Abstract
We present a comprehensive analysis of the human methyltransferasome. Primary sequences, predicted secondary structures, and solved crystal structures of known methyltransferases were analyzed by hidden Markov models, Fisher-based statistical matrices, and fold recognition prediction-based threading algorithms to create a model, or profile, of each methyltransferase superfamily. These profiles were used to scan the human proteome database and detect novel methyltransferases. 208 proteins in the human genome are now identified as known or putative methyltransferases, including 38 proteins that were not annotated previously. To date, 30% of these proteins have been linked to disease states. Possible substrates of methylation for all of the SET domain and SPOUT methyltransferases as well as 100 of the 131 seven-β-strand methyltransferases were surmised from sequence similarity clusters based on alignments of the substrate-specific domains.
A significant percentage of proteins across all organisms are enzymes that catalyze the transfer of a methyl group from the cofactor S-adenosylmethionine to a substrate (1–5). In yeast, these proteins make up about 1.2% of all gene products (6, 7). The ability of methyltransferases to use a variety of different substrates, including RNA, DNA, lipids, small molecules, and proteins, is responsible for their diverse roles in different biological pathways (1–3). Methyltransferases have been shown to be essential in epigenetic control, lipid biosynthesis, protein repair, hormone inactivation, and tissue differentiation (8–14). The identification of new enzymes may allow the delineation of additional pathways and modes of regulation as well as increase our understanding of S-adenosylmethionine metabolism.
Although there are hundreds of known substrates for these reactions, methyltransferases are found in a small number of distinct structural arrangements that are used to classify them into superfamilies (2, 3, 5, 6, 15). Proteins in each superfamily also share conserved amino acid sequences. The seven-β-strand superfamily (also referred to as “Class I” methyltransferases) is the most abundant. These proteins catalyze a wide array of substrates and feature a Rossmann-like structural core (2, 3, 5, 6, 15). The SPOUT methyltransferase superfamily contains a distinctive knot structure and methylates RNA substrates (16). SET domain methyltransferases catalyze the methylation of protein lysine residues with histones and ribosomal proteins as major targets (17–19). Smaller superfamilies with at least one three-dimensional structure available include the precorrin-like methyltransferases (20), the radical SAM1 methyltransferases (21, 22), the MetH activation domain (23), the Tyw3 protein involved in wybutosine synthesis (24), and the homocysteine methyltransferases (25–27). Lastly, an integral membrane methyltransferase family has been defined by sequence alone where no three-dimensional structure is yet available (28, 29).
Advances in computational resources and the availability of increased numbers of three-dimensional structures have allowed the formulation of sequence- and structure-based models describing each methyltransferase superfamily and consequently the prediction of additional members (4, 5, 6, 7, 16). Here, we applied these methods to uncover the entire human methyltransferasome. Statistical profiles were generated from the refined domains of each S-adenosylmethionine-dependent methyltransferase superfamily. Hidden Markov models (HMM) and Fisher-based matrices were created to describe the primary sequence and secondary structures of these methyltransferase domains and were utilized in the computational programs HHpred (30), FHMMER (31), and Multiple Motif Scanning (5) to determine which human proteins align with the models (5, 6). Questionable matches were evaluated by a fold recognition program, PHYRE (32), to create full structural predictions and ensure these proteins share structural homology to solved structures of the known methyltransferases. Additionally, we predicted the substrates of methylation for many of these proteins by cluster analysis utilizing substrate-specific domains.
EXPERIMENTAL PROCEDURES
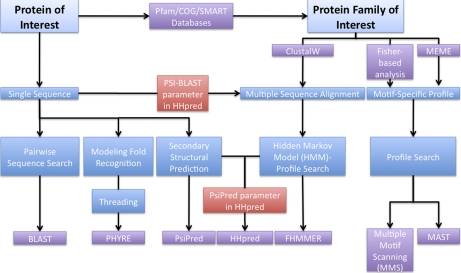
Fig. 1 and supplemental Table I summarize the bioinformatics approaches we used to identify new human methyltransferases. The methodology used for each superfamily depended on the abundance of information known on the specific family. For families represented by only one known protein, its sequence was used to extract other potential members through protein family databases Pfam (33), COG (34), and SMART (35) and/or HHpred using the PSI-BLAST parameter (30) (supplemental Table I). All identified proteins were cross-checked with HHpred against its embedded non-redundant database of proteins from all organisms available to confirm that the nearest homolog was a known or putative methyltransferase (30). All HHpred searches were conducted using the PsiPred parameter to include secondary structural predictions, and all protein alignments of the COG and SMART family databases were done using ClustalW (36).
Fig. 1.
Bioinformatics approaches for searching proteome for methyltransferase family members. Detecting putative methyltransferases requires multiple bioinformatics approaches depending on the existing information for each reference group of superfamily members. An overall scheme of these methods is depicted here; a fuller description is given in the text. Previous bioinformatics searches for novel methyltransferases have used the MEME and MAST programs (4).
Seven-β-strand Methyltransferases
We previously defined a “yeast reference set” profile based on the primary sequence and secondary structural characteristics of the amino acids within the signature motifs of these enzymes (5). This profile was used against the non-redundant human proteome database using the programs HHpred with the PsiPred parameter and FHMMER (supplemental Table I). To confirm that the methyltransferase domain was adequately represented by the inherently non-redundant yeast seven-β-strand methyltransferasome, the “crystal reference set” profile (5) was used as a secondary input with these programs. This profile includes methyltransferases from all non-yeast organisms with solved structures to ensure proper domain/motif identification; these enzymes are diverse in both function and organismal source (5). Both of these reference set profiles were also used as inputs into the Multiple Motif Scanning program (MMS; Ref. 5) against the human proteome.
Two subvariants of the seven-β-strand domain have been described in yeast (37–39). Both variants appear to recognize the adenosine N6 group. One group, including the Ime4 and Kar4 proteins, form a “rearranged motifs” domain (2), whereas a second group, represented by the YGR001C protein, lacks Motif I. These three proteins were independently tested using HHpred with the PSI-BLAST parameter to compile profiles including proteins from all organisms (supplemental Table I). Additionally, the COG database contains a group of the “rearranged domain” proteins from a variety of organisms (COG4725: IME4-transcription activator, adenine-specific DNA methyltransferase); this profile was used as input in the HHpred against the human proteome.
SET Methyltransferases
In addition to the SET domain, these enzymes can contain supplemental domains that vary markedly among the SET proteins. Therefore, the profile used in the bioinformatics search for all SET proteins included only the amino acids within the SET domain. The input profile was obtained from the SMART database (family SM00317), which includes SET domains in proteins from all organisms. HHpred was used to run the profile against the human proteome with the PsiPred secondary structural parameter. Additionally, this profile was used in an FHMMER search to find additional members of the superfamily (supplemental Table I).
