Abstract
Haematopoietic stem cells in umbilical cord blood are an attractive target for gene therapy of inborn errors of metabolism. Three neonates with severe combined immunodeficiency were treated by retroviral-mediated transduction of the CD34+ cells from their umbilical cord blood with a normal human adenosine deaminase complementary DNA followed by autologous transplantation. The continued presence and expression of the introduced gene in leukocytes from bone marrow and peripheral blood for 18 months demonstrates that umbilical cord blood cells may be genetically modified with retroviral vectors and engrafted in neonates for gene therapy.
Inherited deficiency of adenosine deaminase (ADA) is the cause of approximately one-quarter of the cases of severe combined immunodeficiency (SCID)1,2. Based on the successful treatment of ADA-deficient SCID by allogeneic bone marrow transplantation and the cloning of the normal human ADA complementary DNA, ADA-deficiency has been a major target for gene therapy3,4. Theoretically, genetically corrected autologous T-lymphoid precursors should have a selective survival advantage over non-corrected ADA-deficient cells. A broad level of expression by an inserted ADA gene (from one-tenth to above normal) should be beneficial without adverse consequences, although there is no specific information about consequences of vast overexpression of ADA in cells of all haematopoietic lineages. Thus, even with the currently modest capabilities to efficiently transfer genes into primary haematopoietic cells and an inability to control gene expression in a precise manner, ADA-deficient SCID may respond to gene therapy.
The first clinical trial of gene therapy involved two young girls with ADA deficiency, using their peripheral blood T lymphocytes as the target for ADA gene transfer5,6. Multiple courses of leukopheresis, transduction and re-infusion of T lymphocytes led to increased numbers of T lymphocytes and improved T-Iymphocyte function, with attainment in one of the two patients of circulating leukocyte ADA levels equivalent to a quarter of those in normal individuals. Despite these encouraging results with mature T lymphocytes, the genetic correction of haematopoietic stem cells may create a more long-lasting source of ADA-expressing T lymphocytes. Information gained from the treatment of ADA deficiency would be applicable to the use of haematopoietic stem cells for gene therapy for other aetiologic forms of SCID and to other genetic diseases affecting haematopoietic and lymphoid cells.
In the spring of 1993, three women who were known to be heterozygous for ADA deficiency were identified to be carrying ADA-deficient fetuses. After independent genetic counseling, each family elected to carry its respective pregnancy to term. Because of the prenatal diagnosis, the unique opportunity was afforded to attempt gene therapy for the neonates using their umbilical cord blood as a source of haematopoietic stem cells. Extensive information has been accumulated to indicate that umbilical cord blood contains haematopoietic stem cells; more than forty patients have had successful haematopoietic reconstitution by the transplantation of allogeneic cord blood leukocytes following cytoablative conditioning7,8.
Approval to perform gene transfer into the neonates’ umbilical cord blood followed by the transplantation of the cells was obtained from the Institutional Review Boards, the Recombinant DNA Advisory Committee (RAC) of the National Institutes of Health, and the Food and Drug Administration. The parents of each child gave informed consent for the collection, transduction and re-infusion of the umbilical cord blood CD34+ cells. After term delivery of each neonate, umbilical cord blood was collected and processed to isolate the CD34+ population. The CD34+ cells were transduced with the retroviral vector LASN (ref. 9), which carried a normal human ADA cDNA, and returned to their respective donors by intravenous infusion on their fourth day of life. Subsequently, we determined the effectiveness of gene transduction and whether the engraftment of the umbilical cord blood stem cells had occurred. We report here the results of the first use of umbilical cord blood cells for clinical gene therapy.
Clinical course
At birth the prenatal diagnosis of ADA-deficiency was confirmed. Fluorescence-activated cell sorting (FACS) analysis of umbilical cord blood samples demonstrated that the patients had reduced numbers of T lymphocytes (1.5–2%) compared with those in normal umbilical cord blood (56.6% ± 12.5). Their levels of serum deoxyadenosine metabolites (dAXP) were elevated. Following the institution of enzyme replacement therapy with polyethylene glycol (PEG) combined with ADA on days 1–4 of life (30 U kg-1 twice weekly by intramuscular injection), there was correction of the metabolic abnormalities, with dAXP levels in erythrocytes decreasing from 160–302 nmol ml-1 (cord blood red blood cells (RBCs)) to <10 nmol ml-1 within 7–10 weeks. All three patients have remained in good health while receiving PEG–ADA. Growth and development have been normal with no infectious complications except for routine upper respiratory tract infections. Prophylaxis with oral trimethoprim/sulphamethoxazole (TMP/SMX) and intravenous gamma globulin was given for the first six months to patient 1, patient 2 remains on TMP/SMX, and patient 3 continues to receive both intravenous gamma globulin and TMP/SMX.
Gene transduction of umbilical cord blood CD34+ cells
Between 60 and 200 ml of umbilical cord blood was obtained from the three infants. FACS analysis of the umbilical cord leukocytes demonstrated that between 0.7% and 0.9% of the cells were positive for the stem cell/progenitor antigen, CD34. The cord blood samples were mixed with biotinylated monoclonal antibody to CD34 and processed through an avidin immunoaffinlty column. The resulting cell populations were between 32% and 62% CD34 positive, with yields between 24% and 89%.
