Abstract
Knowledge of elaborate structures of protein complexes is fundamental for understanding their functions and regulations. Although cross-linking coupled with mass spectrometry (MS) has been presented as a feasible strategy for structural elucidation of large multisubunit protein complexes, this method has proven challenging because of technical difficulties in unambiguous identification of cross-linked peptides and determination of cross-linked sites by MS analysis. In this work, we developed a novel cross-linking strategy using a newly designed MS-cleavable cross-linker, disuccinimidyl sulfoxide (DSSO). DSSO contains two symmetric collision-induced dissociation (CID)-cleavable sites that allow effective identification of DSSO-cross-linked peptides based on their distinct fragmentation patterns unique to cross-linking types (i.e. interlink, intralink, and dead end). The CID-induced separation of interlinked peptides in MS/MS permits MS3 analysis of single peptide chain fragment ions with defined modifications (due to DSSO remnants) for easy interpretation and unambiguous identification using existing database searching tools. Integration of data analyses from three generated data sets (MS, MS/MS, and MS3) allows high confidence identification of DSSO cross-linked peptides. The efficacy of the newly developed DSSO-based cross-linking strategy was demonstrated using model peptides and proteins. In addition, this method was successfully used for structural characterization of the yeast 20 S proteasome complex. In total, 13 non-redundant interlinked peptides of the 20 S proteasome were identified, representing the first application of an MS-cleavable cross-linker for the characterization of a multisubunit protein complex. Given its effectiveness and simplicity, this cross-linking strategy can find a broad range of applications in elucidating the structural topology of proteins and protein complexes.
Proteins form stable and dynamic multisubunit complexes under different physiological conditions to maintain cell viability and normal cell homeostasis. Detailed knowledge of protein interactions and protein complex structures is fundamental to understanding how individual proteins function within a complex and how the complex functions as a whole. However, structural elucidation of large multisubunit protein complexes has been difficult because of a lack of technologies that can effectively handle their dynamic and heterogeneous nature. Traditional methods such as nuclear magnetic resonance (NMR) analysis and x-ray crystallography can yield detailed information on protein structures; however, NMR spectroscopy requires large quantities of pure protein in a specific solvent, whereas x-ray crystallography is often limited by the crystallization process.
In recent years, chemical cross-linking coupled with mass spectrometry (MS) has become a powerful method for studying protein interactions (1–3). Chemical cross-linking stabilizes protein interactions through the formation of covalent bonds and allows the detection of stable, weak, and/or transient protein-protein interactions in native cells or tissues (4–9). In addition to capturing protein interacting partners, many studies have shown that chemical cross-linking can yield low resolution structural information about the constraints within a molecule (2, 3, 10) or protein complex (11–13). The application of chemical cross-linking, enzymatic digestion, and subsequent mass spectrometric and computational analyses for the elucidation of three-dimensional protein structures offers distinct advantages over traditional methods because of its speed, sensitivity, and versatility. Identification of cross-linked peptides provides distance constraints that aid in constructing the structural topology of proteins and/or protein complexes. Although this approach has been successful, effective detection and accurate identification of cross-linked peptides as well as unambiguous assignment of cross-linked sites remain extremely challenging due to their low abundance and complicated fragmentation behavior in MS analysis (2, 3, 10, 14). Therefore, new reagents and methods are urgently needed to allow unambiguous identification of cross-linked products and to improve the speed and accuracy of data analysis to facilitate its application in structural elucidation of large protein complexes.
