Abstract
The animal immune system provides defence against microbial infection, and the evolution of certain animal–microbial symbioses is predicted to involve adaptive changes in the host immune system to accommodate the microbial partner. For example, the reduced humoral immune system in the pea aphid Acyrthosiphon pisum, including an apparently non-functional immune deficiency (IMD) signalling pathway and absence of peptidoglycan recognition proteins (PGRPs), has been suggested to be an adaptation for the symbiosis with the bacterium Buchnera aphidicola. To investigate this hypothesis, the interaction between Buchnera and non-host cells, specifically cultured Drosophila S2 cells, was investigated. Microarray analysis of the gene expression pattern in S2 cells indicated that Buchnera triggered an immune response, including upregulated expression of genes for antimicrobial peptides via the IMD pathway with the PGRP-LC as receptor. Buchnera cells were readily taken up by S2 cells, but were subsequently eliminated over 1–2 days. These data suggest that Buchnera induces in non-host cells a defensive immune response that is deficient in its host. They support the proposed contribution of the Buchnera symbiosis to the evolution of the apparently reduced immune function in the aphid host.
Keywords: aphid, Buchnera aphidicola, immune deficiency pathway, immune response, symbiosis
1. Introduction
At first sight, it appears paradoxical that animals possess an immune system that protects them from micro-organisms, while simultaneously supporting a large, and often diverse, microbiota that contributes to various host functions, including immune function and nutrition. There is a growing recognition that this paradox is resolved, at least in part, by precisely coordinated interactions between the resident micro-organisms and the host immune system [1–5]; and that these interactions are crucial for the sustained health and wellbeing of the animal host [6–8]. As a consequence, the animal immune system function is predicted to be shaped by the distinct (and possibly conflicting) selection pressures to defend against pathogens and maintain beneficial micro-organisms [9–11].
The possible evolutionary impact of the resident beneficial microbiota on the animal immune system is highlighted by the genetic capacity for immune function in the pea aphid, Acyrthosiphon pisum, revealed by the recent sequencing of its genome [12]. In many insects, the humoral branch of the innate immune system comprises two canonical signalling pathways, immune deficiency (IMD) and Toll, which are activated by DAP-type and lys-type peptidoglycan broadly characteristic of Gram-negative and Gram-positive bacteria, respectively [1,13]. Exceptionally, the pea aphid lacks recognizable genes for peptidoglycan recognition proteins (PGRPs), most genes of the IMD pathway, and also has an apparent dearth of anti-microbial peptides (AMPs; [14]). Most aphids, including the pea aphid, require the vertically transmitted γ-proteobacterium, Buchnera aphidicola, and Gerardo et al. [14] specifically hypothesize that the symbiosis with Buchnera might have selected for the reduction and simplification of the aphid humoral immune system.
One prediction arising from this hypothesis is that Buchnera possesses molecular patterns that are recognized by receptors for the IMD pathway of non-host insect cells. An opportunity to investigate this hypothesis has arisen from the discovery that various bacteria can infect heterologous insect cells in culture. In particular, the Drosophila S2 cell line, an embryo-derived line with properties akin to haemocytes [15], enables dissection of the host–microbial interaction founded on the extensive Drosophila genomic and post-genomic tools and detailed understanding of the molecular basis of the immune system in this species. S2 cells have been used in research on various bacteria, including Mycobacterium, Listeria, Chlamydia, Ehrlichia and Wolbachia [16–18]; and aphid-associated bacteria known as secondary symbionts can also be maintained indefinitely in S2 cell culture [19]. In this study, we investigated the capacity of Buchnera to infect and persist in Drosophila S2 cells, and whether the Drosophila cells mount an immune response to this bacterium.
