Abstract
As host to the genome, the nucleus plays a critical role as modulator of cellular phenotype. To understand the totality of proteins that regulate this organelle, we used proteomics to characterize the components of the cardiac nucleus. Following purification, cardiac nuclei were fractionated into biologically relevant fractions including acid-soluble proteins, chromatin-bound molecules and nucleoplasmic proteins. These distinct subproteomes were characterized by liquid chromatography-tandem MS. We report a cardiac nuclear proteome of 1048 proteins—only 146 of which are shared between the distinct subcompartments of this organelle. Analysis of genomic loci encoding these molecules gives insights into local hotspots for nuclear protein regulation. High mass accuracy and complementary analytical techniques allowed the discrimination of distinct protein isoforms, including 54 total histone variants, 17 of which were distinguished by unique peptide sequences and four of which have never been detected at the protein level. These studies are the first unbiased analysis of cardiac nuclear subcompartments and provide a foundation for exploration of this organelle's proteomes during disease.
The nucleus exists to keep the genetic material separate from the other activities of the eukaryotic cell. It is the final theater of gene regulation and proteins directly controlling gene expression must reside in, or relocate to, this organelle. During cell division, an exquisitely orchestrated processing of the genome and nuclear architecture occurs, facilitating accurate replication. In times of nonreplication, the somatic nucleus exhibits a remarkable set of (somewhat opposing) properties necessary for cellular function: the DNA is discretely packaged; transcription factors and other molecules are shuttled rapidly, and specifically, between the nucleus and cytoplasm; appropriate gene expression for the given cell type is established; and the nucleus remains sufficiently plastic as to enable very rapid changes in transcription. Genes encoding proteins, as well as other biologically active ribonucleotides, clearly play a fundamental role in establishing the features of this organelle. Equally important, and to date less well understood, is how the totality of proteins in the nucleus control nuclear behavior and ultimately cellular phenotype.
The unique functionality of the nucleus is conferred by specialized compartmentalization. The nucleus is bordered by a double membrane, which is impermeable to most molecules, across which nuclear pores transverse, enabling selective transport (1). The outer nuclear membrane is contiguous with the cytoskeleton (2) and the endoplasmic reticulum (3), which is studded with ribosomes, the sites of protein translation. Inside the nucleus, DNA is packaged into chromosomes, continuous pieces of DNA whose selective accessibility controls transcription. After initial production of mRNA, these transcripts are further processed by spliceosome complexes to remove noncoding sequences. These processing events are thought to occur in subcompartments of the nucleus—deemed Cajal bodies, speckles and paraspeckles—prior to their transport to the ribosome (4–6). Although functional ribosomes are located in the cytoplasm and the outer nuclear membrane, these organelles are initially synthesized and assembled from RNA and protein in the nucleolus, a nuclear structure formed around tandem repeats of DNA coding for ribosomal RNA (7, 8). The nuclear lamina (9), a meshwork of structural proteins that provides mechanical support akin to the cytoskeleton, is physically coupled to the nuclear membrane and chromatin, and may participate in gene regulation (10).
Most mammalian cells have a single nucleus and erythrocytes are the only mammalian cell devoid of a nucleus. In contrast, many species of protozoa and fungi have naturally multinucleated cells (11–14). Nonpathological polynucleation in mammals is seen in skeletal muscle, where it results from the fusion of myoblasts (15, 16), and in cardiomyocytes, where it appears to be the result of karyokinesis without cytokinesis (17, 18).
Because heart disease is the leading cause of death in the developed world, understanding control of cardiac phenotype is essential to develop new therapies. Although cardiac gene expression has been studied in detail (19–21), much less is known about the protein make up of the nucleus. In utero, cardiomyocytes grow and divide. Not long after birth these cells exit the cell cycle and become binucleated. In rodents binucleation can be seen as early as 4 days after birth and by 2 weeks of age, 85%–90% of cardiomyocytes have two nuclei (17, 22). Further nuclear division can occur during physiological or pathological hypertrophy, a state in which the cell size increases without cell division. Cell division occurs in the heart, but this is almost exclusively restricted to noncardiomyocytes. As such, the adult cardiomyocyte is an intriguing cell type given its binucleated state and lack of cell division. Furthermore, rapid gene expression changes that must occur to induce changes in cardiac phenotype require efficient packaging of the genetic material. We reason that a major determinant of nuclear function is the proteins that reside in this organelle.
Proteomics has been successfully applied to a number of organelles, however, studies of the mammalian nucleus have been relatively limited (23–27) with most restricted to nuclear protein complexes (28–35). A common challenge for nuclear proteomics studies is the difficulty of isolating this highly interconnected organelle with sufficient purity (36). Studies specific to cardiac nuclei (37) have been further limited by additional experimental difficulties unique to this organ including extensive mitochondria to support high metabolic activity, a rigid contractile apparatus and cytoskeletal network, and heterochromatic DNA organization.
To address the question how the proteome regulates the genome in the heart, we developed a method for isolating cardiac nuclei and subfractionating them into biologically relevant compartments. We carried out exhaustive proteomic dissection of these fractions using high accuracy mass spectrometry. Our findings present the first comprehensive description of the proteomic anatomy of the cardiac nucleus, enabling future work to understand how this anatomy endows normal and diseased physiology of the heart.
EXPERIMENTAL PROCEDURES
Isolation, Subfractionation and Structural Analysis of Cardiac Nuclei
Adult male balb/c mice aged 8–12 weeks (Charles River Laboratories) were used for this study. Neonatal rat ventricular myocytes1 (NRVMs)1 were obtained by enzymatic dissociation from 1-day-old litters and plated in Dulbecco modified Eagle medium (Invitrogen, #11965) containing 1% penicillin, 1% streptomycin, 1% insulin-transferrin-sodium selenite supplement and 10% fetal bovine serum for the first 24 h after which the cells are cultured in serum-free media. 3T3 and Hela cells were obtained from ATCC (Manassas, VA). All protocols involving animals were approved by the University of California Los Angeles Chancellor's Animal Research Committee. All chemicals and reagents, unless otherwise noted, were from Sigma.
Nuclear Purification
To isolate nuclei, hearts were excised, washed thoroughly in PBS, minced with scissors, and homogenized in a glass dounce in buffer containing 10 mm Tris (pH 7.4), 250 mm sucrose, 1 mm EDTA, 0.15% Nonidet P-40, 10 mm sodium butyrate, 0.1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture (Roche) and phosphatase inhibitors (0.2 mm Na3VO4, 0.1 mm NaF). After homogenization the suspension was passed through a 100 μm nylon strainer (BD Falcon, #352360) to remove any large insoluble material. Subcellular fractionation was carried out by centrifuging at 1000 × g to pellet crude nuclear fraction. The crude nuclear pellet was resuspended in homogenization buffer, layered on a 2 m sucrose pad and centrifuged at 7500 × g for 5 min to isolate the enriched nuclear fraction. The first supernatant from the crude nuclear pellet was centrifuged at 5000 × g for 30 min to yield the mitochondria, which were further purified using a 28% Percoll gradient and 14,000 × g spin for 40 min (mitochondria are in the pellet). All steps were carried out at 4 °C. The initial relative purity of these individual fractions was evaluated via Western blotting with histone H2A for nuclei and the adenine nucleotide transporter (ANT) for mitochondria. Nuclei were isolated from NRVMs and 3T3 cells using the same procedure.
Electron Microscopic Analyses
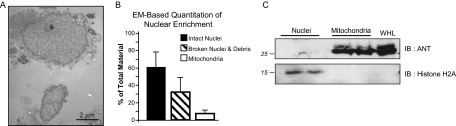
Transmission electron microscopy analyses were performed on isolated nuclei to examine enrichment. Nuclei were fixed in homogenization buffer (detailed above) containing 2% glutaraldehyde at 4 °C, postfixed in osmic acid, dehydrated, and embedded in epoxy resin. A Reichert Ultracut ultramicrotome was used to cut 70 nm slices from embedded samples, which were stained in uranyl acetate followed by lead. Sections were imaged and photographed on a JEOL 100CX Transmission Electron Microscope (JEOL USA, Inc, Peabody, MA). To quantify nuclear enrichment, at least 10 pictures were randomly taken from each EM grid. Two grids were analyzed from each sample block and three blocks were analyzed from two separate nuclei preparations. The area occupied in each picture by intact nuclei, broken nuclei and other membranes, and mitochondria was determined using area measurements in Photoshop. These areas are expressed as a percentage of the total visible material in each picture.
