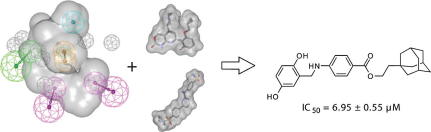
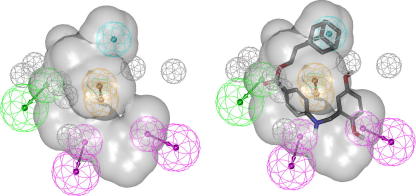
Graphical abstract
Keywords: I kappa B kinase β, IKK-β, Virtual screening, Pharmacophore, Shape-based screening
Abstract
Various inflammatory stimuli that activate the nuclear factor kappa B (NF-κB) signaling pathway converge on a serine/threonine kinase that displays a key role in the activation of NF-κB: the I kappa B kinase β (IKK-β). Therefore, IKK-β is considered an interesting target for combating inflammation and cancer. In our study, we developed a ligand-based pharmacophore model for IKK-β inhibitors. This model was employed to virtually screen commercial databases, giving a focused hit list of candidates. Subsequently, we scored by molecular shape to rank and further prioritized virtual hits by three-dimensional shape-based alignment. One out of ten acquired and biologically tested compounds showed inhibitory activity in the low micromolar range on IKK-β enzymatic activity in vitro and on NF-κB transactivation in intact cells. Compound 8 (2-(1-adamantyl)ethyl 4-[(2,5-dihydroxyphenyl)methylamino]benzoate) represents a novel chemical class of IKK-β inhibitors and shows that the presented model is a valid approach for identification and development of new IKK-β ligands.
Activation of the nuclear factor kappa B (NF-κB) signaling pathway by various stimuli, of which tumor necrosis factor-α (TNF-α) is probably the most extensively studied, induces the transcription of proinflammatory target genes via the activation of a complex intracellular signaling cascade, involving among others TNF receptor-associated factors (TRAFs) 2, 5, and 6, I kappa B kinases (IKKs), and inhibitory κB proteins (IκB). The substantial role of I kappa B kinase β (IKK-β) in NF-κB signaling has been shown by evaluating small molecule IKK-β inhibitors in preclinical models.1,2 The expression of proinflammatory molecules that may promote tumor growth is also facilitated by the IKK/NF-κB signaling pathway.3 IKK-β forming the IKK complex together with IKK-α and the regulatory domain NEMO (IKK-γ) is predominantly contributing to the phosphorylation of the inhibitor of NF-κB protein, IκB-α, at residues Ser32 and Ser36.4 This results in the proteasomal degradation of IκB-α and the release of NF-κB from IκB-α. As inhibition of IKK-β is abolishing NF-κB activation, the development of small molecule IKK-β inhibitors as potential anti-inflammatory and chemosensitizing agents is promising.5–8
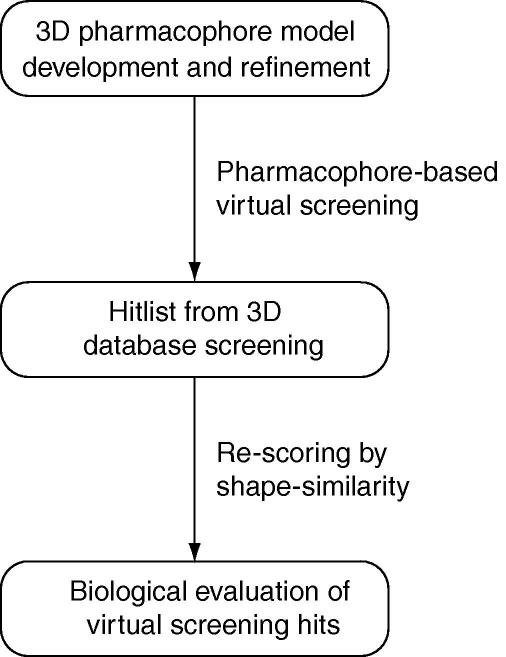
In virtual screening large databases of drug-like molecules are filtered in a knowledge-driven approach to prioritize compounds for biological testing. The use of an X-ray structure or a model of the target receptor to predict the binding mode and the binding affinity by means of high-throughput docking is well established.9 Previous studies on IKK-β inhibitors employed molecular docking of compound libraries into the active site of homology models to investigate binding modes of inhibitors10 and eventually prioritize candidates from a pharmacophore-based screening hitlist for a biological evaluation.11 However, as stated by Muegge, the use of a homology model of the target receptor for the docking campaign in situations, where an X-ray crystal structure is not available, adds a considerable amount of uncertainty to the predictions, accounting that the macromolecule conformation employed in the docking experiment is a predicted conformation, which has to be regarded as putative.12 In our study, we therefore deliberately focused