Abstract
The present study addresses, by transcriptomics and quantitative stable isotope labeling by amino acids in cell culture (SILAC)-based proteomics, the estrogen receptor α (ERα) and β (ERβ)-mediated effects on gene and protein expression in T47D breast cancer cells exposed to the phytoestrogen genistein. Using the T47D human breast cancer cell line with tetracycline-dependent ERβ expression (T47D-ERβ), the effect of a varying intracellular ERα/ERβ ratio on genistein-induced gene and protein expression was characterized. Results obtained reveal that in ERα-expressing T47D-ERβ cells with inhibited ERβ expression genistein induces transcriptomics and proteomics signatures pointing at rapid cell growth and migration by dynamic activation of cytoskeleton remodeling. The data reveal an interplay between integrins, focal adhesion kinase, CDC42, and actin cytoskeleton signaling cascades, occurring upon genistein treatment, in the T47D-ERβ breast cancer cells with low levels of ERα and no expression of ERβ. In addition, data from our study indicate that ERβ-mediated gene and protein expression counteracts ERα-mediated effects because in T47D-ERβ cells expressing ERβ and exposed to genistein transcriptomics and proteomics signatures pointing at a clear down-regulation of cell growth and induction of cell cycle arrest and apoptosis were demonstrated. These results suggest that ERβ decreases cell motility and metastatic potential as well as cell survival of the breast cancer cell line. It is concluded that the effects of genistein on proteomics and transcriptomics end points in the T47D-ERβ cell model are comparable with those reported previously for estradiol with the ultimate estrogenic effect being dependent on the relative affinity for both receptors and on the receptor phenotype (ERα/ERβ ratio) in the cells or tissue of interest.
At present, two main estrogen receptors, ERα1 and ERβ, have been identified in rats, mice, primates, and humans (1). Different biological responses may occur when an estrogen binds to the different ERs. Several studies have shown decreased ERβ expression in malignant cancer tissues as compared with benign tumors or normal tissues where ERα expression persists (2–7). Hence, ERα and ERβ may have different roles in gene regulation, and their relative levels or ratios within the tissues may influence cellular responses to estrogens. To understand the critical role of estrogens in the regulatory cascade involved in the progression of breast cancers, several studies focused on the evaluation of global transcriptomics and showed the association of ERα activation with cell proliferation and the opposing effects for activation of ERβ (8–10). So far, most of these “omics” studies focused on estrogens like estradiol but did not yet widely include so-called phytoestrogens.
Phytoestrogens are a group of plant-derived compounds with estrogenic properties (11). The major types of phytoestrogen are isoflavones, flavones, coumestans, lignans, and stilbenes. Among the most studied flavonoids with respect to antitumor functions are genistein and quercetin. Phytoestrogens have been considered a natural alternative to the hormone replacement therapy because these chemicals are found in the regular diet (12). Genistein is mostly present in soybeans. High consumption of phytoestrogen-rich food correlates with reduced incidence of breast cancer (13, 14). These two flavonoids, genistein and quercetin, have been shown to have potent antiproliferative effects on tumor cells in vitro by halting the cell cycle and inducing apoptosis (15–17). At the same time, these phytoestrogens have been reported to exhibit proliferative effects not only in vitro but also in vivo (18–20). The relative binding affinities of these phytoestrogens for the different estrogen receptor proteins might play a role in these contradictory effects. Indeed, although the phytoestrogens can bind to the ERα protein, they often appear to prefer binding to the ERβ protein (11, 20). However, what effects are induced by phytoestrogens and which mechanisms are activated in cells remain to be elucidated. Therefore, the aim of the present study was to provide omics data on the effects of the phytoestrogen genistein in cells with a variable intracellular ratio of the two estrogen receptors ERα and ERβ and to compare these outcomes qualitatively with those reported earlier for estradiol (8).
The importance of the intracellular ratio of the two estrogen receptors ERα and ERβ for the ultimate potential of estradiol and of the phytoestrogen genistein to stimulate or inhibit cell proliferation was demonstrated previously (20, 21). Using the human T47D breast cancer cell line with tetracycline-dependent ERβ expression (T47D-ERβ), the effect of a varying intracellular ERα/ERβ ratio on estradiol- or genistein-induced cell proliferation was characterized (20). With increased expression of ERβ, estrogen-induced ERα-mediated cell proliferation was reduced. These results point at the importance of the cellular ERα/ERβ ratio for the ultimate effect of (phyto)estrogens on cell proliferation. The results of our previous studies also revealed that in this T47D-ERβ model system the effects of genistein on cell proliferation with varying cellular ERα/ERβ ratios were comparable with the effects induced by estradiol itself (20, 22).
In the present study, we investigated the consequences of the intracellular ERα/ERβ ratio for the effects induced by genistein in more detail using quantitative omics technologies, characterizing both gene and protein expression patterns, and compared the results obtained with those previously reported for estradiol in the same model system (8). To this end, state-of-the-art high throughput methods for systems-wide gene and protein expression analysis were applied. These methods included DNA microarrays, commonly used for global analysis of gene expression changes. In addition, we applied stable isotope labeling by amino acids in cell culture (SILAC), which is a differential and quantitative proteomics technique that uses mass spectrometric analysis (23). The SILAC strategy is based on the metabolic incorporation of “light” (normal) and “heavy” (isotope-labeled) amino acids into the cells, which is a process shown to occur without adverse effects on cellular physiology (24). Full metabolic incorporation of the labeled amino acids into the proteins results in a mass shift of the corresponding peptides. This mass shift can be detected by MS. When two samples are combined, the ratio of peak intensities in the MS reflects the relative protein abundance. Because the metabolic incorporation of the labels does not affect the integrity of genes or proteins, the transcriptomics and SILAC proteomics experiments can be performed on the same cell samples.
The data obtained from our proteomics and transcriptomics studies indicate that genistein induces rapid cell proliferation and migration by dynamic activation of cytoskeleton remodeling in the T47D-ERβ cells expressing low levels of ERα and no ERβ. Interaction between integrins, focal adhesion kinase, CDC42, and actin cytoskeleton signaling cascades occurs upon genistein treatment, supporting the observed cell proliferation. Our results also strengthen the concept that ERβ-mediated gene and protein expression counteracts ERα-mediated effects because in T47D-ERβ cells expressing ERβ and exposed to genistein a clear down-regulation of genes and proteins involved in cell growth and induction of cell cycle arrest and apoptosis was demonstrated.
MATERIALS AND METHODS
Cell Culture
The stably transfected T47D tetracycline-inducible cell line (T47D-ERβ) and the control cell line (T47D-PBI) were made and provided by Ström et al. (21). DMEM SILAC medium lacking arginine (Arg) and lysine (Lys) and supplemented with 5% dialyzed fetal bovine serum and 1000 ng/ml tetracycline (Tet) was used. Arg and Lys, either light or heavy depending on the experimental design, were incorporated in the DMEM SILAC medium. The final concentrations of Arg and Lys were 21 and 48 mg/liter, respectively (25).
During the adaptation phase, cells were grown in light or heavy SILAC medium containing, respectively, [12C6,14N4]arginine and [12C6,14N2]lysine or [13C6,15N2]lysine and [13C6,15N4]arginine until the cells grown in heavy medium had fully incorporated the labeled amino acids. Full incorporation of labels into the cells was checked by MS analysis (26) of cell samples, detecting the time point at which there was no further increase in the amount of label incorporated. Full incorporation of labels for the T47D-ERβ cells without ERβ expression (Tet-containing) was observed after five doubling times or 10 days of cultivation (data not shown). Preparation of the samples after this adaptation phase was performed as follows. Cells in (SILAC) DMEM supplemented with 5% dialyzed fetal bovine serum (FBS) and 1000 ng/ml Tet were seeded in plates for 24 h. After 24 h, DMEM SILAC medium was replaced by the same medium without phenol red, containing the required concentrations of Tet, heavy or light amino acids, and a serum concentration that was reduced to 0.5% dialyzed FBS. To this, 10 nm ICI 182780 was added for cell synchronization (8), and Tet was withdrawn in half of the plates all 12 h before the start of the treatment to allow expression of ERβ. The Tet+ and Tet− cultures were incubated with either a 10 nm concentration of the ERα and ERβ antagonist ICI 182780 (27) or 500 nm genistein for 24 h according to the experimental design (Table I) after which all cells were collected simultaneously for RNA and protein extraction. Three replicates of each sample were prepared in parallel to minimize experimentally induced variability.
Table I. Experimental design of present study based on five cellular samples prepared as indicated.
| Sample | Strain | Tetracycline treatment | ERβ expression | Compound treatment | No. of samples, transcriptomics/proteomics | Label |
|---|---|---|---|---|---|---|
| A | T47D-ERβ | Yes | No | ICI 182780 | 3/3 | Light |
| B | T47D-ERβ | No | Yes | ICI 182780 | 3/3 | Heavy |
| C | T47D-ERβ | Yes | No | Genistein | 3/3 | Heavy |
| D | T47D-ERβ | No | Yes | Genistein | 3/3 | Light |
| E | T47D control | No | No | Genistein | 3/3 | Heavy |
Table I summarizes the experimental design applied to obtain information on the genistein-induced levels of gene and protein expression in T47D cells with variable ERα/ERβ ratios. Table I presents the five different samples prepared, indicating the cell line used, the Tet treatment and resulting ERβ expression, exposure to ICI 182780 or genistein, and the amino acid label (light or heavy) added to the growth medium. In addition to genistein, ICI 182780 was used to block the receptors, thereby suppressing receptor-mediated transcription. A control cell line (T47D-PBI) was used to define genistein-induced non-ERβ-mediated gene expression inherent to the cell system used. Table II presents the pairwise comparisons made using these five samples to analyze (i) genistein-induced ERα-mediated gene and protein expression (C versus A; referred to as sample CA), (ii) genistein-induced ERβ-regulated gene and protein expression in the presence of endogenous levels of ERα (D versus B; referred to as sample DB); and (iii) a second approach to detect genistein-induced ERβ-mediated gene and protein expression in the presence of endogenous levels of ERα (E versus D; referred to as sample ED).
Table II. Experimental design of present study based on their pairwise comparison carried out as indicated.
| Sample | Targets |
|---|---|
| CA | Genistein-modulated ERα-mediated gene and protein expression |
| DB | Genistein-modulated ERβ-mediated gene and protein expression |
| ED | Genistein-modulated ERβ-mediated gene and protein expression (second approach) |
Protein and RNA Preparation
Each sample was split in two, one for RNA and one for protein preparation. RNA was extracted using the TRIzol precipitation method and purified using an RNeasy minikit protocol for second RNA cleanup (RNeasy minikit, Qiagen). Approximately 4 μg of total RNA was collected per replicate.