Purification of Cardiac Chromatin and Nucleoplasm and Acid Extraction of Nuclear Proteins
Following purification of nuclei, further fractionation was carried out to separate nucleoplasm from chromatin. Isolated nuclei were resuspended in buffer (20 mm HEPES, 7.5 mm MgCl2, 30 mm NaCl, 1 m Urea, 1% Nonidet P-40, protease, phosphatase, and deacetylase inhibitors) to solubilize the nuclear membrane and extract soluble proteins in the nucleoplasm. After solubilization samples were centrifuged at 13,000 × g for 10 min to pellet the insoluble chromatin and remove the nucleoplasm fraction. The chromatin pellet was washed with PBS, solubilized in 50 mm Tris (pH 8), 10 mm EDTA, 1% SDS (containing protease, phosphatase, and deacetylase inhibitors) and centrifuged at 13,000 × g to extract proteins (referred to as chromatin fraction). For acid extraction, isolated nuclei or chromatin were treated with 400 μl of 0.4N H2SO4 and placed on a rotator at 4 °C overnight. Samples were centrifuged at 16,000 × g to pellet acid-insoluble material. The pellet was washed with PBS and solubilized in buffer containing 1% SDS, 50 mm Tris and 10 mm EDTA (containing protease, phosphatase, and deacetylase inhibitors) and used as an immunoblotting control (referred to as acid-insoluble fraction). The supernatant was treated with 132 μl of trichloroacetic acid in a dropwise manner, inverting the tube between drops, and placed on ice for 30 min to precipitate the acid extracted proteins. Precipitated proteins were collected by centrifuging at 16,000 × g for 15 min (4 °C). The pellet was washed twice with ice-cold acetone without disturbing the pellet and allowed to air dry. The pellet was resuspended in buffer containing 1% SDS, 50 mm Tris, and 10 mm EDTA (containing protease, phosphatase, and deacetylase inhibitors). Subfractionation of nuclei using this method resulted in ∼15%–20% of the total protein being extracted in the nucleoplasm fraction and ∼80%–85% extracted in the chromatin fraction. Subsequent extraction of the chromatin fraction with acid recovered ∼20% of the total protein in this fraction.
Electrophoresis and Western Blotting
Proteins were separated by standard SDS-PAGE using Laemmli buffer. Gels were stained with SYPRO ruby or Oriole, both from BioRad. For Western blotting, proteins were transferred to nitrocellulose membranes blocked with milk and protein signals detected by enzyme linked chemiluminescence (GE Biosciences), as described (38). Ponceau staining of membranes was used to confirm transfer and protein loading. Antibodies used is this study were as follows, including sources: Histone H2A (Santa Cruz, Santa Cruz, CA; sc-8648); Histone H3 (Abcam, Cambridge, MA; ab1791); Histone H2B (Abcam, ab1790); Histone H4 (Abcam, ab10158); macroH2A.1 (Abcam, ab37264); Histone H1.2 (Abcam, ab4086); Histone H1.3 (Abcam, ab24174); Histone H2A.Z (Abcam, ab4174); adenine nucleotide transporter (Santa Cruz, sc-9299); Nucleoporin P62 (BD Biosciences, 610498); SNRP70 (Abcam, ab51266); Fibrillarin (Cell Signaling, Beverly, MA; C13C3); Nkx2.5 (Santa Cruz, sc-8697); E2F-1 (Thermo Fisher, MS-879); Retinoblastoma (BD Biosciences, 554136); myocyte enhancer factor-2 (Santa Cruz, sc-17785); p300 (Millipore, Billerica, MA, 05–257); serum response factor (Abcam, ab18063); hypoxia inducible factor-1 (Novus Biologicals, Littleton, CO, NB100–469); BiP (Santa Cruz, sc1050); α tubulin (Sigma, T1568). Nuclear lysate control (Nuc) was made by solubilizing intact nuclei in buffer containing 1% SDS, 50 mm Tris, and 10 mm EDTA (containing protease, phosphatase, and deacetylase inhibitors) followed by centrifugation at 13,000 rpm for 15 min to remove insoluble material. Hela lysate control (He) was made by solubilizing Hela cells, isolated by scraping cells from culture dishes, in 20 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, protease, phosphatase, and deacetylase inhibitors. The homogenate was then sonicated three times for 5 s each followed by centrifugation at 13,000rpm to remove insoluble material. Whole heart lysate (WHL) control was made by homogenizing the entire heart in 20 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, protease, phosphatase, and deactylase inhibitors, followed by sonication and centrifugation at 13,000 rpm to remove insoluble material.
Protein Identification by Mass Spectrometry
Enzyme Digestion
Proteins isolated from intact nuclei, nucleoplasm fraction, chromatin fraction, and acid extraction were separated by SDS-PAGE. Each gel lane was cut into ∼25 slices (2 mm each) for protein identification by mass spec. Gel plugs were dehydrated in acetonitrile (ACN) and dried completely in a Speedvac. Samples were reduced and alkylated with 10 mm dithiotreitol and 10 mm Tris(2-carboxyethyl)phosphine hydrochloride solution in 50 mm NH4HCO3 (30 min at 56 °C) and 100 mm iodoacetamide (45 min in dark), respectively. Gels were washed with 50 mm NH4HCO3, dehydrated with ACN, and dried down in a Speedvac. Gel pieces were then swollen in digestion buffer containing 50 mm NH4HCO3, and 20.0 ng/μl of trypsin (37 °C, overnight). Peptides were extracted with 0.1% trifluoroacetic acid in 50% ACN solution, dried down, and resuspended in LC buffer A. Digestion with chymotrypsin (used in a targeted manner for low molecular weight proteins), was performed using the protocol above, except proteins were swollen in buffer containing 50 mm NH4HCO3, and 20.0 ng/μl of chymotrypsin (25 °C, overnight). Biological (de novo preparation of fractions from different animals, protein isolation, digestion, and liquid chromatography (LC)/tandem MS (MS/MS)) and technical (multiple LC/MS/MS experiments on the same preparation) replicates analyzed for each fraction are as follows (biological/technical): intact nuclei (4/2), nucleoplasm (3/2), chromatin (3/2), and acid extraction (5/2).
Mass Spectrometry Analyses and Database Searching
Ten microliters of extracted peptides were analyzed by nano-flow LC/MS/MS on a Thermo Orbitrap with dedicated Eksigent nanopump using a reversed phase column (75 μm i.d. 10 cm, BioBasic C18 5 μm particle size, New Objective). The flow rate was 200 nL/min for separation: mobile phase A is 0.1% formic acid, 2% ACN in water, and mobile phase B is 0.1% formic acid, 20% water in ACN. The gradient used for analyses was linear from 5% B to 50% B over 60 min, then to 95% B over 15 min, and finally keeping constant 95% B for 10 min. Spectra were acquired in data-dependent mode with dynamic exclusion where the instrument selects for fragmentation the top six most abundant ions in the parent spectra. Data were searched against the mouse IPI database (version 3.46; 55,270 entries) using the SEQUEST algorithm in the BioWorks software program version 3.3.1 SP1. False positive rate, which was calculated on several independent datasets within this study by reverse database searching, ranged from 1.4% to 1.7%. All spectra used for identification had deltaCN>0.1, consensus score ≥20 and met the following Xcorr criteria: >2 (+1), >3 (+2), >4 (+3), and >5 (+4); no spectra from singly charged parents were considered in this study. Searches required full cleavage with the given enzyme, ≤4 missed cleavages and were performed with the differential modifications of carbamidomethylation on cysteine and methionine oxidation. Mass tolerance was 2 Da for precursor and 1 Da for product ions. All proteins were identified on the basis of two or more unique peptides; exceptions to this include histone proteins, which were identified on the basis of at least two peptides that did not in all cases allow for distinguishing of isoforms (see Results, Table I and supplemental Table 1 for a detailed description of this point). In addition to these criteria, all individual spectra used to distinguish different isoforms and/or single amino acid differences among proteins were manually examined to ensure correct mass of parent ion, to assign all major peaks and to determine that a diagnostic fragmentation pattern existed to prove the specificity of the peptide. When specific isoforms are reported, at least one peptide unique to that isoform was detected. All reported proteins were identified in at least two biological replicates.
Table I. Histone variants unequivocally identified in cardiac nuclei. Histone variants are organized by family and listed randomly within each family.
Abbreviations: p = Putative; PUP = Putative Uncharacterized Protein; N/A = Not Available. Please see Supplemental Table I for a list of all histone isoforms mapped by the peptides identified in this study.