SPOUT Methyltransferases
Although SPOUT methyltransferases display structural similarity, we were unable to establish one profile based on primary and secondary sequence information alone. Initial tries to generate one profile containing Motifs 1, Post 1, 2, and 3 from either the alignments presented in Ref. 16 or the ClustalW alignments of an updated list of SPOUT proteins were unsuccessful in detecting all SPOUT yeast proteins via HHpred runs against the yeast proteome despite the fact that both reference groups contained those proteins (supplemental Table II). Therefore, COG groups identified as SPOUT methyltransferases in Ref. 16 were independently used in searches via FHMMER and HHpred with the PsiPred parameter with either the exact alignment of the methyltransferase domain presented in Ref. 16 or the ClustalW alignment of the proteins in these COG groups using the full protein sequence (supplemental Table II). To confirm structural homology, all identified human SPOUT methyltransferases were modeled in the fold recognition program PHYRE by threading analysis (supplemental Table I).
Smaller Superfamilies
For families with a single representative, the sequence of the protein was used to develop a profile composed of sequences from all organisms using the PSI-BLAST feature of HHpred. The resulting profile was then used in a search against the human proteome. Each sequence was also entered into the Pfam, COG, or SMART database to determine whether there is a reference set of proteins available to represent the superfamily that could be used as an input in a separate HHpred search against the human proteome. The outputs of protein matches from these searches were evaluated, and proteins with questionable matches were analyzed using the threading program PHYRE to model full structural predictions in a search against all available Protein Data Bank structures. These procedures are summarized in supplemental Table I.
To search for precorrin-like methyltransferases, the program HHpred with the PSI-BLAST parameter was used with the input sequences of the single CbiF sequence of the precorrin-4 C11 methyltransferase from Bacillus megaterium or the multiple alignment of the COG1798 group (DPH5, diphthamide biosynthesis methyltransferase). The input profile for the membrane superfamily was the multiple alignment of PF04191 Pfam group (PEMT, phospholipid methyltransferases). The human family of homocysteine methyltransferases was determined using input sequences of the methionine synthetase from Escherichia coli or the multiple alignment of the COG0646 group (MetH, methionine synthase I (cobalamin-dependent), methyltransferase domain). To search for proteins with the MetH activation domain, the program HHpred with the PSI-BLAST parameter was used with the single input sequence of the C-terminal domain of E. coli MetH. Additionally, use of the fold recognition program PHYRE with the same sequence indicated that no other proteins have significant homology to MetH in the human proteome. Radical SAM methyltransferases were defined by an HHpred search against the human proteome using an input of the multiple alignment generated by ClustalW of proteins identified by the keyword search “radical SAM methyltransferase” in the RefSeq database. Finally, Tyw3-like methyltransferases were identified from the single input sequence of the Saccharomyces cerevisiae Tyw3 wybutosine tRNA methyltransferase using HHpred with the PSI-BLAST parameter. The fold recognition program PHYRE with the same sequence indicated that no other proteins have significant homology to the Tyw3 protein in the human proteome or the Protein Data Bank.
Eliminating Database Redundancy
To ensure a non-redundant human methyltransferasome, all proteins identified in the different search algorithms were matched with Swiss-Prot or TrEMBL accession numbers (40). For the few protein species that lacked these identification numbers, International Protein Index identification numbers were assigned (41). After elimination of duplicate accession numbers, proteins with non-redundant identifiers were then mapped to their chromosomal location through GeneALaCart (www.genecards.org; Refs. 42 and 43). In the event that multiple identifiers were mapped to the same chromosomal location, the species with either the highest UniProt version number and/or the most recent date of sequence was selected. Those species were then identified as members in the non-redundant methyltransferasome.
Protein Subfamily and Ortholog Search of Seven-β-strand Methyltransferases
Protein sequences were entered into the CLANS program for a two- or three-dimensional visualization of sequence similarity clusters (44). The degree of similarity between yeast and human homologs were assessed through BLAST searches of yeast proteins against the non-redundant human protein database set.
Substrate Specificity Search for SET Domain and SPOUT Methyltransferases
Groups of methyltransferases with known substrates were entered in CLANS to test which human protein clusters with the known groups. SET domain methyltransferase groups were obtained from classes described in Ref. 6, proteins from all organisms as described by UniProt (40), subfamilies defined by UniProt (40), and human proteins that were manually identified. SPOUT methyltransferase groups were clustered with yeast SPOUT methyltransferases (6) and with all members within the SPOUT UniProt subfamilies (40).
RESULTS AND DISCUSSION
Human Methyltransferasome
We wanted to identify all of the S-adenosylmethionine-dependent methyltransferases that are encoded in the human genome. This was approached by bioinformatics analyses of the human proteome with respect to each of the known structural superfamilies of these enzymes. As described under “Experimental Procedures,” the specific methods utilized to detect methyltransferases in the human proteome differed for each superfamily depending on the degree of sequence and structural similarity between family members and the availability of three-dimensional structures. Briefly, HMM profiles were created for each superfamily to search the human protein database with the computational program HHpred (30). In certain superfamilies, additional searches were performed using FHMMER (31), and Fisher-based matrices were developed to search using MMS (5). These approaches are summarized in supplemental Table I. The establishment of sets of reference proteins is described under “Experimental Procedures”; seven-β-strand methyltransferases are obtained from Ref. 5, whereas the rearranged and Motif I-less seven-β-strand, SET domain, precorrin-like, membrane, and homocysteine methyltransferases have well defined superfamily databases that are readily available (33–35). Single protein sequences were also used as probes to retrieve additional superfamily members through PSI-BLAST searches within the HHpred search program. Each putative methyltransferase was cross-checked using HHpred against a comprehensive database containing proteins from all organisms to ensure the closest known homolog is a putative or known methyltransferase (30). Proteins that still remained in question were analyzed by PHYRE (32) to predict their structure and ensure their similarity to known methyltransferases. To eliminate redundancy in the output, proteins were mapped to their chromosome location (see “Experimental Procedures”).
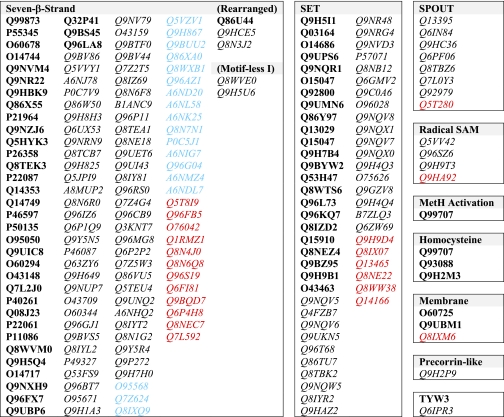
Overall, we found 208 proteins that make up the human methyltransferasome, equating to ∼0.9% of all human gene products. Of these proteins, 31% are currently “known” methyltransferases, whereas 38 proteins have not been annotated previously as methyltransferases. 30% of the methyltransferases are associated with disorders, most frequently cancer and mental disorders (see below). A non-redundant list of all S-adenosylmethionine-dependent methyltransferases in humans is shown in Table I; the functions of the known proteins are given in supplemental Table III. It is possible that additional methyltransferases are present in the human proteome; if so, they may represent new structural families or proteins that have markedly diverged from known enzymes.