For protein extraction, cells were washed twice with PBS and lysed in modified radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 150 mm NaCl, 1% Triton X-100, 0.25% sodium deoxycholate) containing a protease inhibitor mixture (Complete Mini, Roche Applied Science) at 4 °C for 15 min. Samples were sonicated for 1 min and centrifuged at 14,000 × g for 15 min at 4 °C, and the supernatant was collected. Equal amounts of protein (BSA Protein Assay kit, Pierce) from each sample were mixed in a ratio of 1:1 according to the experimental design presented in Tables I and II, and 1 μg of the protein sample thus obtained was separated by 12% Tris-glycine sodium dodecyl sulfate-polyacrylamide gel by electrophoresis (SDS-PAGE) followed by staining of the SDS gel using colloidal Coomassie Blue (Colloidal Blue staining kit, Invitrogen). Then the resulting three gel lanes per replicate (CA, DB, and ED) were excised and cut horizontally into eight equal sections per lane. In-gel digestion was performed as follows. The SDS gel was destained by two washes with water. Cysteine reduction was performed by adding 100 μl of 50 mm dithiothreitol (DTT) in 50 mm NH4HCO3. Samples were sonicated for 1 min and incubated at 60 °C without shaking for 1 h. Alkylation was performed when samples reached room temperature, replacing DDT by 100 μl of 50 mm iodoacetamide in 50 mm NH4HCO3. Samples were sonicated for 1 min and alkylated at room temperature in the dark for 1 h. The gel pieces were washed three times with 100 μl of 50 mm NH4HCO3 at pH 8. For proteolytic digestion, samples were treated overnight with 100 μl of trypsin (10 ng/μl in 50 mm NH4HCO3; sequencing grade, Roche Applied Science) at room temperature. Gel fragments were removed by centrifugation, and the proteolytic peptides were recovered in the supernatant fraction (25 μl). 10% trifluoroacetic acid was added to correct the pH up to 2–4. Finally, all extracts were measured by LC-MS/MS.
Data Acquisition and Mass Spectrometry
The protein samples were analyzed by injecting 18 μl of sample over a 0.10 × 32-mm Prontosil 300-5-C18H (Bischoff) preconcentration column (prepared in house) at a flow of 6 μl/min for 5 min with a Proxeon EASY nLC system. Compounds were eluted from the preconcentration column onto a 0.10 × 250-mm Prontosil 300-3-C18H analytical column (prepared in house) with an acetonitrile gradient at a flow of 0.5 μl/min. The gradient consisted of an increase from 9 to 34% acetonitrile in water with 1 ml/liter formic acid in 50 min followed by a fast increase in the percentage of acetonitrile to 80% (with 20% water and 1 ml/liter formic acid in both the acetonitrile and the water) in 3 min as a column cleaning step. In between the preconcentration and the analytical columns, an electrospray potential of 3.5 kV was applied directly to the eluent via a solid 0.5-mm platinum electrode fitted into a P777 Upchurch MicroCross. Full-scan positive mode FT-MS spectra were measured between m/z 380 and 1400 at a resolution of 60,000 on an LTQ-Orbitrap (Thermo Electron, San Jose, CA). MS/MS scans of the four most abundant doubly or triply charged peaks in the FT-MS scan were recorded in a data-dependent mode in the linear trap (MS/MS threshold, 10,000). Data were acquired using Xcalibur software.
Protein Identification and Quantitation
Mass spectra were analyzed using MaxQuant (version 1.0.13.8), which performs e.g. list generation, ratio heavy/light (H/L) significance A, ratio H/L significance B, SILAC- and extracted ion current-based quantification, false positive rate, and data filtration (28). Data were searched against the human International Protein Index (IPI) database supplemented with frequently observed contaminants and concatenated with reversed copies of all sequences (total sequences, 148,380) using Mascot v2.2 (Matrix Sciences). Spectra determined to result from heavy labeled peptides by presearch MaxQuant analysis were searched with the additional fixed modifications Arg10 and Lys8, whereas spectra with a SILAC state not defined a priori were searched with Arg10 and Lys8 as additional variable modifications. Precursor mass tolerance was set at 10 ppm for the complete peptides and 0.5 Da for peptide fragments as observed in the MS2 spectra. Trypsin/Pro cleavage specificity with up to two missed cleavage and three labeled amino acids (Arg and Lys) were allowed. The required false discovery rate was set to 1% at the protein level, and the minimum required peptide length was set to 6 amino acids. Carbamidomethylcysteine was set as a fixed modification, and methionine oxidation, deamidation (Asn/Gln), and acetylation of the N terminus (protein) were allowed as variable modifications. For protein identification, at least two peptides were required among which at least one peptide was required to be unique in the database. Identified proteins were quantified. Protein ratios calculated by MaxQuant were subjected to manual inspection, and results obtained were compared with those from MSQuant software (version 1.5). Further analysis and plotting were performed using the R statistical and graphic environments.
Microarray
Integrity and quantity of the extracted RNA were assessed by using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA) and the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies Inc.). cRNA was prepared according to the manufacturer's protocol and hybridized to HG-U133 Plus 2.0 GeneChip arrays (Affymetrix, Santa Clara, CA). HG-U133 Plus 2.0 arrays contain 54,675 sets of oligonucleotide probes or probe sets. Data were analyzed based on a mapping to ∼17,500 unique human genes with Entrez Gene annotation.
Calculations and Statistics
Microarray results were processed in R (http://www.r-project.org) using gcRMA and filtered using MAS5 calls. Ratios were calculated using limma in R, applying moderated t tests, and were adjusted for multiple testing. The reported ratios describing biological effects of interest were calculated as indicated in Tables I and II.
Bioinformatics Network Analysis
A cutoff p value ≤0.001 (ratio H/L or L/H) was selected for analysis of differentially expressed proteins, and a multiple testing-adjusted p value ≤0.05 for differentially regulated genes was used. Ingenuity Pathway Analysis 8.5 (Ingenuity Systems Inc.) was used to conduct a knowledge-based network analysis, a molecular and cellular function analysis, and a canonical pathway analysis of the proteomics and transcriptomics data. Ingenuity Pathway Analysis tools rely on curated functional and regulatory interactions extracted from the literature. The biological functions across ER-responsive genes and proteins were identified. Fischer's exact test was used to calculate a p value determining the probability that each biological function assigned to that data set of ER-responsive genes/proteins is due to chance alone.
RESULTS
Optimization of Experimental Protocol and Receptor Expression
T47D cells were previously stably transfected with the ERβ expression plasmid under Tet-responsive promoter regulation (21). Cells were grown in medium containing light or heavy amino acid labels according to the experimental design presented in Table I. After 10 days of preincubation, the MS analysis of cell samples revealed that isotope incorporation did not further increase, reflecting optimal full metabolic incorporation and indicating that the cells were ready for the exposure phase. Cell treatment was performed as indicated by Williams et al. (8) to be able to compare genistein effects with their results for estradiol in the same cell system and under the same conditions. Consequently, after cell synchronization by exposure to ICI 182780, cells were exposed to either ICI 182780 or genistein.
ERβ expression in the T47D control cell lines and the T47D-ERβ cell samples pretreated in the presence or absence of Tet was checked by fluorescence microscopy (enhanced green fluorescent protein co-expression under control of the same Tet-responsive promoter (21)) and mRNA quantification (Fig. 1). ERβ mRNA levels were undetectable in Tet+ samples or the control cell line (no endogenous or exogenous ERβ). LC-MS/MS measurements detected high levels of ERβ protein in all Tet− samples (see below).
Fig. 1.
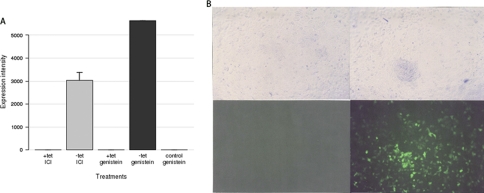
ERβ expression. A, mean expression intensities of ERβ at the mRNA level based on microarray normalized results under five different conditions: Tet+ and ICI (sample A; Table I), Tet− and ICI (sample B; Table I), Tet+ and genistein (sample C; Table I), Tet− and genistein (sample D; Table I), and Tet− in control cell line (T47D-PBI) and genistein (sample E; Table I). Each data point represents the mean of triplicate exposure ± standard deviation. B, picture of cells (upper) and green fluorescent protein (enhanced GFP) detection (lower) of Tet+ (left)- and Tet− (right)-cultured cells treated with genistein.
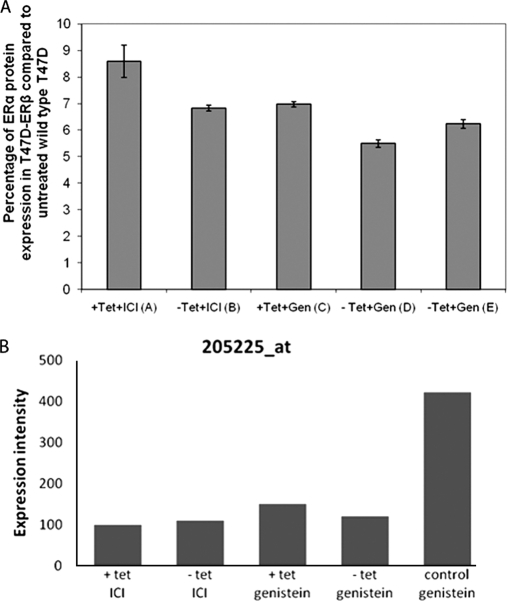
Expression of ERα was not found at the protein level using LC-MS/MS. However, using an ERα ELISA (ActiveMotif sandwich ELISA), ERα could be detected in all the experimental samples (Fig. 2A) at expression levels of 6–10% of the levels observed in the wild type T47D cells (not treated with ICI 182780). ERα (Affymetrix probe 205225_at) mRNA was detected in all samples at different expression levels (Fig. 2B).
Fig. 2.
A, ActiveMotif sandwich ERα ELISA. Amounts of intact ERα from SILAC samples were compared with the ERα from untreated wild type T47D. Shown is the percentage of expression of three measurements of ERα protein under five different conditions: Tet+ and ICI (sample A; Table I), Tet− and ICI (sample B; Table I), Tet+ and genistein (sample C; Table I), Tet− and genistein (sample D; Table I), and Tet− in control cell line (T47D-PBI) and genistein (sample E; Table I). B, mean expression intensities of ERα at the mRNA probe level based on microarray normalized results under the five different conditions. Each data point represents the mean of triplicate exposure ± standard deviation.
Protein Identification and Quantitation
Corresponding light and heavy samples were mixed in a 1:1 ratio according to the experimental design, providing three data sets (CA, DB, and ED; Table II). On average, 2600 proteins were identified per data set (2616 in sample CA, 2543 in DB, and 2627 in ED). From these, for about 1950 proteins, relative quantification data with at least two identified peptides per experiment were obtained (1994 in sample CA, 1905 in DB, and 1995 in ED). For proteins for which corresponding peptides were only detected in the heavy data set but not in the light data set, hampering calculation of an H/L ratio, the H/L ratio was set to the largest H/L ratio determined in the whole data set. Likewise, for proteins for which corresponding peptides were only detected in the light data set but not in the heavy data set, also hampering calculation of an H/L ratio, the H/L ratio was set to the smallest H/L ratio in the whole data set. This enabled comparison of the proteomics results with results obtained with the microarray or finding de novo regulated proteins.
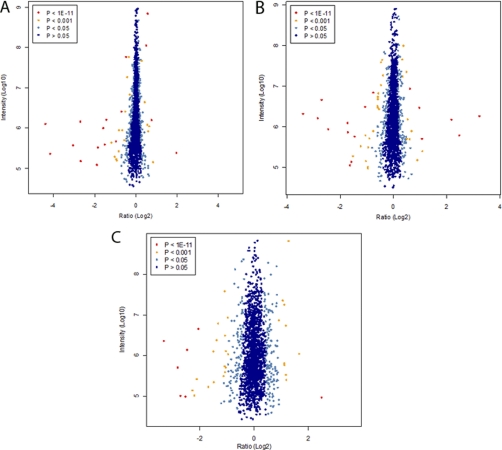
The ratio distributions obtained from each experiment (CA, DB, and ED) are displayed in Fig. 3, A, B, and C. The spread of the cloud is lower at high protein abundance (higher intensity on y axis), which indicates higher precision in the quantification. Notably, higher spread of the cloud in the x direction (ratio (log2)) was observed during ERβ expression, indicating larger -fold changes in protein expression levels.