The enriched CD34+ cells were transduced with three rounds of LASN retroviral vector-containing supernatant in the presence of recombinant haematopoietic growth factors (interleukin-3 (IL-3), IL-6 and stem cell factor) to enhance gene transfer. After three days of culture, the total numbers of leukocytes increased by 1.3- to 2.4-fold. The LASN-transduced umbilical cord blood cells were then infused intravenously into each patient on his fourth day of life. No toxicities were noted during or after the infusions.
To assess the efficiency of retroviral-mediated transduction of clonogenic myeloid progenitors contained within the cord blood cells before transplantation, samples were plated in a colony-forming unit granulocyte-macrophage (CFU-GM) assay, with and without the selective neomycin analogue, G418. The expression of the neo gene contained in the LASN vector permits colonies derived from transduced progenitors to survive in G418. From the three patients, 21.5%, 12.5% and 19.4% of the clonogenic progenitors were G418-resistant.
Peripheral blood leukocytes
To determine whether the transduced CD34+ cells had engrafted and whether they contributed to circulating leukocyte populations, we obtained serial peripheral blood samples at monthly intervals after transplantation. The frequency of peripheral blood mononuclear cells and granulocytes that contained vector DNA sequences was determined by semiquantitative polymerase chain reaction (PCR) analysis. PCR primers were designed to specifically amplify only vector-derived sequences; one primer was complementary to sequences of the Moloney murine leukaemia virus backbone of the vector and one was complementary to the human ADA cDNA (Fig. 1a). The intensities of the signals from leukocyte samples were compared with those of a standard curve to allow an estimation of the frequency of circulating leukocytes containing the integrated LASN provirus sequences.
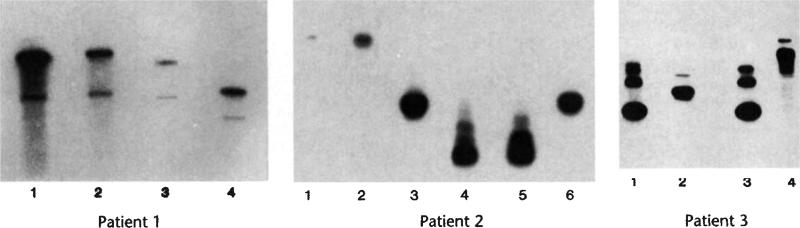
Fig. 1.
Analysis of peripheral blood leukocytes. a, Map of LASN retroviral vector. LTR, Moloney murine leukaemia virus long-terminal repeat; Φ+, extended packaging signal; ADA, human ADA cDNA; SV, SV40 early region promoter; NEO, neomycin resistance gene. Position of PCR primers used to detect proviral DNA sequences (half arrows) and probe (p) used to detect PCR products by hybridization are shown. b, Semiquantitative PCR analysis of leukocyte DNA to detect cells transduced by the LASN retroviral vector. DNA from representative leukocyte samples (PBM, mononuclear cells; gran, granulocytes) from each patient were assayed for the frequency of vector-containing cells by semiquantitative PCR as described in Methods. Samples from the untreated umbilical cord blood (0 months) were tested from each patient as negative controls. A standard curve for the frequency of vector-containing cells was produced by diluting cells containing the LASN vector with non-transduced cells. c, Temporal analYSis of semiquantitative PCR of leukocyte DNA. The results from all peripheral blood leukocyte samples are shown.
An example of a semiquantitative DNA PCR blot is shown in Fig. 1b. The standard curve showed that the intensity of autoradiography signal decreases as the LASN-containing cells are diluted, with a detection limit of 1 vector-containing cell per 100,000 cells. Samples of both mononuclear cells and granulocytes collected over the course of 18 months documented the sustained presence of circulating vector-transduced cells (Fig. 1c). Vector sequences are present in both the mononuclear cell and granulocyte fractions. Although there was some variability in the level of vector-containing cells, circulating leukocytes containing the LASN sequences have persisted in all three patients at frequencies between 1/3,000 and 1/100,000 for at least 18 months following transplantation.
Bone marrow leukocytes
Frequency of transduced cells
Bone marrow aspirates were obtained from each patient at 1 year of age to determine the frequency of bone marrow leukocytes containing the LASN vector. Samples of mononuclear and granulocytic cells were separately assayed by semiquantitative DNA PCR analysis, as described for the peripheral blood samples. All three patients showed the presence of bone marrow granulocytes and mononuclear cells containing the LASN vector, at approximately 1/10,000 cells, a frequency similar to that seen in peripheral blood leukocytes (data not shown).
Bone marrow cells were also analysed to detect the presence of clonogenic myeloid progenitors containing the vector. Colonies were grown in methylcellulose, with and without G418, to test for expression of the neo gene of the LASN vector. Between 4% and 6% of the CFU-GM colonies from all three patients were G418-resistant, whereas minimal numbers of G418-resistant colonies grew from normal bone marrow (Table 1). The higher numbers of colonies grown in the absence of G418 from the patients’ marrow compared with those from the control donors reflects the higher colony-forming efficiency seen in marrow from young (l-year-old) subjects compared with that obtained from adults who were the sources of the control samples.
Table 1.