| Variant | IPI Number | UniProt | % Coverage | # of Peptides (Unique peptides) | Gene loci |
|---|---|---|---|---|---|
| H1.1 | IPI00228616 | P43275; Q5SZ98 | 18 | 2(2) | Chr 13 |
| H1.2a | IPI00223713 | P15864; Q3UXH2; Q5SZA3 | 32 | 6(0) | Chr 13 |
| H1.3a | IPI00331597 | P43277; Q149Z9; Q3U292 | 30 | 6(0) | Chr 13 |
| H1.4 | IPI00223714 | P43274 | 30 | 6(2) | Chr 13 |
| H1.5 | IPI00230133 | P43276; Q1WWK3; Q5T003 | 29 | 4(1) | Chr 13 |
| H1.0 | IPI00467914 | P10922 | 26 | 4(4) | Chr 13 |
| H1.0 | IPI00404590 | Q8C1Y3 | 28 | 4(4) | Chr 15 |
| H1.X | IPI00118590 | Q80ZM5 | 12 | 2(2) | Chr 6 |
| macroH2A.1a | |||||
| isoform 2 | IPI00378480 | Q9QZQ8–2 | 29 | 6(1) | Chr 13 |
| H2A | IPI00665557 | Q8CA90 | 48 | 6(1)b | Chr 13 |
| H2A.Za | IPI00331734 | P0C0S6 | 26 | 5(0) | Chr 3 |
| H2B.1A | IPI00111957 | P70696; A0JNS9 | 46 | 5(1) | Chr 13 |
| PUP (H2B) | IPI00459318 | Q8C622 | 17 | 2(1)b | Chr 13 |
| H3.1 | IPI00553538 | P68433; A1L0V4 | 71 | 9(1) | Chr 13 |
| Similar to H3 (H3.1t) | IPI00277753 | N/A | 71 | 9(2)b | Chr 11 |
| H3.2 | IPI00230730 | P84228; B9EI85; A1L0U3 | 71 | 9(1) | Chr 3, 13 |
| H3.3 | IPI00785343 | P84244; A2A852 | 71 | 9(1) | Chr 1, 11 |
| H3-like 4 | IPI00379693c | N/A | 71 | 9(1)b | Chr 3,13 |
| P. H3-like | IPI00108200 | P02301 | 33 | 5(1)b | Chr 13 |
| H4 | IPI00407339 | P62806; B2RTM0; Q61667; Q6B822 | 62 | 11(11) | Chr 3, 6, 13 |
a Indicates histone variants also confirmed by western blot analysis.
b Denotes identification of novel histone variants (See spectra in Figure 5 and Supplemental Figure 4E).
c Designates an IPI number listed in v3.46 of the mouse IPI database which is no longer available in the online EBI database.
Bioinformatics and Protein Annotation
All physical-chemical properties of proteins, including molecular weight, isoelectric point, and hydropathy were calculated using in-house software based on the algorithms in the Swiss-Prot Protparam tool. Analysis of disorder was carried out using the DISOPRED2 program (39). Redundancy in proteins was eliminated at the primary sequence level by manual inspection using CLUSTAL to compare the sequences in UniProt. Genome analysis was performed from the NCBI genome browser following conversion of IPI to ref-seq accession numbers. All Gene Ontology (GO) annotations were extracted from Uniprot. IPI to Uniprot accession number linkage was performed in an automated manner from the EBI website followed by manual annotation. GO annotation clustering was performed using the GO Term Mapper program available through Princeton University (http://go.princeton.edu); UniProt numbers were converted to MGI terms for clustering. The ScanProSite tool associated with the Swiss-Prot website was utilized to identify proteins containing nuclear localization sequences and DNA- and RNA-binding domains (please see Supplemental Methods for a list of ProSite identifiers used in this study).
RESULTS
Nuclear Isolation and Subfractionation
To characterize the cardiac nuclear proteome we isolated intact nuclei from the heart using differential centrifugation and sucrose density enrichment. Other subcellular fractions (including mitochondria) were isolated and used as controls in immunoblotting and electron microscopy analyses. Transmission electron microscopy analysis of cardiac nuclei demonstrated enrichment of this organelle (Fig. 1A); manual evaluation of EM images to quantify organelles revealed between 60% and 80% (Fig. 1B) of the nuclei were intact with reproducibly low contamination from nonnuclear membranes. Western blotting with antibodies against organelle-specific proteins further confirmed enrichment and purity of the nuclei (Fig. 1C, supplemental Fig. 8).
Fig. 1.
Purification of cardiac nuclei. Following centrifugation-based enrichment and sucrose gradient purification, cardiac nuclear fractions were examined by transmission electron microscopy. As shown in image (A) and quantified in graph (B), nuclear preparations were enriched in intact nuclei and had minimal mitochondrial contamination. C, Fractions were further analyzed by Western blotting, demonstrating enrichment of nuclear proteins and depletion of mitochondrial proteins (using antibodies against adenine nucleotide transporter (ANT) and histone H2A); 30 μg protein loaded per well).
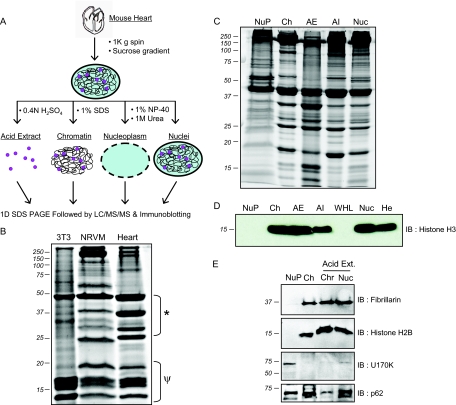
Subfractionation of nuclei utilized a mild salt and detergent buffer to extract the nuclear membrane and soluble nucleoplasmic proteins, followed by subsequent extraction of chromatin bound proteins by 1% SDS. In addition, DNA- and RNA-binding proteins were separately enriched by acid extraction (Fig. 2A). Examination of chromatin-bound proteins from rapidly dividing cells (3T3) in comparison to isolated ventricular myocytes or heart revealed that 3T3 cell chromatin is dominated by histone and other low molecular weight chromatin structural proteins, whereas cardiac myocytes and heart have a number of prominent higher molecular weight proteins (Fig. 2B).
Fig. 2.
Unique protein anatomy of cardiac nuclei. A, Isolation strategy for cardiac chromatin, nucleoplasm and acid soluble proteins. B, Optimized chromatin isolation protocol reveals a distinct protein complement in this subcompartment of the nucleus in heart as compared with NIH 3T3 fibroblasts or neonatal rat ventricular myocytes (NRVMs); * and ψ indicate areas of marked difference in protein components (15 μg protein loaded per lane). C, One dimensional separation of the distinct fractions generated using protocol in panel A demonstrates the distinct protein character of each compartment. D, Histone H3 (as well as other histones examined, see supplemental Fig. 4) was substantially enriched in chromatin and acid extracted fractions and not present in nucleoplasm. E, To validate the functional make-up of the biochemical fractions, Western blotting was performed for proteins with functions restricted to nucleoplasm or chromatin, U170K and H3 respectively. Likewise, the chromatin-bound nucleolar protein fibrillarin was present in total nucleus and chromatin fractions, whereas the splicing factor U170K was confined to the nucleoplasm. The nuclear pore structural protein p62 was present in all fractions. Abbreviations: NuP, nucleoplasm; Ch, chromatin; AE, acid extracted fraction; AI, acid insoluble fraction; Nuc, total nucleus; WHL, whole heart lysate (no fractionation/enrichment); He, Hela cell lysate.
In an attempt to preserve transient protein-DNA interactions throughout the subfractionation process, cross linking with 1% formaldehyde was utilized prior to nuclear enrichment. Although standard in other cell types and before different analyses (such as PCR to detect target genes), cross linking DNA to protein resulted in extensive loss of recoverable proteins in the heart (supplemental Fig. 1). Because cardiac chromatin is extremely heterochromatic, we reason that cross linking produced insoluble aggregates that prevented extraction of proteins. This was visualized by examining the recovery of histones in the presence or absence of formaldehyde cross linking (supplemental Fig. 1), demonstrating poor recovery after cross linking. Based on these observations, formaldehyde cross linking was not incorporated into the fractionation procedure.
SDS-PAGE separation (Fig. 2C) of the proteins from each nuclear fraction demonstrates that these subcompartments harbor distinct subsets of proteins. Also notable from this experiment is the additional enrichment of chromatin structural proteins, as evidenced by the strong histone protein bands (∼10–18kDa), following acid extraction. Western blotting for nuclear compartment-specific proteins further confirmed subfractionation of the organelle (Figs. 2D and E). As expected histone H3 and H2B were exclusively identified in chromatin and further enriched in acid extracted fractions, whereas the soluble spliceosomal factor, U170K, was restricted to the nucleoplasm. Western blotting for other nuclear proteins, including the nucleolus specific methyltransferase fibrillarin and the nuclear pore component p62, indicate distinct features of the subnuclear fractions.
Mass Spectrometric Identification of Nuclear Proteins
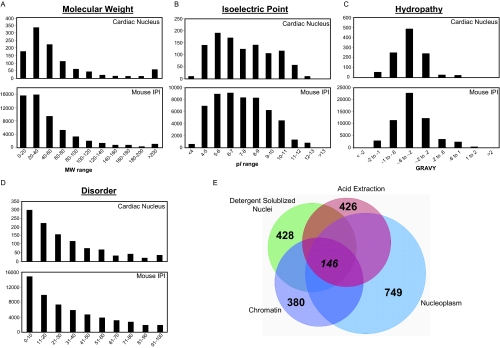
Mass spectrometric analysis of proteins from nucleoplasm, chromatin and acid extracted fractions, as well as from detergent-solubilized nuclei, identified a total of 1048 proteins (supplemental Table 2) represented by 246,845 peptide events (supplemental Table 3). By fraction, 749 protein were identified from the nucleoplasm, 380 from chromatin, 426 from the acid extracted fraction, and 428 from intact nuclei. Despite the large number of proteins identified in each population, only 146 proteins were common to all four preparations (Fig. 3E), suggesting that these fractions host functionally distinct subproteomes and serving as posthoc affirmation for the experimental approach. Additionally, pair-wise comparisons between these fractions (supplemental Fig. 2) reveal proteins involved in multiple different functions in the nucleus (for example proteins residing in the nucleoplasm and regulating chromatin). To determine the physical and chemical properties (molecular weight, isoelectric point, and hydropathy) of the 1048 identified proteins, we examined these features using in-house software based on the tools available from EBI. The cardiac nuclear proteins displayed similar distributions of physical-chemical properties as compared with the entire mouse IPI database (Fig. 3A–C), suggesting that the technical approach is not biased for separation/identification of a certain type of protein. To explore structure/function features within this subset of proteins, we evaluated protein disorder using the DISOPRED algorithm (39), which predicts regions of proteins that lack defined secondary structure. In contrast to what we observe in other specialized subproteomes (supplemental Fig. 3), the distribution of disordered proteins in our nuclear dataset exhibited a distribution indistinguishable from that of the entire mouse IPI database (Fig. 3D).