on ligand-based virtual screening techniques to identify novel compounds with activity for IKK-β. We started the development of ligand-based pharmacophore models based on training set compounds with high activity (IC50 <100 nM) and several-fold selectivity for IKK-β over IKK-α. Afterward, the model was carefully and iteratively refined with exclusion volume spheres (XVOLs) and shape constraints. The final model was used to virtually screen the database from the National Cancer Institute (NCI). For post-filtering, the OMEGA/ROCS13,14 software packages from OpenEye (www.eyesopen.com) were employed to re-score the NCI hitlist by comparing the query molecules (NCI hits) with two highly active and structurally diverse IKK-β inhibitors as reference molecules. In ROCS, a different alignment algorithm accounting shape/physicochemical properties for the query molecule orientation gives a complementary filter for the hitlist from pharmacophore-based screening. Figure 1 shows a graphical summary of the workflow.
Figure 1.
Overview of the molecular modeling workflow.
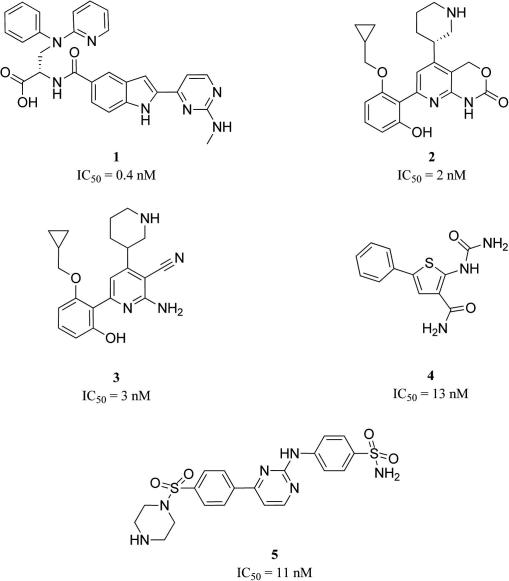
In order to implement this modeling work-flow, a dataset of 128 active and 44 biologically confirmed inactive compounds was collected from the literature.15–31 It is well-known that kinase inhibitors tend to form hydrogen-bonds to the hinge region, which is a highly conserved segment among protein kinases.32 Therefore, the IKK-β inhibitors are also supposed to interact with the hinge motif, which in case of IKK-β is composed of Glu97 and Cys99, to stabilize inhibitor binding and contribute to the reduction of catalytic enzyme activity.10,11 The heterogeneous dataset of IKK-β inhibitors is in all cases composed of chemical moieties to form these interactions. Furthermore, the molecules contain an aromatic ring and a hydrophobic moiety in most cases (Fig. 2).
Figure 2.
Heterogeneous dataset of IKK-β inhibitors.15–19
For pharmacophore model generation conformational models of the training set compounds were generated with a maximum number of 255 conformations per molecule and ‘BEST’ quality within DiscoveryStudio 2.1. The ‘HipHop algorithm’ of Catalyst 4.11 from Accelrys (accelrys.com), which now is incorporated into DiscoveryStudio, was used to build the common feature hypotheses.33 The results from experiments with different training sets showed that the four training set compounds 1, 2, 3, and 4 (Fig. 2) are most appropriate for ligand-based pharmacophore generation. Pharmacophore model generation based on these highly active molecules resulted in ten five-feature hypotheses. Out of these models, the highest ranked 3D pharmacophore displayed the best results in an initial model validation and was selected for a step-wise model refinement. The ‘Refine algorithm’—in DiscoveryStudio available as ‘Steric refinement with excluded volumes’ protocol—was used to strategically place XVOLs in approximated ‘forbidden areas’ from steric hindrance around the ligand-based model.34 This refinement was based on information from confirmed biologically inactive compounds 6 and 7 (Fig. 3). The final model (Fig. 4) consisted of two hydrogen-bond donor (HBD), one hydrogen-bond acceptor (HBA), one ring aromatic (RA), and one hydrophobic feature (H), respectively. In model refinement, XVOLs and a molecular shape constraint derived from the highly active compound 1 in its conformation that optimally fit the highest ranked model were added as steric restrictions. The complete training set of our HipHop refine model is shown in Table 1.