A number of approaches have been developed to facilitate MS detection of low abundance cross-linked peptides from complex mixtures. These include selective enrichment using affinity purification with biotinylated cross-linkers (15–17) and click chemistry with alkyne-tagged (18) or azide-tagged (19, 20) cross-linkers. In addition, Staudinger ligation has recently been shown to be effective for selective enrichment of azide-tagged cross-linked peptides (21). Apart from enrichment, detection of cross-linked peptides can be achieved by isotope-labeled (22–24), fluorescently labeled (25), and mass tag-labeled cross-linking reagents (16, 26). These methods can identify cross-linked peptides with MS analysis, but interpretation of the data generated from interlinked peptides (two peptides connected with the cross-link) by automated database searching remains difficult. Several bioinformatics tools have thus been developed to interpret MS/MS data and determine interlinked peptide sequences from complex mixtures (12, 14, 27–32). Although promising, further developments are still needed to make such data analyses as robust and reliable as analyzing MS/MS data of single peptide sequences using existing database searching tools (e.g. Protein Prospector, Mascot, or SEQUEST).
Various types of cleavable cross-linkers with distinct chemical properties have been developed to facilitate MS identification and characterization of cross-linked peptides. These include UV photocleavable (33), chemical cleavable (19), isotopically coded cleavable (24), and MS-cleavable reagents (16, 26, 34–38). MS-cleavable cross-linkers have received considerable attention because the resulting cross-linked products can be identified based on their characteristic fragmentation behavior observed during MS analysis. Gas-phase cleavage sites result in the detection of a “reporter” ion (26), single peptide chain fragment ions (35–38), or both reporter and fragment ions (16, 34). In each case, further structural characterization of the peptide product ions generated during the cleavage reaction can be accomplished by subsequent MSn1 analysis. Among these linkers, the “fixed charge” sulfonium ion-containing cross-linker developed by Lu et al. (37) appears to be the most attractive as it allows specific and selective fragmentation of cross-linked peptides regardless of their charge and amino acid composition based on their studies with model peptides.
Despite the availability of multiple types of cleavable cross-linkers, most of the applications have been limited to the study of model peptides and single proteins. Additionally, complicated synthesis and fragmentation patterns have impeded most of the known MS-cleavable cross-linkers from wide adaptation by the community. Here we describe the design and characterization of a novel and simple MS-cleavable cross-linker, DSSO, and its application to model peptides and proteins and the yeast 20 S proteasome complex. In combination with new software developed for data integration, we were able to identify DSSO-cross-linked peptides from complex peptide mixtures with speed and accuracy. Given its effectiveness and simplicity, we anticipate a broader application of this MS-cleavable cross-linker in the study of structural topology of other protein complexes using cross-linking and mass spectrometry.
EXPERIMENTAL PROCEDURES
Materials and Reagents
General chemicals were purchased from Fisher Scientific or VWR International (West Chester, PA). Bovine heart cytochrome c (98% purity) and bovine erythrocyte ubiquitin (98% purity) were purchased from Sigma-Aldrich. Synthetic peptide Ac-IR7 (Ac-IEAEKGR; 98.1% purity) was synthesized by GL Biochem (Shanghai, China). Sequencing grade modified trypsin was purchased from Promega (Fitchburg, WI). The 20 S proteasome core particle was affinity-purified using a Pre1-TAP-expressing yeast strain as described previously (39).
