Chiao & Blizinsky's [1] recently published study, ‘Culture-gene coevolution of individualism–collectivism and the serotonin transporter gene’, addressed an important and under-studied area. It stands in contrast to the dogma that, despite the dramatic cultural and biological diversity of contemporary humans, adaptive genetic changes and especially adaptive population differences have been limited or non-existent over the past 10 000 years. This belief probably stems from a reluctance to apply evolutionary theory to humans and human behaviour. This reluctance dates back at least to Alfred Russel Wallace (co-discoverer along with Darwin of evolution via natural selection) disbelieving that natural selection could have led to the formation of the human mind [2]. Contrary to this, evidence shows that adaptive evolution has accelerated in humans (e.g. [3,4]) and that genes related to psychiatric conditions are highly differentiated across human populations [5]. This is consistent with the notion that cultural context is a salient determinant of human behaviour and the local fitness value of behaviours.
While it is a pleasure to see Chiao and Blizinsky test hypotheses relating to the evolutionary origins of behaviourally implicated genes, there are a number of flaws in their methodology which call their findings into question. In particular, neutral evolutionary processes that might account for the observed patterns in serotonin transporter gene (5-HTTLPR) allele frequencies are not adequately explored and rejected. In the absence of this, the case for natural selection (including more complex gene-culture co-evolutionary processes) changing national-level allele frequencies is weak, and a null hypothesis of no selection, or neutrality, should be retained. We make this argument by reviewing relevant literature and re-analysing Chiao and Blizinsky's data together with published human population genetic data.
Comparative studies, which attempt to explain the origins of variation between populations or species, are fundamental to evolutionary research and are considered key to leading to the very discovery of evolution via natural selection by both Alfred Russel Wallace and Charles Darwin. However, comparative studies are also notoriously susceptible to confounding and other difficulties. Chief among these problems are defining what constitutes an independent observation and determining how to accommodate non-independent observations. That is, ‘when historical connection between two phenomena can be proved, they must not be admitted as independent evidence’ ([6], p. 907). When comparing species, it is recognized that one must control for the degree of relatedness between species ([7]; cf. [8]). When considering population-level comparisons in humans, this adjustment is more problematic, since the cultural and genetic connections between groups often do not follow simple discrete tree-like patterning [9,10]. While Chiao and Blizinsky do briefly acknowledge these problems and perform a corrective analysis, it appears that the correction is incorrectly applied. A lack of proper control for non-independence issues can not only attenuate the ability to detect significant effects, but also cause the detection of effects in the ‘opposite direction of the true evolutionary relationship’ [7, p. 82].
In addition to the 22 primarily Eurasian populations sampled by Chiao and Blizinsky, seven non-Eurasian countries were sampled (South Africa, Brazil, Australia, Argentina, New Zealand, the USA and Mexico). These ostensibly provide independent confirmation of the trend observed in Eurasia. However, this seemingly independent confirmation fails to account for the post-colonial movements of human populations which add considerable complexity to comparative studies. Upon examination of the original sources cited by Chiao and Blizinsky from which 5-HTTLPR frequencies were derived, the above non-Eurasian locales exhibit considerable European cultural and genetic influence. The sampled South Africans from Saunders et al. [11], Brazilians from Grevet et al. [12], Australians from Jorm et al. [13] and Argentineans from Sookoian et al. [14] were of Caucasian/European descent. Additionally, the New Zealand sample is predominantly of European descent, with fewer than 7% Maori or Polynesian individuals [15,16].
Chiao and Blizinsky used world regions (n = 6) and cultural clusters (n = 10) as well as countries (n = 29) as units of analysis. However, these larger units of analysis do not provide confirmation of their hypothesis as they suggest. Rather, as outlined in the last paragraph, these different world regions illustrate the global dispersal of European culture and genes rather than independent data points. To be independent confirmations, they would have had to sample groups that are independent with respect to both their genes and culture. The effective sample size of their study is probably much closer to 2 (Europe versus East Asia) than to 29 (or 6 or 10). This can be observed in their fig. 2, which shows that all the East Asian populations (and only the East Asian populations) show 5-HTTLPR S allele frequencies greater than 60 per cent.