Table I. The human methyltransferasome.
Proteins are listed by their UniProtKB identification number and categorized by their methyltransferase superfamily. Proteins with confirmed functional evidence are in bold, proteins that are already designated as putative methyltransferases are in italics, and proteins that are now annotated as methyltransferases are in color. Specifically, proteins in blue are designated as “unknown” in UniProtKB yet contain suspected methyltransferases through previous domain searches, whereas proteins in red are newly discovered as methyltransferases.
Identification of Human Methyltransferase Superfamily Members
In previous work, we developed profiles of the seven-β-strand methyltransferase motifs in the yeast reference set based on redefined primary and secondary structural analysis of the motifs (5). We used these aligned motif sequences to search for novel superfamily members in a non-redundant human proteome database using FHMMER (31) and HHpred (30). The “crystal and yeast matrix” sets were analyzed by the MMS (5) to identify additional candidate seven-β-strand methyltransferases. The known and putative seven-β-strand members are now presented in Table I; this list includes 30 previously unannotated proteins.
To our surprise, this analysis led to the discovery of an additional yeast protein (Dre2) that contains seven-β-strand signature methyltransferase motifs. The scan of the human proteome for methyltransferases brought up CIAPIN1, the anamorsin protein that is a cytokine-induced apoptosis inhibitor (UniProtKB accession number Q6FI81). This protein is a homolog of yeast Dre2. The HHpred search of the N-terminal region of Dre2 and CIAPIN1 showed that it has sequence similarity to Motifs II and III of the seven-β-strand methyltransferase domain but lacks Motifs I and Post I. A PHYRE search confirmed that this structural prediction is consistent with the sequence-based search. However, it remains to be seen whether Dre2 is an active methyltransferase.
A variant type of seven-β-strand methyltransferase having a distinct sequence pattern although maintaining the overall three-dimensional structure has been described in yeast (2, 3, 6, 15). One subgroup is characterized by the yeast Ime4 and Kar4 proteins where Motif I follows Motif III (37, 38). A second subgroup is characterized by the yeast YGR001C protein, which lacks Motif I (39). To assess the relationship between these proteins and discover novel human members, the sequences of yeast proteins Ime4, Kar4, and the methyltransferase domain of YGR001C were independently used in a PSI-BLAST search to gather more members in each of these subgroups. Each of these three profiles was searched against the human database using HHpred. Ime4 and Kar4 retrieved the same group of human proteins and are now defined as the “rearranged” seven-β-strand methyltransferases (Table I). These results were confirmed by an HHpred search using the corresponding COG group COG4725. The YGR001C-built profile search in HHpred resulted in a separate group of proteins that are now defined as the “Motif I-less” seven-β-strand proteins (Table I).
Although the SPOUT superfamily methyltransferases have similar structural folds, sequence similarity within the SPOUT domain itself is very weak (16). We were unable to output a single, comprehensive list of SPOUT methyltransferases from one profile search using primary and secondary sequence information alone. Initial tries to generate this profile began with the full sequence ClustalW alignments of every COG member identified as SPOUT methyltransferases (16). However, the HHpred and FHMMER searches using this profile against the human proteome were unsuccessful in detecting all human SPOUT proteins. Therefore, an alternative profile was developed with these proteins using only the sequences within the methyltransferase domain that are cited as being common throughout all superfamily members. However, the same results were observed; this analysis only retrieved three of the eight human SPOUT methyltransferases: TARBP1 (UniProtKB accession number Q13395), MRM1 (UniProtKB accession number Q6IN84), and RNMTL1 (UniProtKB accession number Q9HC36) (supplemental Table II). To solve this problem, we performed several profiles searches, each derived from a SPOUT methyltransferase COG family. Through this analysis, we were able to collect all of the human SPOUT methyltransferase members, including the newly identified C9orf114 (UniProtKB accession number Q5T280). All of these human SPOUT methyltransferases contain the SPOUT structural folding as predicted by the fold recognition program PHYRE (32) (supplemental Table II). Interestingly, the SPOUT superfamily is the only methyltransferase superfamily where a comprehensive list of enzymes within a superfamily cannot be generated from a single sequence-based methyltransferase domain search.
Unlike the situation for the SPOUT methyltransferases, the sequences for the SET domain methyltransferases are well conserved (6, 17, 45). A reference group of these SET domain sequences (SM00317) was obtained from the SMART database (35) to search the human proteome using HHpred and FHMMER. The 57 human SET methyltransferases discovered are presented in Table I and supplemental Table III; this group includes six species not annotated previously as methyltransferases.
The remaining six superfamilies are presently not as well defined as the five groups described above and make up less than 10% of the total number of putative methyltransferases (Table I and supplemental Table III).
The single sequence of CbiF (UniProtKB accession number O87696) was used as a probe to build the reference group of precorrin-like methyltransferases through the PSI-BLAST parameter in HHpred. Through this search, DPH5 (UniProtKB accession number Q9H2P9) was the only human protein classified in the precorrin-like methyltransferases superfamily. This was additionally confirmed by the HHpred search using the multiple alignment of sequences corresponding to COG group COG1798 (DPH5, diphthamide biosynthesis methyltransferase) against the human proteome.
Three human proteins of the membrane-bound methyltransferase superfamily were detected from the HHpred scan of the human proteome using the multiple alignment of the Pfam PEMT reference group (Pfam family PF04191). These proteins are ICMT (UniProtKB accession number O60725), PEMT (UniProtKB accession number Q9UBM1), and NRM (UniProtKB accession number Q8IXM6) (Table I). Interestingly, NRM has not been described previously as a potential methyltransferase, although it is very similar to ICMT (HHpred p value = 9.8 × 10−13).
As expected, BHMT (UniProtKB accession number Q93088), BHMT2 (UniProtKB accession number Q9H2M3), and MTR (UniProtKB accession number Q99707) were detected as homocysteine methyltransferase family members. These three proteins were found by the HHpred search of the single sequence of the N-terminal sequence of MetH (UniProtKB accession number P13009 (residues 2–325)) using the PSI-BLAST parameter. These proteins were confirmed by the HHpred search using the multiple alignment of its corresponding COG group COG0646 (MetH, methionine synthase I (cobalamin-dependent), methyltransferase domain).