Fig. 3.
Proteome-wide accurate quantification significance in ERα- (A) and ERβ (B)-mediated proteins expressed as a result of genistein treatment. A, ERα-mediated proteins expressed as a result of genistein treatment (sample CA; Table II). Normalized protein ratios (H/L) are plotted against summed peptide intensities. The left side of the plot shows proteins up-regulated in the sample cultured in light amino isotope conditions: ERα(+), ERβ(−), and ICI 182780; the right side of the plot shows proteins up-regulated in the sample cultured in heavy amino acid isotope conditions: ERα(+), ERβ(−), and genistein. B, ERβ-mediated proteins expressed as a result of genistein treatment (sample DB; Table II). Normalized protein ratios (H/L) are plotted against summed peptide intensities. The left side of the plot shows proteins up-regulated in the sample culture in light amino isotope conditions: ERα(+), ERβ(+), and genistein; the right side of the plot shows proteins up-regulated in the sample culture in heavy amino isotope conditions: ERα(+), ERβ(+), and ICI 182780. C, second approach to detect ERβ-mediated proteins expressed as a result of genistein treatment (sample ED; Table II). Normalized protein ratios (H/L) are plotted against summed peptide intensities. The left side of the plot shows proteins up-regulated in the sample culture in light amino isotope conditions: ERα(+), ERβ(+), and genistein; The right side of the plot shows proteins up-regulated in the sample culture in heavy amino isotope conditions: ERα(+), ERβ(−), and genistein in the T47D-PBI cell line. The spread cloud is lower at high abundance, indicating that quantification is more precise. The data points are colored by their “significance B” (intensity-dependent p value) with dark blue circles having values >0.05, light blue circles having values between 0.05 and 0.001, yellow circles having values between 0.001 and 1e−11, and red circles having values <1e−11.
Differentially expressed proteins were selected using a cutoff confidence level of ratio H/L or L/H significance B ≤0.001. In the next step, we used canonical pathways and molecular and cellular function classification from Ingenuity Pathway Analysis (IPA) to identify overrepresented biological themes among these differentially expressed genes and proteins.
Genistein Modulated Protein Expression in Absence of ERβ (Sample CA)
To determine the genistein-induced ERα-mediated effects on protein expression levels, cells grown in heavy amino acid isotope-containing medium were treated for 24 h with 500 nm genistein, whereas cells grown in light amino acid isotope-containing medium were treated for 24 h with a 10 nm concentration of the ER antagonist ICI 182780 in both cases in the presence of Tet and therefore without the expression of ERβ (sample CA; Tables I and II). This process resulted in the identification of 1994 proteins with at least two unique peptides. The results obtained showed small changes for most proteins (Fig. 3A). Based on the statistical selection criteria (significance B ≤0.001), the data revealed 59 SILAC proteins to be significantly regulated (Table III).
Table III. Most significant ERα-mediated proteins as a result of genistein treatment (sample CA; Table II).
Only H/L SILAC pair proteins are listed.
| UniProt name | Gene name | Protein name | Ratio H/L, normalized | Ratio significance | Ratio H/L count | Sequence coverage |
|---|---|---|---|---|---|---|
| % | ||||||
| A2RRB4_HUMAN | NFAT5 | Nuclear factor of activated T-cells 5 protein | 3.9151 | 8.32e−28 | 2 | 1.5 |
| NBN_HUMAN | NBN | Nibrin | 1.7846 | 1.02e−06 | 2 | 5.3 |
| Q8IWY7_HUMAN | TTBK2 | Tau-tubulin kinase | 1.7012 | 1.26e−16 | 2 | 2.9 |
| MYH10_HUMAN | MYH10 | Myosin-10 | 1.5769 | 2.11e−07 | 14 | 15.5 |
| CCD25_HUMAN | CCDC25 | Coiled coil domain-containing protein 25 | 1.5499 | 0.00013085 | 2 | 11.1 |
| MGP_HUMAN | MGP | Matrix Gla protein | 1.521 | 1.53e−06 | 5 | 23.3 |
| T120A_HUMAN | TMEM120A | Transmembrane protein 120A | 1.5178 | 0.00024373 | 2 | 3.2 |
| MYH14_HUMAN | MYH14 | Myosin-14 | 1.5152 | 0.00025613 | 3 | 4.6 |
| RM20_HUMAN | MRPL20 | 39 S ribosomal protein L20, mitochondrial | 1.5095 | 0.0002857 | 2 | 12.8 |
| K1C18_HUMAN | KRT18 | Keratin, type I cytoskeletal 18 | 1.4982 | 2.90e−18 | 230 | 91.9 |
| Q53HL1_HUMAN | MYL12B | Myosin regulatory light chain MRCL3 variant | 1.4819 | 5.81e−06 | 8 | 37.3 |
| K2C8_HUMAN | KRT8 | Keratin, type II cytoskeletal 8 | 1.4635 | 2.30e−16 | 545 | 86.1 |
| K1C19_HUMAN | KRT19 | Keratin, type I cytoskeletal 19 | 1.4508 | 1.10e−15 | 246 | 95.5 |
| MYH9_HUMAN | MYH9 | Myosin-9 | 1.4135 | 9.37e−14 | 269 | 63.9 |
| MYL6_HUMAN | MYL6 | Myosin light polypeptide 6 | 1.3405 | 3.34e−06 | 22 | 61.6 |
| ASNS_HUMAN | ASNS | Asparagine synthetase (glutamine-hydrolyzing) | 1.3256 | 0.00074994 | 4 | 8.9 |
| LAT1_HUMAN | SLC7A5 | Large neutral amino acids transporter small subunit 1 | 1.2556 | 0.00024142 | 6 | 6.7 |
| NIBAN_HUMAN | FAM129A | Protein Niban | 1.2546 | 0.00025301 | 6 | 9.2 |
| RCN1_HUMAN | RCN1 | Reticulocalbin-1 | 1.2251 | 0.00093813 | 7 | 18.4 |
| NPM_HUMAN | NPM1 | Nucleophosmin | 1.2138 | 3.04e−05 | 71 | 64.6 |
| IPI: IPI00041625.1 | LOC130773 | Similar to 60 S ribosomal protein L23a | 1.1925 | 0.00014684 | 5 | 30.6 |
| 4F2_HUMAN | SLC3A2 | 4F2 cell surface antigen heavy chain | 1.1855 | 0.00024133 | 54 | 28.7 |
| H12_HUMAN | HIST1H1C | Histone H1.2 | 1.1649 | 0.00095077 | 48 | 39.9 |
| RS25_HUMAN | RPS25 | 40 S ribosomal protein S25 | 0.85955 | 0.0001805 | 24 | 55.2 |
| RS16_HUMAN | RPS16 | 40 S ribosomal protein S16 | 0.81669 | 1.37e−06 | 41 | 61 |
| TPR_HUMAN | TPR | Nucleoprotein translocated promoter region (to activated MET oncogene) | 0.81623 | 0.00058563 | 22 | 9.7 |
| NPC2_HUMAN | NPC2 | Epididymal secretory protein E1 | 0.80913 | 0.00035735 | 16 | 35.3 |
| VAT1_HUMAN | VAT1 | Synaptic vesicle membrane protein VAT-1 homolog | 0.80802 | 0.00032994 | 16 | 35.1 |
| Q6ZSA3_HUMAN | BCKDHA | cDNA FLJ45695 fis, clone FEBRA2013570, highly similar to 2-oxoisovalerate (lipoate) dehydrogenase α subunit, mitochondrial (EC 1.2.4.4) | 0.80718 | 0.00031066 | 8 | 16.1 |
| ACTBL_HUMAN | ACTBL2 | β-Actin-like protein 2 | 0.78565 | 1.53e−08 | 9 | 21.8 |
| RS18_HUMAN | RPS18 | 40 S ribosomal protein S18 | 0.75035 | 2.95e−11 | 40 | 67.1 |
| NFS1_HUMAN | NFS1 | Cysteine desulfurase, mitochondrial | 0.73481 | 0.00073737 | 6 | 9.4 |
| IPI: IPI00903046.1 | Putative uncharacterized protein ENSP00000384045 (fragment) | 0.73352 | 0.00069096 | 2 | 13.5 | |
| Q53H03_HUMAN | NASP | Nuclear autoantigenic sperm protein isoform 2 variant | 0.73307 | 0.00067556 | 3 | 7.5 |
| LMNA_HUMAN | LMNA | Lamin-A/C | 0.71605 | 1.83e−14 | 102 | 54.8 |
| RS29_HUMAN | RPS29 | 40 S ribosomal protein S29 | 0.70426 | 1.21e−08 | 5 | 47.4 |
| LPP_HUMAN | LPP | Lipoma-preferred partner | 0.6822 | 3.54e−05 | 3 | 8.2 |
| MAP4_HUMAN | MAP4 | Microtubule-associated protein 4 | 0.63442 | 1.02e−06 | 9 | 12.7 |
| THOC2_HUMAN | THOC2 | THO complex subunit 2 | 0.62342 | 0.00051663 | 2 | 1.4 |
| NEUL_HUMAN | NLN | Neurolysin, mitochondrial | 0.62126 | 0.00047125 | 2 | 3 |
| CHCH1_HUMAN | CHCHD1 | Coiled coil-helix-coiled coil-helix domain-containing protein 1 | 0.61855 | 0.0004196 | 2 | 25.4 |
| CHM2B_HUMAN | CHMP2B | Charged multivesicular body protein 2b | 0.61376 | 0.00034025 | 2 | 4.7 |
| TOM40_HUMAN | TOMM40 | Mitochondrial import receptor subunit TOM40 homolog | 0.61264 | 3.56e−15 | 4 | 10.5 |
| % | ||||||
| LMNB1_HUMAN | LMNB1 | Lamin-B1 | 0.56768 | 1.45e−09 | 4 | 12.1 |
| APLP2_HUMAN | APLP2 | Amyloid-like protein 2 | 0.56042 | 2.33e−05 | 2 | 4.7 |
| PIP_HUMAN | PIP | Prolactin-inducible protein | 0.52576 | 2.73e−06 | 4 | 26 |
| HMGN1_HUMAN | HMGN | Non-histone chromosomal protein HMG-14 | 0.51956 | 1.79e−06 | 3 | 14 |
| SYAP1_HUMAN | SYAP1 | Synapse-associated protein 1 | 0.51352 | 1.20e−12 | 2 | 7.4 |
| MOGS_HUMAN | MOGS | Mannosyl-oligosaccharide glucosidase | 0.49431 | 2.80e−07 | 2 | 3.8 |
| HNRPC_HUMAN | HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 0.44009 | 2.22e−09 | 2 | 13.4 |
| SPS1_HUMAN | SEPHS1 | Selenide, water dikinase 1 | 0.37038 | 4.08e−56 | 4 | 13.3 |
| OFUT1_HUMAN | POFUT1 | GDP-fucose protein O-fucosyltransferase 1 | 0.35029 | 1.99e−14 | 2 | 4.1 |
| MYO15_HUMAN | MYO15A | Myosin-XV | 0.33393 | 1.87e−31 | 2 | 0.9 |
| S10A8_HUMAN | S100A8 | Protein S100-A8 | 0.28004 | 1.44e−20 | 2 | 20.4 |
| KPRP_HUMAN | KPRP | Keratinocyte proline-rich protein | 0.2706 | 1.29e−21 | 2 | 1.6 |
| TIM9B_HUMAN | FXC1 | Mitochondrial import inner membrane translocase subunit Tim9 B | 0.15658 | 4.51e−42 | 2 | 24.3 |
| S10A9_HUMAN | S100A9 | Protein S100-A9 | 0.15405 | 5.78e−193 | 5 | 26.3 |
| LEG7_HUMAN | LGALS7 | Galectin-7 | 0.12058 | 2.35e−54 | 2 | 19.9 |
| PNKD_HUMAN | PNKD | Probable hydrolase paroxysmal nonkinesigenic dyskinesia | 0.05602 | 8.55e−100 | 3 | 30.3 |
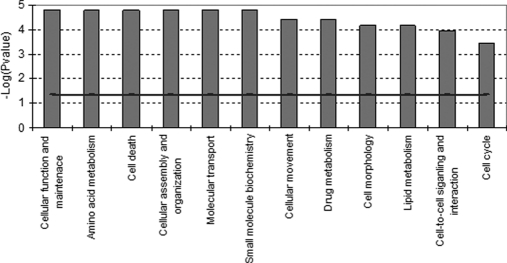
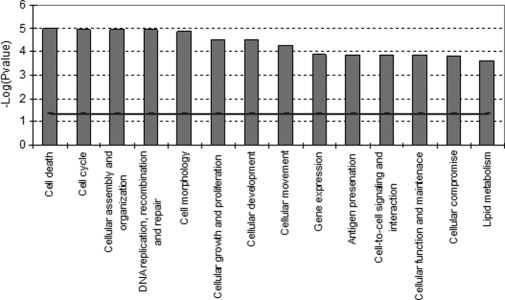
Functional analysis (see “Materials and Methods”) of the proteins in this data set revealed that the major biological functions that were affected (increased or decreased) by genistein included cellular function and maintenance, cell death, cellular assembly and organization, cell movement, cell morphology, lipid metabolism, and cell cycle (Fig. 4). Most of the proteins belonging to these functional classes were up-regulated, whereas down-regulated proteins were categorized in the classes representing cell death and lipid metabolism (Table III). Notably, five of the most up-regulated proteins upon genistein-induced ERα-mediated protein expression were myosins (MYH10, MYH14, MYL12B, MYH9, and MYL6). The majority of these myosins are actively involved in cell assembly and organization or cytoskeleton reorganization. Among the several significantly down-regulated proteins, we found at least three proteins, S100A8, S100A9, and PIP, of specific interest because their expression is associated with a decrease of cell proliferation and induction of apoptosis (29, 30). The two S100 EF-hand calcium-binding proteins S100A8/A9 induce apoptosis in various cells, especially tumor cells like MCF-7 (29).