Synthesis and Characterization of DSSO
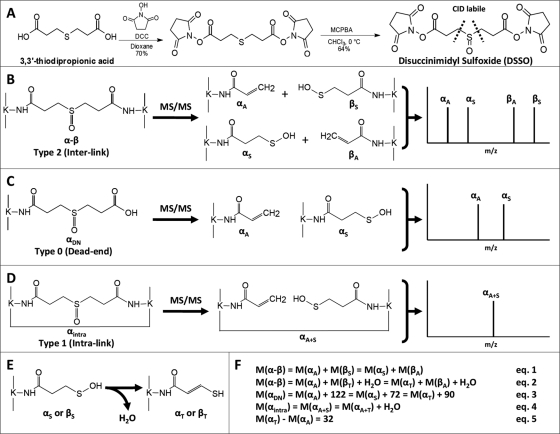
Fig. 1A displays a two-step synthesis scheme of DSSO with an extended spacer length of 10.1 Å. Briefly, sulfide S-1 was first synthesized by mixing 3,3′-thiodipropionic acid (2.50 g; 14.0 mmol) with N-hydroxysuccinimide (3.30 g; 28.6 mmol) in dioxane (60 ml). The reaction mixture was stirred under an atmosphere of argon, and a solution of N,N′-dicyclohexylcarbodiimide (5.79 g; 28.1 mmol) in dioxane (20 ml) was added dropwise. After 12 h, the insoluble urea was filtered from the reaction. The filtrate was concentrated to form a white solid. The solid residue was washed with cold diethyl ether followed by cold hexanes. After drying under reduced pressure, 5.20 g (70%) of sulfide S-1 was recovered and used without further purification: 1H (500 MHz, DMSO-d6) δ 3.02 (t, J = 7.0 Hz, 4H), 2.86 (t, J = 7.0 Hz, 4H), 2.81 (s, 8H); 13C (125 MHz, DMSO-d6) δ 170.1, 167.8, 31.4, 25.6, 25.4; IR (KBr pellet) 1801, 1732 cm−1; HRMS (ES/MeOH) m/z calculated for C14H16N2O8SNa [M + Na]+: 395.0525, found: 395.0531.
Fig. 1.
Proposed fragmentation schemes of DSSO-cross-linked peptides. A, DSSO synthesis and structure. B–D, MS/MS fragmentation patterns of the three types of DSSO-cross-linked peptides: interlinked (B), dead end (C), and intralinked (D). E, conversion of a sulfenic acid-modified fragment to an unsaturated thiol-modified fragment after a water loss. F, mass relationships between MS/MS fragment ions shown in B–D and their parent ions. DCC, N,N′-dicyclohexylcarbodiimide; MCPBA, m-chloroperbenzoic acid.
To synthesize DSSO, a solution of sulfide S-1 (0.600 g; 1.61 mmol) in CHCl3 (30 ml) at 0 °C was mixed with a solution of m-chloroperbenzoic acid (0.371 g; 1.61 mmol) in CHCl3 (10 ml). The reaction product was filtered and washed with cold CHCl3 (10 ml) and cold MeOH (10 ml). The filtrate was cooled to −10 °C for 1 h, washed again with CHCl3 and MeOH, and dried under reduced pressure to yield 0.400 g (64%) of DSSO: 1H (600 MHz, DMSO-d6) δ 3.28–3.21 (m, 2H), 3.17–3.13 (m, 4H), 3.08–2.99 (m, 2H), 2.88–2.75 (s, 8H); 13C (125 MHz, DMSO-d6) δ 170.08, 167.74, 44.62, 25.46, 23.41; IR (KBr pellet) 2943, 1786, 1720 cm−1; HRMS (ES/MeOH) m/z calculated for C14H16N2O9Na [M + Na]+: 411.0474, found: 411.0471.
Cross-linking of Synthetic Peptides with DSSO
Synthetic peptides Ac-IR7, Ac-myelin, and substance P were dissolved in DMSO to 1 mm and cross-linked with DSSO dissolved in DMSO in a ratio of 1:1 in the presence of 1 eq of diisopropylethylamine similarly as described (21). The cross-linked peptide solution was then diluted to 1 pmol/μl in 4% ACN, 0.1% formic acid for liquid chromatography multistage tandem mass spectrometry (LC MSn) analysis.
Cross-linking of Cytochrome c and Ubiquitin with DSSO
Lyophilized bovine cytochrome c or ubiquitin was reconstituted in 1× PBS (pH 7.5) to 200 μm, 20 μl of which was mixed with 2 μl of 20 mm DSSO (in DMSO) in a molar ratio of 1:10 (protein:cross-linker) for the cross-linking reaction as described (21). The cross-linked protein was digested with trypsin (1%, w/w) overnight at 37 °C. The cross-linked peptide digest was then diluted to 1 pmol/μl in 4% ACN, 0.1% formic acid for LC MSn analysis.