The only human protein detected to have a “MetH activation domain” is MTR (UniProtKB accession number Q99707). Several different bioinformatics approaches were used to confirm this conclusion. Initially, the single sequence of the C terminus of MetH (UniProtKB accession number P13009 (residues 897–1227)) was used to build its superfamily profile through PSI-BLAST and scan the human proteome in HHpred. To search for proteins that may exhibit homology only on the structural level, the fold recognition program PHYRE (32) was tested using the same sequence of MetH indicated above. The results of these methods confirmed that the C-terminal domain of MetH is unique by both sequence and structure from any other protein in the human proteome.
Gathering a reference database of the radical SAM methyltransferase family was difficult because the sequence similarity between these enzymes and radical SAM non-methyltransferases is very high; thus, most family databases lump these proteins into one group. Therefore, the keywords radical SAM methyltransferase were searched in the RefSeq database (46) to create our desired superfamily reference group, and a ClustalW alignment of proteins was used to search against the human proteome by HHpred. Four proteins were identified as most similar to this radical SAM methyltransferase profile (Table I and supplemental Table III).
The search for additional human protein members in the TYW3 superfamily still only retrieved the human ortholog (UniProtKB accession number Q6IPR3). This HHpred search was performed by creating a superfamily profile from yeast Tyw3 protein with the PSI-BLAST parameter. Additionally, we used this superfamily profile to search against the Protein Data Bank to see whether any protein structures have been solved to date that match the description of this superfamily. This search flagged four unannotated proteins: PH1069 in Pyrococcus horikoshii (UniProtKB accession number O58796; Protein Data Bank code 2IT2), AF2059 in Archaeoglobus fulgidus (UniProtKB accession number O28220; Protein Data Bank code 2QG3), UPF0130 in Aeropyrum pernix (UniProtKB accession number Q9YDV3; Protein Data Bank code 2DVK), and SSO0622 in Sulfolobus solfataricus (UniProtKB accession number Q9UX16; Protein Data Bank code 1TLJ). The similarity value of these proteins compared with the profile equaled 0, indicating that they are orthologs of Tyw3. To confirm that the tRNA wybutosine-synthesizing protein is structurally unique from all other methyltransferases, a PHYRE search using the human TYW3 protein resulted in no matches with any other proteins.
The complete non-redundant list of all S-adenosylmethionine-dependent methyltransferases in humans is shown in Table I.
Comparison of Yeast and Human Methyltransferasomes
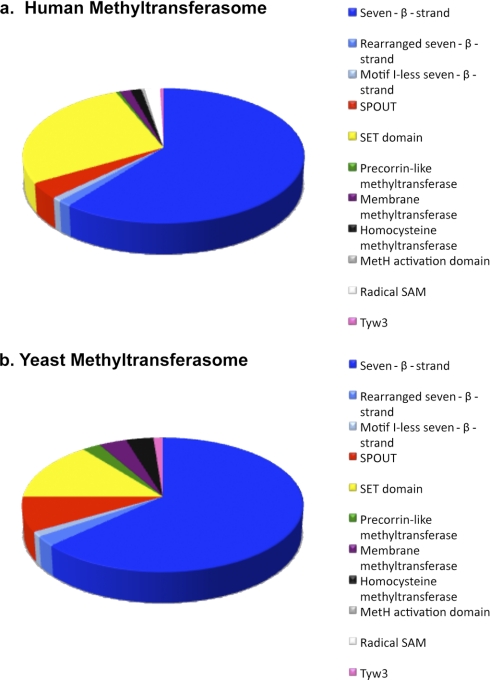
The human methyltransferasome identified here includes 208 known and putative members, comprising about 0.9% of protein open reading frames. In contrast, the yeast methyltransferasome includes some 81 species, or about 1.2% of open reading frames (6). The distribution of these proteins among superfamilies, however, differs between the two species (7) (Fig. 2). In both organisms, the majority of the methyltransferases fall into the seven-β-strand family (60% in human and 63% in yeast). However, the second most abundant superfamily, SET domain methyltransferases, makes up 27% of the human methyltransferasome compared with only 14% of the yeast methyltransferasome. This is due to the presence of a large partially redundant group of histone methyltransferases in humans as well as to the presence of subfamilies in humans not found in yeast (see below). The increase in histone methyltransferases may reflect the greater importance of epigenesis in humans; yeast does not have DNA methylation, although histone methylation does occur (6, 47–49).
Fig. 2.
Composition of methyltransferasome. Proteins in the human (a) and yeast (b) methyltransferasomes are sorted into their superfamilies. The composition of the human methyltransferasome was taken from Table I; the yeast methyltransferasome was obtained from Ref. 6. The radical SAM proteins identified previously in yeast (6) were not included here because they are more closely related to radical SAM non-methyltransferases than to radical SAM methyltransferases.
There are about 4 times the number of open reading frames in humans than in yeast (50). This increase is reflected in an increase in the number of seven-β-strand and SET domain methyltransferases. However, the number of human methyltransferases in the other superfamilies is often not much greater or is even less than those of the corresponding yeast superfamilies (7) (Fig. 2). For example, there is only one more methyltransferase in the human SPOUT and precorrin-like superfamilies than in yeast, and there are an equal number of membrane and homocysteine methyltransferase superfamily members in both organisms. Additionally, there is no evidence for members of the radical SAM methyltransferase superfamily in yeast (6, 7). Although there were four proteins that were detected with a radical SAM domain, all of them have homologs in other organisms that had non-methyltransferase functions. Additionally, sequence searches and fold recognition programs did not detect any proteins with a MetH activation domain in yeast (6, 7).
Many of the yeast and human methyltransferases share the same substrates. The human methyltransferases in the smaller superfamilies (not including the seven-β-strand, SPOUT, and SET domain groups) have at least one well defined ortholog where the function has been demonstrated. The exceptions to this include the radical SAM methyltransferases that have functionally defined homologs in prokaryotes and the membrane methyltransferase nurim protein (UniProtKB accession number Q8IXM6) with an unknown function (51). As for the larger methyltransferase superfamilies, further bioinformatics analysis (described below) was used to determine more information about the substrates of methylation and degree of similarity between the yeast and human proteins.
Analysis of Potential Substrates for Human Seven-β-Strand Species
To date, little insight is available into the substrates or physiological roles of many of the proteins already designated as “putative” methyltransferases. Here, we used sequence and structural similarity of unknown and known methyltransferases in multiple organisms to reveal possible similarities in functions. Specifically, human methyltransferases were grouped with yeast proteins in sequence similarity networks to provide this additional information.