Fig. 4.
Molecular and cellular functional classes affected by genistein-induced ERα-mediated effects on protein expression (sample CA; Table II). IPA molecular and cellular functions of the H/L proteins with a p value <0.001 are shown. The selected scoring method was based on Fisher's exact test p value. The high level functional categories that are involved in this analysis are displayed along the x axis in decreasing order of significance. The y axis displays the −(log) significance. The horizontal line denotes the cutoff for significance (p value of 0.05).
Genistein Modulated Protein Expression in Presence of ERβ (Sample DB)
Proteins induced by genistein in ERα- and ERβ-expressing cells could be identified by comparing cells grown in light amino acid isotope-containing medium treated with 500 nm genistein with cells grown in heavy isotope-containing medium exposed to 10 nm pure ER antagonist ICI 182780, both grown in the absence of Tet and thus expressing ERβ (sample DB; Tables I and II). This resulted in the identification of 1905 proteins with at least two unique peptides. Significant changes observed in the protein level after 24 h of treatment are depicted in Fig. 3B. Based on the statistical selection criteria (significance B ≤0.001), the data revealed 66 SILAC proteins to be significantly regulated (Table IV). For IPA and further analysis, inverse ratios were used to allow additional interexperiment comparisons.
Table IV. Most significant ERβ-mediated proteins as a result of genistein treatment (sample DB; Table II).
Only L/H SILAC pair proteins are listed.
| UniProt name | Gene name | Protein name | Ratio L/H, normalized | Ratio significance | Ratio H/L count | Sequence coverage |
|---|---|---|---|---|---|---|
| % | ||||||
| FGFR2_HUMAN | FGFR2 | Fibroblast growth factor receptor 2 | 10.47724 | 1.33e−124 | 2 | 2.1 |
| H4_HUMAN | HIST1H4A | Histone H4 | 7.141837 | 5.69e−88 | 7 | 51.5 |
| B2R5B6_HUMAN | HIST1H2AG | Histone H2A | 5.420936 | 8.26e−47 | 2 | 27.2 |
| TR112_HUMAN | AD-001 | TRM112-like protein | 3.275038 | 5.13e−24 | 4 | 32 |
| SPS1_HUMAN | SEPHS1 | Selenide, water dikinase 1 | 3.242227 | 1.22e−23 | 2 | 9.2 |
| ELOV1_HUMAN | ELOVL1 | Elongation of very long chain fatty acids protein 1 | 3.09272 | 1.72e−12 | 2 | 4.3 |
| PIP_HUMAN | PIP | Prolactin-inducible protein | 2.97274 | 9.65e−12 | 4 | 26 |
| ARM10_HUMAN | ARMC10 | Armadillo repeat-containing protein 10 | 2.851521 | 5.55e−11 | 2 | 4.7 |
| K0753_HUMAN | KIAA0753 | Uncharacterized protein KIAA0753 | 2.74198 | 7.31e−18 | 2 | 3.1 |
| DHB8_HUMAN | HSD17B8 | Estradiol 17β-dehydrogenase 8 | 2.345051 | 8.49e−08 | 2 | 10.7 |
| HNRPC_HUMAN | HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 2.083811 | 5.49e−14 | 11 | 21.9 |
| ANM1_HUMAN | PRMT1 | Protein arginine N-methyltransferase 1 | 2.078829 | 3.24e−10 | 2 | 10.8 |
| A8MXP9_HUMAN | MATR3 | Putative uncharacterized protein MATR3 | 2.051366 | 5.57e−06 | 2 | 3 |
| RMTL1_HUMAN | RNMTL1 | RNA methyltransferase-like protein 1 | 1.955225 | 2.12e−05 | 2 | 2.9 |
| S10A9_HUMAN | S100A9 | Protein S100-A9 | 1.939827 | 2.62e−05 | 2 | 13.2 |
| ESR2_HUMAN | ESR2 | Estrogen receptor β | 1.93304 | 2.88e−05 | 3 | 7.9 |
| HYES_HUMAN | EPHX2 | Epoxide hydrolase 2 | 1.871608 | 6.24e−08 | 2 | 5.2 |
| A6NKB8_HUMAN | RNPEP | Putative uncharacterized protein RNPEP | 1.750271 | 0.0003341 | 2 | 36.5 |
| H10_HUMAN | H1F0 | Histone H1.0 | 1.687564 | 5.23e−06 | 11 | 19.6 |
| H2A1B_HUMAN | HIST1H2AB | Histone H2A type 1-B/E | 1.686369 | 1.54e−12 | 13 | 35.4 |
| Q6Q3G8_HUMAN | LAMP2 | Lysosomal-associated membrane protein 2C | 1.520751 | 0.0002132 | 2 | 1.9 |
| H2B1N_HUMAN | HIST1H2BN | Histone H2B type 1-N | 1.518603 | 1.13e−05 | 10 | 25.3 |
| LMNA_HUMAN | LMNA | Lamin-A/C | 1.511624 | 0.0002583 | 2 | 51.2 |
| INO1_HUMAN | ISYNA1 | Inositol-3-phosphate synthase 1 | 1.504959 | 1.68e−05 | 14 | 24.2 |
| HNRL2_HUMAN | HNRNPUL2 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 | 1.49997 | 0.0003293 | 3 | 3.1 |
| NHRF1_HUMAN | SLC9A3R1 | Na+/H+ exchange regulatory cofactor NHE-RF1 | 1.497253 | 2.11e−05 | 18 | 34.9 |
| B4DWK6_HUMAN | MUC1 | cDNA FLJ60927, highly similar to Mucin-1 | 1.496468 | 2.16e−05 | 16 | 19.1 |
| CD9_HUMAN | CD9 | CD9 antigen | 1.494098 | 0.0003718 | 3 | 14.5 |
| NUMA1_HUMAN | NUMA1 | Nuclear mitotic apparatus protein 1 | 1.491402 | 3.76e−08 | 71 | 35.5 |
| H14_HUMAN | HIST1H1E | Histone H1.4 | 1.45588 | 6.87e−05 | 11 | 39.7 |
| TFR1_HUMAN | TFRC | Transferrin receptor protein 1 | 1.386155 | 4.75e−06 | 62 | 37.2 |
| H15_HUMAN | HIST1H1B | Histone H1.5 | 1.385003 | 5.00e−06 | 40 | 38.1 |
| H12_HUMAN | HIST1H1C | Histone H1.2 | 1.314389 | 9.28e−05 | 45 | 45.1 |
| PYGB_HUMAN | PYGB | Glycogen phosphorylase, brain form | 1.298937 | 0.0001691 | 24 | 21.9 |
| ROA2_HUMAN | HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 1.272961 | 0.000447 | 97 | 74.5 |
| ACLY_HUMAN | ACLY | ATP-citrate synthase | 0.820075 | 0.0007347 | 65 | 35.6 |
| COTL1_HUMAN | COTL1 | Coactosin-like protein | 0.818666 | 0.0006644 | 22 | 55.6 |
| IF4G1_HUMAN | EIF4G1 | Eukaryotic translation initiation factor 4 γ1 | 0.811886 | 0.0003983 | 60 | 29.8 |
| B4DUT8_HUMAN | CNN2 | cDNA FLJ52765, highly similar to Calponin-2 | 0.808211 | 0.0002993 | 27 | 36.7 |
| A7E2F8_HUMAN | TNKS1BP1 | Tankyrase 1-binding protein 1, 182 kDa | 0.798276 | 0.0001335 | 63 | 31.9 |
| VINC_HUMAN | VCL | Vinculin | 0.792896 | 8.39e−05 | 98 | 61 |
| REEP5_HUMAN | REEP5 | Receptor expression-enhancing protein 5 | 0.786225 | 4.62e−05 | 25 | 34.4 |
| CYB5_HUMAN | CYB5A | Cytochrome b5 | 0.770594 | 0.0006788 | 10 | 47 |
| FLNA_HUMAN | FLNA | Filamin-A | 0.768285 | 8.22e−06 | 239 | 53.9 |
| PEPL_HUMAN | PPL | Periplakin | 0.761325 | 0.0003987 | 34 | 17.3 |
| NFKB2_HUMAN | NFKB2 | Nuclear factor NF-κB p100 subunit | 0.753977 | 0.0009489 | 5 | 5.1 |
| MAP4_HUMAN | MAP4 | Microtubule-associated protein 4 | 0.742666 | 0.0005389 | 4 | 6 |
| PUR8_HUMAN | ADSL | Adenylosuccinate lyase | 0.720305 | 0.0001592 | 2 | 5.6 |
| UBF1_HUMAN | UBTF | Nucleolar transcription factor 1 | 0.717051 | 0.0001318 | 4 | 4.6 |
| CSRP1_HUMAN | CSRP1 | Cysteine- and glycine-rich protein 1 | 0.709673 | 6.73e−09 | 30 | 67.9 |
| DDRGK_HUMAN | DDRGK1 | DDRGK domain-containing protein 1 | 0.701361 | 5.04e−05 | 2 | 8.6 |
| ANXA1_HUMAN | ANXA1 | Annexin A1 | 0.691324 | 2.67e−06 | 9 | 20.2 |
| % | ||||||
| CSN1_HUMAN | GPS1 | COP9 signalosome complex subunit 1 | 0.654407 | 1.75e−06 | 2 | 6.8 |
| ANXA3_HUMAN | ANXA3 | Annexin A3 | 0.647249 | 1.76e−13 | 22 | 37.8 |
| TOP2B_HUMAN | TOP2B | DNA topoisomerase 2-β | 0.643791 | 0.0004078 | 4 | 2.1 |
| SLK_HUMAN | SLK | STE20-like serine/threonine-protein kinase | 0.62692 | 0.0001979 | 2 | 1.9 |
| DNPEP_HUMAN | DNPEP | Aspartyl aminopeptidase | 0.621157 | 9.79e−08 | 2 | 4.9 |
| UB2L6_HUMAN | UBE2L6 | Ubiquitin/ISG15-conjugating enzyme E2 L6 | 0.534416 | 1.20e−06 | 2 | 11.8 |
| STAT1_HUMAN | STAT1 | Signal transducer and activator of transcription 1-α/β | 0.511326 | 5.44e−17 | 9 | 15.1 |
| Q8WYY7_HUMAN | TTC9C | Tetratricopeptide repeat protein 9C (TPR repeat protein 9C) | 0.474518 | 2.35e−16 | 3 | 6.5 |
| A8K5J7_HUMAN | BAG5 | cDNA FLJ77290, highly similar to Homo sapiens BCL2-associated athanogene 5 (BAG5), mRNA | 0.470389 | 7.84e−09 | 2 | 6.6 |
| MX1_HUMAN | MX1 | Interferon-induced GTP-binding protein Mx1 | 0.221029 | 2.22e−78 | 4 | 11.9 |
| UCRP_HUMAN | ISG15 | Interferon-induced 17-kDa protein | 0.181045 | 3.92e−77 | 4 | 29.7 |
| PRAF2_HUMAN | PRAF2 | PRA1 family protein 2 | 0.106702 | 9.82e−170 | 2 | 16.9 |
As reported previously (20), in T47D-ERβ cells grown in the absence of Tet and thus expressing both ERα and ERβ, genistein-induced cell proliferation was notably reduced as compared with genistein-induced cell proliferation in T47D cells expressing only ERα. The SILAC data of the sample reflect genistein-induced up-regulation of nine histone-related proteins (HIST1H4A, HIST1H2AG, H1F0, HIST1H2AB, HIST1H2BN, HIST1H1E, HIST1H1B, HIST1H1C, and NUMA1), which points at a general down-regulation of gene transcription (31, 32). In addition, down-regulation of TOP2B (Topoisomerase IIβ) was significant. Topoisomerases are enzymes that play important roles in transcription, DNA synthesis, and chromosome segregation (33). This result is in line with a previously published inhibition of Topoisomerase IIβ by genistein influencing cell growth (34). Important proteins belonging to the same signaling network, i.e. interferon signaling pathways (35), were STAT1, MX1, ANXA1, UBE2L6, and ISG15. Also, ANXA3, ANXA1, and VCL, proteins important for actin cytoskeleton signaling, cell migration, and anti-inflammatory processes (36, 37), were found to be down-regulated by genistein in T47D cells expressing both ERα and ERβ.