Figure 3.
Confirmed biologically inactive compounds 6 and 7.20,21
Figure 4.
Ligand-based pharmacophore model for IKK-β inhibitors. The model consists of five features, two HBDs, one HBA, one RA, and one H feature. Chemical features are color-coded: HBD—magenta, HBA—green, H—cyan, aromatic ring—brown, shape—shaded volume, XVOL—gray (left). Compound 8 fitted into the model (right).
Table 1.
Training set for HipHop refine model
Due to the availability of only 44 confirmed biologically inactive compounds on IKK-β, a diverse virtual library consisting of 12,775 diverse random compounds (‘decoy set’) was generated using the program ilib diverse35–37 for model validation. For the theoretical assessment of predictability, active literature dataset compounds that were not included in the training set and the virtual decoy database were combined to a validation dataset. The enrichment of active compounds in the resulting hit lists was evaluated by calculating enrichment factors (EF) using the equation
where TP is the number of active compounds fitting into the model, n is the total number of molecules (active compounds and decoys) that were returned as hits by the pharmacophore-based screening, A is the number of active compounds in the entire validation database, and N is the number of all compounds in the validation database.38
The training set composed of the highly active compounds 1–4 resulted in ten pharmacophore models, where the 1st-ranked model showed the best enrichment rate. This model was iteratively refined by addition of XVOLs and shape constraints, which improved the prediction quality from an initial enrichment factor of 15.7–25.8 in the final model (see Table 2 for enrichment factors and percentage of actives, inactives and decoys identified).
Table 2.
Results from the theoretical model validation by screening the literature dataset and the virtual library decoys
| Literature dataset |
Virtual library decoys | EF | ||
|---|---|---|---|---|
| Active compounds | Confirmed inactives | |||
| Hypo1 | 34 | 6 | 191 | 15.7 |
| (27.4%) | (13.6%) | (1.5%) | ||
| Hypo1-R1 | 33 | 1 | 115 | 23.2 |
| (26.6%) | (2.3%) | (0.9%) | ||
| Hypo1-R1-S1 | 31 | 0 | 94 | 25.8 |
| (25.0%) | (0.0%) | (0.7%) | ||
3D resulting pharmacophore model ‘Hypo-1-R1-S1’ was used to screen the compound database of the National Cancer Institute (NCI) for new compounds that fulfill the required chemical features and steric constraints. First, conformers for all NCI compounds were generated using a maximum of 100 conformations per molecule and the Discovery Studio ‘FAST’ algorithm. Virtual screening was conducted using the ‘fast flexible search’ method and 1860 compounds out of 247,041 entries (0.8%).
In order to select the most promising compounds from the large number of hits, the pharmacophore hit list was ranked using shape-based scoring employing ROCS—Rapid Overlay of Chemical Structures. The algorithm is based on the idea that the molecular shape of compounds is similar if the molecules overlay well and any volume mismatch is resulting from shape dissimilarity. For the superimposition of molecules, a smooth Gaussian function is used to represent the molecular volume. Subsequently, the overlay of the molecules is corrected by simple matching of chemical functionalities.39 A recent study40 shows that the enrichment of shape-based screening does not necessarily depend on the availability of a bioactive conformation as reference molecule. These findings encouraged us to employ this complementary technique as a post-processing of the retrieved NCI hit list.