Forty of the 56 yeast seven-β-strand methyltransferases have human orthologs as detected through sequence similarity clusters using CLANS (44) (Fig. 4 and supplemental Table IV). Proteins clustered in the “convex” mode with standard deviations of more than 8 were used to identify similar species. When the standard deviation “cutoff” value was relaxed to 4, most of the proteins clustered to their protein subfamilies, which all have related substrates in common. Protein families with both yeast and human proteins are shown in supplemental Table V. Interestingly, almost all of the Group J proteins as described in yeast (5) were found using the “network” clustering setting in CLANS; in fact, yeast proteins YIL110W and YLR137W were not clustered with this group by the convex 4 setting alone. The only missing yeast Group J protein, YJR129C, was found to group with six other human proteins using the convex 8 setting. Many proteins clustered into unknown functional families, including the yeast methyltransferase “Group J,” which now includes 10 additional human proteins (supplemental Table V).
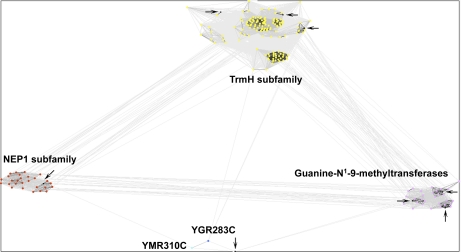
Fig. 4.
Sequence similarity cluster of SPOUT methyltransferases. All of the known and putative SPOUT methyltransferases were analyzed by CLANS with reference proteins to infer substrate specificity. All human SPOUT methyltransferases are in black and identified with arrows. Reference proteins in the NEP1 subfamily are in red, and those in the TrmH subfamily are in yellow. Yeast proteins YGR283C and YMR310C are in dark and light blue, respectively, and guanine-N1-9-methyltransferase reference proteins are in pink. BLAST correlations are shown with gray lines; lighter shades of gray represent BLAST correlations closer to an E-value of 1, whereas darker shades of gray represent BLAST correlations that are closer to an E-value of 0.
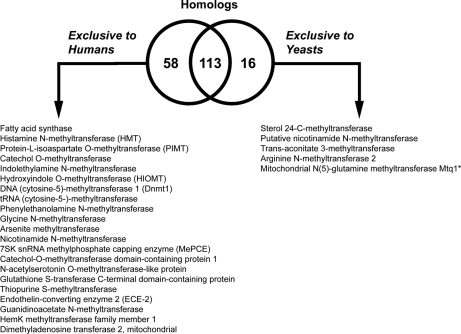
58 of the human methyltransferases do not have an orthologous partner in yeast; these proteins are known to function in human-specific catalytic reactions, including protein isoaspartate methyltransferase, catechol methyltransferase, DNA methyltransferase, glycine N-methyltransferase, arsenite methyltransferase, meP capping, MraW, and acetylserotonin O-methyltransferase (Fig. 3 and supplemental Table VI). Protein families containing solely human methyltransferases are shown in supplemental Table VII. Additionally, we confirmed that there are several yeast-specific methyltransferase reactions, including those catalyzed by sterol methyltransferase (Erg6) and trans-aconitate methyltransferase (Tmt1) (Fig. 3 and supplemental Table VI). Interestingly, although Nnt1 is annotated as “putative nicotinamide N-methyltransferase” (52), by sequence analysis, it does not seem to be orthologous to human NNT1.
Fig. 3.
Homologs between yeast and human seven-β-strand methyltransferases. The Venn diagram represents the numbers of proteins that have homologs in both human and yeast or are exclusive to each organism (see supplemental Tables IV and V). Proteins that are exclusive either to human or to yeasts are described by function. Although there is poor sequence similarity, yeast Mtq1 is the functional homolog of human HemK.
A potential limitation of this analysis comes from the poor sequence similarity of yeast Mtq1 and human HemK, although it is clear that they are functionally identical in methylating translational release factors (53). Some orthologs, such as yeast Abp140, show homology to the human protein only in limited regions. A PSI-BLAST of this protein against the human proteome revealed similarity with Q6P1Q9 (E-value = 7 × 10−45) in the Abp140 C-terminal region; the N-terminal region of the yeast protein may include an additional function. Other putative methyltransferases in humans and yeast that appear to be unique to each species are given in supplemental Table VI.
Some of the methyltransferases reveal a predicted structure similar to the seven-β-strand core domain yet are composed from rearranged motifs. The known rearranged seven-β-strand proteins in yeast have homologs that function as RNA (2′-O-methyladenosine-N6-)-methyltransferases (37, 38). These proteins are similar to the non-rearranged, Motif I-less DNA N6-methylating enzymes that do have solved crystal structures (40). The sequence similarity of the RNA and DNA methylating species are within the Motif II-DPPY-Motif III-β-strand of Motif I, confirming their relationship.
Interestingly, two putative human methyltransferase enzymes were identified as species with additional catalytic activities. Fatty-acid synthase (UniProtKB accession number P49327) has been identified as a seven-β-strand putative methyltransferase in Table I. This methyltransferase-like sequence occurs in a unique domain whose experimental three-dimensional structure is similar to seven-β-strand enzymes (54). This result suggests that fatty-acid synthase may catalyze a previously unidentified methylation reaction. It has been assumed that this is not a functioning domain (54); however, further experimentation will be needed to confirm those predictions. Additionally, we found that the human GSTCD glutathione S-transferase (UniProtKB accession number Q8NEC7) has a domain of the seven-β-strand methyltransferase superfamily. The target of this potential methyltransferase remains to be determined. We note that glutathione S-transferase itself is methylated; perhaps the reaction catalyzed is automethylation (55).
It is important to note that a few enzymes have a conserved seven-β-strand methyltransferase domain but do not catalyze a methyl transfer reaction (family 1a in supplemental Table III). Spermidine synthase and spermine synthase catalyze aminopropyl transfer from decarboxylated AdoMet (56), whereas TRMT12 catalyzes an aminobutyryltransferase reaction with AdoMet in wybutosine synthesis (24). In each of these cases, nucleophilic attack on bound AdoMet or its decarboxylated derivative occurs on the γ carbon of the methionine moiety rather than the methyl group.
Substrate Specificity of Human SPOUT Proteins
Although there is one more SPOUT methyltransferase present in humans than in yeast, the human enzymes methylate a less diverse group of substrates because no human homolog was detected for yeast protein YOR021C (Table II). Reference groups of SPOUT proteins with known substrates were clustered together with the eight putative human SPOUT methyltransferases in Table I via CLANS (see “Experimental Procedures”). This analysis revealed sequence similarity clusters of proteins that shared similar substrates. This is likely due to the additional, conserved domains within these proteins, such as THUMP, OB-fold, L30e, and PUA domains, that are responsible for substrate specificity (6, 16). The CLANS analysis revealed that six of the eight SPOUT human methyltransferases likely methylate guanosine, half of which methylate 2′-O-guanosine, whereas the remaining show homology to Trm10 and methylate N1-guanine (Table II). Substrates are currently unknown for human Nep1, which is homologous to yeast Emg1, and for human CI114. Human CI114 clustered very closely to both yeast YMR310C and YGR283C, indicating that they all possibly share the same substrate (Fig. 4).
