 2008;15(6):319–331. doi: 10.1097/PAP.0b013e31818a5c19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 3.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10(2):77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Balter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Gronberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 5.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Sun J, Kader AK, Lindstrom S, Wiklund F, Hsu FC, Johansson JE, Zheng SL, Thomas G, Hayes RB, Kraft P, Hunter DJ, Chanock SJ, Isaacs WB, Gronberg H. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69(14):1565–1572. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ioannidis JP, Gwinn M, Little J, Higgins JP, Bernstein JL, Boffetta P, Bondy M, Bray MS, Brenchley PE, Buffler PA, Casas JP, Chokkalingam A, Danesh J, Smith GD, Dolan S, Duncan R, Gruis NA, Hartge P, Hashibe M, Hunter DJ, Jarvelin MR, Malmer B, Maraganore DM, Newton-Bishop JA, O'Brien TR, Petersen G, Riboli E, Salanti G, Seminara D, Smeeth L, Taioli E, Timpson N, Uitterlinden AG, Vineis P, Wareham N, Winn DM, Zimmern R, Khoury MJ. Human Genome Epidemiology Network and the Network of Investigator Networks. A road map for efficient and reliable human genome epidemiology. Nat Genet. 2006 Jan;38(1):3–5. doi: 10.1038/ng0106-3. [DOI] [PubMed] [Google Scholar]
- 8.de Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17(2):R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86(1):6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O'Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G, Easton DF. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41(10):1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindstrom S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40(3):281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40(3):316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 14.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, Hopper JL, Southey MC, Muir KR, English DR, Dearnaley DP, Ardern-Jones AT, Hall AL, O'Brien LT, Wilkinson RA, Sawyer E, Lophatananon A, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper C, Donovan JL, Hamdy FC, Neal DE, Eeles RA, Easton DF. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 15.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 16.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni JF, Jr, Hunter DJ, Thomas G, Chanock SJ. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41(10):1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 18.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 19.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38(6):652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Zheng SL, Wiklund F, Isaacs SD, Purcell LD, Gao Z, Hsu FC, Kim ST, Liu W, Zhu Y, Stattin P, Adami HO, Wiley KE, Dimitrov L, Li T, Turner AR, Adams TS, Adolfsson J, Johansson JE, Lowey J, Trock BJ, Partin AW, Walsh PC, Trent JM, Duggan D, Carpten J, Chang BL, Gronberg H, Isaacs WB, Xu J. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40(10):1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 22.Hsu FC, Sun J, Wiklund F, Isaacs SD, Wiley KE, Purcell LD, Gao Z, Stattin P, Zhu Y, Kim ST, Zhang Z, Liu W, Chang BL, Walsh PC, Duggan D, Carpten JD, Isaacs WB, Gronberg H, Xu J, Zheng SL. A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 2009;69(7):2720–2723. doi: 10.1158/0008-5472.CAN-08-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genet Epidemiol. 2010;34(1):60–66. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 25.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]