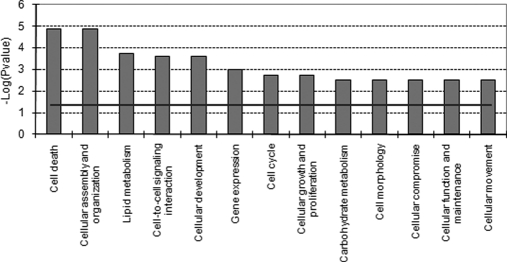
Molecular and cellular function profiling of these data (Fig. 5) points at significant effects on pathways involved in cell death, cell cycle, cellular assembly and organization, DNA replication, recombination and repair, cell growth and proliferation, and cellular movement. Proteins up-regulated by genistein were directly involved in the first three categories, whereas the down-regulated proteins were part of the categories cell movement and cell death (Table IV).
Fig. 5.
Molecular and cellular functional classes affected by genistein-induced ERβ-mediated effects on protein expression (sample DB; Table II). IPA molecular and cellular functions of the L/H proteins with a p value <0.001 are shown. The selected scoring method was based on Fisher's exact test p value. The high level functional categories that are involved in this analysis are displayed along the x axis in decreasing order of significance. The y axis displays the −(log) significance. The horizontal line denotes the cutoff for significance (p value of 0.05).
Genistein Modulated Protein Expression in Presence of ERβ (Sample ED)
Genistein-modulated ERβ-regulated proteins could also be identified by comparing T47D-ERβ cells grown in light amino acid isotope-containing medium treated with 500 nm genistein to mock-transfected T47D control cells grown in heavy isotope-containing medium treated with 500 nm genistein, both grown in the absence of Tet with the T47D-ERβ cells expressing ERβ (sample ED; Tables I and II). Significant changes observed in protein levels after 24 h of treatment are depicted in Fig. 3C. The mock control cell line does not express ERβ. This comparison resulted in the identification of 1995 proteins with at least two unique peptides that were differentially expressed upon genistein exposure in the presence of ERβ.
Based on the statistical selection criteria (significance B ≤0.001) the data revealed 58 SILAC proteins that appeared to be significantly regulated (Fig. 3C and Table V). Genistein-induced ERβ-mediated protein expression causes the most significant changes in proteins involved in processes like cell death (ERβ, FHL2, LMNA, and CYB5A), lipid metabolism (ERβ, ACOX1, COLT1, and CYB5A), gene expression (ERβ, HIST1H2AG, HIST1H4A, H2AFZ, and HNRNPC), and cell cycle (LMNA, ITGAV, and ERβ) (Fig. 6 and Table V).
Table V. Most significant ERβ-mediated proteins as a result of genistein treatment (sample ED; Table II).
Only L/H SILAC pair proteins are listed.
| UniProt name | Gene name | Protein name | Ratio H/L, normalized | Ratio significance | Ratio H/L count | Sequence coverage |
|---|---|---|---|---|---|---|
| % | ||||||
| U634C_HUMAN | ANKRD36B | UPF0634 protein C | 5.594 | 1.04e−17 | 2 | 1.5 |
| MGP_HUMAN | MGP | Matrix Gla protein | 3.1457 | 1.26e−09 | 7 | 23.3 |
| K1C18_HUMAN | KRT18 | Keratin, type I cytoskeletal 18 | 2.4136 | 8.26e−09 | 215 | 94.4 |
| GIPC1_HUMAN | GIPC1 | PDZ domain-containing protein GIPC1 | 2.2777 | 1.76e−05 | 2 | 9.3 |
| QORX_HUMAN | TP53I3 | Putative quinone oxidoreductase | 2.245 | 1.11e−07 | 21 | 40.4 |
| CLD1_HUMAN | CLDN1 | Claudin-1 | 2.2442 | 1.38e−06 | 5 | 19 |
| EFHD1_HUMAN | EFHD1 | EF-hand domain-containing protein D1 | 2.1675 | 3.63e−06 | 8 | 18.4 |
| K1107_HUMAN | KIAA1107 | Uncharacterized protein KIAA1107 | 2.1623 | 3.87e−06 | 2 | 2.1 |
| REEP5_HUMAN | REEP5 | Receptor expression-enhancing protein 5 | 2.1537 | 4.50e−07 | 32 | 34.9 |
| AATM_HUMAN | GOT2 | Aspartate aminotransferase, mitochondrial | 2.0696 | 1.61e−06 | 36 | 41.6 |
| K2C8_HUMAN | KRT8 | Keratin, type II cytoskeletal 8 | 2.0237 | 3.21e−06 | 535 | 85.7 |
| B4DY64_HUMAN | COMTD1 | cDNA FLJ52132, highly similar to catechol O-methyltransferase (EC 2.1.1.6) domain-containing protein 1 | 1.89 | 0.00061374 | 2 | 2.8 |
| CRIP1_HUMAN | CRIP1 | Cysteine-rich protein 1 | 1.8892 | 2.35e−05 | 7 | 79.2 |
| B4DG34_HUMAN | VPS24 | cDNA FLJ58988, highly similar to charged multivesicular body protein 3 | 1.8736 | 0.00071107 | 2 | 4.8 |
| MET7A_HUMAN | METTL7A | Methyltransferase-like protein 7A | 1.8288 | 0.00023505 | 6 | 17.2 |
| RAB18_HUMAN | RAB18 | Ras-related protein Rab-18 | 1.8235 | 0.00025018 | 12 | 34.5 |
| CNPY2_HUMAN | CNPY2 | Protein canopy homolog 2 | 1.8106 | 0.00094353 | 14 | 37.9 |
| K1C19_HUMAN | KRT19 | Keratin, type I cytoskeletal 19 | 1.8021 | 8.29e−05 | 240 | 93.8 |
| A8MPQ9_HUMAN | LYRM7 | Putative uncharacterized protein LYRM7 | 1.7869 | 0.00038649 | 4 | 24 |
| NPM_HUMAN | NPM1 | Nucleophosmin | 1.7869 | 0.00010282 | 68 | 71.4 |
| APT_HUMAN | APRT | Adenine phosphoribosyltransferase | 1.7725 | 0.00012622 | 36 | 78.3 |
| GRP75_HUMAN | HSPA9 | Stress-70 protein, mitochondrial | 1.7722 | 0.00012673 | 142 | 60.8 |
| PTGR2_HUMAN | PTGR2 | Prostaglandin reductase 2 | 1.7713 | 0.00046415 | 3 | 15.7 |
| CALX_HUMAN | CANX | Calnexin | 1.7616 | 0.0001473 | 31 | 28.2 |
| CBX5_HUMAN | CBX5 | Chromobox protein homolog 5 | 1.7157 | 0.00088702 | 5 | 19.4 |
| ACY1_HUMAN | ACY1 | Aminoacylase-1 | 1.7126 | 0.00091913 | 6 | 10.6 |
| FLNA_HUMAN | FLNA | Filamin-A | 0.60289 | 0.00070352 | 269 | 51.8 |
| INO1_HUMAN | ISYNA1 | Inositol-3-phosphate synthase 1 | 0.60177 | 0.00067576 | 17 | 23.8 |
| COTL1_HUMAN | COTL1 | Coactosin-like protein | 0.56046 | 0.00013071 | 22 | 54.2 |
| PRSS8_HUMAN | PRSS8 | Prostasin | 0.54848 | 0.00087159 | 2 | 12.8 |
| CYB5_HUMAN | CYB5A | Cytochrome b5 | 0.52474 | 0.00012112 | 13 | 56 |
| A8MZH9_HUMAN | COX7A2L | Putative uncharacterized protein COX7A2L | 0.51931 | 0.00031952 | 3 | 34.8 |
| S10AG_HUMAN | S100A16 | Protein S100-A16 | 0.5155 | 8.08e−05 | 9 | 68 |
| RL38_HUMAN | RPL38 | 60 S ribosomal protein L38 | 0.51546 | 0.00027718 | 5 | 50 |
| B2R5B6_HUMAN | HIST1H2AG | Histone H2A | 0.5063 | 9.01e−06 | 15 | 27.2 |
| Q2I5I4_HUMAN | FHL2 | Four and a half LIM domain protein 2 | 0.48592 | 8.52e−05 | 6 | 26 |
| ACOX1_HUMAN | ACOX1 | Peroxisomal acyl-coenzyme A oxidase 1 | 0.48194 | 1.59e−05 | 14 | 20 |
| SRPR_HUMAN | SRPR | Signal recognition particle receptor subunit α | 0.47482 | 5.24e−05 | 2 | 3.3 |
| ANXA3_HUMAN | ANXA3 | Annexin A3 | 0.47299 | 9.83e−06 | 14 | 33.7 |
| LMNA_HUMAN | LMNA | Lamin-A/C | 0.47133 | 1.09e−06 | 113 | 61.1 |
| ITAV_HUMAN | ITGAV | Integrin αV | 0.46812 | 3.86e−05 | 5 | 4.8 |
| S10A8_HUMAN | S100A8 | Protein S100-A8 | 0.46418 | 0.00078094 | 2 | 32.3 |
| ARMX3_HUMAN | ARMCX3 | Armadillo repeat-containing X-linked protein 3 | 0.45982 | 0.00067904 | 3 | 12.1 |
| PREX1_HUMAN | PREX1 | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | 0.45533 | 2.10e−05 | 11 | 7.1 |
| DHSB_HUMAN | SDHB | Succinate dehydrogenase (ubiquinone) iron-sulfur subunit, mitochondrial | 0.42786 | 0.00022185 | 2 | 7.1 |
| H4_HUMAN | HIST1H4A | Histone H4 | 0.39614 | 2.83e−09 | 9 | 44.7 |
| HNRPC_HUMAN | HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 0.37985 | 1.58e−08 | 11 | 25.2 |
| % | ||||||
| H2AZ_HUMAN | H2AFZ | Histone H2A.Z | 0.35593 | 8.43e−06 | 2 | 20.3 |
| GABT_HUMAN | ABAT | 4-Aminobutyrate aminotransferase, mitochondrial | 0.35076 | 1.04e−09 | 9 | 13 |
| LMNB2_HUMAN | LMNB2 | Lamin-B2 | 0.31103 | 5.26e−07 | 2 | 4.5 |
| UCRP_HUMAN | ISG15 | Interferon-induced 17-kDa protein | 0.22984 | 3.20e−10 | 5 | 29.7 |
| UGDH_HUMAN | UGDH | UDP-glucose 6-dehydrogenase | 0.21775 | 7.17e−11 | 2 | 6.1 |
| MX1_HUMAN | MX1 | Interferon-induced GTP-binding protein Mx1 | 0.17993 | 3.85e−23 | 12 | 17.1 |
| A8MXP9_HUMAN | MATR3 | Putative uncharacterized protein MATR3 | 0.17222 | 5.90e−14 | 2 | 3.6 |
| ESR2_HUMAN | ESR2 | Estrogen receptor β | 0.15132 | 7.65e−16 | 2 | 5.7 |
| DMD_HUMAN | DMD | Dystrophin | 0.14177 | 1.63e−24 | 4 | 0.3 |
| LDHB_HUMAN | LDHB | l-Lactate dehydrogenase B chain | 0.099128 | 1.64e−40 | 6 | 33.2 |
Fig. 6.