Total and G418-resistant colony-forming units in bone marrow aspirates one year after infusion of transduced umbilical cord blood cells
| No. CFU per 104 cells plated | ||||
|---|---|---|---|---|
| Patient No. | +G418 | –G418 | % G418-R | PCR(+)/testeda |
| 1 | 2.1 | 38.3 | 5.5 | 6/10 |
| Controlb | 0.07 | 4.7 | 1.5 | |
| 2 | 1.8 | 45.4 | 4.0 | 7/7 |
| Control | 0.02 | 7.1 | 0.3 | |
| 3 | 2.6 | 43.3 | 6.0 | 12/12 |
| Control | 0 | 5.2 | 0 | |
G418-resistant colonies were isolated from methylcellulose and assayed for the presence of the LASN vector by PCR as described in Methods.
For the control, normal, untransduced bone marrow mononuclear cells were assayed simultaneously.
To confirm that the G418-resistant colonies that grew from the patients’ bone marrow samples were transduced, individual G418-resistant colonies were isolated from the methylcellulose and assayed for LASN vector sequences by DNA PCR. A representative blot is shown in Fig. 2a. From patients 2 and 3, each G418-resistant colony showed the presence of vector sequences, whereas six out of ten of the colonies analysed from patient 1 were PCR-positive for the LASN vector.
Fig. 2.
Analysis of bone marrow cells. a, PCR analysis of G418-resistant bone marrow CFU-GM to determine the presence of the LASN vector provirus. +, LASN vector-producing PA317 cell line; –, PA317/LNc11 negative control vector packaging cell line; 1–7, individual G418-resistant CFU-GM. b, Semiquantitative DNA PCR of bone marrow-derived cells. Left, lane 1, bone marrow-derived mononuclear cells; lane 2, bone marrow-derived CD34+ cells. Right, lane 1, pooled cells from CFU grown from marrow mononuclear cells without G418 selection; lane 2, pooled cells from CFU grown from marrow CD34+ cells without G418 selection.
The relatively high frequency of colony-forming cells that contained the LASN vector (4–6%) contrasted with the frequency of vector-containing cells in the peripheral blood and bone marrow mononuclear cell fractions (0.03–0.001%) (Fig. 2b, left, lane 1). To determine whether the frequency of vector-containing cells was higher in bone marrow progenitor cells than in the general mononuclear cell population, CD34+ cells were directly isolated from the marrow mononuclear cells by immunomagnetic bead selection. Semiquantitative DNA PCR analysis of the CD34+ cells showed a 1% frequency of LASN-containing cells, a frequency similar to that detected by in vitro colony growth (Fig. 2b, left, lane 2). Pooled CFU-GM colonies grown without G418 selection from both the unseparated bone marrow mononuclear cells and the CD34-enriched cells also contained approximately 1% transduced cells (Fig. 2b, right, lanes 1 and 2). Thus, transduced cells were present among the bone marrow progenitor cells at a higher frequency than in the peripheral leukocytes.
Analysis of clonality of transduced, engrafted progenitor cells
Because retroviral vectors integrate into chromosomal DNA at random sites, each vector integrant is expected to be at a unique chromosomal position. Thus, integrated retroviral vectors can be used as clonal tags, with all cells derived from a transduced progenitor containing the same vector integrant10,11. The individual vector integrants can be detected from extremely small cell samples using inverse PCR12–14.
The vector integrants of individual bone marrow-derived, G418-resistant CFU-GM were examined by inverse PCR. The CFU-GM shown in each panel of Fig. 3 demonstrate three different integrant patterns for each patient. Overall, each patient had between three and five distinct inverse PCR patterns of vector integrants from among 9–10 CFU-GM (Table 2). These results demonstrate that at least three to five separate transduced progenitor cells were contributing to the CFU-GM colony-forming cells present in the bone marrow one year after transplantation.
Fig. 3.
Clonal analysis by inverse PCR of G418-resistant bone marrow CFU-GM for patients 1, 2 and 3.
Table 2.
Clonal integration patterns from bone marrow CFU-GM
| Distinct clonal integrant (no. colonies with pattern) | |||
|---|---|---|---|
| Patient No. | 1 | 2 | 3 |
| 1A-(4) | 2A-(4) | 3A-(4) | |
| 1B-(2) | 2B-(4) | 3B-(3) | |
| 1C-(2) | 2C-(2) | 3C-(1) | |
| 1D-(1) | 3D-(1) | ||
| 1E-(1) | |||
| Total colonies | 10 | 10 | 9 |
ADA expression
The low frequency of vector-containing leukocytes in blood or marrow samples precluded direct measurements of vector-conferred ADA enzymatic activity. Instead, we used reverse-transcriptase (RT)-linked PCR to detect vector-derived RNA transcripts. Following reverse transcription of cellular RNA into cDNA, we used the PCR primers that are specific for the vector-derived sequences for amplification.
We showed the presence of the vector-derived transcripts in RNA extracted directly from peripheral blood mononuclear cells (Fig. 4). The intensity of the signal from RT-PCR of patient mononuclear cells was intermediate to the intensity of signals produced from standards containing 0.03% and 0.3% LASN-transduced cells, a level consistent with the measured frequency of transduced leukocytes in peripheral blood samples. The specificity of this assay for vector RNA and not DNA is shown by the absence of a PCR product in samples in which the reverse transcription step was omitted (Fig. 4, bottom row). Thus, the LASN vector is expressed in vivo at the level of RNA.
Fig. 4.