Cross-linking of Yeast 20 S Proteasome with DSSO
Affinity-purified yeast 20 S proteasome complex was concentrated by Microcon (Billerica, MA) to ∼1.2 μm in 1× PBS buffer (pH 7.5). Typically, 50 μl of the 20 S proteasome was cross-linked with 3 μl of DSSO (20 mm) dissolved in DMSO (final concentration, ∼1 mm) at a molar ratio of 1:1000 (protein:cross-linker). Cross-linking was performed for 0.5 h or overnight and quenched with excess ammonium bicarbonate buffer. Cysteine residues were reduced with 5 mm DTT at 56 °C for 30 min and alkylated with 10 mm chloroacetamide for 30 min at room temperature. The cross-linked protein complex was digested with trypsin (2%, w/w) overnight at 37 °C. Digested peptides were desalted by C18 OMIX ZipTip (Varian, Palo Alto, CA) prior to LC MSn analysis.
For some analyses, two-dimensional LC MSn analysis was carried out. Off-line strong cation exchange chromatography was performed as the first dimension of separation using an ÄKTA HPLC system (GE Healthcare) as described (9). Each fraction was desalted by ZipTip prior to LC MSn analysis.
LC MSn Analysis
LC MSn analysis of DSSO-cross-linked peptides was performed using an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA) with an on-line Eksigent NanoLC system (Eksigent, Dublin, CA). The LC separation was the same as described previously (21). The MSn method was set specifically for analyzing DSSO-cross-linked peptides. Each acquisition cycle of an MSn experiment includes one MS scan in FT mode (350–1800 m/z, resolution of 60,000 at m/z 400) followed by two data-dependent MS/MS scans with normalized collision energy at 10 or 15% on the top two peaks from the MS scan and then three MS3 scans operated in the LTQ with normalized collision energy at 29% on the top three peaks from each of the MS/MS scans. For initial analyses, MS/MS spectra were acquired in the LTQ in LC MSn experiments. For automated data analysis, MS/MS spectra were obtained in FT mode (resolution of 7500).
Data Analysis of DSSO-cross-linked Peptides
Monoisotopic masses of parent ions and corresponding fragment ions, parent ion charge states, and ion intensities from LC MS/MS and LC MS3 spectra were extracted using in-house software based on the Raw_Extract script from Xcalibur v2.4 (Thermo Scientific). Database searching was performed with a developmental version of Protein Prospector (v5.5.0, University of California San Francisco) (http://prospector.ucsf.edu/prospector/mshome.htm) using its software suite, i.e. Batch-Tag and MS-Bridge, as described (27). Using in-house scripts, extracted MS3 data were reformatted such that MS3 fragment ions were directly linked to their MS/MS parent ions. For cytochrome c (P62894) and ubiquitin (P62990) analyses, database searching of MS3 spectra was performed using Batch-Tag against their accession numbers in Swiss-Prot September 1, 2009 database. For the 20 S proteasome, a Batch-Tag search of MS3 data was performed against a decoy database consisting of a normal Saccharomyces Genome Database concatenated with its reversed version (13,490 total protein entries). The mass tolerances for parent ions and fragment ions were set as ±20 ppm and 0.6 Da, respectively. Trypsin was set as the enzyme, and a maximum of two missed cleavages was allowed. Protein N-terminal acetylation, methionine oxidation, and N-terminal conversion of glutamine to pyroglutamic acid were selected as variable modifications. In addition, three defined modifications on uncleaved lysines were chosen, including alkene (C3H2O; +54 Da), sulfenic acid (C3H4O2S; +104 Da), and thiol (C3H2SO; +86 Da) modifications due to remnants of the cross-linker (Fig. 1). Initial acceptance criteria for peptide identification required a reported expectation value ≤0.05. For the 20 S proteasome analysis, the false positive rate for peptide identification was less than 1%.