2. Material and methods
(a). Aphids and cells
The pea aphid Acyrthosiphon pisum clone LL01 was reared on preflowering Vicia faba cv. Sutton at 20°C with 18 L : 6 D. It was confirmed to bear no detectable internal bacteria other than Buchnera by microscopical examination, PCR assays with general 16S rRNA gene primers and microbiological cultivation experiments. Aposymbiotic aphids, i.e. lacking the symbiotic bacterium Buchnera, were generated by oral administration of the antibiotic rifampicin at 50 µg ml−1 diet over 2 days after birth; and aphids were used when 7 days old, at which time Buchnera were undetectable microscopically and by Buchnera-specific PCR assay [20]. The Drosophila S2 cells were cultured aseptically in Schneider's Drosophila medium (SDM) + 10% foetal calf serum (FCS) (Invitrogen) at 20°C and maintained in exponential phase by passaging every fourth day.
To isolate Buchnera for administration to Drosophila S2 cells, ca 0.4 g aphids were surface-sterilized by immersion in 70 per cent ethanol for 30 s, rinsed in sterile water and then homogenized in 2 ml SDM + 10% FCS (Invitrogen) under aseptic conditions. The homogenate was passed slowly through a 5 µm filter (Sartorius), the filtrate was centrifuged at 1600g for 3 min, and the pellet was washed twice in SDM + 10% FCS with centrifugation. As a control, aposymbiotic aphids were subjected to the same treatment, and the equivalent extract was administered to S2 cells.
For Buchnera infections, S2 cells were brought to 8 × 106 cells ml−1 and incubated overnight in 1.9 ml aliquots in 6-well plates. Buchnera (ca 108 bacterial cells in 0.1 ml SDM + 10% FCS) were added to each culture of S2 cells (which had proliferated overnight to ca 107 cells ml−1), the plate was swirled gently, centrifuged at 125g for 10 min and returned to standard culture conditions. Buchnera in S2 cells were quantified by quantitative real-time PCR (qRT-PCR) of the dnaK gene by TaqMan in 96-well reaction plates, exactly as in Douglas et al. [21].
(b). Analysis of S2 cell gene expression
A microarray experiment was conducted on four replicate S2 cell cultures that had been maintained separately for at least 20 passages prior to analysis. Six replicate 2 ml volumes of each culture were transferred to wells of a 6 well plate allocated randomly as three experimental samples (infected with Buchnera) and three uninfected control samples; and one experimental and control samples from each culture were harvested at 1 h, 6 h and 24 h. After RNA extraction and labelling, the control samples from all four cultures were pooled to give a single common reference sample. Each of the 12 experimental samples (four cultures × three time points) was hybridized against the common reference sample, with dye swap, to give a total of 24 hybridizations. Standard protocols of FlyChip were used for RNA extraction and labelling, hybridization on a FL002 microarray (14 400 genes), and fluorescent spot finding (electronic supplementary material). Data were normalized using the package VSN (the variance stabilizing normalization library [22] within Bioconductor [23], and differentially expressed genes identified by Microarray Significant Profiles [24] in Bioconducter (MaSigPro: single time-series analysis with parameters α = 0.05/3; degree = 2; Q = 0.05; step.method = ‘backward’; rsq = 0.7; min.obs = 3; see the electronic supplementary material). The microarray experiment is summarized at GEO (http://www.ncbi.nlm.nih.gov/ geo/) under accession ID: GSE11 012.
As a control, the expression of the genes attB CG18 372, cec A2 CG1367 and dipt CG12 763 in S2 cells challenged with Buchnera cells and the equivalent extract from aposymbiotic cells was quantified by qRT-PCR (see methods in electronic supplementary material).
(c). Microscopical analysis
The vitality of Buchnera cells was assessed by BacLight RedoxSensor CTC vitality kit (Invitrogen), using cells incubated with 5 mM 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) for 0.5–2 h and counterstained with the green-fluorescent 0.5 µM SYTO24 nucleic acid stain. Their viability was assessed by BacLight bacterial viability kit (Invitrogen), by incubating cells with 0.5 µM SYTO9 and 30 µM propidium iodide for 15 mins. (These concentrations are lower than advised by the manufacturer; at the recommended concentrations, the concentration of dimethyl sulphoxide in the propidium iodide preparation is deleterious to Buchnera, causing an artefactual reduction in scored viability.) At least 600 Buchnera cells per sample were scored.