Analysis of gene expression in peripheral blood mononuclear cells by the LASN retroviral vector by RT-PCR. RNA from mononuclear cells from patient 1 obtained 6 months (12/93) and 17 months (10/94) after gene therapy were assayed for the frequency of vector-containing cells by semiquantitative RT-PCR as described in the Methods. A standard curve for the frequency of vector-containing cells was produced by diluting cells containing the LASN vector with non-transduced cells. +RT, reversed transcriptase used to copy RNA to DNA; –RT, no reverse transcriptase added.
To demonstrate that the LASN vector produces enzymatically active ADA, G418-resistant CFU-GM colonies from the bone marrow of patient 1 were expanded by in vitro culture for two weeks, and the ADA enzymatic activity was measured. Because these colonies grew in vitro in the presence of G418, they all contained the LASN vector and were expressing the neo gene. The level of ADA present in a pool of six CFU-GM exceeded the ADA levels seen in normal phytohaemagglutinin (PHA)-stimulated T lymphocytes and in G418-resistant CFU-GM grown from LN-transduced normal bone marrow (Table 3). Purine nucleoside phosphorylase (PNP) is unaffected by ADA-deficiency and serves as a control for the amount and quality of cell lysate assayed. At the same time, pools of peripheral blood T lymphocytes grown from the patients by ex vivo stimulation with PHA and IL-2 did not contain ADA enzymatic activity above the background level, consistent with the low frequency of gene-containing cells.
Table 3.
Cellular ADA activity at 18 months after gene transduction
| ADAa | PNPa | ADA/PNP | |
|---|---|---|---|
| G418-resistant CFU-GM colonies | |||
| Patient 1 | 24,051.0 | 28,038.0 | 0.86 |
| Normalb | 4,694.0 | 7,730.0 | 0.61 |
| Unselected T lymphocytes c | |||
| Patient 1 | 99.0 | 4,262.0 | 0.02 |
| Patient 2 | 65.7 | 2,366.0 | 0.03 |
| Patient 3 | 29.3 | 1,996.0 | 0.01 |
| ADA- brother of Patient 2c | 60.6 | 2,212.0 | 0.03 |
| ADA- SCID patients (n = 13)c | 41.3 ± 33.4 | 3,710 ± 1,678 | 0.01 |
| Normal | 2,047 ± 1,360 | 3,007 ± 824 | 0.68 |
U, nmol hr-1 per mg protein; PNP, purine nucleoside phosphorylase.
CD34+ cells from the bone marrow of a normal donor were transduced with the LN retroviral vector and grown in CFU-GM assay with 0.9 mg ml-1 G418.
T lymphocytes were cultured by a modification of Arredondo-Vega31; blood mononuclear cells were grown for 2 days in RPMI 1640, 15% heat-inactivated fetal bovine serum containing 5 μg ml-1 PHA, then in medium lacking PHA, containing 50 U ml-1 IL-2. After 10–14 days, 107 cells were harvested for enzyme assay.
dUnder treatment with PEG-ADA, but not with ADA-transduced cells.
Discussion
These findings attest to the successful introduction of the retroviral vector into long-lived haematopoietic progenitors present in the umbilical cord blood. The ability of haematopoietic stem cells to engraft in vivo after clinical bone marrow transplantation without ‘making space’ with cytoablative therapy has long been controversial. Recent experiments with murine bone marrow transplantation models indicate that haematopoietic stem cell engraftment is possible without the use of cytoablative therapy15–17. Our results demonstrate that long-lived human haematopoietic progenitor cells present in umbilical cord blood are capable of engrafting in neonates without the administration of cytoablative therapy.
The absolute frequency of circulating leukocytes containing the LASN vector (0.03–0.001%) is in accord with theoretical expectations. In preclinical studies of gene transfer/bone marrow transplantation in large animals and retroviral-mediated gene marking of human bone marrow from patients with cancer, a maximum of 1% of the haematopoietic stem cells have been transduced18-20. The efficiency of gene transfer into the umbilical cord haematopoietic stem cells was, therefore, most probably in the 1% range. Because the infants did not receive cytoablative therapy, the endogenous bone marrow mass was still present. Assuming that the infused umbilical cord blood cells represent 1% of the endogenous marrow mass, dilution of the infused cord blood cells would result in a final frequency of gene-transduced haematopoietic stem cells in the range of 0.01%. To achieve higher levels of vector-containing stem cells, it would be necessary to increase gene transfer efficiency and/or to administer cytoablative therapy before transplantation of the transduced cells.
Evaluation of the patients’ bone marrow one year after treatment was highly informative. The frequency of vector-containing progenitor cells exceeds by 100-fold the frequency of vector-containing cells in the mature haematopoietic cell compartments. The explanation for this dichotomy is unknown. Potentially, the expression of the ADA gene is beneficial for progenitor cell proliferation and allows expansion of the committed CD34+ progenitor pool in a fashion similar to that expected for T-Iymphoid progenitors. However, the relatively high frequency of progenitor cells containing the vector is not reflected in mature leukocytes. In studies in which normal mice were transplanted with bone marrow without prior cytoablation, higher frequencies of donor cells were seen in the early haematopoietic progenitor cells than in the mature peripheral leukocytes17. This observation suggests that although primitive progenitor cells . may engraft without cytoablative therapy, they fail to undergo complete maturation in vivo. Alternatively, the presence of the vector may interfere with mature haematopoietic cell production. Some reports have suggested that the neomycin phosphotransferase gene (neo) may impair haematopoietic cell function21,22. The high level of ADA expression seen in progenitor-derived colonies could also be harmful to developing cells.