The Link-Finder program (http://www.ics.uci.edu/∼baldig/Link-Finder/) was developed to search MS/MS data and identify the list of putative DSSO-interlinked and dead end products based on their unique MS fragmentation patterns as illustrated in Fig. 1 (for details, see “Results”). Monoisotopic masses and charges of parent ions measured in MS scans for those putative cross-linked peptides identified by the Link-Finder program were subsequently submitted to MS-Bridge to determine cross-linked peptide sequences by mass mapping with a given cross-linker (i.e. DSSO) and protein sequences (27). The parent mass error for MS-Bridge search was set as ±10 ppm, and only one cross-link was allowed in the cross-linked peptides for general search. All of the three types of the cross-linked peptides (14), i.e. interlinked (type 2), intralinked (type 1), and dead end-modified (type 0), can be computed and matched in MS-Bridge (27).
The search results from Link-Finder, Batch-Tag, and MS-Bridge programs were integrated together using in-house scripts to compile a list of cross-linked peptides identified with high confidence. The final results were validated manually by examining MS/MS spectra and MS3 spectra, respectively.
RESULTS
Development of Novel Sulfoxide-containing MS-cleavable Cross-linker
To develop a robust MS-cleavable cross-linking reagent, the incorporated MS-labile bond must have the ability to selectively and preferentially fragment prior to peptide backbone breakage independently of peptide charges and sequences. It is well documented that methionine sulfoxide-containing peptides have preferential fragmentation at the C–S bond adjacent to the sulfoxide during collision-induced dissociation (CID) analysis (40), and this fragmentation is dominant and much more labile than peptide bonds. Such labile fragmentation has often been observed as the loss of 64 Da (−SOCH4) from oxidized methionine-containing peptides in our routine peptide analysis. Therefore, we expect that if a sulfoxide is incorporated in the spacer region of a NHS ester, the C–S bond adjacent to the sulfoxide will be MS-labile and prone to preferential fragmentation. To test this, we designed and synthesized a CID-cleavable cross-linker, namely DSSO, which contains two NHS ester functional groups and two symmetric MS-labile C–S bonds adjacent to the sulfoxide (Fig. 1A). DSSO has a spacer length of 10.1 Å, making it well suited for detecting protein interaction interfaces of protein complexes and generating highly informative distance constraints. In comparison with existing MS-cleavable cross-linkers, DSSO can be easily synthesized in a two-step process as shown in Fig. 1A.
Proposed CID Fragmentation Pattern of DSSO-cross-linked Peptides
Three types of cross-linked peptides can be formed during the cross-linking reaction: interlinked (type 2), intralinked (type 1), and dead end (type 0)-modified peptides (14), among which interlinked peptides are the most informative for generating distance constraints. Fig. 1, B–D, show the proposed fragmentation schemes of DSSO-cross-linked peptides. As shown in Fig. 1B, during CID analysis of a DSSO-interlinked peptide α-β, the cleavage of one C–S bond next to the sulfoxide separates the interlinked peptide into a pair of peptide fragments, i.e. αA/βS in which the α peptide fragment is modified with the alkene (A) moiety (+54 Da) and the β peptide fragment is modified with the sulfenic acid (S) moiety (+104 Da). If peptides α and β have different sequences, two possible pairs of fragments (i.e. αA/βS and αS/βA) will be observed due to the breakage of either of the two symmetric C–S bonds next to the sulfoxide in the spacer region of DSSO (Fig. 1B), thus resulting in four individual peaks in the MS/MS spectrum. But if peptides α and β have the same sequences, only one fragment pair, i.e. two peaks, will be detected in the MS/MS spectrum. To determine sequences of interlinked peptides and assign the cross-linking site, the resulting peptide fragments (i.e. αA, βS, αS, or βA) generated in MS/MS can be further subjected to MS3 analysis. Because these fragments represent single peptide sequences, the interpretation of the MS3 spectra by the Batch-Tag program in Protein Prospector is identical to the identification of a single peptide with a defined modification (in this case, remnant of the cross-linker). This will dramatically simplify data interpretation and improve the identification accuracy of cross-linked products.