Fig. 3.
Global features of mouse nuclear cardiac proteome. A–C, Physical-chemical properties of nuclear cardiac proteins were examined and compared with the mouse IPI database (version 3.46). D, Protein disorder was calculated for cardiac proteins and exhibited a similar distribution in comparison to the entire IPI database, in contrast to what we observed in other specialized subproteomes (supplemental Fig. 3). E, Venn diagram displaying shared and distinct components of nuclear subproteomes analyzed in this study. Each of the totals from the individual fractions are in parentheses and are the result of at least two technical and two biological replicates. Only 146 proteins are shared between these fractions, emphasizing the compartmentalization of functions in this organelle and illustrating the need for the comprehensive fractionation approach utilized.
Primary Sequence Motifs
After synthesis on the ribosome, nuclear proteins must be selectively transported across the nuclear membrane in a process often governed by primary sequence motifs (i.e. nuclear localization sequences). A wide array of proteins have been found to contain primary sequence motifs necessary for their transport into the nucleus; however, these motifs have for the most part been identified on an individual protein basis and there is no consensus of an exact nuclear localization sequence. Nevertheless, shared features of these sequences have been shown to include two clusters of basic residues (usually K or R) separated by ∼10 amino acids, a so-called bipartite motif (40). Using the ScanProsite tool (ExPASy), which examines primary sequence for bipartite nuclear localization sequences, we found that 71 of the 1048 proteins (6.8%) contain these sequences. Interestingly only 31% (17/54) of all histone proteins, the quintessential nuclear protein, were found to contain a nuclear localization sequence via this method.
We also examined the proteins in our dataset for the presence of DNA- and RNA-binding domains using the ScanProsite tool. Although this mapping tool allows the user to specify individual domains or classes of domains to search for, it is limited by its ability to only report proteins whose UniProt entry is annotated with the exact sequence and location of the binding domain. Proteins known to bind DNA or RNA in which the exact sequence has not been identified or annotated are thus not included. Using the ScanProsite tool 62 proteins were identified with DNA-binding domains and 40 identified with RNA binding domains; 3 of which were common to both populations (contained both DNA- and RNA-binding domains). As a complementary technique, all proteins were evaluated for GO-annotations indicating known or hypothesized DNA- or RNA- binding. Evaluation of GO-annotations identified 111 proteins with DNA-binding annotations, 72 identified with RNA-binding annotations, and 13 with general nucleic-acid binding (NAB) annotations. Collectively, the proteins in our dataset with known or hypothesized DNA- and RNA-binding domains are 133 and 83, respectively.
Genomic Origins of Nuclear Proteins
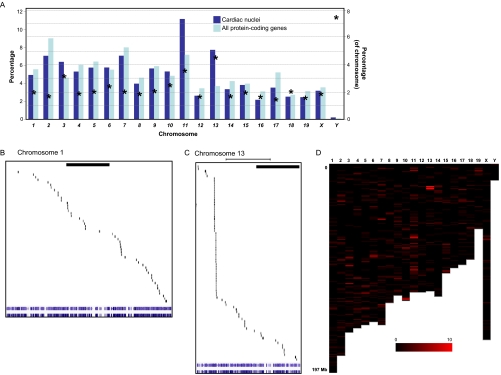
To evaluate the source of nuclear proteins from a genomic structure standpoint, we examined the localization of genes from cardiac nuclear proteins across each of the 21 (1 through 19, plus X and Y) mouse chromosomes in the context of the background of all protein-coding genes (Fig. 4). Among the 1048 proteins identified in this study, 964 had direct counterparts in Ensembl. Because some IPI numbers have more than one UCSC gene ID (and vice versa), the total number of genes mapped in this analysis was 1051. The number of genes for proteins identified in this study, plus the total number of known genes, are quantified for each chromosome in Figs. 4A and 4D. This analyses uncovered differential contribution of proteins from different chromosomes to the nuclear proteome, with chromosomes 11 and 13 housing the largest fractions of genes encoding nuclear proteins. This can be partially attributed to the clusters of histone genes found on these chromosomes (supplemental Table 1). Mapping of the gene loci for all proteins in this study resulted in two distinct patterns, the first being a relatively random distribution of genes (Fig. 4B; for the example of chromosome 1) and the second being a clustering of genes within specific regions (Fig. 4C; for the example of chromosome 13).
Fig. 4.
Genomic origin of cardiac nuclear proteome. A, Summary of data from analysis of all genomic loci for nuclear cardiac proteins shows large representation from chromosomes 11 and 13. Dark blue bars indicate proteins identified on a given chromosome as a percentage of the total number identified in this study, whereas light blue bars are all protein-coding genes in Ensembl on a given chromosome (using left y axis labels). Asterisks indicate the per chromosome number of proteins identified in this study as a percentage of the total number of proteins encoded by the respective chromosome (right y axis labels), indicating a proportionately larger representation from chromosomes Y, 13 and 11, in that order. Ref-seq/UCSC gene identifiers from Ensembl were determined for cardiac nuclear proteins and these genes mapped to the physical structure of the individual chromosomes. Shown in expanded format are the example of chromosomes 1 (B) and 13 (C), which display markedly different distributions of genes encoding nuclear proteins across the individual chromosomes (bar indicates 50 megabases). D, Distribution of all nuclear protein-coding genes from the mouse genome; color intensity represents the number of expressed genes in a given region based on proteomic data in this study (≥10 genes are displayed with maximum intensity in this display; 1 bin = 1 megabase). See supplemental Fig. 7 for an enlargement of panels B, C, and D, including a list of the specific proteins mapping to these chromosomes.
Histone Proteins
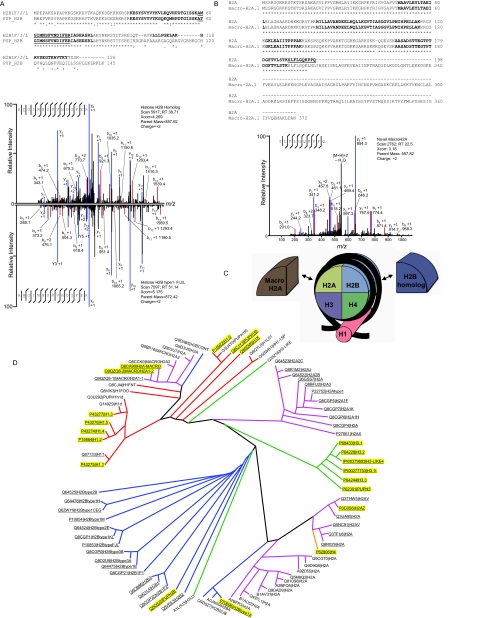
As histone proteins play a prominent role in mediating DNA packaging and transcriptional regulation within the nucleus, we specifically focused on identifying cardiac histone variants. We identified peptides that mapped to a total of 54 histone proteins (supplemental Table 1), including a number of canonical, inducible and novel variants. Seven of these variants were also confirmed by Western blotting (supplemental Fig. 4). Because a significant degree of sequence similarity exists between variants within the same family, many of the peptides used for identification map to multiple variants. These minor variations in histone primary sequence also results in proteins that differed by as little as a single amino acid. The reproducibility of our chromatin fraction enrichment and the mass accuracy of the Orbitrap enabled us to differentiate between such peptides in many cases, bringing the number of individual histone variants uniquely identified to 17 (Fig. 5, Table I, supplemental Table 1 and supplemental Fig. 4). This included the identification of the histone variants H2B homolog and H2B type 1-F/J/L which differ by a single amino acid (T→M) at position 60, as seen in Fig. 5A. We also identified peptides which mapped to a specific subset of inducible variants, as in the case of histone H2A.Z and H2A.V. For these two variants, whose sequences differ by three amino acids, three peptides were identified that mapped uniquely to both of these variants but did not allow us to differentiate between the two (supplemental Fig. 4). By comparative analysis of our data, we were able to conclude that a minimum of 25 different histone variants are present in the cardiac nucleus (this number is the 17 isoforms to which at least one peptide was identified that mapped to only that isoform, plus six additional isoforms for which peptides were detected that map to only a small subset of histone variants and two detected only by Western blotting; please see supplemental Table 1 for more details), although we believe the actual number to be much higher. Among the histone proteins identified are five variants novel to the cardiac system, the existence (at the protein level) of four that we report for the first time in any cell type. Although these five variants have been hypothesized to exist, only one has previously been identified at the protein level; two have previously been identified in high-throughput mRNA screens, and the remaining two defined by genome sequence analysis. Clustal-based sequence alignment and phylogenetic mapping revealed that we identified isoforms from all major branches of the histone family (Fig. 5D).