The genetic relationships between human populations show clear clines and clusters with respect to geographical distance and geographical barriers, respectively (e.g. [17]). Individuals from more distant populations are less likely to interbreed with each other than individuals from less distant populations (a concept known as ‘isolation-by-distance’). Similarly, members of populations that have large social pressures or geographical barriers separating them are less likely to mate. Oceans, deserts or mountain ranges decrease the rate of interbreeding. Social norms promoting endogamy, as well as propensities to violate these norms, probably play a role as well (see [18]). These genetic relationships between human populations, or underlying population substructure, have long been acknowledged as important confounders in genetic association studies. For example, while a strong association between a lactase gene polymorphism and height has been found among Europeans, controlling for confounding due to population substructure eliminates this association [19].
Eurasian human genetics, as with human genetics globally, are characterized by clear isolation-by-distance and clustering due to geographical barriers (figure 1a; [17,20]). Europeans are, on average, more closely genetically related to Middle-Easterners than to Central Asians, and more related to Central Asians than to East Asians. This is seen by looking at allele frequencies of primarily neutral alleles at many loci across the genome. This means that even with no natural selection, the historical relationship between populations is expected to create a cline in allele frequencies moving from Western Europe into East Asia. The pattern that Chiao and Blizinsky show mimics such a cline (their fig. 2).
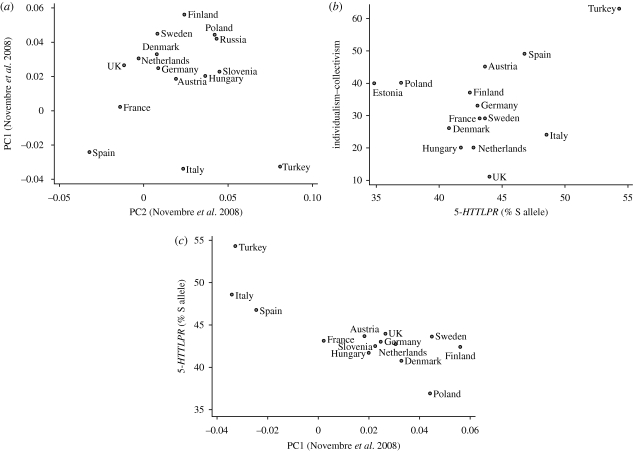
Figure 1.
(a) Population mean principal component 1 (PC1) versus PC2 of 197 146 loci from 1387 individuals across Europe from Novembre et al. [20] including only those overlapping populations used in Chiao & Blizinsky [1]. Genetic structure clearly mirrors geography. (b) Association of 5-HTTLPR allele frequency with individualism–collectivism in the European subgroup from Chiao and Blizinsky (r(14) = 0.32). If Turkey is dropped from the analysis, the relationship switches direction (r(13) = −0.18). (c) PC1 (from figure 1a) has a strong association with 5-HTTLPR allele frequency (r(14) = −0.81). The relationship remains strong without Turkey (r(13) = −0.76).
Further, since the differences between Europe and Asia in both 5-HTTLPR allele frequency and collectivism were observed a priori by Chiao and Blizinsky to make their hypothesis that 5-HTTLPR S alleles were selected for within more collectivist cultures, this East–West difference cannot be admitted as evidence for their hypothesis. Since the data outside of Eurasia are primarily of Europeans, these samples cannot be considered independent evidence either. In an effort to evaluate Chiao and Blizinksy's hypothesis further, but to control for the a priori knowledge, a multivariate regression model predicting 5-HTTLPR allele frequency in Eurasia based upon individualism–collectivism but including a dummy variable for continent found no significant effect of individualism collectivism (n = 21; p = 0.15), but a significant effect of continent (p = 0.01; see electronic supplementary material 1 for dataset). With clustering by continent included in the model, neither coefficient was significant. It must be noted that, while p-values are given above and below for comparison purposes with the original study, they must be interpreted with caution since the data remain non-independent.
We then proceeded to examine the association between individualism–collectivism and 5-HTTLPR allele frequency in Eurasia inside Europe and Asia separately. No association between 5-HTTLPR and individualism–collectivism in the sample of 14 European countries is evident (figure 1b; r(14) = 0.32, p = 0.26). Since this lack of association could be due to reduced sample sizes, we took 10 000 different random samples of 14 countries from the original dataset of 29 and examined the distribution of correlation coefficients. Correlations 0.32 or less were found less than 3 per cent of the time, suggesting that the small correlation in the European only analysis was not due to the limited sample size in Europe. In fact, if Turkey is dropped from the analysis, the relationship switches to the opposite direction of predicted (r(13) = −0.18, p = 0.55).