There have been no adverse effects from the administration of gene-modified umbilical cord blood cells. The continued administration of ADA enzyme replacement therapy has allowed the patients to develop normal immune function and to remain free of infections. To be successful, the PEG-ADA therapy must lower the levels of toxic deoxyadenosine metabolites, and would, therefore, blunt any selective survival advantage afforded to T-lymphocyte precursors by vector-derived ADA expression. Based on the stable presence of vector-containing cells, we have recently decreased the dosage of PEG-ADA administered from 60 U kg–1 per week to 30 U kg-1 per week.
ADA deficiency has been the focus for the initial attempts of gene therapy with haematopoietic stem cells, because transduced T-lymphoid progenitors are expected to have a selective survival advantage in vivo, similar to that seen in allogeneic BMT for ADA deficiency. The present studies demonstrate that primitive haematopoietic cells present in umbilical cord blood can be genetically modified and engrafted without the administration of cytoablative therapy. A similar approach may be used in the future to treat other genetic diseases. Clinical benefits may be expected even with the present inefficient gene transfer techniques if the progeny of the transduced progenitor cells have a selective advantage in vivo. The successful application of gene therapy to other haematologic disorders where the transduced progenitors do not have a selective advantage, such as haemoglobinopathies, lysosomal storage disorders, and AIDS, will require more efficient gene transfer. These advances will require better understanding of the biology of haematopoietic stem cells and the cytokines that regulate their proliferation.
Methods
Umbilical cord blood collection
Umbilical cord blood was collected into CPD anti-coagulant, either by gravity drainage from the umbilical cord stump immediately after clamping of the umbilical cord or by cannulization of the umbilical vessels. Cord blood samples were transported from the sites of neonatal delivery to the laboratory of the Division of Research Immunology and Bone Marrow Transplantation at Childrens Hospital, Los Angeles, California, at room temperature within 1–8 hours of birth.
CD34 isolation
CD34+ cells were isolated from the umbilical cord blood samples by using the CellPro Ceprate system (Bothell, Washington). Biotinylated monoclonal antibody to CD34 (12.8) was added directly to the unseparated cord blood, which was then processed as recommended by the manufacturer.
LASN retroviral vector
The retroviral vector LASN was constructed and packaged in the PA317 cell line in the laboratory of A.D. Miller (Fred Hutchinson Cancer Center, Seattle, Washington)9,23. LASN contains a normal human ADA cDNA under transcriptional control of the MoMuLV long-terminal repeat (LTR) and the bacterial neomycin resistance gene (neo) encoding resistance to the aminoglycoside, G418, under the transcriptional control of the SV40 promoter. The LASN vector-containing supernatant was manufactured and provided by Genetic Therapy Inc. (Gaithersburg, Maryland) under the guidelines of the Food and Drug Administration and had a titre of 1.3 × 105 infectious units per millilitre. The vector supernatant was free of replication competent retrovirus, as determined by S+, L– assay24. LASN supernatant was stored at –70 °C until immediately before use, when it was thawed in a water bath at 37 °C.
Retroviral-mediated transduction
CD34+ cells were transduced by three cycles of exposure to LASN vector-containing supernatant as described25. For each cycle, the CD34+ cells were suspended at 5 × 104 cells ml-1 in 2× transduction medium mixed with an equal volume of LASN vector-containing supernatant. This medium consists of IMDM (Irvine Scientific, Santa Ana, California) with 30% heat-inactivated fetal calf serum (Gemini Bioproducts, Calabasas, California) with recombinant human haematopoietic growth factors (interleukin-3 (IL-3), Sandoz, Piscataway, New Jersey), interleukin-6 (IL-6, Sandoz) and stem cell factor (Amgen, Thousand Oaks, California), plus penicillin (100 U ml-1), streptomycin (10 μg ml-1), and l-glutamine (2 mM). The final concentrations of growth factors were IL-3, 20 ng ml-1; IL-6, 50 ng ml-1; and stem cell factor, 100 ng ml-1. Protamine sulphate was added to a final concentration of 4 μg ml-1 to enhance retroviral-mediated transduction. The cells and vector-containing supernatant were incubated for 24 hours at 37 °C in 5% CO2, and then the cells were pelleted by centrifugation at 800g for 10 min. The cells were resuspended in 2× transduction medium mixed 1:1 with freshly thawed LASN supernatant and incubated for 24 hours for two further cycles on successive days. After the third transduction cycle, the cells were pelleted and washed three times with phosphate-buffered saline and once with RPMI 1640 (Irvine Scientific). The cells were then resuspended in 5 ml RPMI for intravenous infusion. Aliquots were saved for CFU-GM assay and FACS analysis. Daily bacterial cultures and Gram stains performed on the cultured cells were negative.
Analysis of peripheral blood leukocytes
Peripheral blood samples collected by venipuncture into preservative-free heparin were separated by centrifugation on Ficoll-Hypaque gradients (Pharmacia Pharmaceuticals, Uppsala, Sweden). Cells from the mononuclear cell and granulocyte fractions were collected separately and frozen as cell pellets for subsequent DNA analysis and in guanidinium isothiocyante (GITC) for subsequent RNA analysis. We measured PEG-ADA activity in plasma, the level of dAdo nucleotides (dAXP) in erythrocytes, and ADA activity in CFU-GM cultured from bone marrow and T cells cultured from peripheral blood mononuclear cells as described26.