Buchnera in S2 cells were scored by fluorescence in situ hybridization (FISH). S2 cells infected with Buchnera and uninfected control cells were washed once in phosphate-buffer saline (PBS) and transferred to glass slides by centrifugation in a CytoFuge (Stat-spin, Westwood, USA) at 22g for 4 min. They were fixed sequentially in 4 per cent paraformaldehyde and 70 per cent ethanol, each for 10 min, rinsed in three changes of PBS and hybridized overnight with the DNA-specific counterstain 4′, 6-diamidino-2-phenylindole (DAPI) at 1 µg ml−1 and 5′-FITC-labelled Apis-P (5′-FITC-TCTTTTGGGTAGATCC-3′), a DNA probe (HPLC-purified: MWG Biotech, GmbH) specific to the 16S rRNA of Buchnera at 70 pmole ml−1 [25]. All experiments included control treatments comprising probe-free controls, RNase-treated slides and competitive suppression with excess (10 µg ml−1) unlabelled probe.
Confocal microscopy was performed with a Zeiss LSM 510 Meta attached to a Zeiss Axiovert 200M fitted with a Plan-Apochromat 63x oil immersion lens (N.A. 1.4). DAPI was excited with the 405 nm diode laser line and emissions collected via a 420–480 nm band pass filter. SYTO9, SYTO24 and FITC were excited with the laser at 488 nm with emissions collected via a 505–530 nm band pass filter (SYTO dyes) and 505–570 nm band pass filter (FITC). CTC was excited with a 458 nm argon laser and emission was collected with a 650–710 nm band pass filter. Propidium iodide was excited with the HeNe laser line at 543 nm and emissions collected via a 560 nm long pass filter. To prevent bleed-through in the experiments using BacLIGHT stain and DAPI/FITC staining, images were acquired sequentially.
3. Results
(a). Fate of Buchnera in Drosophila S2 cell culture
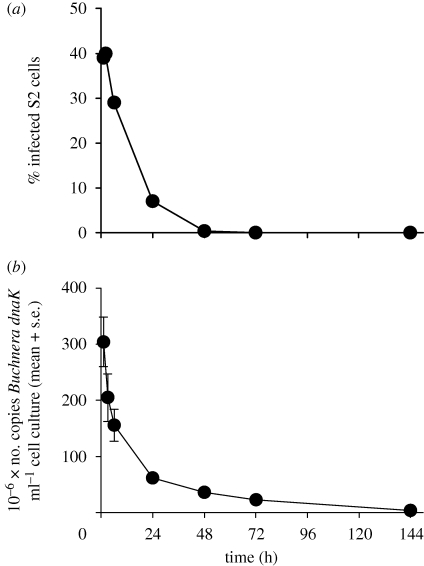
Cells of the symbiotic bacterium Buchnera freshly isolated from their native aphid host were incorporated into cultured Drosophila S2 cells, as revealed by confocal microscopical examination (figure 1a). Optical sectioning confirmed that the bacterial cells were internalized and not borne on the S2 cell surface. Up to 30–40% of the S2 cells were infected by Buchnera within an hour, but the abundance of Buchnera in the cells subsequently declined. The infection was no longer detectable at 72 h as determined microscopically (figure 2a) and at 144 h as determined by qRT-PCR of the Buchnera gene dnaK (figure 2b).
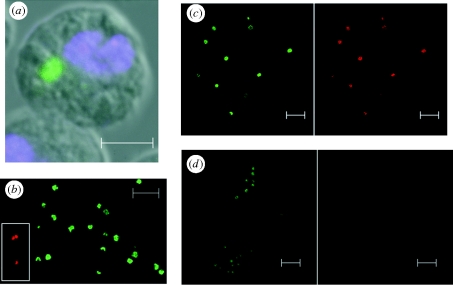
Figure 1.