Fig. 5.
Features of cardiac histone proteins. High mass accuracy detection of peptides on the Orbitrap allowed for differentiation of histone isoforms in the heart. Shown are spectra for the examples of histone H2B homolog (UniProt:Q8C622) and H2B type 1 F/J/L (UniProt:P10853;inverted; A), which differ by a single amino acid, as well as a single spectra allowing the identification of a novel form of histone macro H2A (UniProt:Q9QZQ8, compared with canonical macro H2A, Uniprot:Q8CA90; B). In the sequence comparisons (determined by Clustal 2.0.12 from EBI), bold letters indicate MS/MS coverage from tryptic and chymotryptic peptides; * indicates identical residues, . conserved substitutions and: semi-conserved substitutions. In the spectra, assigned y and b ions are blue and red, respectively, and m/z values are listed for daughter ions that distinguish the single amino acid difference. supplemental Fig. 4 reports additional spectra for the identification of novel histone variants, confirmatory data from Western blotting, and sequence-based comparison of the four core histone families. C, Diagrammatic representation of the structural assembly of the nucleosome. Two copies each of four core histones (H2A, H2B, H3 and H4) form a nucleosome, around which wraps ∼147 bp of genomic DNA (black strand). Histone H1 is peripheral to the nucleosome core and facilitates quaternary structure of chromatin. D, Phylogenetic representation (created with Dendroscope (87) based on neighbor-joining alignment from Clustal) of all 80 known histone isoforms (confirmed or predicted) from UniProt; peptides identified in this study map to 54 variants (underlined) with 20 of these variants unequivocally confirmed (yellow highlight). Edges to different families are colored as follows: H1 (red), H2A (pink), H2B (blue), H3 (green), and H4 (orange).
Transcription Factors
Another class of proteins, which we were interested in because of their nuclear-specific function, were cardiac transcription factors. Although a variety of proteins are involved in gene regulation the defining feature of transcription factors is their ability to bind DNA. However, as mentioned above when querying protein databases for DNA-binding domains this information may be excluded because the exact location and sequence of binding is not known or may be inferred (thereby lacking experimental validation). We identified 15 known and 6 hypothesized transcription factors from our proteomic analysis, not including transcriptional associated proteins such as chromatin remodelers and RNA polymerases. Of these 15 known transcription factors, 11 have annotated DNA binding domains (as determined by ScanProsite mapping) whereas only 7 have GO-annotations listing DNA-binding. In addition to mass spectrometry analyses, we used Western blotting to confirm expression levels and subnuclear localization of six well-characterized cardiac transcription factors, five of which were undetectable by mass spectrometry presumably because of their low abundance (supplemental Fig. 5).
Gene Ontology
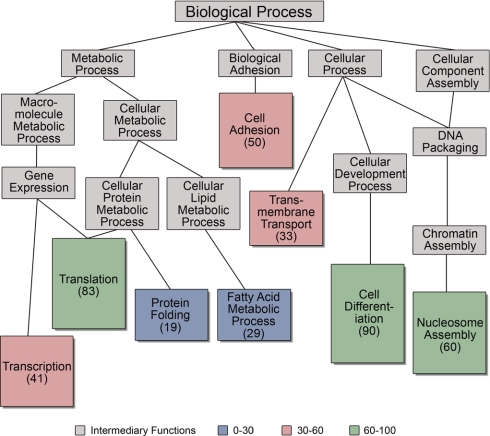
To capture additional information regarding preconceived gene-based annotation of the proteome we define in this study, the 1) cellular component, 2) the molecular function and 3) the biological process that were extracted from Gene Ontology (GO) annotations in the UniProt database. When possible, all IPI numbers were mapped to individual UniProt numbers. However, many IPI numbers map to multiple UniProt numbers, and therefore all GO annotations for all UniProt entries mapping to an individual IPI number were collected and appear in supplemental Table 2. The genes for 43 of the proteins identified in this study lacked GO annotations. The GO annotations for individual proteins were clustered based on annotation hierarchy and reveal subsets of proteins whose genes are annotated with similar functionality and/or localization. Grouping of the molecular function annotations resulted in the following eight prominent clusters: binding, protein binding, nucleotide binding, RNA binding, DNA binding, metal ion binding, ATP binding, and actin binding (Fig. 6). The nuclear proteins had cellular component annotations including nucleus, ribonucleoprotein complex, nucleosome, chromosome, ribosome, and endoplasmic reticulum (supplemental Fig. 6). Component annotations also included cytoplasm and mitochondria (due in part to the fact that most proteins have multiple annotations for each GO category), emphasizing that gene-based annotation is only a weak indicator of endogenous protein localization. There were also eight enriched biological processes: translation, transcription, cell adhesion, nucleosome assembly, membrane transport, cell differentiation, protein folding, and fatty acid metabolic process (supplemental Fig. 6). Overall, these annotations emphasize the importance of proteomic detection of proteins and biochemical characterization of function, rather than sole reliance on gene annotation.
Fig. 6.
Gene ontology-based annotation. GO terms for molecular function, biological process and cellular component were retrieved for all cardiac nuclear proteins and clustering performed using GO Term Mapper as described in Methods. Shown is the diagram for biological process. Node colors correspond to the number of proteins with a given annotation, whereas edges represent relationships between the different processes according to the GO hierarchy. supplemental Fig. 6 has similar clustering results for molecular function and cellular component. For each GO category, individual proteins often have multiple annotations.
DISCUSSION
The rigid and extensive contractile apparatus of the cardiomyocyte, which is anchored to both the plasma membrane and the nuclear envelope, as well as the abundance of mitochondria in these cells (∼40% of the cell volume (41, 42)) have provided significant hurdles to nuclear enrichment. Our goal for this study was to establish such an enrichment protocol for the study of cardiac nuclei, but in the process, it became apparent that subfractionation of the organelle was an essential step to comprehensive analysis. Thus, the present paper presents the methodology for enrichment of nuclei from heart suitable for proteomic analyses, along with subfractionation protocols to obtain chromatin, nucleoplasm, and acid-soluble proteins. That said, it is important to note that it is indeed enrichment that is carried out—not purification—because remnants of other organelles are still present in the preparation. Electron microscopy and Western blotting clearly lack the sensitivity (and are accompanied by their own experimental biases) to completely rule out the presence of other cellular components.
Nuclei were subfractioned into nucleoplasm, chromatin and acid-extracted fractions and, along with analysis of detergent solublized nuclei in toto. Not surprisingly, these fractions have shared and distinct constituents. One example of the strength of combining these approaches can be seen by examining the heterogeneous nuclear ribonucleoprotein (hnRNP) family of mRNA processing proteins. This family of proteins is mainly involved in mRNA protection, stability, splicing, and transport, requiring some members to remain in the nucleoplasm whereas others shuttle between nucleoplasm and cytoplasm (43–45). Recently some family members have also been found to participate in DNA-specific functions including DNA repair, telomere elongation, chromatin remodeling, and transcription (46–48). Of the 17 hnRNP proteins identified, 8 were exclusively identified in the nucleoplasm and 6 were identified in both chromatin and nucleoplasm (3 identified in AE and/or I). Five of the six proteins identified in both fractions have been reported to participate in RNA- and DNA-specific functions, constituting all variants identified with known DNA-regulatory roles. This unique compartmentalization supports the reported DNA- and RNA-regulatory functions of this family and suggests specific roles for the heretofore uncharacterized members.
The nuclear membrane is composed of an outer membrane, which is continuous with and thought to be indistinguishable from the rough endoplasmic reticulum, and an inner membrane, which houses a number of proteins facilitating interactions with lamins and chromatin. The nuclear membrane is perforated by nuclear pores, which have also been shown to interact with the nuclear lamina, enabling connection to the nuclear matrix and facilitating shuttling of factors imported through these pores. Until recently the role of nuclear pore proteins has been limited to membrane transport, however, recent investigations support a role of this complex to influence local gene expression (49). This functionality in the cardiomyocyte is supported by the presence of the nuclear pore protein, p62, in both nucleoplasm and chromatin fractions. In our proteomic analysis we identified 142 integral membrane proteins, the majority being exclusive to the nucleoplasm fraction (73%), however, 29 proteins were identified in both chromatin and nucleoplasm fractions, indicative of interactions between the nuclear envelope and chromatin.
Cardiac nuclei are intimately regulated by the cytoskeleton. The outer nuclear membrane is contiguous with the cytoskeleton that participates in movement of molecules from specialized domains of the cytoplasm into the nucleus. In addition, the myocyte contractile apparatus, which dominates the regions of the cell not dedicated to energy production (mitochondria) or calcium handling (sarcoplasmic reticulum), functionally couples to the nucleus to regulate gene expression by mechanical perturbation, referred to by some investigators (50) as “excitation-transcription coupling.” In agreement with these specialized functions of the nucleus, our proteomic dissection reveals 89 and 37 proteins known to also participate in cytoskeletal and contractile processes, respectively.