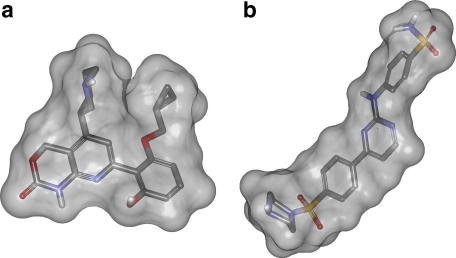
The 3D similarity-based ranking with ROCS was performed using a combined scoring method, using the shape Tanimoto coefficient and the score retrieved from the ROCS ‘color force field’, which includes rough information about chemical functions. As both scores can vary between 0 and 1, the combo score as the sum of both varies between 0 and 2.34 For the shape-based screening we selected compounds 2 and 5 from the literature dataset—two structural dissimilar and highly active reference molecules (Figs. 2 and 5). The reference molecules were submitted to 3D geometry generation using CORINA41 from Molecular Networks (www.molecular-networks.com). The option for conformational sampling of ring moieties was enabled, giving ten conformations for each reference molecule. The conformational models of the different query molecules were calculated using omega (default settings, with a maximum of 400 conformations per molecule). In the subsequently carried out shape-based similarity screening, the query molecules were ranked using the combo score. We accounted the highest similarity score for each query molecule, which was retrieved from virtually screening the ensemble of conformations for each reference molecule against all single query molecule conformations. This highest scoring conformation was determined using the multi-conformer query option in ROCS. The re-scored hit list was visually inspected and ten compounds were chosen for biological testing (Fig. 6). Compounds with a high score were preferred in the selection. However, we emphasized on compounds, which were (i) available at the time of our study, and (ii) showed sufficient structural diversity among each other.
Figure 5.
Compounds 2 and 5 were selected as reference molecules for the shape-based scoring. Shown is the 3D illustration of compounds 2 (a) and 5 (b) with solvent accessible surface.
Figure 6.
Compounds selected for biological evaluation.
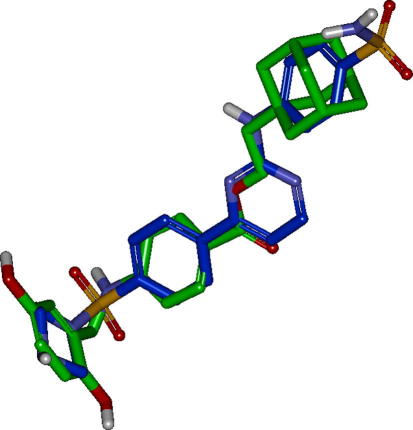
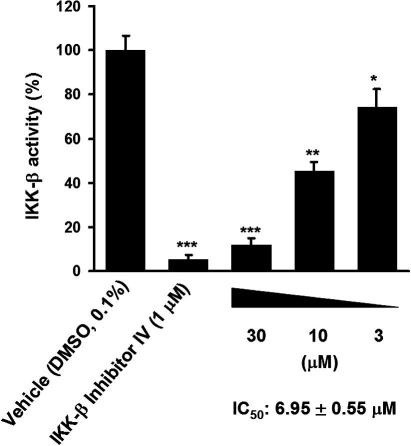
Out of these ten virtual screening hits, compound NSC 719177 (compound 8, Figs. 4, 6 and 7), showed inhibitory activity against IKK-β in a cell-free in vitro assay42 with an IC50 of 6.95 ± 0.55 μM (Fig. 8).
Figure 7.
Alignment of the biologically active compound 8 (green) to the ROCS query molecule compound 5 (blue).
Figure 8.
IKK-β inhibitory activity of the compound identified by the virtual screening approach. IKK-β activity is determined by ELISA as described in detail in the experimental section. The enzymatic activity of human recombinant IKK-β is quantified for 30 min at 30 °C in the presence of DMSO vehicle, different concentrations of 8, or 1 μM IKK-β inhibitor IV that was used as a control. The color development of the substrate was quantified on a GeniosPro plate reader (Tecan, Austria), and the IKK-β activity is presented as percentage of the activity of the vehicle treated control. The graph represents the mean of three independent experiments ± SD (∗p <0.05 ∗∗p <0.01 and ∗∗∗p <0.001, two-tailed paired t-test).