Buchnera aphidicola in cell culture. (a) Incorporation into Drosophila S2 cells (at 1 h after challenge): the cells were probed with FITC-ApisP (green: specific to Buchnera 16S rRNA) with DAPI (blue: DNA stain) counterstain. (b). Integrity of isolated Buchnera cells assessed by BacLight viability assay, displayed as merged dual colour images; inset, cells killed by incubation at 70°C for 30 min before addition of stain. (c,d) Metabolic vitality of isolated Buchnera cells assessed by CTC assay. Cells were incubated for 30 min at room temperature (c) and at 70°C (d) before incubation with CTC stain for 1 h. Split images are displayed. For (b) green is viable and red is dead; for (c,d), all cells stain green and metabolically active cells additionally stain red. Scale bars, (a) 5 µm, (b,c,d) 10 µm.
Figure 2.
Time course of Buchnera-infection of S2 cells. (a) determined by FISH (604–842 cells scored) and (b) determined by qRT-PCR of the Buchnera gene dnaK (n = 3).
The condition of Buchnera cells used to infect the S2 cells was assessed with the BacLight stain for viability and the RedoxSensor CTC stain for respiratory activity via the electron transport chain. On isolation, 98 per cent of the Buchnera cells were both viable (figure 1b) and metabolically active (figure 1c); control experiments using killed cells confirmed the validity of the two stains (figure 1b inset and figure 1d). The incidence of viable Buchnera cells in the culture medium was stable over 6 h of incubation but declined subsequently to 60–65% at 24 h (electronic supplementary material, figure S1). These results are consistent with published evidence that Buchnera cells remain viable and metabolically active for some hours after isolation [26], but cannot be brought into long-term culture.
(b). Impact of bacterial infection on gene expression of S2 cells
When S2 cells were challenged with Buchnera, 795 genes were identified with expression significantly different from the control S2 cells by least-squares regression for multiple comparisons in MaSigPro. These genes represent 13 per cent of the 6137 genes on the microarray with signal higher than background. The differentially expressed genes include 13 genes with median upregulation by greater than threefold (table 1) at one or more of the three time points of the experiment; no genes were downregulated by twofold or more in Buchnera-infected cells. Four genes were upregulated greater than threefold at all times: the antimicrobial peptide (AMP) genes attacin A and attacin B induced by the IMD pathway and with activity principally against Gram-negative bacteria [27]; PGRP-LB, coding a peptidoglycan amidase, which regulates IMD signalling [28] and CG32 185, also implicated in defence against bacteria [29]. Of the further five genes in table 1 with a described function, four are implicated in bacterial detection and defence (cecropin A2, defensin, diptericin and PGRP-SD) and one (omega) has a role in cell surface receptor-linked signal transduction.
Table 1.
Genes upregulated by greater than threefold in Buchnera-infected S2 cells (instances of greater than threefold upregulation (log2 >2) are shown in bold).