Observations on the Properties of the Cardiac Nuclear Proteome
Although informative and essential, no annotation strategy or bioinformatic approach can supplant experimentation. Our knowledge of proteomes, therefore, must ultimately yield to the results of well controlled experiments that measure proteins. Nevertheless, several critical observations emerge from bioinformatic analyses of the proteomic data obtained in this study. It has been postulated that proteins localized to the nucleus contain nuclear localization sequences necessary for their selective transport across the nuclear membrane. Many experimental examples of such domains exist (40, 51–53). Domain mapping of nuclear localization sequences in our data indicate that only 6.8% of proteins contain bipartite motifs, which are two regions of basic residues separated by ∼10 amino acids. A similar frequency of these sequences was observed in previous studies (37) of nuclei from heart (11.4%) as well as from brain (13.3%) and liver (17.9%) nuclei and was only slight higher than that seen in two studies of cardiac mitochondria (4.1% in (37) and 3.1% in (54)). These observations highlight the paucity of understanding how nuclear transport is controlled on a global level, i.e. whether (and if so, how) it is coded in the primary sequence.
Although only a small fraction of proteins identified in our study contained bipartite nuclear localization sequences (as mapped by ScanProsite) the identity of several of these proteins is intriguing. Three myosin isoforms (9, 10, and 11) contained possible nuclear localization sequences. Of the three isoforms only myosin-10 has been reported to have a nuclear-specific function, that of spindle assembly and nuclei positioning during meiosis as observed in Xenopus embryos (55). A nuclear specific myosin I isoform (identified in our study) has been found to contain an N-terminal motif necessary for its localization to the nuclear interior (56, 57).
Cardiac DNA is exceptionally heterochromatic, a feature reflected in various aspects of our analyses. A number of proteins involved in cellular differentiation and the inhibition of cell proliferation were identified in this study, including the transcriptional regulator p204 (58) and the histone methyltransferase Smyd1 (59), which have both been separately shown to be necessary for differentiation of embryonic progenitor cells into functional cardiomyocytes. A number of proteins involved in heterochromatin formation and transcriptional silencing were identified, including heterochromatin protein 1-binding protein 3 (60), methyl-CpG-binding protein 2 (61), and transcription intermediary factor 1-β (62). Although these three proteins have established roles in maintaining transcriptionally silenced chromatin regions, their roles in the cardiomyocyte have only begun to be explored (Bissonnette and colleagues examined Mecp2-null mice and revealed no alterations in cardiac phenotype (63)).
As discussed above, the heterochromatic state of cardiac DNA makes the execution of rapid and precise transcriptional responses particularly important: genes to be expressed must be rendered structurally accessible for transcriptional machinery and genes to be repressed must be maintained structurally inaccessible to prevent aberrant expression. Cross-referencing protein identification numbers (IPI) with gene identifiers (ref-seq/UCSC) allowed us to explore which regions of the genome contribute proteins to the nucleus (Fig. 4). Although our understanding of the structural constraints affecting global gene expression on a genome-wide scale is in its infancy, this type of analysis will contribute insights into the bidirectional relationship between the expressed proteome and genome structure, and how this in turn influences phenotype in different cell types.
Novel Histone Variants
Once thought to be inert chromatin structural proteins, histones have more recently been shown to function as transcriptional regulators. Several variants of the canonical histone family have been described—which are incorporated into chromatin during cell division—with additional inducible variants found to be expressed (and integrated into chromatin) in a replication-independent manner (64–66). Tissue and species specific variants have also been identified (67, 68). However, there remain a number of histone genes which have been cataloged in the mouse and human genome with no corresponding evidence for their expression at the protein level. This lack of identification may be because of low expression levels, unexplored tissue- or disease-specific expression, or the challenges associated with identifying histones by mass spectrometry. In general the identification of histones (at the protein level) has been impeded by their small size, which results in fewer peptides to detect in a data-dependent LC-MS/MS experiment, extensive modification (for example, detection of unmodified peptides from cardiac histone H3 is exceedingly rare in our hands, hence the reporting of modified peptides in supplemental Fig. 4) and the abundance of lysine and arginine residues, which interfere with enzymatic digestion prior to mass spectrometry. Using a combination of digestion protocols subsequent to the aforementioned subfractionation, we were able to identify five histone variants never before reported in the heart (four of which had never been detected at the protein level in any cell type, in addition to 12 other isoforms): H2A (IPI00665557), H2B (IPI00459318), H3.1t (IPI00277753), H3 (IPI00379693), and H3 (IPI00108200). Although the cataloging of their genetic loci predicted their existence, only H3.1t has ever been identified at the protein level (and this identification was in testes (69)) with an additional two having been previously identified in high-throughput mRNA screens. The human homolog of H3.1t was originally reported to exhibit testis-specific expression by RNA hybridization (70), but the transcript has since been identified in mouse brain and embryo by rt-PCR (71), although its function remains unknown. Additional variants, originally reported to have testis-specific expression (including H1t), have also been identified in somatic tissues. The implications of expression of these protein variants in the adult mouse heart open intriguing possibilities for DNA packaging and transcriptional regulation, to be further explored in future studies.
Proteomics has been employed to characterize whole cardiac tissue (72–76) as well as some cardiac organelles, including mitochondria (54, 77–84), and myofilaments (85, 86). Only one study of which we are aware has characterized intact nuclei from heart (37), and none have examined subnuclear compartments. Kislinger et al. performed a large-scale proteomic analysis of four organelles from six mouse tissues, including the cardiac nucleus, from which 1044 proteins were identified. UniProt-based comparison indicated 367 proteins in common between the Kislinger et al. paper and the current study. We attribute these differences primarily to the use of subfractionation in our study. Other methodological differences between our studies include trypsin and chymotrypsin separately (present study) versus trypsin and Lys-C combined (37), SDS-PAGE plus C18 for separation (present study) versus C18 and SCX (37) and nuances in peptide/protein identification.
Commercial kits have been marketed for enrichment of nuclei; however, when followed by rigorous biochemical and proteomic analyses, these kits exhibited a poor ability to enrich nuclear proteins (36). Furthermore, previous investigations showed that classical density centrifugation-based approaches (which were the basis for our work in the current study) performed better to enrich substructures of the nucleus (36). Whether hearts are fresh (as in the present study) or frozen (36) also plays a fundamental role in the ability to enrich individual organelles and should be taken into account when designing any proteomic investigation. Both approaches (fresh versus frozen) have relative strengths and weaknesses.
Summary and Outlook
In summary, we report proteomic dissection of the cardiac nucleus using a combination of subfractionation, multiple digestion enzymes, and high mass accuracy mass spectrometry. Our analyses identified proteins from all major compartments of the nucleus: outer nuclear membrane (ribosomes), inner nuclear membrane (importin, exportin), nuclear structural proteins (laminins, myosin), chromatin (both structural and regulatory; i.e. histones, chromatin remodeling), transcriptional machinery (transcription factors and regulators), splicing machinery (hnRNP, sNRNP), and inner nuclear structures including nucleolus, paraspeckles, and cajal bodies.
A consideration for the interpretation of this study is the use of intact hearts, rather than isolated cardiac myocytes, as the source of nuclei. Although cardiomyocytes make up 50%–70% of cardiac mass, the nuclear proteins we map are undoubtedly a mixture of proteins from cardiomyocytes and other cells present in the heart (including but not limited to fibroblasts, endothelial cells, and smooth muscle cells). Because the heart functions as an organ, and not as isolated cells, it can be viewed as an advantage to capture the proteomic information contained in all cells in the organ, rather than biasing for the contractile cells. Our rationale for examining the entire heart, then, was that we wanted to capture the nuclear proteome in a state as close as possible to that in vivo, which would be impossible were we to isolate adult cells by enzymatic dissociation. The reality is that both approaches (intact organ and isolated cell) must be taken to overcome the limitations of both. This discovery-based dissection of the basal cardiac nucleus will enable future investigations into the changes in proteome dynamics that accompany heart disease.
Acknowledgments
We thank Robb MacLellan for isolation of NRVMs and Marianne Cilluffo for assistance with EM analyses, both at UCLA.
Footnotes
* This study was supported by NHLBI grants and the Laubisch Endowment at UCLA (to TMV). SF is the recipient of an NHLBI Ruth Kirschstein Post-doctoral Fellowship.
 This article contains supplemental Figs. 1–8 and Tables 1–3.
This article contains supplemental Figs. 1–8 and Tables 1–3.
1 The abbreviations used are:
- NRVM
- neonatal rat ventricular myocytes
- LC-MS/MS
- liquid chromatography-tandem MS
- GO
- Gene Ontology
- ACN
- acetonitrile.