Molecular and cellular functional classes affected in second approach to detect genistein-induced ERβ-mediated effects on protein expression (sample ED; Table II). IPA molecular and cellular functions of the H/L proteins with a p value <0.001 are shown. The selected scoring method was based on Fisher's exact test p value. The high level functional categories that are involved in this analysis are displayed along the x axis in decreasing order of significance. The y axis displays the −(log) significance. The horizontal line denotes the cutoff for significance (p value of 0.05).
Both ERβ target approaches (samples DB and ED) found comparable results. In both experiments, genistein-mediated ERβ activation regulated molecular functions including cell death, cell cycle, cell movements, and lipid metabolism. Among the most significant ERβ targets identified were proteins mediating gene transcription such as HNRNPC and histone-related proteins.
Transcriptomics Data
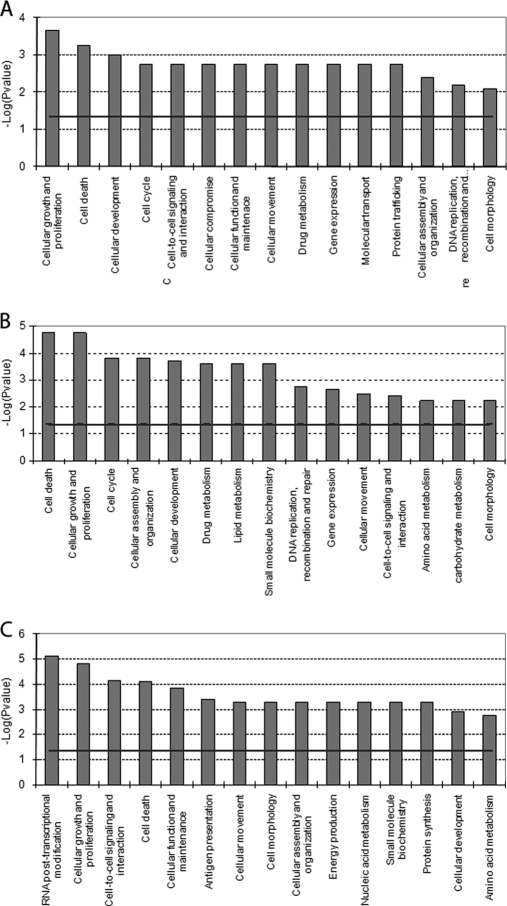
In addition to the proteomics, transcriptomics also was performed. Using the same samples as for the SILAC protein analysis, differentially expressed genes for each experiment (CA, DB, and ED) were obtained. The comparison of gene expression levels between genistein- and ICI-induced T47D-ERβ cells in the absence of ERβ (sample CA) did not identify significant differentially expressed genes. This is different from the study of Williams et al. (8). However, it is in agreement with the relatively low levels of ERα present. The presence of more subtle gene regulations that could indeed be ERα-mediated was investigated. A functional analysis of the top 30 genes as ranked by p value (Fig. 7A) showed that these genes are involved in biological processes that match ERα-mediated effects. Cellular function and maintenance, cell-to-cell signaling and interaction, cellular movement, cell death, and cellular assembly and organization were enhanced at the mRNA level as already seen at the protein level. The majority of the top 30 gene set were up-regulated as a result of genistein treatment, resulting in stimulation of pathways for cellular growth and proliferation, cell death, and cellular development.
Fig. 7.
Most significant molecular and cellular functional classes affected as result of genistein-induced ERα-mediated effects on gene expression (sample CA; Table II) (A), genistein-induced ERβ-mediated effects on gene expression (sample DB; Table II) (B), and a second approach to detect genistein-induced ERβ-mediated effects on gene expression (sample ED; Table II) (C). IPA molecular and cellular functions of the genes with a p value <0.05 are shown. The selected scoring method was based on Fisher's exact test p value. The high level functional categories that are involved in this analysis are displayed along the x axis in decreasing order of significance. The y axis displays the −(log) significance. The horizontal line denotes the cutoff for significance (p value of 0.05).
The results of the genistein treatment effects in the presence of ERβ (sample DB) contained a set of 73 differentially regulated genes (p ≤ 0.05). Functional analysis of this gene set showed up-regulation of cell death and down-regulation of cellular growth and proliferation and cell cycle regulation. These effects (Fig. 7B) were similar to those observed at the protein level, pointing at increases in expression of genes and proteins involved in for example cellular growth and proliferation, cell cycle, and cellular assembly and organization.
The second approach to detect genistein-modulated ERβ-mediated gene expression (sample ED) revealed 1011 differentially regulated genes (p ≤ 0.05). Genistein-modulated ERβ-mediated effects (Fig. 7C) were related to RNA post-transcriptional modification, cellular growth and proliferation, cell-to-cell signaling and interaction, cell death, and cellular function and maintenance.
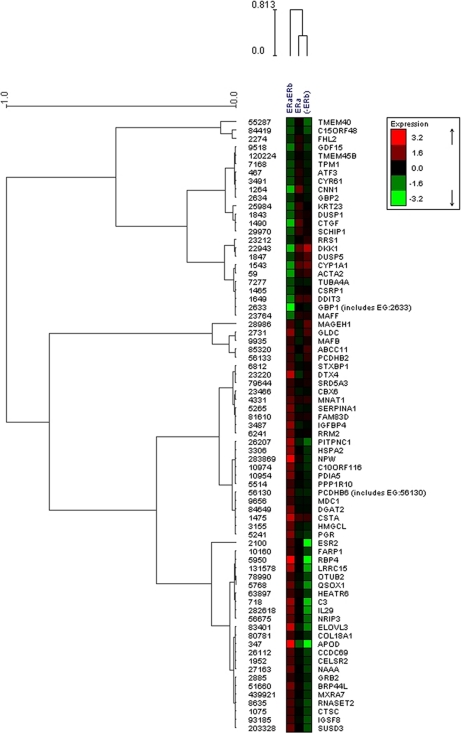
Furthermore, hierarchical cluster analysis of the gene expression of the 73 genes differentially regulated by ERβ was used to identify three main clusters in this gene set (Fig. 8). Genes present in the upper part of the dendrogram correspond to those genes down-regulated in genistein-exposed T47D-ERβ cells with ERβ expression. Pathway and functional analysis shows the involvement of these genes with cellular growth, proliferation, and cell cycle. Genes clustered in the lower part of the dendrogram correspond to those genes up-regulated in genistein-exposed T47D-ERβ cells expressing ERβ and belong to the functional group of cell death. APOD, RBP4, IL29, NRIP3, and LRRC15 genes were found to be clearly up-regulated by genistein-ERβ (see supplemental Fig. S1). It was also observed that negative regulators of ERα transcription are clustered together with ERβ (ESR2) and progesterone receptor (PGR). In addition, down-regulation of genes involved in cellular growth and proliferation was seen in the last cluster of genes in the center of the dendrogram. Most of the genes belonging to this group are negative regulators of colony formation of cancer cell lines.
Fig. 8.
Hierarchical cluster analysis of 73 differentially regulated genes using complete linkage and Pearson correlation similarity. Gene symbols and Entrez Gene identity are shown.
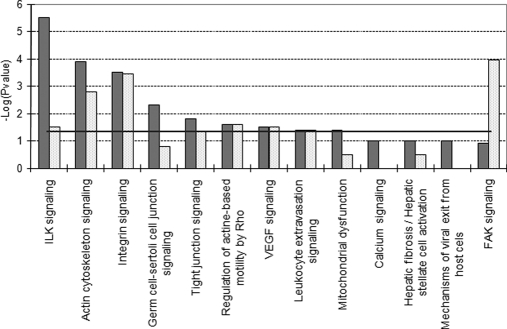
Finally, to have a global view of the pathways affected by genistein treatment, we conducted pathway enrichment analysis using IPA. The transcriptome and the proteome (Fig. 9) data were used to compare genistein-exposed cells expressing ERβ with those not expressing ERβ. Integrin-linked kinase signaling, actin cytoskeleton signaling, and integrin signaling were the most significant pathways influenced by genistein in the presence of low levels of ERα and the absence of ERβ, whereas focal adhesion kinase signaling, actin cytoskeleton signaling, and integrin signaling were affected by the presence of ERβ.