| gene name | gene symbol | median log2 expression |
||
|---|---|---|---|---|
| 1 h | 6 h | 24 h | ||
| Attacin A | CG10 146 | 3.124 | 3.530 | 2.656 |
| Attacin B | CG18 372 | 3.126 | 3.723 | 2.834 |
| Cecropin A2 | CG1367 | 2.635 | 2.527 | 1.067 |
| Defensin | CG1385 | 0.932 | 2.937 | 2.557 |
| Diptericin | CG12 763 | 1.380 | 2.361 | 3.192 |
| Omega | CG32 145 | 1.181 | 2.954 | 2.295 |
| PGRP-LB | CG14 704 | 2.110 | 2.813 | 3.164 |
| PGRP-SD | CG7496 | 1.074 | 1.621 | 2.282 |
| — | CG4267 | 2.616 | 1.619 | 0.199 |
| — | CG6231 | 1.302 | 2.212 | 1.474 |
| — | CG7510 | 2.018 | 1.161 | 0.652 |
| — | CG9932 | 2.061 | 0.546 | 0.533 |
| — | CG32 185 | 2.748 | 3.433 | 3.506 |
Further analysis revealed that other genes in the IMD pathway were expressed in the S2 cells. These included the receptor gene PGRP-LC [30,31], components of the IMD pathway (IMD/CG5576, TAB/CG7417, ird5/CG4201, key/CG16910, dredd/CG7486) and the NFκB transcriptional factor relish/CG11992. Genes for the putative intracellular receptor of the IMD pathway, PGRP-LE (CG8995) and two alternative phagocytic receptors implicated in the binding and uptake of bacteria, Sr-Cl (scavenger receptor class C, type 1: CG4099) and Eater (CG6124) [32,33], were not expressed at levels above background. The gene PGRP-SD (CG11 709), coding one receptor for the Toll pathway (the second principal signalling pathway in the humoral response of the Drosophila immune system) was also upregulated, suggesting that the Toll cascade may also have been activated. Genes coding other defensive functions (e.g. lysozymes, cathepsins, phenoloxidases) were not upregulated (electronic supplementary material, table S3a).
The qRT-PCR analysis of expression of three S2 cell genes challenged with Buchnera preparation and an equivalent extract from aposymbiotic aphids lacking Buchnera revealed significant upregulation of attacin B, cecropin A2 and diptericin exclusively in Buchnera-challenged cells (electronic supplementary material, figure S2). This experiment provides both validation of the microarray data and confirmation that the S2 cell response is not mounted against non-Buchnera products that may be present in the Buchnera preparations.
These results indicate that the genes with a defensive function, especially those regulated by the IMD pathway, make an important contribution to the response of S2 cells to Buchnera. This interpretation was confirmed by global analysis of the patterns of gene expression over the time course of the experiment (electronic supplementary material), which identified defence against micro-organisms as one of five broad functional groupings of differentially expressed genes. The other groups, genes implicated in uptake and intracellular transport, cytoskeleton organization, carbohydrate metabolism and rRNA processing are considered in electronic supplementary material, figure S3 and table S3).
4. Discussion
The upregulated expression of genes encoding AMPs in Drosophila S2 cells challenged with Buchnera (table 1) demonstrates that Buchnera possesses molecular patterns generally detectable by the innate immune system of insects. The gene expression pattern in the S2 cells suggests that Buchnera principally triggers the IMD pathway of the humoral immune system via the cell membrane receptor PGRP-LC. The ligand for this receptor is extracellular peptidoglycan. Consistent with this interpretation, Buchnera has the genetic capacity to synthesize the peptidoglycan backbone, despite the reduction and simplification of its cell wall and outer membrane architecture associated with its very small genome size [34,35].
These data are compatible with the proposal that the Buchnera symbiosis may have selected for reduced functionality of the host immune system, including loss of genes for the IMD pathway and all PGRPs (see §1). A further important test for this hypothesis, the phylogenetic distribution of IMD pathway genes in aphids and related insect groups, will become increasingly possible as the genome sequences for relevant insects become available.
A striking result of this study was that Buchnera cells fail to persist in S2 cells over the timescale that the isolated bacteria are viable. Other bacteria, including the facultative secondary symbionts of aphids Hamiltonella defensa and Regiella insecticola, can persist and proliferate in S2 cells [19]. This difference suggests that Buchnera has limited capacity to tolerate or suppress the immune responses of non-host cells; an interpretation that is consistent with the proposal (above) that the aphid host immune system may be reduced to accommodate the Buchnera symbiosis. Even so, aphids are not without immunological protection, including phagocytic haemocytes and lysozyme and phenoloxidase activities [14,36]. This raises the possibility that the localization to one specialized cell type, the bacteriocyte, in the aphid protects Buchnera cells from some elements of the host immune system. Nevertheless, bacteriocytes have considerable potential competence against bacteria, as is suggested by their expression of genes for lysozyme (ACYPI002 175) and cathepsin L (ACYPI006 974) [37,38], and histochemical evidence for high levels of nitric oxide synthetase [39], indicative of nitrosative stress in these cells. The evidence here that expression of Drosophila genes for lysozyme, cathepsin L and nitric oxide were not upregulated in S2 cells challenged by Buchnera further illustrates the differences between the defensive capabilities of aphid bacteriocytes and Drosophila S2 cells. It remains to be established whether the poised anti-microbial capability of the bacteriocyte is functional against Buchnera and how this capability is kept in check.