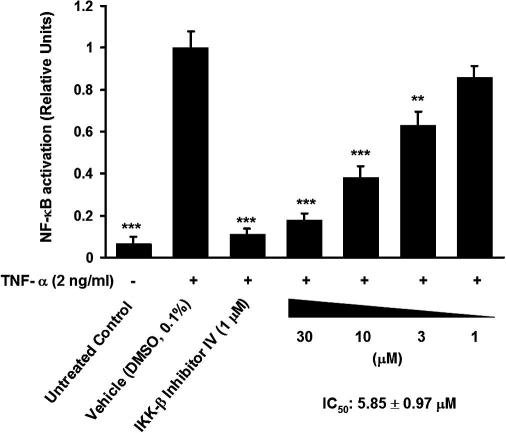
In order to verify the effectiveness of 8 in intact cells, we tested its ability to block NF-κB activation induced by TNF-α in stably transfected HEK293 cells carrying a luciferase reporter gene driven by a promoter containing multiple copies of the NF-κB response element.43 Similar to the control IKK-β inhibitor (IKK-β inhibitor IV),44 compound 8 inhibited TNF-α-induced luciferase activity dose-dependently. The inhibitory effect (IC50: 5.85 ± 0.97 μM, Fig. 9) appeared in a concentration range similar to that needed for the inhibition of IKK-β enzyme activity in vitro.
Figure 9.
HEK293 cells stably transfected with a NF-κB luciferase reporter were treated as indicated and stimulated with 2 ng/ml TNF-α for 6 h before cell lysates were subjected to detection of luciferase activity. The data shown are means ± SD from three independent experiments. (∗∗p <0.01 and ∗∗∗p <0.001, two-tailed paired t-test).
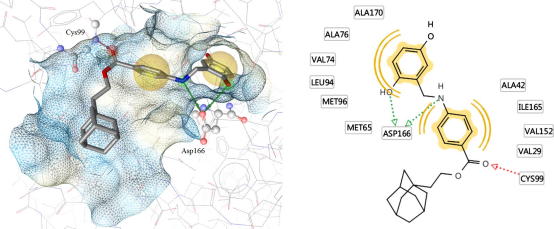
In order to get hints about possible binding modes of compound 8, it was submitted to molecular docking using a homology model of IKK-β.45 The docking results suggest a contact of the inhibitor with the hinge region by forming a hydrogen bond between Cys99 and the ester carbonyl group of the ligand. Additionally, two hydrogen bonds can be surmised with the residue Asp166 by one phenolic hydroxyl group and by the amine in the linker chain. The two aromatic moieties of the ligand are positioned in hydrophobic pockets of the assumed binding site (Fig. 10).
Figure 10.
Predicted binding pose of compound 8 docked into a homology model of IKK-β. The 3D representation of the ligand binding pose is shown with the receptor-binding surface (color-coded by aggregated hydrophilicity/hydrophobicity: blue/gray, respectively). In the 2D representation predicted protein–ligand interactions are given. Chemical features are color-coded: red/green arrow—hydrogen-bond acceptor/donor; yellow spheres—hydrophobic interactions.
In brief, we report the development of a ligand-based pharmacophore model for IKK-β inhibitors, as well as the application of pharmacophore-based virtual screening techniques combined with 3D shape-based re-scoring. Biological testing of 10 virtual screening hits resulted in the identification of compound 8, which has inhibitory activity in the low micromolar range, both in a cell-free IKK-β in vitro assay and a cell-based NF-κB transactivation assay. Therefore, compound 8 is a promising candidate for further medicinal chemistry optimization in order to obtain novel drugs against inflammation and cancer.
Acknowledgements
This work was financed by the NFN-project ‘Drugs from Nature Targeting Inflammation—DNTI’, Grant Nos. S10702-B03, S10704-B03, and S10703-B03 from the Austrian Science Foundation (FWF) by the Austrian Federal Ministry for Science and Research (to S.M.N., D.S., and N.F.) [Technologiestipendien Südostasien Doktorat ACM-2007-00178, ACM-2008-00857 and ACM-2009-01206] and a Young Talents Grant to D.S. from the University of Innsbruck, Austria. Test compounds were provided free of charge by the National Cancer Institute.
We also thank E. Geiger (University of Vienna) for excellent technical assistance.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmcl.2010.10.051.