Table II. SPOUT methyltransferases and their substrates.
Human SPOUT proteins are listed by their UniProtKB identification and official name along with their yeast homolog and inferred substrate specificity.
| Protein | Yeast homolog | Substrate specificity |
|---|---|---|
| Q13395 TARB1 | Trm3 | Guanosine-2′-O-methyltransferase |
| Q6IN84 MRM1 | Mrm1 | Guanosine-2′-O-methyltransferase |
| Q9HC36 RMTL1 | Mrm1 | Guanosine-2′-O-methyltransferase |
| Q6PF06 Rg9D3 | Trm10 | Guanine-N1-9-methyltransferase |
| Q8TBZ6 Rg9D2 | Trm10 | Guanine-N1-9-methyltransferase |
| Q7L0Y3 MRRP1 | Trm10 | Guanine-N1-9-methyltransferase |
| Q92979 NEP1 | Emg1 | |
| Q5T280 CI114 | YMR310C/YGR283C |
We wanted to test the nature of the SPOUT methyltransferase domain and whether the amino acids within the SPOUT domain also have substrate-specific characteristics. We developed HMM profiles from each individual substrate-specific COG group using only the sequences within the domain (16). Interestingly, we found that these “common” domains still flagged only substrate-specific proteins (supplemental Table II). This indicates that the SPOUT domain may itself help recognize specific substrates (supplemental Table II).
Substrate Specificity of Human SET Proteins
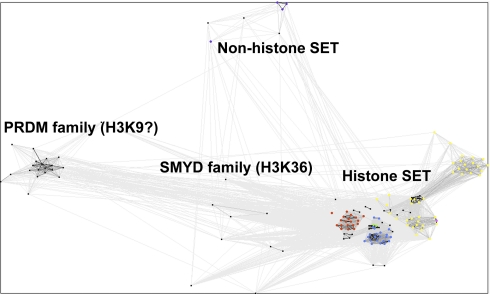
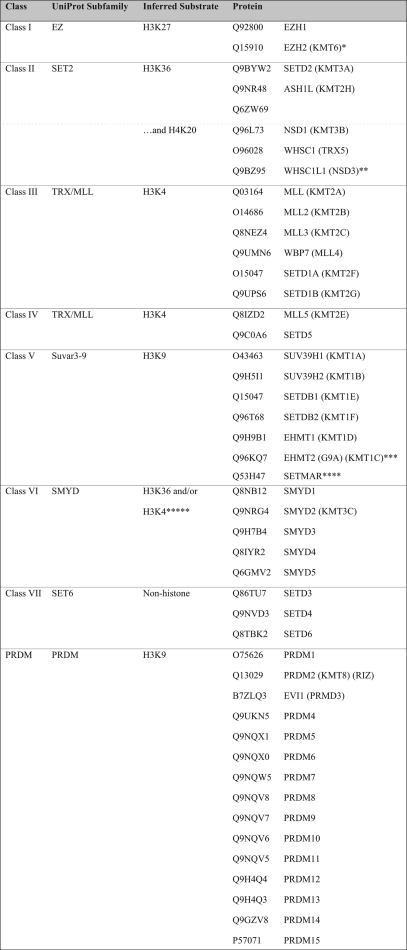
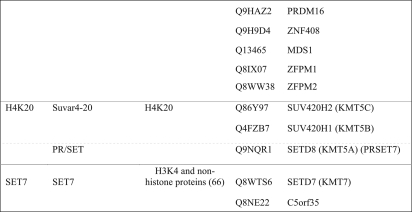
SET domain methyltransferases, like SPOUT methyltransferases, contain supplemental domains that can bind specifically to their methyl-accepting substrates. The human SET domain methyltransferases presented in Table I were clustered alongside a reference database of SET proteins through CLANS reiterative PSI-BLAST searches (see “Experimental Procedures” and Fig. 5). We were able to group all of the SET proteins into 10 classes (Classes I–VII, PRDM, H4K20, and SET7) that can help define their substrate specificity (Table III).
Fig. 5.
Sequence similarity clusters of SET domain methyltransferases. All of the known and putative SET domain methyltransferases were analyzed by CLANS with reference proteins to infer substrate specificity. All human SET domain methyltransferases are in black. Reference proteins methylating only H3K4 are in red; those methylating H3K9 are in yellow; those methylating H3K36 are in blue; those methylating both H3K36 and H4K20 are in green; those methylating H3K9, H3K27, and H4K20 are in pink; and those methylating Rubisco are in purple. BLAST correlations are shown with gray lines; lighter shades of gray represent BLAST correlations closer to an E-value of 1, whereas darker shades of gray represent BLAST correlations that are closer to an E-value of 0.
Table III. Classification of human SET domain methyltransferases.
Proteins are listed by their UniProtKB identification and are categorized by SET class, UniProt subfamily, and inferred substrate specificity (see text). Class members are separated by lines; subclass members are separated by dotted lines.
* Protein has been shown to also methylate H1K26 depending on its complex of proteins (57).
** Although UniProt also describes this as SET2, the function of this protein does not match its subfamily description; it has H3K4 and H3K27 activity (58).
*** This protein has also been shown to have H3K27 activity (59).
**** This protein actually has been shown to have H3K4 and H3K36 activity. The N terminus matches the Class III SET methyltransferases, whereas the C terminus matches the mariner transposase family and may be responsible for this change in specificity (60).
Many of the SET methyltransferase classes have been well described by their homologs in S. cerevisiae and/or Drosophila melanogaster (6). Class I proteins (UniProt EZ subfamily) methylate lysine 27 on histone H3 (denoted H3K27). Interestingly, the EZH2 protein can selectively methylate H1K26 depending on its complex partners (57). Class II proteins (UniProt SET2) methylate the H3K36 substrate. A new putative SET methyltransferase protein (UniProtKB accession number Q6ZW69) clusters with this group of enzymes. Class II proteins NSD1 and WHSC1 have also been shown to methylate H4K20. The WHSC1L1 protein, a homolog of WHSC1, is exceptional because it displays both H3K4 and H3K27 activity (58). Proteins in the UniProt TRX/MLL subfamily, which methylate H3K4, fall into Classes III and IV. Although they methylate the same substrate, Class IV members only share a common PHD region with its Class III counterparts (6).
SUV39H1, EHMT, and SETDB proteins are Class V enzymes that methylate H3K9. Although the protein SETMAR is found in this class, it has H3K4 and H3K36 activity (60). SMYD proteins fall into Class VI SET domain methyltransferases. Although there are no SMYD proteins in yeast, previous analysis of Class VI proteins included yeast proteins Set5 and Set6 (6). SMYD3 is H3K4-specific (61), whereas SMYD2 is H3K4- and H3K36-specific (62, 63). Therefore, our additional knowledge of the H3K4 activity within this class of proteins allows us to predict that other class members may methylate H3K4 as well.