These considerations indicate that specific adaptations of the host immune system may have played a key role in the evolution and maintenance of the aphid–Buchnera symbiosis. In this respect, the pea aphid–Buchnera symbiosis appears to be distinct from some other associations, notably the tsetse fly symbiosis with the bacterium Wigglesworthia and Sitophilus weevil symbiosis with a Sodalis-allied bacterium, in which the IMD pathway of the insect host is intact but downregulated in bacteriocytes through the action of a peptidoglycan amidase, PGRP-LB [40,41]. The emerging evidence that the interaction between the host immune system and microbial symbionts varies among insect groups raises the possibility that symbiosis with micro-organisms may have played a role in the evolutionary diversification of immune system function among insects.
Acknowledgements
We thank Laura Briggs (University of York) for advice on S2 cell culture, Neil Boonham (CSL, York) for advice on qRT-PCR, Bettina Fisher (University of Cambridge) for assistance in conducting the microarray assays, Steve Russell (University of Cambridge) for advice on the design of the microarray experiments and colleagues in the Imaging and Cytometry Laboratory of the University of York Technology Facility for assistance with microscopy experiments. David Schneider (Stanford University), Sean Sweeney, Paul Pryor and Adam Peltan (University of York) provided valuable discussions. This research was supported by a BBSRC Research Fellowship BB/C520 898, FlyChip grant and the Sarkaria Institute of Insect Physiology and Toxicology (to A.E.D.).
References
- 1.Lemaitre B., Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 10.1146/annurev.immunol.25.022106.141615 (doi:10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- 2.Mazmanian S. K., Round J. L., Kasper D. L. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 10.1038/nature07008 (doi:10.1038/nature07008) [DOI] [PubMed] [Google Scholar]
- 3.Ryu J.-H., Ha E.-M., Lee W.-J. 2009. Innate immunity and gut-microbe mutualism in Drosophila. Dev. Comp. Immunol. 34, 369–376 10.1016/j.dci.2009.11.010 (doi:10.1016/j.dci.2009.11.010) [DOI] [PubMed] [Google Scholar]
- 4.Salzman N. H., et al. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–82 10.1038/ni.1825 (doi:10.1038/ni.1825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei B., Wingender G., Fujiwara D., Chen D. Y., McPherson M., Brewer S., Borneman J., Kronenberg M., Braun J. 2010. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J. Immunol. 184, 1218–1226 10.4049/jimmunol.0902620 (doi:10.4049/jimmunol.0902620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillon R. J., Dillon V. M. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92 10.1146/annurev.ento.49.061802.123416 (doi:10.1146/annurev.ento.49.061802.123416) [DOI] [PubMed] [Google Scholar]
- 7.Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 10.1038/4441022a (doi:10.1038/4441022a) [DOI] [PubMed] [Google Scholar]
- 8.Wen L., et al. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 10.1038/nature07336 (doi:10.1038/nature07336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas A. E. 2010. The symbiotic habit. Princeton, NJ: Princeton University Press [Google Scholar]
- 10.Feldhaar H., Gross R. 2008. Immune reactions of insects on bacterial pathogens and mutualists. Microbes Infect. 10, 1082–1088 10.1016/j.micinf.2008.07.010 (doi:10.1016/j.micinf.2008.07.010) [DOI] [PubMed] [Google Scholar]