Fig. 9.
Top IPA canonical pathways regulated by ERα-mediated effects (dark columns) and ERβ-mediated effects (light columns) as based on Fisher's exact test p value using larger protein data set (p < 0. 05). The horizontal line denotes the cutoff for significance (p value of 0.05). ILK, integrin-linked kinase; FAK, focal adhesion kinase.
In summary, both at the gene and protein levels, genistein induced signatures of cell proliferation and negative regulation of cell death in the absence of ERβ (sample CA), whereas signatures of up-regulation of cell death, negative regulation of cell cycle, and cell proliferation were observed during genistein treatment in the presence of ERβ (contrast DB and ED).
DISCUSSION
Various molecular profiling technologies have been developed recently to identify and quantify proteins and/or genes in biological samples. From these studies, it was observed that mRNA levels are only a partial reflection of the functional state of an organism and that a comprehensive understanding of the genomic information will require means of analyzing quantitative differences in protein expression on a proteome-wide scale (38–41). In general, an overall positive correlation between protein and mRNA abundance has been observed in many organisms, but simple correlations are insufficient to assess regulatory patterns of gene and protein expression (41). Therefore, in this study, we combined SILAC-based proteomics and microarray analysis of identical samples to address the signaling effects of estrogen receptor subtypes in the T47D-ERβ breast cancer cells exposed to the phytoestrogen genistein.
In ERα-positive breast cancer cells, growth and proliferation are preserved by active transcription of ER targets (42, 43), whereas ERβ expression results in a significant decrease in cell proliferation (21, 22). In previous studies, we observed that genistein, a phytoestrogen with relatively higher affinity for ERβ (11, 44) than for ERα, increased cell proliferation in ERα-positive cells at physiologically relevant concentrations (20). Furthermore, genistein did not induce cell proliferation in ER-negative breast cancer cells like MDA-MB-231 (17, 45).
In the present study, ERα expression levels in the ICI-synchronized T47D-ERβ cell line were 6–10% compared with the wild type T47D cells as measured by ELISA (Fig. 2A). However, ERα mRNA was detected in all samples (Fig. 2B) albeit at levels 30-fold lower than the maximum ERβ mRNA levels detected in the T47D-ERβ cells in the absence of tetracycline. The low levels of ERα protein observed are in accordance with previous studies where it was shown that (i) the ERα protein displays a shorter half-life after ligand binding (46), (ii) ERα is subject to degradation induced by its antagonist ICI 182780 (47), and (iii) ERα may be down-regulated by the genistein treatment. It has been proposed that proteasomal degradation of ERα protein is increased in the presence of the co-regulated proteins KRT18 and KRT8 (48, 49). Up-regulation of these cytokeratins (Tables III and V) attracts the receptor into close proximity to nuclear matrix-associated proteasomes for degradation as has been described after treatment with ICI 182780 (50). In the samples treated with genistein, expression of ERα, after ICI withdrawal, could return to low basal levels (46, 48, 49) because genistein influences the expression of ERα. This observation is supported by the co-regulation of ERα and c-MYC proteins. The proto-oncogene c-MYC is normally up-regulated by ERα in response to the presence of estradiol (51, 52). On the other hand, ERβ is able to repress c-MYC expression (5, 8, 50). In our proteome data set, the differential regulation of most of the ERα-regulated proteins appeared to be similar to that of c-MYC (53), including up-regulation of KRT18, MYH10, HNRPC, MYH9, and MYL6.
It has been proposed that epigenetic mechanisms including DNA methylation and histone modifications might contribute to ERβ-mediated ERα silencing (44, 45, 54, 55). Furthermore, genistein has been shown to alter DNA methylation patterns in mice (56). A possible increase of DNA and protein methylation is supported by regulation of methyltransferases, histones, and histone deacetylases as observed in our SILAC studies. Additional studies investigating epigenetic mechanisms in the presence of genistein would be of interest.
Our transcriptomics results showed some overlap with those reported by Williams et al. (8) for the effect of estradiol on ERβ-regulated genes. We used the same experimental conditions but with a different ligand (genistein instead of estradiol) and a different technology platform (Affymetrix gene chip versus Operon's oligomer spotted array). Comparing expression data, similar ERβ targets are induced in both studies, e.g. LRRC15, APOD, HMGCL, and NRIP3. Expression of APOD is absent in proliferating cells and induced in cells that undergo growth arrest and senescence (57). APOD protein levels were only detected in one SILAC experiment, namely in cells co-expressing both receptors during genistein treatment. Both studies confirmed that estradiol and genistein induce changes at the gene and protein expression levels leading to stimulation of cell proliferation in the absence of ERβ expression, whereas expression of ERβ drives the cell to protein and gene expression leading to negative regulation of cell cycle and induction of apoptosis. Moreover, our results appear to be similar to the genistein effects described by Chang et al. (10) in a similar model system with MCF-7 cells. At a similar concentration of genistein (300 nm), co-expression of both ERα and ERβ resulted in a markedly reduced ability to stimulate cell proliferation. The MCF-7 cells of Chang et al. (10) contained relatively high levels of ERα, whereas our cells contain relatively low amounts of ERα, yet at the concentration of genistein used in both studies the transcriptomic and proteomic changes are very similar to those of 17β-estradiol, suggesting that the ultimate estrogenic effect is dependent on the relative affinity of a ligand for both receptors and on the receptor phenotype (ERα/ERβ ratio) in the cells or tissue of interest.
Functional analysis of the genistein-induced ERα-activated proteome data revealed that genistein was able to induce growth of the breast epithelial cells as indicated by the activation of cellular reorganization and maintenance. Cytoskeletal rearrangement, which is organized by microtubules, actin-containing microfilaments, and mechanochemical molecules, is a sensitive indicator of cell growth. Expression of myosins is important for proliferation and migration of breast cancer cells controlled by Rho and ERK signaling (58, 59). Consequently, genistein has similar effects on estrogen-dependent tumor growth as has been shown in in vivo mouse studies (19).
ERβ decreases the cell motility and cell proliferation, two of the most common components of breast cancer progression. This was observed during functional analysis of the genistein-induced ERβ-activated proteome (samples DB and ED). Cellular organization and cell movement (migration) were processes down-regulated by ERβ expression. More importantly and in accordance with gene expression experiments, ERβ activity induces negative regulation of cell proliferation and induction of cell death. An increase of histone-related proteins is thought to result in a general repression of ERα transcription (31, 32), which will alter cell proliferation in breast cancer cell lines. Repression of transcription was also observed by the down-regulation of TOP2B after genistein treatment (34, 49).
Finally, enrichment of canonical pathways (Fig. 9) comparing genistein-exposed cells expressing ERβ with those not containing ERβ agreed with similar findings in genistein-exposed epithelial cells (59), indicating that cells not containing ERβ lose their cell-cell interactions and cell polarity and undergo a major change in their actin cytoskeleton, enabling them to acquire an increased motility and invasiveness. Expression of ERβ halts genistein-induced cell migration and cytoskeleton reorganization by inactivating the same signaling pathways.
In summary, we have demonstrated by using proteomics and transcriptomics that even in the presence of low ERα levels genistein induces effects on gene and protein expression in the T47D-ERβ cells comparable with those previously reported for estradiol. It is concluded that genistein can act as an estrogen and that its ultimate estrogenic effect on cells and tissues is dependent on the receptor phenotype and the ratio between the receptor subtypes within these cells or tissues, a phenotype that may potentially be modified upon genistein exposure.
Acknowledgments
We thank Dr. Henk W. P. van der Toorn and Dr. Engineer Bas van Breukelen from Biomolecular Mass Spectrometry department (Utrecht University) for help with Mascot. All proteomics LC-MS/MS measurements were done at Biqualys Wageningen (www.biqualys.nl).
Footnotes
* This work was partially funded by the Graduate School of Voeding, Levensmiddelentechnologie, Agrobiotechnologie en Gezondheid (VLAG) (Project 61.61.100.040) and the Swedish Cancer Society.
 This article contains supplemental Fig. S1.
This article contains supplemental Fig. S1.
1 The abbreviations used are:
- ER
- estrogen receptor
- SILAC
- stable isotope labeling by amino acids in cell culture
- Tet
- tetracycline
- Arg10
- [13C6,15N4]arginine
- Lys8
- [13C6,15N2]lysine
- IPA
- Ingenuity Pathway Analysis
- ICI
- ICI 182780
- H/L
- heavy/light
- IPI
- International Protein Index
- PIP
- prolactin-inducible protein
- HNRNPC
- heterogeneous nuclear ribonucleoproteins C1/C2
- LMNA
- lamin-A/C
- ITGAV
- integrin αV.