DSSO dead end-modified peptides have a defined mass modification (+176 Da) due to the half-hydrolyzed DSSO (Fig. 1C). MS/MS analysis of a dead end-modified peptide αDN would result in two possible fragment ions, i.e. αA and αS, due to the cleavage of the C–S bond on either side of the sulfoxide. We name the αA and αS fragments as the dead end fragment pair, and the mass difference between these fragments correlates to the difference between the remnants of DSSO attached to the fragments. Similarly, intralinked peptides (e.g. αintra) also have a defined mass modification (+158 Da) due to DSSO cross-linking of two distinct lysines in the same peptide sequence (Fig. 1D). The cleavage of the C–S bond will result in only one fragment peak in MS/MS with the same mass as the parent ion observed in MS. MS3 analysis of fragment ions detected in MS/MS will lead to the detection of y or b ions containing either A or S modifications.
As shown in Fig. 1E, the sulfenic acid-containing fragment (e.g. αS, βS, or αA+S) may undergo further fragmentation and lose a water molecule (−18 Da) to generate a new fragment containing an unsaturated thiol (T) moiety (+86 Da) (e.g. αT, βT, or αA+T). We do not expect any complication with data analysis as the thiol-containing fragment ion will become the dominant ion instead of the sulfenic acid-modified fragment ion in the MS/MS spectrum. Thus, we anticipate that the total number of pairs and peaks will remain similar as shown in Fig. 1, B–D. Because of the specific and unique MS/MS fragmentation patterns for different types of DSSO-cross-linked peptides, there are fixed mass relationships between parent ions and their fragment ions as listed in Fig. 1F. For DSSO-interlinked peptides (α-β), the mass sum of each fragment pair (αA/βS or αS/βA) is equivalent to the mass of the parent ion (Fig. 1F, Equation 1). If αS or βS loses a water and becomes αT or βT, respectively, the fragment pairs will be αA/βT and αT/βA, and the mass sum of each fragment pair plus a water will be the same as the parent mass (Fig. 1F, Equation 2). As for the dead end (DN)-modified peptide αDN, each fragment (i.e. αA, αS, or αT) has a distinct mass difference from the parent ion (Fig. 1F, Equation 3). For the intralink peptide αintra, the fragment mass could be either the same as the parent mass (i.e. αA+S) or 18 Da less than the parent mass (i.e. αA+T) (Fig. 1F, Equation 4). Moreover, there is a defined mass difference (Δ32 Da) between the T- and A-modified forms of the same sequence (Fig. 1F, Equation 5). These characteristic mass relationships were incorporated into the Link-Finder program to identify DSSO-cross-linked peptides.
Characterization of DSSO-cross-linked Model Peptides by MSn Analysis
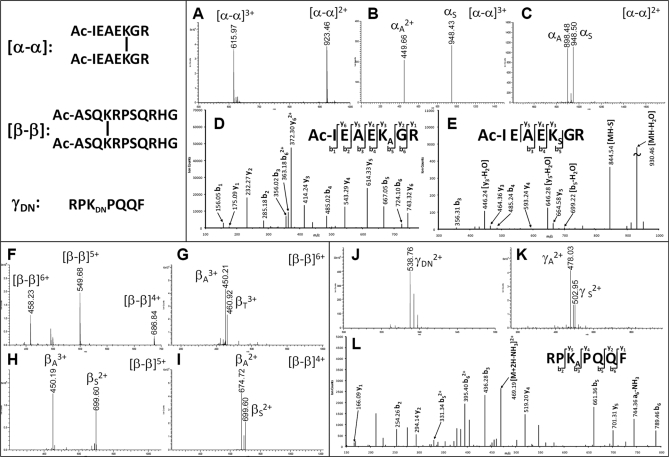