Colony-forming unit granulocyte-macrophage (CFU-GM) assay
Samples of bone marrow were obtained one year after treatment by aspiration of 2–5 ml of marrow from the posterior iliac crest into preservative-free heparin. The marrow cells were separated into granulocyte and mononuclear cell fractions by centrifugation on Ficoll-Hypaque; portions of each were saved for subsequent DNA and RNA assays.
Haematopoietic progenitor-derived colonies of the myelomonocytic lineages (CFU-GM) were grown in a methylcellulose-based assay27. CD34+ enriched cells from umbilical cord blood were plated following transduction at 1-, 2-, 3- and 5 × 103 cells per dish. To measure CFU-GM from the mononuclear cells of the bone marrow aspirates, cells were plated at densities of 1-, 2.5-, 5- and 10 × 104 cells per dish. Quadruplicate dishes were made at each cell plating density, and G418 was added to two dishes at each cell density at a concentration of 0.9 mg ml-1 of active drug. Umbilical cord blood CD34+ cells or marrow mononuclear cells from normal donors were processed and assayed in parallel, with and without G418, to detect the breakthrough growth of non-transduced cells in G418. After 14 days, colonies containing at least 50 cells were enumerated, and the percentages that were G418-resistant calculated. Three to seven days later, well-separated individual colonies growing in the presence of G418 were isolated using a micropipettor for subsequent PCR assays.
CD34+ cells from bone marrow mononuclear cells of patient 1 were isolated using an anti-CD34 monoclonal antibody (9C5 a generous gift of Ping Law, Baxter Immunotherapeutics, Santa Ana, California) and goat anti-mouse immunoglobulin-coated immunomagnetic beads (Dynal, Oslo, Norway). Portions of the unseparated marrow mononuclear cells and the isolated CD34+ cells were plated in the CFU assay with and without G418. After 14 days, colonies were enumerated. The cells growing in the absence of G418 were pooled for semiquantitative DNA PCR. CFU-GM growing in the presence of G418 were isolated from the methylcellulose and grown in culture for two weeks to expand the numbers of cells available for ADA enzyme assay, by culture on irradiated autologous bone marrow stromal cells as described28. The remainder of the unseparated mononuclear cells and CD34+ cells were snap frozen for DNA preparation.
PCR analysis
1. Semiquantitative PCR of leukocyte DNA
PCR was performed to determine the frequency of peripheral blood and bone marrow leukocytes containing the LASN vector. DNA was extracted from frozen leukocyte pellets by solubilization in TNE buffer, pH = 8.0 with 1% SDS and proteinase K at 100 μg ml-1 for 16 hours at 37 °C, followed by two extractions with phenol:chloroform:isoamyl alcohol (25:24:1), one extraction with chloroform:isoamyl alcohol (24:1), and ethanol precipitation. DNA was resuspended in TE buffer (10 mM tris hydrochloride, pH = 8.0, 1 mM EDTA) and quantified with a microfluorometer (Hoefer Scientific Instruments, San Francisco, California). Five hundred nanograms of genomic DNA were used for each PCR reaction. Vector copy number per cell standards were prepared by diluting cells from an HTLV-1 transformed T-lymphocyte line from an ADA-deficient patient, which contained a single copy of the LASN provirus with cells of the same T-cell line that lacked the LASN provirus (10%, 3%, 1%, 0.3%, 0.1%, 0.03%, 0.01%, 0.003% and 0.001% LASN-positive cells). DNA was extracted from the cell mixtures and used as concomitant standards for each PCR reaction. For every assay, samples of DNA extracted from cells not exposed to LASN vector were tested in parallel to detect contamination of reagents with PCR product.
PCR was performed using a primer pair derived to specifically detect vector provirus DNA, with the 5′ (sense) primer (5′-GGAGCAGGCTCATCGAGAAG-3′) in the Φ region of the MoMuLV vector backbone and the 3′ (antisense) primer (5′-ACCGCACCTCCACATACACC-3′) in the third exon of the ADA cDNA. PCR reactions were performed in buffer containing 10 mM tris, pH = 8.4; 50 mM KCI, 1.5 mM MgCl2, 0.01% Triton X-100 and 0.01% gelatin. PCR reactions conditions were 1 cycle of denaturation at 94 °C for five minutes, annealing at 65 °C for 2 minutes and polymerization at 75 °C for 2 minutes followed by 30 cycles with denaturation at 94 °C for 1 minute with the same annealing and polymerization steps. PCR products were separated on 1.5% agarose gels, transferred to nylon membranes by capillary transfer, and cross-linked with UV light. Membranes were hybridized with an oligonucleotide probe from the 5′ end of the ADA cDNA (5′–CCTAGACGGATCCATCAAGCCTGAA–3′) labelled with polynucleotide kinase and [γ-32P]ATP as described29. Membranes were washed with 2× SSPE buffer with 0.1% SDS for 5 minutes, two to four times, and used to expose Kodak XAR film for 18–72 hours at –70 °C. Blots were analysed using a SciScan 5000 densitometer (U.S. Biochemicals, Cleveland, Ohio) reading the signals from the patients’ cell samples and simultaneously performed standard curves.