REFERENCES
- 1. Kuiper G. G., Enmark E., Pelto-Huikko M., Nilsson S., Gustafsson J. A. (1996) Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. U.S.A. 93, 5925–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leygue E., Dotzlaw H., Watson P. H., Murphy L. C. (1998) Altered estrogen receptor {alpha} and {beta} messenger RNA expression during human breast tumorigenesis. Cancer Res. 58, 3197–3201 [PubMed] [Google Scholar]
- 3. Rutherford T., Brown W. D., Sapi E., Aschkenazi S., Muñoz A., Mor G. (2000) Absence of estrogen receptor-[beta] expression in metastatic ovarian cancer. Obstet. Gynecol. 96, 417–421 [DOI] [PubMed] [Google Scholar]
- 4. Roger P., Sahla M. E., Mäkelä S., Gustafsson J. A., Baldet P., Rochefort H. (2001) Decreased expression of estrogen receptor beta protein in proliferative preinvasive mammary tumors. Cancer Res. 61, 2537–2541 [PubMed] [Google Scholar]
- 5. Lazennec G., Bresson D., Lucas A., Chauveau C., Vignon F. (2001) ER beta inhibits proliferation and invasion of breast cancer cells. Endocrinology 142, 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fixemer T., Remberger K., Bonkhoff H. (2003) Differential expression of the estrogen receptor beta (ERbeta) in human prostate tissue, premalignant changes, and in primary, metastatic, and recurrent prostatic adenocarcinoma. Prostate 54, 79–87 [DOI] [PubMed] [Google Scholar]
- 7. Bardin A., Boulle N., Lazennec G., Vignon F., Pujol P. (2004) Loss of ER{beta} expression as a common step in estrogen-dependent tumor progression. Endocr. Relat. Cancer 11, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams C., Edvardsson K., Lewandowski S. A., Ström A., Gustafsson J. A. (2008) A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene 27, 1019–1032 [DOI] [PubMed] [Google Scholar]
- 9. Lin C. Y., Ström A., Li Kong S., Kietz S., Thomsen J. S., Tee J. B., Vega V. B., Miller L. D., Smeds J., Bergh J., Gustafsson J. A., Liu E. T. (2007) Inhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancer. Breast Cancer Res. 9, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang E. C., Charn T. H., Park S. H., Helferich W. G., Komm B., Katzenellenbogen J. A., Katzenellenbogen B. S. (2008) Estrogen receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol. Endocrinol. 22, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner J. V., Agatonovic-Kustrin S., Glass B. D. (2007) Molecular aspects of phytoestrogen selective binding at estrogen receptors. J. Pharm. Sci. 96, 1879–1885 [DOI] [PubMed] [Google Scholar]
- 12. Galluzzo P., Marino M. (2006) Nutritional flavonoids impact on nuclear and extracellular estrogen receptor activities. Genes Nutr. 1, 161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarkar F. H., Li Y. (2003) Soy isoflavones and cancer prevention—clinical sciences review. Cancer Invest. 21, 744–757 [DOI] [PubMed] [Google Scholar]
- 14. Messina M., McCaskill-Stevens W., Lampe J. W. (2006) Addressing the soy and breast cancer relationship: review, commentary, and workshop proceedings. J. Natl. Cancer Inst. 98, 1275–1284 [DOI] [PubMed] [Google Scholar]
- 15. McCarty M. F. (2006) Isoflavones made simple—genistein's agonist activity for the beta-type estrogen receptor mediates their health benefits. Med. Hypotheses 66, 1093–1114 [DOI] [PubMed] [Google Scholar]
- 16. Nebe B., Peters A., Duske K., Richter D. U., Briese V. (2006) Influence of phytoestrogens on the proliferation and expression of adhesion receptors in human mammary epithelial cells in vitro. Eur. J. Cancer Prev. 15, 405–415 [DOI] [PubMed] [Google Scholar]
- 17. Conklin C. M., Bechberger J. F., MacFabe D., Guthrie N., Kurowska E. M., Naus C. C. (2007) Genistein and quercetin increase connexin43 and suppress growth of breast cancer cells. Carcinogenesis 28, 93–100 [DOI] [PubMed] [Google Scholar]
- 18. Hsieh C. Y., Santell R. C., Haslam S. Z., Helferich W. G. (1998) Estrogenic effects of genistein on the growth of estrogen receptor-positive human breast cancer (MCF-7) cells in vitro and in vivo. Cancer Res. 58, 3833–3838 [PubMed] [Google Scholar]
- 19. Allred C. D., Allred K. F., Ju Y. H., Virant S. M., Helferich W. G. (2001) Soy diets containing varying amounts of genistein stimulate growth of estrogen-dependent (MCF-7) tumors in a dose-dependent manner. Cancer Res. 61, 5045–5050 [PubMed] [Google Scholar]
- 20. Sotoca A. M., Ratman D., van der Saag P., Ström A., Gustafsson J. A., Vervoort J., Rietjens I. M., Murk A. J. (2008) Phytoestrogen-mediated inhibition of proliferation of the human T47D breast cancer cells depends on the ER[alpha]/ER[beta] ratio. J. Steroid Biochem. Mol. Biol. 112, 171–178 [DOI] [PubMed] [Google Scholar]
- 21. Ström A., Hartman J., Foster J. S., Kietz S., Wimalasena J., Gustafsson J. A. (2004) Estrogen receptor beta inhibits 17beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc. Natl. Acad. Sci. U.S.A. 101, 1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sotoca A. M., van den Berg H., Vervoort J., van der Saag P., Ström A., Gustafsson J. A., Rietjens I., Murk A. J. (2008) Influence of cellular ER{alpha}/ER{beta} ratio on the ER{alpha}-agonist induced proliferation of human T47D breast cancer cells. Toxicol. Sci. 105, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 24. Everley P. A., Krijgsveld J., Zetter B. R., Gygi S. P. (2004) Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Mol. Cell. Proteomics 3, 729–735 [DOI] [PubMed] [Google Scholar]
- 25. Blagoev B., Mann M. (2006) Quantitative proteomics to study mitogen-activated protein kinases. Methods 40, 243–250 [DOI] [PubMed] [Google Scholar]
- 26. Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 27. Wakeling A. E., Dukes M., Bowler J. (1991) A potent specific pure antiestrogen with clinical potential. Cancer Res. 51, 3867–3873 [PubMed] [Google Scholar]
- 28. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 29. Ghavami S., Kerkhoff C., Chazin W. J., Kadkhoda K., Xiao W., Zuse A., Hashemi M., Eshraghi M., Schulze-Osthoff K., Klonisch T., Los M. (2008) S100A8/9 induces cell death via a novel, RAGE-independent pathway that involves selective release of Smac/DIABLO and Omi/HtrA2. Biochim. Biophys. Acta 1783, 297–311 [DOI] [PubMed] [Google Scholar]
- 30. Debily M. A., Marhomy S. E., Boulanger V., Eveno E., Mariage-Samson R., Camarca A., Auffray C., Piatier-Tonneau D., Imbeaud S. (2009) A functional and regulatory network associated with PIP expression in human breast cancer. PLoS One 4, e4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheung E., Zarifyan A. S., Kraus W. L. (2002) Histone H1 represses estrogen receptor {alpha} transcriptional activity by selectively inhibiting receptor-mediated transcription initiation. Mol. Cell. Biol. 22, 2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vani G., Devi C. S. (2005) Effect of histone H1 on estrogen receptor status of human breast cancer MCF 7 cells. Mol. Cell. Biochem. 272, 151–155 [DOI] [PubMed] [Google Scholar]
- 33. Wang J. C. (1991) DNA topoisomerases: why so many? J. Biol. Chem. 266, 6659–6662 [PubMed] [Google Scholar]
- 34. López-Lazaro M., Willmore E., Austin C. A. (2007) Cells lacking DNA topoisomerase IIbeta are resistant to genistein. J. Nat. Prod. 70, 763–767 [DOI] [PubMed] [Google Scholar]
- 35. Weichselbaum R. R., Ishwaran H., Yoon T., Nuyten D. S., Baker S. W., Khodarev N., Su A. W., Shaikh A. Y., Roach P., Kreike B., Roizman B., Bergh J., Pawitan Y., van de Vijver M. J., Minn A. J. (2008) An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. 105, 18490–18495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DePasquale J. A., Samsonoff W. A., Gierthy J. F. (1994) 17-beta-Estradiol induced alterations of cell-matrix and intercellular adhesions in a human mammary carcinoma cell line. J. Cell Sci. 107, 1241–1254 [DOI] [PubMed] [Google Scholar]
- 37. Perretti M., Gavins F. N. (2003) Annexin 1: an endogenous anti-inflammatory protein. News Physiol. Sci. 18, 60–64 [DOI] [PubMed] [Google Scholar]
- 38. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hegde P. S., White I. R., Debouck C. (2003) Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 14, 647–651 [DOI] [PubMed] [Google Scholar]
- 40. Dechering K. J. (2005) The transcriptome's drugable frequenters. Drug Discov. Today 10, 857–864 [DOI] [PubMed] [Google Scholar]
- 41. Schmidt M. W., Houseman A., Ivanov A. R., Wolf D. A. (2007) Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe. Mol. Syst. Biol. 3, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katzenellenbogen B. S., Katzenellenbogen J. A. (2000) Estrogen receptor transcription and transactivation: estrogen receptor alpha and estrogen receptor beta—regulation by selective estrogen receptor modulators and importance in breast cancer. Breast Cancer Res. 2, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Power K. A., Thompson L. U. (2003) Ligand-induced regulation of ERα and ERβ is indicative of human breast cancer cell proliferation. Breast Cancer Res. Treat. 81, 209–221 [DOI] [PubMed] [Google Scholar]
- 44. Bovee T. F., Helsdingen R. J., Rietjens I. M., Keijer J., Hoogenboom R. L. (2004) Rapid yeast estrogen bioassays stably expressing human estrogen receptors [alpha] and [beta], and green fluorescent protein: a comparison of different compounds with both receptor types. J. Steroid Biochem. Mol. Biol. 91, 99–109 [DOI] [PubMed] [Google Scholar]
- 45. Nomoto S., Arao Y., Horiguchi H., Ikeda K., Kayama F. (2002) Oestrogen causes G2/M arrest and apoptosis in breast cancer cells MDA-MB-231. Oncol. Rep. 9, 773–776 [PubMed] [Google Scholar]
- 46. Ascenzi P., Bocedi A., Marino M. (2006) Structure-function relationship of estrogen receptor [alpha] and [beta]: Impact on human health. Mol. Aspects Med. 27, 299–402 [DOI] [PubMed] [Google Scholar]
- 47. Long X., Nephew K. P. (2006) Fulvestrant (ICI 182,780)-dependent interacting proteins mediate immobilization and degradation of estrogen receptor-alpha. J. Biol. Chem. 281, 9607–9615 [DOI] [PubMed] [Google Scholar]
- 48. Cotroneo M. S., Wang J., Eltoum I. A., Lamartiniere C. A. (2001) Sex steroid receptor regulation by genistein in the prepubertal rat uterus. Mol. Cell. Endocrinol. 173, 135–145 [DOI] [PubMed] [Google Scholar]
- 49. Fritz W. A., Wang J., Eltoum I. E., Lamartiniere C. A. (2002) Dietary genistein down-regulates androgen and estrogen receptor expression in the rat prostate. Mol. Cell. Endocrinol. 186, 89–99 [DOI] [PubMed] [Google Scholar]
- 50. Zilli M., Grassadonia A., Tinari N., Di Giacobbe A., Gildetti S., Giampietro J., Natoli C., Iacobelli S. (2009) Molecular mechanisms of endocrine resistance and their implication in the therapy of breast cancer. Biochim. Biophys. Acta 1795, 62–81 [DOI] [PubMed] [Google Scholar]
- 51. Cheng A. S., Jin V. X., Fan M., Smith L. T., Liyanarachchi S., Yan P. S., Leu Y. W., Chan M. W., Plass C., Nephew K. P., Davuluri R. V., Huang T. H. (2006) Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-[alpha] responsive promoters. Mol. Cell 21, 393–404 [DOI] [PubMed] [Google Scholar]
- 52. Hua S., Kallen C. B., Dhar R., Baquero M. T., Mason C. E., Russell B. A., Shah P. K., Liu J., Khramtsov A., Tretiakova M. S., Krausz T. N., Olopade O. I., Rimm D. L., White K. P. (2008) Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Syst. Biol. 4, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng P. C., Chang H. K., Chen S. H. (2010) Quantitative nanoproteomics for protein complexes (QNanoPX) related to estrogen transcriptional action. Mol. Cell. Proteomics 9, 209–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang S., Litt M., Felsenfeld G. (2005) Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 19, 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou Q., Shaw P. G., Davidson N. E. (2009) Epigenetics meets estrogen receptor: regulation of estrogen receptor by direct lysine methylation. Endocr.-Relat. Cancer 16, 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Day J. K., Bauer A. M., DesBordes C., Zhuang Y., Kim B. E., Newton L. G., Nehra V., Forsee K. M., MacDonald R. S., Besch-Williford C., Huang T. H., Lubahn D. B. (2002) Genistein alters methylation patterns in Mice. J. Nutr. 132, 2419S–2423S [DOI] [PubMed] [Google Scholar]
- 57. Do Carmo S., Séguin D., Milne R., Rassart E. (2002) Modulation of apolipoprotein D and apolipoprotein E mRNA expression by growth arrest and identification of key elements in the promoter. J. Biol. Chem. 277, 5514–5523 [DOI] [PubMed] [Google Scholar]
- 58. Zhou X., Liu Y., You J., Zhang H., Zhang X., Ye L. (2008) Myosin light-chain kinase contributes to the proliferation and migration of breast cancer cells through cross-talk with activated ERK1/2. Cancer Lett. 270, 312–327 [DOI] [PubMed] [Google Scholar]
- 59. Giretti M. S., Fu X. D., De Rosa G., Sarotto I., Baldacci C., Garibaldi S., Mannella P., Biglia N., Sismondi P., Genazzani A. R., Simoncini T. (2008) Extra-nuclear signalling of estrogen receptor to breast cancer cytoskeletal remodelling, migration and invasion. PLoS One 3, e2238. [DOI] [PMC free article] [PubMed] [Google Scholar]