A. Supplementary data
References and notes
- 1.Sommers C.D., Thompson J.M., Guzova J.A., Bonar S.L., Rader R.K., Mathialagan S., Venkatraman N., Holway V.W., Kahn L.E., Hu G., Garner D.S., Huang H.C., Chiang P.C., Schindler J.F., Hu Y., Meyer D.M., Kishore N.N. J. Pharmacol. Exp. Ther. 2009;330:377. doi: 10.1124/jpet.108.147538. [DOI] [PubMed] [Google Scholar]
- 2.Hacker H., Karin M. Sci. STKE. 2006:1. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 3.Greten F.R., Eckmann L., Greten T.F., Park J.M., Li Z.W., Egan L.J., Kagnoff M.F., Karin M. Cell. 2004;118:285. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh S., Hayden M.S. Nat. Rev. Immunol. 2008;8:837. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 5.Karin M., Yamamoto Y., Wang Q.M. Nat. Rev. Drug. Disc. 2004;3:17. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 6.Strnad J., Burke J.R. Trends Pharmacol. Sci. 2007;28:142. doi: 10.1016/j.tips.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Gavriil M., Lucas J., Mandiyan S., Follettie M., Diesl V., Sum F.W., Powell D., Haney S., Abraham R., Arndt K. Cancer Res. 2008;68:9519. doi: 10.1158/0008-5472.CAN-08-1549. [DOI] [PubMed] [Google Scholar]
- 8.Schon M., Wienrich B.G., Kneitz S., Sennefelder H., Amschler K., Vohringer V., Weber O., Stiewe T., Ziegelbauer K., Schon M.P. J. Natl. Cancer Inst. 2008;100:862. doi: 10.1093/jnci/djn174. [DOI] [PubMed] [Google Scholar]
- 9.Waszkowycz B. Drug Discovery Today. 2008;13:219. doi: 10.1016/j.drudis.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Lauria A., Ippolito M., Fazzari M., Tutone M., Di Blasi F., Mingoia F., Almerico A.M. J. Mol. Graphics Modell. 2010;29:72. doi: 10.1016/j.jmgm.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Nagarajan S., Choo H., Cho Y.S., Oh K.S., Lee B.H., Shin K.J., Pae A.N. Bioorg. Med. Chem. 2010;18:3951. doi: 10.1016/j.bmc.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Muegge I. Mini-Rev. Med. Chem. 2008;8:927. doi: 10.2174/138955708785132792. [DOI] [PubMed] [Google Scholar]
- 13.omega version 2.0, OpenEye Scientific Software, Santa Fe, NM, USA.
- 14.ROCS version 2.3.1, OpenEye Scientific Software, Santa Fe, NM, USA.
- 15.Haddad, E.-B.; Ritzeler, O.; Aldous, D. J.; Cox, P. J. United States Patent US 2007/0142417 A1, Jun. 21, 2007; Int. Pat. Appl. US 2007/0142417 A1, 2005.
- 16.Ziegelbauer K., Gantner F., Lukacs N.W., Berlin A., Fuchikami K., Niki T., Sakai K., Inbe H., Takeshita K., Ishimori M., Komura H., Murata T., Lowinger T., Bacon K.B. Br. J. Pharmacol. 2005;145:178. doi: 10.1038/sj.bjp.0706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata T., Shimada M., Sakakibara S., Yoshino T., Masuda T., Shintani T., Sato H., Koriyama Y., Fukushima K., Nunami N., Yamauchi M., Fuchikami K., Komura H., Watanabe A., Ziegelbauer K.B., Bacon K.B., Lowinger T.B. Bioorg. Med. Chem. Lett. 2004;14:4019. doi: 10.1016/j.bmcl.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 18.Baxter A., Brough S., Cooper A., Floettmann E., Foster S., Harding C., Kettle J., McInally T., Martin C., Mobbs M., Needham M., Newham P., Paine S., St-Gallay S., Salter S., Unitt J., Xue Y. Bioorg. Med. Chem. Lett. 2004;14:2817. doi: 10.1016/j.bmcl.2004.03.058. [DOI] [PubMed] [Google Scholar]