2. DNA PCR of individual bone marrow CFU-GM
Individual CFU-GM grown from marrow aspirates in the presence of G418 were picked from the methylcellulose for PCR confirmation of the presence of the vector and determination of clonal origin. Single, well-isolated colonies were plucked from the dish using a micropipette tip and incubated for 1 hour at room temperature in 1 ml of PBS. The sample was then pelleted and lysed for 1 hour at 56 °C in 20 μl of whole-cell lysis buffer (50 mM KCI, 10 mM Tris-HCI (pH = 8.3), 1.5 mM MgCl2, 0.1 mg ml-1 gelatin, 0.45% Nonidet P-40, and 0.45% Tween 20) with 3 μl proteinase K (10 mg ml-1, Gibco BRL). Lysates were boiled for 10 minutes to inactivate proteinase K before amplification. PCR amplification to detect LASN sequences was then performed as described above, using 1 μl of the whole-cell lysate.
3. Inverse PCR to detect clonality of bone marrow CFU-GM
The clonal identity of vector proviral integrants was determined using inverse PCR by a modification of the method previously described (ref. 14 and J.A. Nolta et al., manuscript submitted). Briefly, genomic DNA from individual CFU-GM was digested with the TaqI restriction endonuclease, ligated using T4 DNA ligase, and amplified by serial rounds of nested PCR with primers designed to extend from the vector LTR outward through the flanking cellular DNA sequences. The PCR products were fractionated by electrophoresis on a 2% agarose gel, transferred to nylon membrane and probed with an oligonucleotide probe complementary to the LTR as described14. This method will yield two PCR products of unique size (one from each LTR of the vector) for each clonal vector integrant. To verify that PCR bands from distinct CFU-GM with the same size represent identical integration sites, the bands were subjected to DNA sequence analysis. Clones with the same vector integrant are presumed to be derived from a common precursor haematopoietic stem cell.
4. Reverse transcriptase-linked PCR (RT-PCR) to detect expression of vector RNA
RT-PCR was performed to assess expression of the vector in peripheral blood cells. RNA was prepared from 500,000 cells preserved in GITC as described30. The cell suspensions were passed through a 27fi gauge needle 20 times to shear genomic DNA to minimize its contamination of the RNA preparations. RNA from 100,000 cells was copied into cDNA using an antisense oligonucleotide primer (5′-ACCACCTCGTCT-3′), which lies 84 bp downstream of the 3′ antisense DNA PCR primer described above. AMV reverse transcriptase (25 U) (Boehringer-Mannheim) was used in 20 μl of the supplied buffer at 42 °C for 60 minutes. The entire product of the RT reaction was used as template for PCR, as described above for genomic DNA, in a final volume of 100 μl. To verify that only RNA is detected by this assay, duplicate samples were prepared without reverse transcriptase.
Acknowledgements
We wish to thank the patients and their families for their cooperation with these studies. Jay Greenblatt and MaryEllen Franko of the Cancer Treatment Evaluation Program of the National Cancer Institute were invaluable in development of the protocol. Paul Tolstoshev (Genetic Therapy, Inc.), Sherrie Brown (Amgen), Randi Issacs (Sandoz), and Ronald Berenson (CellPro) each generously provided essential reagents and helpful advice for the treatment of the umbilical cord blood cells. The gene transfer reported here was made possible by the prior arduous efforts of French Anderson, Michael Blaese and Kenneth Culver in developing their ADA T-lymphocyte gene transfer protocol, to which this protocol was approved by the NIH RAC as an amendment.
References
- 1.Giblett ER, Anderson JE, Cohen F, Pollara B, Meuwissen HJ. Adenosine-deaminase deficiency in two patients with severely impaired cellular immunity. Lancet. 1972;2:1067–1069. doi: 10.1016/s0140-6736(72)92345-8. [DOI] [PubMed] [Google Scholar]
- 2.Hershfield MS, Mitchell BS. Immunodeficiency diseases caused by adenosine deaminase deficiency and purine nucleoside phosphorylase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 1994. pp. 1725–1768. [Google Scholar]
- 3.Parkman R, Gelfand EW, Rosen FS, Sanderson A, Hirschhorn R. Severe combined immunodeficiency and adenosine deaminase. New Engl. J. Med. 1975;292:714–719. doi: 10.1056/NEJM197504032921402. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg KI, Kohn DB. Somatic gene therapy for severe combined immunodeficiency (SCID). In: Chang P, editor. Somatic Gene Therapy. CRC Press; Boca Raton: 1995. pp. 31–47. [Google Scholar]
- 5.Blaese RM. Development of gene therapy for immunodeficiency: adenosine deaminase deficiency. Pediatr. Res. 1993;33(suppl.):S49–S55. doi: 10.1203/00006450-199305001-00278. [DOI] [PubMed] [Google Scholar]
- 6.Hershfield MS, Chaffee S, Sorensen RU. Enzyme replacement therapy with PEG-ADA in adenosine deaminase deficiency: Overview and case report of three patients, including two now receiving gene therapy. Pediatr. Res. 1993;33(suppl.):S42–S48. doi: 10.1203/00006450-199305001-00236. [DOI] [PubMed] [Google Scholar]