REFERENCES
- 1. Dauer W. T., Worman H. J. (2009) The nuclear envelope as a signaling node in development and disease. Dev. Cell 17, 626–638 [DOI] [PubMed] [Google Scholar]
- 2. Wang N., Tytell J. D., Ingber D. E. (2009) Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 [DOI] [PubMed] [Google Scholar]
- 3. Webster M., Witkin K. L., Cohen-Fix O. (2009) Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122, 1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cioce M., Lamond A. I. (2005) Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21, 105–131 [DOI] [PubMed] [Google Scholar]
- 5. Bond C. S., Fox A. H. (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Handwerger K. E., Gall J. G. (2006) Subnuclear organelles: new insights into form and function. Trends Cell Biol. 16, 19–26 [DOI] [PubMed] [Google Scholar]
- 7. Raska I., Shaw P. J., Cmarko D. (2006) Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 18, 325–334 [DOI] [PubMed] [Google Scholar]
- 8. Carmo-Fonseca M., Mendes-Soares L., Campos I. (2000) To be or not to be in the nucleolus. Nat Cell Biol. 2, E107–112 [DOI] [PubMed] [Google Scholar]
- 9. Stewart C. L., Roux K. J., Burke B. (2007) Blurring the boundary: the nuclear envelope extends its reach. Science 318, 1408–1412 [DOI] [PubMed] [Google Scholar]
- 10. Shaklai S., Amariglio N., Rechavi G., Simon A. J. (2007) Gene silencing at the nuclear periphery. FEBS J 274, 1383–1392 [DOI] [PubMed] [Google Scholar]
- 11. Walker C. A., Köppe M., Grenville-Briggs L. J., Avrova A. O., Horner N. R., McKinnon A. D., Whisson S. C., Birch P. R., van West P. (2008) A putative DEAD-box RNA-helicase is required for normal zoospore development in the late blight pathogen Phytophthora infestans. Fungal. Genet. Biol. 45, 954–962 [DOI] [PubMed] [Google Scholar]
- 12. Kostka M., Hampl V., Cepicka I., Flegr J. (2004) Phylogenetic position of Protoopalina intestinalis based on SSU rRNA gene sequence. Mol. Phylogene. Evol. 33, 220–224 [DOI] [PubMed] [Google Scholar]
- 13. Gladfelter A. S. (2006) Nuclear anarchy: asynchronous mitosis in multinucleated fungal hyphae. Curr. Opin. Microbiol. 9, 547–552 [DOI] [PubMed] [Google Scholar]
- 14. Philippsen P., Kaufmann A., Schmitz H. P. (2005) Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 8, 370–377 [DOI] [PubMed] [Google Scholar]
- 15. Richardson B. E., Nowak S. J., Baylies M. K. (2008) Myoblast fusion in fly and vertebrates: new genes, new processes and new perspectives. Traffic 9, 1050–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nowak S. J., Nahirney P. C., Hadjantonakis A. K., Baylies M. K. (2009) Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J. Cell Sci. 122, 3282–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li F., Wang X., Capasso J. M., Gerdes A. M. (1996) Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J. Mol. Cell Cardiol. 28, 1737–1746 [DOI] [PubMed] [Google Scholar]
- 18. MacLellan W. R., Schneider M. D. (2000) Genetic dissection of cardiac growth control pathways. Annu. Rev. Physiol. 62, 289–319 [DOI] [PubMed] [Google Scholar]
- 19. Olson E. N., Schneider M. D. (2003) Sizing up the heart: development redux in disease. Genes Dev. 17, 1937–1956 [DOI] [PubMed] [Google Scholar]
- 20. Molkentin J. D., Dorn G. W., 2nd (2001) Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 63, 391–426 [DOI] [PubMed] [Google Scholar]
- 21. Fishman M. C., Chien K. R. (1997) Fashioning the vertebrate heart: earliest embryonic decisions. Development 124, 2099–2117 [DOI] [PubMed] [Google Scholar]
- 22. Clubb F. J., Jr., Bishop S. P. (1984) Formation of binucleated myocardial cells in the neonatal rat. An index for growth hypertrophy. Lab. Invest. 50, 571–577 [PubMed] [Google Scholar]
- 23. Hwang S. I., Lundgren D. H., Mayya V., Rezaul K., Cowan A. E., Eng J. K., Han D. K. (2006) Systematic characterization of nuclear proteome during apoptosis: a quantitative proteomic study by differential extraction and stable isotope labeling. Mol. Cell. Proteomics 5, 1131–1145 [DOI] [PubMed] [Google Scholar]
- 24. Escobar M. A., Hoelz D. J., Sandoval J. A., Hickey R. J., Grosfeld J. L., Malkas L. H. (2005) Profiling of nuclear extract proteins from human neuroblastoma cell lines: the search for fingerprints. J. Pediatr. Surg. 40, 349–358 [DOI] [PubMed] [Google Scholar]
- 25. Buhr N., Carapito C., Schaeffer C., Kieffer E., Van Dorsselaer A., Viville S. (2008) Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. Electrophoresis 29, 2381–2390 [DOI] [PubMed] [Google Scholar]
- 26. Salzano A. M., Paron I., Pines A., Bachi A., Talamo F., Bivi N., Vascotto C., Damante G., Quadrifoglio F., Scaloni A., Tell G. (2006) Differential proteomic analysis of nuclear extracts from thyroid cell lines. J. Chromatogr. B Analyt. Technol Biomed. Life Sci. 833, 41–50 [DOI] [PubMed] [Google Scholar]
- 27. Turck N., Richert S., Gendry P., Stutzmann J., Kedinger M., Leize E., Simon-Assmann P., Van Dorsselaer A., Launay J. F. (2004) Proteomic analysis of nuclear proteins from proliferative and differentiated human colonic intestinal epithelial cells. Proteomics 4, 93–105 [DOI] [PubMed] [Google Scholar]
- 28. Barboro P., D'Arrigo C., Repaci E., Bagnasco L., Orecchia P., Carnemolla B., Patrone E., Balbi C. (2009) Proteomic analysis of the nuclear matrix in the early stages of rat liver carcinogenesis: identification of differentially expressed and MAR-binding proteins. Exp. Cell Res. 315, 226–239 [DOI] [PubMed] [Google Scholar]
- 29. Dreger M., Bengtsson L., Schöneberg T., Otto H., Hucho F. (2001) Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. U.S.A. 98, 11943–11948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 31. Lambert J. P., Mitchell L., Rudner A., Baetz K., Figeys D. (2009) A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell Proteomics 8, 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malik P., Korfali N., Srsen V., Lazou V., Batrakou D. G., Zuleger N., Kavanagh D. M., Wilkie G. S., Goldberg M. W., Schirmer E. C. (2010) Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell Mol. Life Sci. 67, 1353–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tweedie-Cullen R. Y., Reck J. M., Mansuy I. M. (2009) Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J. Proteome Res. 8, 4966–4982 [DOI] [PubMed] [Google Scholar]
- 34. Chu D. S., Liu H., Nix P., Wu T. F., Ralston E. J., Yates J. R., 3rd, Meyer B. J. (2006) Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature 443, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uchiyama S., Kobayashi S., Takata H., Ishihara T., Hori N., Higashi T., Hayashihara K., Sone T., Higo D., Nirasawa T., Takao T., Matsunaga S., Fukui K. (2005) Proteome analysis of human metaphase chromosomes. J. Biol. Chem. 280, 16994–17004 [DOI] [PubMed] [Google Scholar]
- 36. Murray C. I., Barrett M., Van Eyk J. E. (2009) Assessment of ProteoExtract subcellular fractionation kit reveals limited and incomplete enrichment of nuclear subproteome from frozen liver and heart tissue. Proteomics 9, 3934–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 38. Vondriska T. M., Zhang J., Song C., Tang X. L., Cao X., Baines C. P., Pass J. M., Wang S., Bolli R., Ping P. (2001) Protein kinase C epsilon-Src modules direct signal transduction in nitric oxide-induced cardioprotection: complex formation as a means for cardioprotective signaling. Circ. Res. 88, 1306–1313 [DOI] [PubMed] [Google Scholar]
- 39. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 40. Cokol M., Nair R., Rost B. (2000) Finding nuclear localization signals. EMBO Rep. 1, 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sommer JR, J. E (1979) Ultrastructure of cardiac muscle. In: Handbook of Physiololgy: The Cardiovascular System, pp. 113–186, Am Physiol Soc, Bethesda, MD [Google Scholar]
- 42. Anversa P., Vitali-Mazza L., Visioli O., Marchetti G. (1971) Experimental cardiac hypertrophy: a quantitative ultrastructural study in the compensatory stage. J. Mol. Cell Cardiol 3, 213–227 [DOI] [PubMed] [Google Scholar]
- 43. Brewer G. (1991) An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11, 2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Min H., Chan R. C., Black D. L. (1995) The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9, 2659–2671 [DOI] [PubMed] [Google Scholar]
- 45. Vautier D., Chesné P., Cunha C., Calado A., Renard J. P., Carmo-Fonseca M. (2001) Transcription-dependent nucleocytoplasmic distribution of hnRNP A1 protein in early mouse embryos. J. Cell Sci. 114, 1521–1531 [DOI] [PubMed] [Google Scholar]
- 46. LaBranche H., Dupuis S., Ben-David Y., Bani M. R., Wellinger R. J., Chabot B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19, 199–202 [DOI] [PubMed] [Google Scholar]