- 19.Bingham A.H., Davenport R.J., Gowers L., Knight R.L., Lowe C., Owen D.A., Parry D.M., Pitt W.R. Bioorg. Med. Chem. Lett. 2004;14:409. doi: 10.1016/j.bmcl.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 20.Waelchli R., Bollbuck B., Bruns C., Buhl T., Eder J., Feifel R., Hersperger R., Janser P., Revesz L., Zerwes H.G., Schlapbach A. Bioorg. Med. Chem. Lett. 2006;16:108. doi: 10.1016/j.bmcl.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 21.Murata T., Shimada M., Sakakibara S., Yoshino T., Kadono H., Masuda T., Shimazaki M., Shintani T., Fuchikami K., Sakai K., Inbe H., Takeshita K., Niki T., Umeda M., Bacon K.B., Ziegelbauer K.B., Lowinger T.B. Bioorg. Med. Chem. Lett. 2003;13:913. doi: 10.1016/s0960-894x(02)01046-6. [DOI] [PubMed] [Google Scholar]
- 22.Murata T., Shimada M., Kadono H., Sakakibara S., Yoshino T., Masuda T., Shimazaki M., Shintani T., Fuchikami K., Bacon K.B., Ziegelbauer K.B., Lowinger T.B. Bioorg. Med. Chem. Lett. 2004;14:4013. doi: 10.1016/j.bmcl.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 23.Bingham A.H., Davenport R.J., Fosbeary R., Gowers L., Knight R.L., Lowe C., Owen D.A., Parry D.M., Pitt W.R. Bioorg. Med. Chem. Lett. 2008;18:3622. doi: 10.1016/j.bmcl.2008.04.062. [DOI] [PubMed] [Google Scholar]
- 24.Castro A.C., Dang L.C., Soucy F., Grenier L., Mazdiyasni H., Hottelet M., Parent L., Pien C., Palombella V., Adams J. Bioorg. Med. Chem. Lett. 2003;13:2419. doi: 10.1016/s0960-894x(03)00408-6. [DOI] [PubMed] [Google Scholar]
- 25.Palanki M.S., Erdman P.E., Ren M., Suto M., Bennett B.L., Manning A., Ransone L., Spooner C., Desai S., Ow A., Totsuka R., Tsao P., Toriumi W. Bioorg. Med. Chem. Lett. 2003;13:4077. doi: 10.1016/j.bmcl.2003.08.047. [DOI] [PubMed] [Google Scholar]
- 26.Olsen L.S., Hjarnaa P.J., Latini S., Holm P.K., Larsson R., Bramm E., Binderup L., Madsen M.W. Int. J. Cancer. 2004;111:198. doi: 10.1002/ijc.20255. [DOI] [PubMed] [Google Scholar]
- 27.Podolin P.L., Callahan J.F., Bolognese B.J., Li Y.H., Carlson K., Davis T.G., Mellor G.W., Evans C., Roshak A.K. J. Pharmacol. Exp. Ther. 2005;312:373. doi: 10.1124/jpet.104.074484. [DOI] [PubMed] [Google Scholar]
- 28.Bonafoux D., Bonar S., Christine L., Clare M., Donnelly A., Guzova J., Kishore N., Lennon P., Libby A., Mathialagan S., McGhee W., Rouw S., Sommers C., Tollefson M., Tripp C., Weier R., Wolfson S., Min Y. Bioorg. Med. Chem. Lett. 2005;15:2870. doi: 10.1016/j.bmcl.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 29.Wen D., Nong Y., Morgan J.G., Gangurde P., Bielecki A., Dasilva J., Keaveney M., Cheng H., Fraser C., Schopf L., Hepperle M., Harriman G., Jaffee B.D., Ocain T.D., Xu Y. J. Pharmacol. Exp. Ther. 2006;317:989. doi: 10.1124/jpet.105.097584. [DOI] [PubMed] [Google Scholar]
- 30.Morwick T., Berry A., Brickwood J., Cardozo M., Catron K., DeTuri M., Emeigh J., Homon C., Hrapchak M., Jacober S., Jakes S., Kaplita P., Kelly T.A., Ksiazek J., Liuzzi M., Magolda R., Mao C., Marshall D., McNeil D., Prokopowicz A., 3rd, Sarko C., Scouten E., Sledziona C., Sun S., Watrous J., Wu J.P., Cywin C.L. J. Med. Chem. 2006;49:2898. doi: 10.1021/jm0510979. [DOI] [PubMed] [Google Scholar]
- 31.Newton R., Holden N.S., Catley M.C., Oyelusi W., Leigh R., Proud D., Barnes P.J. J. Pharmacol. Exp. Ther. 2007;321:734. doi: 10.1124/jpet.106.118125. [DOI] [PubMed] [Google Scholar]
- 32.Ghose A.K., Herbertz T., Pippin D.A., Salvino J.M., Mallamo J.P. J. Med. Chem. 2008;51:5149. doi: 10.1021/jm800475y. [DOI] [PubMed] [Google Scholar]
- 33.Clement O.O., Mehl A.T., editors. HipHop: Pharmacophores Based on Multiple Common-feature Alignments. International University Line; La Jolla, CA, USA: 2000. [Google Scholar]