Three additional classes of human SET domain methyltransferases are not defined by the traditional SET class categorization (64). The PRDM class of enzymes is the largest group in the human proteome. Interestingly, there are no yeast homologs of this class. These proteins are proposed to methylate H3K9 based on the activity in PRDM2 (65). There are four new putative methyltransferases in this class: MDS1, ZFPM1, ZFPM2, and ZNF408 (Table III). The H4K20 class of proteins contains the SUV420 proteins and the protein SETD8, which has been described as a PR/SET subfamily member in the UniProt database. The SET7 class of proteins is defined by SETD7, a protein that methylates H3K4 as well as a number of additional proteins (66). We found an additional member of this class, C5orf35 (UniProtKB accession number Q8NE22) (Table III); it will be of interest to see whether this protein may also catalyze non-histone protein methylation.
Class VII represents classical non-histone SET domain methyltransferases. In humans, three proteins fall in this category, half the number that is present in yeast (Table III and Ref. 6). The human methyltransferasome does not contain orthologs to the six Class VII yeast enzymes that modify cytochrome c, ribosomal large subunit proteins, and elongation factor 1A (67). In humans, the sequences of SETD3 and SETD4 are most similar to the plant Rubisco methyltransferase, whereas the SETD6 sequence is closest to the yeast ribosomal methyltransferase Rkm4 (19). The substrates of these putative methyltransferases remain to be identified. This class of SET domain proteins represents a case where there is a contraction of methyltransferase species in the human proteome compared with the yeast proteome. Overall, the grouping of SET domain methyltransferases in the 10 classes shown in Table III now provides an opportunity to more rapidly discover the specific substrates of each of the uncharacterized proteins.
Disease Correlations
All 208 human methyltransferase proteins were entered in GeneALaCart (http://www.genecards.org/cgi-bin/BatchQueries/Batch.pl) and searched against the Online Mendelian Inheritance in Man (OMIM) disorder, UniProt disorder, Novoseek disorder, and Mouse Genome Informatics mutant phenotype databases to find the proteins that are known to be correlated to diseases (42, 43). We found that 63 of these species (30%) were associated with disease (supplemental Table VIII). These species were largely in the seven-β-strand (27 species) and the SET domain (28 species) superfamilies. We found that 49% of SET domain species are disease-related, whereas only 22% of seven-β-strand species are associated with disease.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant GM026020 and Training Grant GM008496 (through the UCLA Chemistry-Biology Interface to T. C. P.). This work was also supported by a senior scholar award in aging from the Ellison Medical Foundation (Grant AG-SS-2076-08).
 This article contains supplemental Tables I–VIII.
This article contains supplemental Tables I–VIII.
1 The abbreviations used are:
- SAM or AdoMet
- S-adenosylmethionine
- HMM
- hidden Markov model
- COG
- clusters of orthologous groups
- SMART
- simple modular architecture research tool
- MMS
- Multiple Motif Scanning program
- PEMT
- phosphatidylethanolamine N-methyltransferase
- ICMT
- protein-S-isoprenylcysteine O-methyltransferase
- NRM
- nuclear rim protein
- BHMT
- betaine-homocysteine S-methyltransferase
- MTR
- methionine synthase
- GSTCD
- glutathione S-transferase C-terminal domain-containing protein
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- BLAST
- basic local alignment search tool.
REFERENCES
- 1. Cheng X., Blumenthal R. M. (eds) (1999) S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions, World Scientific, Singapore [Google Scholar]
- 2. Martin J. L., McMillan F. M. (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 [DOI] [PubMed] [Google Scholar]
- 3. Schubert H. L., Blumenthal R. M., Cheng X. (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katz J. E., Dlakić M., Clarke S. (2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteomics 2, 525–540 [DOI] [PubMed] [Google Scholar]
- 5. Petrossian T. C., Clarke S. G. (2009) Multiple Motif Scanning to identify methyltransferases from the yeast proteome. Mol. Cell. Proteomics 8, 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petrossian T., Clarke S. (2009) Bioinformatic identification of novel methyltransferases. Epigenomics 1, 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petrossian T. C., Clarke S. G. (2009) Computational methods to identify novel methyltransferases. BMC Bioinformatics 10, Suppl. 13, P7 [Google Scholar]
- 8. Feng J., Fan G. (2009) The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 89, 67–84 [DOI] [PubMed] [Google Scholar]
- 9. Poleshko A., Einarson M. B., Shalginskikh N., Zhang R., Adams P. D., Skalka A. M., Katz R. A. (2010) Identification of a functional network of human epigenetic silencing factors. J. Biol. Chem. 285, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Albert M., Helin K. (2010) Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 21, 209–220 [DOI] [PubMed] [Google Scholar]
- 11. Baba S. W., Belogrudov G. I., Lee J. C., Lee P. T., Strahan J., Shepherd J. N., Clarke C. F. (2004) Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279, 10052–10059 [DOI] [PubMed] [Google Scholar]
- 12. Clarke S. (2003) Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res. Rev. 2, 263–285 [DOI] [PubMed] [Google Scholar]
- 13. Lehmann L., Jiang L., Wagner J. (2008) Soy isoflavones decrease the catechol-O-methyltransferase-mediated inactivation of 4-hydroxyestradiol in cultured MCF-7 cells. Carcinogenesis 29, 363–370 [DOI] [PubMed] [Google Scholar]
- 14. Yadav N., Cheng D., Richard S., Morel M., Iyer V. R., Aldaz C. M., Bedford M. T. (2008) CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 9, 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozbial P. Z., Mushegian A. R. (2005) Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tkaczuk K. L., Dunin-Horkawicz S., Purta E., Bujnicki J. M. (2007) Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dillon S. C., Zhang X., Trievel R. C., Cheng X. (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao B., Gamblin S. J., Wilson J. R. (2006) Structure of set domain protein lysine methyltransferases, in The Enzymes: Protein Methyltransferases (Clarke S. G., Tamanoi F. eds) pp. 155–178, Elsevier, Amsterdam, The Netherlands: [DOI] [PubMed] [Google Scholar]
- 19. Webb K. J., Laganowsky A., Whitelegge J. P., Clarke S. G. (2008) Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. J. Biol. Chem. 283, 35561–35568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schubert H. L., Wilson K. S., Raux E., Woodcock S. C., Warren M. J. (1998) The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase. Nat. Struct. Biol. 5, 585–592 [DOI] [PubMed] [Google Scholar]
- 21. Berkovitch F., Nicolet Y., Wan J. T., Jarrett J. T., Drennan C. L. (2004) Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan F., LaMarre J. M., Röhrich R., Wiesner J., Jomaa H., Mankin A. S., Fujimori D. G. (2010) RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J. Am. Chem. Soc. 132, 3953–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dixon M. M., Huang S., Matthews R. G., Ludwig M. (1996) The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure 4, 1263–1275 [DOI] [PubMed] [Google Scholar]