- 11.Gross R., Vavre F., Heddi A., Hurst G. D., Zchori-Fein E., Bourtzis K. 2009. Immunity and symbiosis. Mol. Microbiol. 73, 751–759 10.1111/j.1365-2958.2009.06820.x (doi:10.1111/j.1365-2958.2009.06820.x) [DOI] [PubMed] [Google Scholar]
- 12.Richards S. & The International Aphid Genomics Consortium 2010. Genome sequence of the pea aphid, Acyrthosiphon pisum. PLoS Biol. 8, e1000313. 10.1371/journal.pbio.1000313 (doi:10.1371/journal.pbio.1000313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutros M., Agaisse H., Perrimon N. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3, 711–722 [DOI] [PubMed] [Google Scholar]
- 14.Gerardo N. M., et al. 2010. Immunity and other defence in pea aphids, Acyrthosiphon pisum. Genome Biol. 11, R21. 10.1186/gb-2010-11-2-r21 (doi:10.1186/gb-2010-11-2-r21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27, 353–365 [PubMed] [Google Scholar]
- 16.Elwell C., Engel J. N. 2005. Drosophila melanogaster S2 cells: a model system to study Chlamydia interaction with host cells. Cell Microbiol. 7, 725–739 10.1111/j.1462-5822.2005.00508.x (doi:10.1111/j.1462-5822.2005.00508.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luce-Fedrow A., Von Ohlen T., Boyle D., Ganta R. R., Chapes S. K. 2008. Use of Drosophila S2 cells as a model for studying Ehrlichia chaffeensis infections. Appl. Env. Microbiol. 74, 1886–1891 10.1128/AEM.02467-07 (doi:10.1128/AEM.02467-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi Z., Gavotte L., Xie Y., Dobson S. L. 2008. Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genomics 9, 1. 10.1186/1471-2164-9-1 (doi:10.1186/1471-2164-9-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darby A. C., Chandler S. M., Welburn S. C., Douglas A. E. 2005. Aphid-symbiotic bacteria cultured in insect cell lines. Appl. Env. Microbiol. 71, 4833–4839 10.1128/Aem.71.8.4833-4839.2005 (doi:10.1128/Aem.71.8.4833-4839.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas A. E., Minto L. B., Wilkinson T. L. 2001. Quantifying nutrient production by the microbial symbiosis in an aphid. J. Exp. Biol. 204, 349–358 [DOI] [PubMed] [Google Scholar]
- 21.Douglas A. E., Price D. R. G., Minto L. B., Jones E., Pescod K. V., Francois C. L. M. J., Pritchard J., Boonham N. 2006. Sweet problems: insect traits defining the limits to dietary sugar utilisation by the pea aphid, Acyrthosiphon pisum. J. Exp. Biol. 209, 1395–1403 10.1242/Jeb.02148 (doi:10.1242/Jeb.02148) [DOI] [PubMed] [Google Scholar]
- 22.Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl. 1), S96–S104 [DOI] [PubMed] [Google Scholar]
- 23.Gentleman R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. 10.1186/gb-2004-5-10-r80 (doi:10.1186/gb-2004-5-10-r80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conesa A., Nueda M. J., Ferrer A., Talon M. 2006. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 22, 1096–1102 10.1093/bioinformatics/btl056 (doi:10.1093/bioinformatics/btl056) [DOI] [PubMed] [Google Scholar]
- 25.Fukatsu T., Nikoh N., Kawai R., Koga R. 2000. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: homoptera). Appl. Env. Microbiol. 66, 2748–2758 10.1128/AEM.66.7.2748-2758.2000 (doi:10.1128/AEM.66.7.2748-2758.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehead L. F., Douglas A. E. 1993. A metabolic study of Buchnera, the intracellular bacterial symbionts of the pea aphid Acyrthosiphon pisum. J. Gen. Microbiol. 139, 821–826 [Google Scholar]