- 7.Broxmeyer HE, Cooper S, Yoder M, Hangoc G. Human umbilical cord blood as a source of transplantable hematopoietic stem and progenitor cells. Curr. Topics Microbial. Immunol. 1992;177:195–204. doi: 10.1007/978-3-642-76912-2_15. [DOI] [PubMed] [Google Scholar]
- 8.Wagner JE, et al. Transplantation of umbilical cord blood after myeloablative therapy: Analysis of engraftment. Blood. 1992;79:1874–1881. [PubMed] [Google Scholar]
- 9.Hock RA, Miller AD, Osborne WR. Expression of human adenosine deaminase from various strong promoters after gene transfer into human hematopoietic cell lines. Blood. 1989;74:876–881. [PubMed] [Google Scholar]
- 10.Williams DA, Lemischka IR, Nathan DG, Mulligan RC. Introduction of new genetic material into pluripotent haematopoietic stem cells of the mouse. Nature. 1984;310:476–480. doi: 10.1038/310476a0. [DOI] [PubMed] [Google Scholar]
- 11.Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 12.Triglia T, Peterson MG, Kemp DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rill DR, et al. Direct demonstration that autologous bone marrow transplantation for solid tumors can return a multiplicity of tumorigenic cells. Blood. 1994;84:380–383. [PubMed] [Google Scholar]
- 15.Harrison DE. Competitive repopulation in unirradiated normal recipients. Blood. 1993;81:2473–2474. [PubMed] [Google Scholar]
- 16.Stewart FM, Crittenden RB, Lowry PA, Pearson-White S, Quesenberry PJ. Long-term engraftment of normal and post-5-fluorouracil murine marrow into normal nonmyeloablated mice. Blood. 1993;81:2566–2571. [PubMed] [Google Scholar]
- 17.Wu DD, Keating A. Hematopoietic stem cells engraft in untreated transplant recipients. Expl. Hematol. 1993;21:251–256. [PubMed] [Google Scholar]
- 18.Van Beusechem VW, Kakler A, Meidt PJ, Valerio D. Long-term expression of human adenosine deaminase in rhesus monkeys transplanted with retrovirus-infected bone marrow cells. Proc. natn. Acad. Sci. U.S.A. 1992;79:7640–7644. doi: 10.1073/pnas.89.16.7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodine DM, et al. Long-term in vivo expression of a murine adenosine deaminase gene in rhesus monkey hematopoietic cells of multiple lineages after retroviral mediated gene transfer into CD34+ bone marrow cells. Blood. 1993;82:1975–1980. [PubMed] [Google Scholar]
- 20.Brenner MK, et al. Gene marking to determine whether autologous marrow infusion restores long-term haemopoiesis in cancer patients. Lancet. 1993;342:1134–1137. doi: 10.1016/0140-6736(93)92122-a. [DOI] [PubMed] [Google Scholar]
- 21.Valera A, Perales JC, Hatzoglou M, Bosch F. Expression of the neomycin-resistance (neo) gene induces alterations in gene expression and metabolism. Hum. Gene Ther. 1994;5:449–456. doi: 10.1089/hum.1994.5.4-449. [DOI] [PubMed] [Google Scholar]
- 22.von Melchner H, Housman DE. The expression of neomycin phosphotransferase in human promyelocytic leukemia cells (HL-60) delays their differentiation. Oncogene. 1988;2:137–140. [PubMed] [Google Scholar]
- 23.Miller AD, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Malec. cell. Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassin RH, Tuttle N, Fischinger PJ. Isolation of murine sarcoma virus-transformed mouse cells which are negative for leukemia virus from agar suspension cultures. Int. J. Cancer. 1970;6:95–107. doi: 10.1002/ijc.2910060114. [DOI] [PubMed] [Google Scholar]
- 25.Hanley ME, Nolta JA, Parkman R, Kohn DB. Umbilical cord blood cell transduction by retrovlral vectors: Pre-clinical studies to optimize gene transfer. Blood Cells. 1994;20:539–546. [PubMed] [Google Scholar]
- 26.Hershfield MS, et al. Treatment of adenosine deaminase deficiency with polyethylene glycol-modified adenosine deaminase. New Engl. J. Med. 1987;316:589–596. doi: 10.1056/NEJM198703053161005. [DOI] [PubMed] [Google Scholar]
- 27.Nolta JA, Kohn DB. Comparison of the effects of growth factors on retroviral vector-mediated gene transfer and the proliferative status of human hematopoietic progenitor cells. Hum. Gene Ther. 1990;1:257–268. doi: 10.1089/hum.1990.1.3-257. [DOI] [PubMed] [Google Scholar]
- 28.Nolta JA, Yu XJ, Bahner I, Kohn DB. Retroviral mediated transfer of the human glucocerebrosidase gene into cultured Gaucher bone marrow. J. clin. Invest. 1992;90:343–348. doi: 10.1172/JCI115868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinberg K, Parkman R. Severe combined immunodeficiency due to a specific defect in the production of interleukin-2. New Engl. J. Med. 1990;322:1718–1723. doi: 10.1056/NEJM199006143222406. [DOI] [PubMed] [Google Scholar]
- 30.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyante-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 31.Arredondo-Vega FX, et al. Paradoxical expression of adenosine deaminase in T cell cultured from a patient with adenosine deaminase deficiency and combined immunodeficiency. J. clin. Invest. 1990;86:444–452. doi: 10.1172/JCI114730. [DOI] [PMC free article] [PubMed] [Google Scholar]