- 24. Noma A., Kirino Y., Ikeuchi Y., Suzuki T. (2006) Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li F., Feng Q., Lee C., Wang S., Pelleymounter L. L., Moon I., Eckloff B. W., Wieben E. D., Schaid D. J., Yee V., Weinshilboum R. M. (2008) Human betaine-homocysteine methyltransferase (BHMT) and BHMT2: common gene sequence variation and functional characterization. Mol. Genet. Metab. 94, 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vinci C. R., Clarke S. G. (2007) Recognition of age-damaged (R,S)-adenosyl-L-methionine by two methyltransferases in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 27. Szegedi S. S., Castro C. C., Koutmos M., Garrow T. A. (2008) Betaine-homocysteine S-methyltransferase-2 is an S-methylmethionine-homocysteine methyltransferase. J. Biol. Chem. 283, 8939–8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Romano J. D., Michaelis S. (2001) Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol. Biol. Cell 12, 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griggs A. M., Hahne K., Hrycyna C. A. (2010) Functional oligomerization of the Saccharomyces cerevisiae isoprenylcysteine carboxyl methyltransferase, Ste14p. J. Biol. Chem. 285, 13380–13387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eddy S. R. (2009) A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211 [PubMed] [Google Scholar]
- 32. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 33. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tatusov R. L., Koonin E. V., Lipman D. J. (1997) A genomic perspective on protein families. Science 278, 631–637 [DOI] [PubMed] [Google Scholar]
- 35. Letunic I., Doerks T., Bork P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res. 37, D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clancy M. J., Shambaugh M. E., Timpte C. S., Bokar J. A. (2002) Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bujnicki J. M., Feder M., Radlinska M., Blumenthal R. M. (2002) Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J. Mol. Evol. 55, 431–444 [DOI] [PubMed] [Google Scholar]
- 39. Albrecht M., Lengauer T. (2004) Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 569, 18–26 [DOI] [PubMed] [Google Scholar]
- 40. The UniProt Consortium (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 42. Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. (1997) GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 13, 163. [DOI] [PubMed] [Google Scholar]
- 43. Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E., Adato A., Peter I., Khen M., Atarot T., Groner Y., Lancet D. (2003) Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 31, 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frickey T., Lupas A. (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 45. Qian C., Zhou M. M. (2006) SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell. Mol. Life Sci. 63, 2755–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pruitt K. D., Tatusova T., Maglott D. R. (2007) NCBI reference sequences (RefSeq): a curated nonredundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Proffitt J. H., Davie J. R., Swinton D., Hattman S. (1984) 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol. Cell. Biol. 4, 985–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boa S., Coert C., Patterton H. G. (2003) Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast 20, 827–835 [DOI] [PubMed] [Google Scholar]
- 49. Strahl B. D., Grant P. A., Briggs S. D., Sun Z. W., Bone J. R., Caldwell J. A., Mollah S., Cook R. G., Shabanowitz J., Hunt D. F., Allis C. D. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22, 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Flicek P., Aken B. L., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Gräf S., Haider S., Hammond M., Howe K., Jenkinson A., Johnson N., Kähäri A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Koscielny G., Kulesha E., Lawson D., Longden I., Massingham T., McLaren W., Megy K., Overduin B., Pritchard B., Rios D., Ruffier M., Schuster M., Slater G., Smedley D., Spudich G., Tang Y. A., Trevanion S., Vilella A., Vogel J., White S., Wilder S. P., Zadissa A., Birney E., Cunningham F., Dunham I., Durbin R., Fernández-Suarez X. M., Herrero J., Hubbard T. J., Parker A., Proctor G., Smith J., Searle S. M. (2010) Ensembl's 10th year. Nucleic Acids Res. 38, D557–D562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofemeister H., O'Hare P. (2005) Analysis of the localization and topology of nurim, a polytopic protein tightly associated with the inner nuclear membrane. J. Biol. Chem. 280, 2512–2521 [DOI] [PubMed] [Google Scholar]
- 52. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003) Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Polevoda B., Span L., Sherman F. (2006) The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 281, 2562–2571 [DOI] [PubMed] [Google Scholar]
- 54. Maier T., Leibundgut M., Ban N. (2008) The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 55. Johnson J. A., Finn K. A., Siegel F. L. (1992) Tissue distribution of enzymic methylation of glutathione S-transferase and its effects on catalytic activity. Methylation of glutathione S-transferase 11-11 inhibits conjugating activity towards 1-chloro-2,4-dinitrobenzene. Biochem. J. 282, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. White W. H., Gunyuzlu P. L., Toyn J. H. (2001) Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J. Biol. Chem. 276, 10794–10800 [DOI] [PubMed] [Google Scholar]
- 57. Kuzmichev A., Jenuwein T., Tempst P., Reinberg D. (2004) Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14, 183–193 [DOI] [PubMed] [Google Scholar]
- 58. Kim S. M., Kee H. J., Eom G. H., Choe N. W., Kim J. Y., Kim Y. S., Kim S. K., Kook H., Kook H., Seo S. B. (2006) Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem. Biophys. Res. Commun. 345, 318–323 [DOI] [PubMed] [Google Scholar]
- 59. Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317 [DOI] [PubMed] [Google Scholar]
- 60. Lee S. H., Oshige M., Durant S. T., Rasila K. K., Williamson E. A., Ramsey H., Kwan L., Nickoloff J. A., Hromas R. (2005) The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl. Acad. Sci. U.S.A. 102, 18075–18080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F. P., Li M., Yagyu R., Nakamura Y. (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6, 731–740 [DOI] [PubMed] [Google Scholar]
- 62. Abu-Farha M., Lambert J. P., Al-Madhoun A. S., Elisma F., Skerjanc I. S., Figeys D. (2008) The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell. Proteomics 7, 560–572 [DOI] [PubMed] [Google Scholar]
- 63. Brown M. A., Sims R. J., 3rd, Gottlieb P. D., Tucker P. W. (2006) Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ng D. W., Wang T., Chandrasekharan M. B., Aramayo R., Kertbundit S., Hall T. C. (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta 1769, 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim K. C., Geng L., Huang S. (2003) Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 63, 7619–7623 [PubMed] [Google Scholar]
- 66. Pradhan S., Chin H. G., Estève P. O., Jacobsen S. E. (2009) SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics 4, 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lipson R. S., Webb K. J., Clarke S. G. (2010) Two novel methyltransferases acting upon eukaryotic elongation factor 1A in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 500, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]