In the Asian populations, we found the correlation between individualism–collectivism and 5-HTTLPR to be 0.42 (n = 7, p = 0.35)—a value which occurs less than 15 per cent of the time in 10 000 random samples of the same size taken from Chiao and Blizinsky's original study. If India and Russia were excluded from the Asian analysis due to their high degree of European admixture [17,21], the association reversed (r(5) = −0.61, p = 0.28). Using 10 000 random samples of five from the larger dataset, a negative correlation as strong as or stronger than this is expected to occur by chance less than 3 per cent of the time.
Novembre et al. [20] took data from 197 146 loci from 1387 European individuals and derived two principal components (PC1 and PC2) of genetic variation. PC1 and PC2 of genetic variation show that genetic distance clearly mirrors geographical distance (figure 1a). Combining Novembre et al.'s data with those of Chiao and Blizinsky, we see that PC1, which parallels the North–South geographical axis (figure 1a), is strongly correlated with 5-HTTLPR allele frequency in Europe (figure 1c; r(14) = −0.81, p = 0.0005; without Turkey: r(13) = −0.76, p = 0.0026). Population structure is apparently the primary determinant of 5-HTTLPR allele frequencies in Europe. The principal components are mostly based upon neutral genetic polymorphisms and thought to primarily reflect non-selective differences between populations due to genetic drift and gene flow between populations.
While Chiao and Blizinsky's hypothesis is compelling and deserves further examination, we feel that it is not supported by the current data in light of these analyses. Further study would benefit from a larger number of independent samples and controls for underlying population structure (e.g. [22]). In addition, since selection probably works on multiple loci in related pathways simultaneously (e.g. [23,24]), expanding to other loci that are correlated with similar phenotypes would further strengthen the case. Through more careful cross-population analysis, we predict that cases of gene-culture coevolution will be found to be prevalent (see [25] for a recent review).
Acknowledgements
Thanks to Chris Kuzawa, Benjamin Campbell, Jared Bragg and three anonymous reviewers for feedback on earlier drafts. D.T.A.E. is supported by a National Science Foundation Graduate Research Fellowship.
References
- 1.Chiao J. Y., Blizinsky K. D. 2009. Culture-gene coevolution of individualism–collectivism and the serotonin transporter gene. Proc. R. Soc. B 277, 529–537 10.1098/rspb.2009.1650 (doi:10.1098/rspb.2009.1650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace A. R. 1869. Sir Charles Lyell on geological climates and the origin of species. Q. Rev. 126, 359–394 [Google Scholar]
- 3.Hawks J., Wang E. T., Cochran G. M., Harpending H. C., Moyzis R. K. 2007. Recent acceleration of human adaptive evolution. Proc. Natl Acad. Sci. USA 104, 20 753–20 758 10.1073/pnas.0707650104 (doi:10.1073/pnas.0707650104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cochran G., Harpending H. 2009. The 10 000 year explosion: how civilization accelerated human evolution. New York, NY: Basic Books [Google Scholar]
- 5.Amato R., Pinelli M., Monticelli A., Marino D., Miele G., Cocozza S. 2009. Genome-wide scan for signatures of human population differentiation and their relationship with natural selection, functional pathways and diseases. PLoS ONE 4, e7927. 10.1371/journal.pone.0007927 (doi:10.1371/journal.pone.0007927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boas F. 1896. The limitations of the comparative method of anthropology. Science 4, 901–908 10.1126/science.4.103.901 (doi:10.1126/science.4.103.901) [DOI] [PubMed] [Google Scholar]
- 7.Nunn C. L., Barton R. A. 2001. Comparative methods for studying primate adaptation and allometry. Evol. Anthropol. 10, 81–98 10.1002/evan.1019 (doi:10.1002/evan.1019) [DOI] [Google Scholar]
- 8.Martin R. D., Genoud M., Hemelrijk C. K. 2005. Problems of allometric scaling analysis: examples from mammalian reproductive biology. J. Exp. Biol. 208, 1731–1747 10.1242/jeb.01566 (doi:10.1242/jeb.01566) [DOI] [PubMed] [Google Scholar]