- 47. Ritchie S. A., Pasha M. K., Batten D. J., Sharma R. K., Olson D. J., Ross A. R., Bonham K. (2003) Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: implications in the regulation of SRC1A transcription. Nucleic Acids Res. 31, 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iwanaga K., Sueoka N., Sato A., Hayashi S., Sueoka E. (2005) Heterogeneous nuclear ribonucleoprotein B1 protein impairs DNA repair mediated through the inhibition of DNA-dependent protein kinase activity. Biochem. Biophys. Res. Commun. 333, 888–895 [DOI] [PubMed] [Google Scholar]
- 49. Taddei A. (2007) Active genes at the nuclear pore complex. Curr. Opin. Cell Biol. 19, 305–310 [DOI] [PubMed] [Google Scholar]
- 50. Wu X., Bers D. M. (2006) Sarcoplasmic reticulum and nuclear envelope are one highly interconnected Ca2+ store throughout cardiac myocyte. Circ. Res. 99, 283–291 [DOI] [PubMed] [Google Scholar]
- 51. Felin J. E., Mayo J. L., Loos T. J., Jensen J. D., Sperry D. K., Gaufin S. L., Meinhart C. A., Moss J. B., Bridgewater L. C. (2010) Nuclear variants of bone morphogenetic proteins. BMC Cell Biol. 11, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jerke U., Tkachuk S., Kiyan J., Stepanova V., Kusch A., Hinz M., Dietz R., Haller H., Fuhrman B., Dumler I. (2009) Stat1 nuclear translocation by nucleolin upon monocyte differentiation. PLoS One 4, e8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Christophe D., Christophe-Hobertus C., Pichon B. (2000) Nuclear targeting of proteins: how many different signals? Cell Signal.12, 337–341 [DOI] [PubMed] [Google Scholar]
- 54. Zhang J., Li X., Mueller M., Wang Y., Zong C., Deng N., Vondriska T. M., Liem D. A., Yang J. I., Korge P., Honda H., Weiss J. N., Apweiler R., Ping P. (2008) Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics 8, 1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weber K. L., Sokac A. M., Berg J. S., Cheney R. E., Bement W. M. (2004) A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature 431, 325–329 [DOI] [PubMed] [Google Scholar]
- 56. Pestic-Dragovich L., Stojiljkovic L., Philimonenko A. A., Nowak G., Ke Y., Settlage R. E., Shabanowitz J., Hunt D. F., Hozak P., de Lanerolle P. (2000) A myosin I isoform in the nucleus. Science 290, 337–341 [DOI] [PubMed] [Google Scholar]
- 57. Nowak G., Pestic-Dragovich L., Hozák P., Philimonenko A., Simerly C., Schatten G., de Lanerolle P. (1997) Evidence for the presence of myosin I in the nucleus. J. Biol. Chem. 272, 17176–17181 [DOI] [PubMed] [Google Scholar]
- 58. Ding B., Liu C. J., Huang Y., Hickey R. P., Yu J., Kong W., Lengyel P. (2006) p204 is required for the differentiation of P19 murine embryonal carcinoma cells to beating cardiac myocytes: its expression is activated by the cardiac Gata4, Nkx2.5, and Tbx5 proteins. J. Biol. Chem. 281, 14882–14892 [DOI] [PubMed] [Google Scholar]
- 59. Gottlieb P. D., Pierce S. A., Sims R. J., Yamagishi H., Weihe E. K., Harriss J. V., Maika S. D., Kuziel W. A., King H. L., Olson E. N., Nakagawa O., Srivastava D. (2002) Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 31, 25–32 [DOI] [PubMed] [Google Scholar]
- 60. Smallwood A., Black J. C., Tanese N., Pradhan S., Carey M. (2008) HP1-mediated silencing targets Pol II coactivator complexes. Nat. Struct. Mol. Biol. 15, 318–320 [DOI] [PubMed] [Google Scholar]
- 61. Skene P. J., Illingworth R. S., Webb S., Kerr A. R., James K. D., Turner D. J., Andrews R., Bird A. P. (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Riclet R., Chendeb M., Vonesch J. L., Koczan D., Thiesen H. J., Losson R., Cammas F. (2009) Disruption of the interaction between transcriptional intermediary factor 1{beta} and heterochromatin protein 1 leads to a switch from DNA hyper- to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol. Biol. Cell 20, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bissonnette J. M., Knopp S. J., Maylie J., Thong T. (2007) Autonomic cardiovascular control in methyl-CpG-binding protein 2 (Mecp2) deficient mice. Auton. Neurosci. 136, 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mannironi C., Bonner W. M., Hatch C. L. (1989) H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3′ processing signals. Nucleic Acids Res. 17, 9113–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hatch C. L., Bonner W. M. (1990) The human histone H2A.Z gene. Sequence and regulation. J. Biol. Chem. 265, 15211–15218 [PubMed] [Google Scholar]
- 66. Brush D., Dodgson J. B., Choi O. R., Stevens P. W., Engel J. D. (1985) Replacement variant histone genes contain intervening sequences. Mol. Cell. Biol. 5, 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Krieg P. A., Robins A. J., D'Andrea R., Wells J. R. (1983) The chicken H5 gene is unlinked to core and H1 histone genes. Nucleic Acids Res. 11, 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Busslinger M., Barberis A. (1985) Synthesis of sperm and late histone cDNAs of the sea urchin with a primer complementary to the conserved 3′ terminal palindrome: evidence for tissue-specific and more general histone gene variants. Proc. Natl. Acad. Sci. U.S.A. 82, 5676–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Garcia B. A., Thomas C. E., Kelleher N. L., Mizzen C. A. (2008) Tissue-specific expression and post-translational modification of histone H3 variants. J. Proteome Res. 7, 4225–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Albig W., Ebentheuer J., Klobeck G., Kunz J., Doenecke D. (1996) A solitary human H3 histone gene on chromosome 1. Hum. Genet. 97, 486–491 [DOI] [PubMed] [Google Scholar]
- 71. Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., Thévenon J., Catena R., Davidson I., Garin J., Khochbin S., Caron C. (2007) Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Isserlin R., Merico D., Alikhani-Koupaei R., Gramolini A., Bader G. D., Emili A. Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics 10, 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gramolini A. O., Kislinger T., Alikhani-Koopaei R., Fong V., Thompson N. J., Isserlin R., Sharma P., Oudit G. Y., Trivieri M. G., Fagan A., Kannan A., Higgins D. G., Huedig H., Hess G., Arab S., Seidman J. G., Seidman C. E., Frey B., Perry M., Backx P. H., Liu P. P., MacLennan D. H., Emili A. (2008) Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Mol. Cell Proteomics 7, 519–533 [DOI] [PubMed] [Google Scholar]
- 74. Cutillas P. R., Vanhaesebroeck B. (2007) Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol. Cell Proteomics 6, 1560–1573 [DOI] [PubMed] [Google Scholar]
- 75. Cagney G., Park S., Chung C., Tong B., O'Dushlaine C., Shields D. C., Emili A. (2005) Human tissue profiling with multidimensional protein identification technology. J. Proteome Res. 4, 1757–1767 [DOI] [PubMed] [Google Scholar]
- 76. Bousette N., Kislinger T., Fong V., Isserlin R., Hewel J., Emili A., Gramolini A. (2009) Large scale characterization and analysis of the murine cardiac proteome. J. Proteome Res. 8, 1887–1901 [DOI] [PubMed] [Google Scholar]
- 77. Bugger H., Chen D., Riehle C., Soto J., Theobald H. A., Hu X. X., Ganesan B., Weimer B. C., Abel E. D. (2009) Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58, 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Boja E. S., Phillips D., French S. A., Harris R. A., Balaban R. S. (2009) Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J. Proteome Res. 8, 4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Agnetti G., Kaludercic N., Kane L. A., Elliott S. T., Guo Y., Chakir K., Samantapudi D., Paolocci N., Tomaselli G. F., Kass D. A., Van Eyk J. E. (2010) Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ. Cardiovasc. Genet. 3, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wong R., Aponte A. M., Steenbergen C., Murphy E. Cardioprotection leads to novel changes in the mitochondrial proteome. Am. J. Physiol. Heart Circ Physiol 298, H75–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang J., Liem D. A., Mueller M., Wang Y., Zong C., Deng N., Vondriska T. M., Korge P., Drews O., Maclellan W. R., Honda H., Weiss J. N., Apweiler R., Ping P. (2008) Altered proteome biology of cardiac mitochondria under stress conditions. J. Proteome Res. 7, 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 83. Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 84. Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. (2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 [DOI] [PubMed] [Google Scholar]
- 85. Yin X., Cuello F., Mayr U., Hao Z., Hornshaw M., Ehler E., Avkiran M., Mayr M. (2010) Proteomics analysis of the cardiac myofilament subproteome reveals dynamic alterations in phosphatase subunit distribution. Mol. Cell Proteomics 9, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yuan C., Sheng Q., Tang H., Li Y., Zeng R., Solaro R. J. (2008) Quantitative comparison of sarcomeric phosphoproteomes of neonatal and adult rat hearts. Am. J. Physiol. Heart Circ Physiol 295, H647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]