- 27.Hoffmann J. A. 2003. The immune response of Drosophila. Nature 426, 33–38 10.1038/nature02021 (doi:10.1038/nature02021) [DOI] [PubMed] [Google Scholar]
- 28.Zaidman-Remy A., et al. 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473 10.1016/j.immuni.2006.02.012 (doi:10.1016/j.immuni.2006.02.012) [DOI] [PubMed] [Google Scholar]
- 29.Verleyen P., Baggerman G., D'Hertog W., Vierstraete E., Husson S. J., Schoofs L. 2006. Identification of new immune induced molecules in the haemolymph of Drosophila melanogaster by 2D-nanoLC MS/MS. J. Insect Physiol. 52, 379–388 10.1016/j.jinsphys.2005.12.007 (doi:10.1016/j.jinsphys.2005.12.007) [DOI] [PubMed] [Google Scholar]
- 30.Gottar M., Gobert V., Michel T., Belvin M., Duyk G., Hoffmann J. A., Ferrandon D., Royet J. 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644 10.1038/nature734 (doi:10.1038/nature734) [DOI] [PubMed] [Google Scholar]
- 31.Rämet M., Manfruelli P., Pearson A., Mathey-Prevot B., Ezekowitz R. A. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for. E. coli. Nature 416, 644–648 10.1038/nature735 (doi:10.1038/nature735) [DOI] [PubMed] [Google Scholar]
- 32.Kocks C., et al. 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123, 335–346 10.1016/j.cell.2005.08.034 (doi:10.1016/j.cell.2005.08.034) [DOI] [PubMed] [Google Scholar]
- 33.Rämet M., Pearson A., Manfruelli P., Li X., Koziel H., Gobel V., Chung E., Krieger M., Ezekowitz R. A. B. 2001. Drosophila scavenger receptor Cl is a pattern recognition receptor for bacteria. Immunity 15, 1027–1038 10.1016/S1074-7613(01)00249-7 (doi:10.1016/S1074-7613(01)00249-7) [DOI] [PubMed] [Google Scholar]
- 34.Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 10.1038/35024074 (doi:10.1038/35024074) [DOI] [PubMed] [Google Scholar]
- 35.Zientz E., Dandekar T., Gross R. 2004. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68, 745–770 10.1128/MMBR.68.4.745-770.2004 (doi:10.1128/MMBR.68.4.745-770.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altincicek B., Gross J., Vilcinskas A. 2008. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 17, 711–716 10.1111/j.1365-2583.2008.00835.x (doi:10.1111/j.1365-2583.2008.00835.x) [DOI] [PubMed] [Google Scholar]
- 37.Nakabachi A., Shigenobu S., Sakazume N., Shirak I. T., Hayashizaki Y., Carninci P., Ishikawa H., Kudo T., Fukatsu T. 2005. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl Acad. Sci. USA 102, 5477–5482 10.1073/pnas.0409034102 (doi:10.1073/pnas.0409034102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikori K., Morioka K., Kubo T., Morioka M. 2009. Age- and morph-dependent activation of the lysosomal system and Buchnera degradation in aphid endosymbiosis. J. Insect Physiol. 55, 351–357 10.1016/j.jinsphys.2009.01.001 (doi:10.1016/j.jinsphys.2009.01.001) [DOI] [PubMed] [Google Scholar]
- 39.Ganassi S., Tagliazucchi D., Mola L. 2005. Occurrence of nitric oxide synthase in Megoura viciae Buckton (Homoptera, Aphididae): an histochemical and immunohistochemical localisation. Eur. J. Histochem. 49, 385–393 [DOI] [PubMed] [Google Scholar]
- 40.Anselme C., Vallier A., Balmand S., Fauvarque M. O., Heddi A. 2006. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl. Environ. Microbiol. 72, 6766–6772 10.1128/AEM.00942-06 (doi:10.1128/AEM.00942-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Wu Y., Yang G., Aksoy S. 2009. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl Acad. Sci. USA 106, 12 133–12 138 10.1073/pnas.0901226106 (doi:10.1073/pnas.0901226106) [DOI] [PMC free article] [PubMed] [Google Scholar]