- 9.Cavalli-Sforza L. L., Feldman M. W. 1973. Cultural versus biological inheritance: phenotypic transmission from parents to children. (A theory of the effect of parental phenotypes on children's phenotypes.) Am. J. Hum. Genet. 25, 618–637 [PMC free article] [PubMed] [Google Scholar]
- 10.Borgerhoff Mulder M. 2001. Using phylogenetically based comparative methods in anthropology: more questions than answers. Evol. Anthropol. Issues News Rev. 10, 99–111 10.1002/evan.1020 (doi:10.1002/evan.1020) [DOI] [Google Scholar]
- 11.Saunders C. J., de Milander L., Hew-Butler T., Xenophontos S. L., Cariolou M. A., Anastassiades L. C., Noakes T. D., Collins M. 2006. Dipsogenic genes associated with weight changes during Ironman Triathlons. Hum. Mol. Genet. 15, 2980–2987 10.1093/hmg/ddl240 (doi:10.1093/hmg/ddl240) [DOI] [PubMed] [Google Scholar]
- 12.Grevet E. H., et al. 2007. Serotonin transporter gene polymorphism and the phenotypic heterogeneity of adult ADHD. J. Neural Transm. 114, 1631–1636 10.1007/s00702-007-0797-2 (doi:10.1007/s00702-007-0797-2) [DOI] [PubMed] [Google Scholar]
- 13.Jorm A. F., Henderson A. S., Jacomb P. A., Christensen H., Korten A. E., Rodgers B., Tan X., Easteal S. 1998. An association study of a functional polymorphism of the serotonin transporter gene with personality and psychiatric symptoms. Mol. Psychiatry 3, 449–451 10.1038/sj.mp.4000424 (doi:10.1038/sj.mp.4000424) [DOI] [PubMed] [Google Scholar]
- 14.Sookoian S., Gianotti T. F., Gemma C., Burgueno A., Pirola C. J. 2008. Contribution of the functional 5-HTTLPR variant of the SLC6A4 gene to obesity risk in male adults. Obesity (Silver Spring) 16, 488–491 10.1038/oby.2007.64 (doi:10.1038/oby.2007.64) [DOI] [PubMed] [Google Scholar]
- 15.Caspi A., Moffitt T., Silva P., Stouthamer-Loeber M. 1994. Are some people crime-prone-replications of the personality–crime relationship across countries, genders, races, and methods. Criminology 32, 163. 10.1111/j.1745-9125.1994.tb01151.x (doi:10.1111/j.1745-9125.1994.tb01151.x) [DOI] [Google Scholar]
- 16.Caspi A., et al. 2003. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301, 386–389 10.1126/science.1083968 (doi:10.1126/science.1083968) [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg N. A., Mahajan S., Ramachandran S., Zhao C., Pritchard J. K., Feldman M. W. 2005. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1, e70. 10.1371/journal.pgen.0010070 (doi:10.1371/journal.pgen.0010070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenberg D. T. A., Apicella C., Campbell B., Dreber A., Garcia J., Lum J. K. 2010. Assortative human pair-bonding for partner ancestry and allelic variation of the dopamine receptor D4 (DRD4) gene. Social Cogn. Affect. Neurosci. 5, 194–202 (doi:10.1093/scan/nsp026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price A. L., et al. 2008. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 4, e236. 10.1371/journal.pgen.0030236 (doi:10.1371/journal.pgen.0030236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novembre J., et al. 2008. Genes mirror geography within Europe. Nature 456, 98–101 10.1038/nature07331 (doi:10.1038/nature07331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg N. A., et al. 2006. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2, e215. 10.1371/journal.pgen.0020215 (doi:10.1371/journal.pgen.0020215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenberg D. T. A., Kuzawa C. W., Hayes M. G. 2010. Worldwide allele frequencies of the human apoliprotein E (APOE) gene: climate, local adaptations and evolutionary history. Am. J. Phys. Anthropol. 143, 100–111 (doi:10.1002/ajpa.21298) [DOI] [PubMed] [Google Scholar]
- 23.Edelman G. M., Gally J. A. 2001. Degeneracy and complexity in biological systems. Proc. Natl Acad. Sci. USA 98, 13 763–13 768 10.1073/pnas.231499798 (doi:10.1073/pnas.231499798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harpending H., Cochran G. 2006. Genetic diversity and genetic burden in humans. Infect. Genet. Evol. 6, 154–162 10.1016/j.meegid.2005.04.002 (doi:10.1016/j.meegid.2005.04.002) [DOI] [PubMed] [Google Scholar]
- 25.Laland K. N., Odling-Smee J., Myles S. 2010. How culture shaped the human genome: bringing genetics and the human sciences together. Nat. Rev. Genet. 11, 137–148 10.1038/nrg2734 (doi:10.1038/nrg2734) [DOI] [PubMed] [Google Scholar]