- 34.Schuster D., Spetea M., Music M., Rief S., Fink M., Kirchmair J., Schutz J., Wolber G., Langer T., Stuppner H., Schmidhammer H., Rollinger J.M. Bioorg. Med. Chem. 2010;18:5071. doi: 10.1016/j.bmc.2010.05.071. [DOI] [PubMed] [Google Scholar]
- 35.Wolber G., Langer T. In: Rational Approaches to Drug Design. Sippl H.-D.H.A.W., editor. Prous Science; 2000. p. 390. [Google Scholar]
- 36.Wolber G., Langer T. J. Chem. Inf. Model. 2005;45:160. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 37.Schuster D., Laggner C., Steindl T.M., Palusczak A., Hartmann R.W., Langer T. J. Chem. Inf. Model. 2006;46:1301. doi: 10.1021/ci050237k. [DOI] [PubMed] [Google Scholar]
- 38.Güner O.F., Waldman M., Hoffmann R., Kim J.H., editors. Strategies for Database Mining and Pharmacophore Development. International University Line; La Jolla, CA, USA: 2000. [Google Scholar]
- 39.Rush T.S., Grant J.A., Mosyak L., Nicholls A. J. Med. Chem. 2005;48:1489. doi: 10.1021/jm040163o. [DOI] [PubMed] [Google Scholar]
- 40.Kirchmair J., Distinto S., Markt P., Schuster D., Spitzer G.M., Liedl K.R., Wolber G. J. Chem. Inf. Model. 2009;49:678. doi: 10.1021/ci8004226. [DOI] [PubMed] [Google Scholar]
- 41.CORINA—Generation of 3D coordinates, version 3.0, Molecular Networks GmbH, Erlangen, Germany.
- 42.IKK-β activity: To measure the IKK-β activity we utilized the ELISA-based (K-LISA™) IKK-β activity assay (Calbiochem, V., Austria) with the conditions recommended by the manufacturer. The GST-IκBα 50-amino acid peptide that includes the Ser32 and Ser36 IKK-β phosphorylation sites is used as a substrate and incubated for 30 min at 30 °C with human recombinant IKK-β in a glutathione-coated 96-well plate, which allows substrate phosphorylation and capture in a single step. The phosphorylated GST-IκBα substrate was subsequently detected using anti-phospho IκBα (Ser32/Ser36) as first antibody, followed by the HRP-conjugated secondary antibody. The color development of the HRP substrate was monitored at 450 nm on a GeniosPro plate reader (Tecan, Austria) and the absorbance intensity was used as a measure for the IKK-β activity.
- 43.NF-κB transactivation activity: HEK293 cells stably transfected with a NF-κB luciferase reporter (293/NFκB-luc cells, P., RC0014) were seeded in Dulbecco’s Modified Eagle Medium at a density of 6 × 106 cells/10 cm dish, incubated for 18 h to allow adherence, and transfected with 5 μg pEGFP-C1 (Clontech, France). Six hours later the cells were harvested and re-seeded in serum-free Dulbecco’s Modified Eagle Medium in a 96-well plate, followed by incubation at 37 °C and 5% CO2 overnight. On the next day, the cells were treated with the respective test compounds for 30 minutes, and further stimulated with 2 ng/ml human recombinant TNF-α for 6 h. After a lysis step, the luminescence of the firefly luciferase and the fluorescence of EGFP were quantified on a GeniosPro plate reader (Tecan, Austria). The luciferase signals derived from the NF-κB reporter were normalized by the EGFP-derived fluorescence to account for differences in the cell number and/or transfection efficiency.
- 44.Gavriil M., Tsao C.C., Mandiyan S., Arndt K., Abraham R., Zhang Y. Mol. Carcinog. 2009;48:678. doi: 10.1002/mc.20550. [DOI] [PubMed] [Google Scholar]
- 45.http://swissmodel.expasy.org/repository/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.