To characterize the new DSSO linker, we first cross-linked several model peptides, including Ac-IR7, Ac-myelin, and substance P. Under our experimental conditions, the major cross-linked products for Ac-IR7 and Ac-myelin are interlinked, whereas substance P mostly formed dead end-modified peptides. All of the cross-linked model peptides were subjected to LC MSn analysis. The interlinked Ac-IR7 peptide (α-α) was detected as doubly charged (m/z 923.462+) and triply charged (m/z 615.973+) ions (Fig. 2A). MS/MS analyses of the two differently charged parent ions resulted in two dominant fragment ions, respectively (Fig. 2, B and C). Because the two interlinked sequences are identical, only one fragment pair (i.e. αA/αS) was observed as expected. The results suggest that MS/MS fragmentation of interlinked peptides is independent of peptide charges. It should be noted that besides unique mass relationships the fragment ions in each pair have a defined charge relationship associated to the charge of the parent ion. In other words, the sum of the observed charges for each fragment in a pair equals the charge of the parent ion. For example, the triply charged parent ion (m/z 615.973+) generated the fragment pair with one doubly charged (αA2+) ion and one singly charged (αS1+) ion, whereas the doubly charge parent ion (m/z 923.462+) only produced a fragment pair with two singly charged (αA1+ and αS1+) ions. This information can be used to validate the fragment pairs identified by masses. The respective MS3 analysis of αA and αS ions (Fig. 2, D and E) allowed unambiguous identification of the peptide sequence and cross-linked site based on a series of y and b ions. Similar analysis was carried out for interlinked Ac-myelin (β-β), and a characteristic fragment pair was observed in MS/MS spectra of the parent ion (β-β) at three different charge states (m/z 458.236+, 549.685+, and 686.844+), respectively (Fig. 2, F–I), which represent the expected fragmentation of two identical interlinked peptides. Whereas the fragment pair βA/βS was detected in MS/MS spectra of quintuply and quadruply charged interlinked Ac-myelin (β-β) (m/z 549.685+ and 686.844+) (Fig. 2, H and I), the fragment pair βA/βT was observed in the MS/MS spectrum of sextuply charged interlinked Ac-myelin (β-β) (458.236+) (Fig. 2G). The βT fragment, namely the β peptide fragment containing an unsaturated T moiety (+86 Da), was generated due to the loss of H2O from the sulfenic acid moiety on the βS fragment (Fig. 1E). This is likely due to excess collision energy deposited on the highest charged species as the collision energy chosen for CID analysis in the LTQ-Orbitrap XL mass spectrometer does not change with peptide charges during LC MSn runs.
Fig. 2.
MSn analysis of DSSO-cross-linked model peptides. A–E, MSn analysis of the DSSO-interlinked Ac-IR7 (α-α). A, MS spectrum of α-α: [α-α]3+ (m/z 615.973+) and [α-α]2+ (m/z 923.462+). B and C, MS/MS spectra of [α-α]3+ (B) and [α-α]2+ (C) in which alkene (αA) and sulfenic acid (αS) fragments were detected. D and E, MS3 spectra of αA (m/z 449.662+) (D) and αS (m/z 948.43) (E). F–I, MSn analysis of DSSO-interlinked Ac-myelin (β-β). F, MS spectrum of β-β: [β-β]6+ (m/z 458.236+), [β-β]5+ (m/z 549.685+), and [β-β]4+ (m/z 686.844+). G–I, MS/MS spectra of [β-β]6+ in which βA/βT pair was observed (G), [β-β]5+ in which the βA/βS pair was observed (H), and [β-β]4+ in which βA/βS pair was observed (I). J–L, MSn analysis of DSSO dead end-modified substance P peptide γDN. J, MS spectrum of γDN (m/z 538.762+). K, MS/MS spectrum of γDN in which two fragments, γA (m/z 478.032+) and γS (m/z 502.952+), were detected. L, MS3 spectrum of γA (m/z 478.032+). Sequences of Ac-IR7, Ac-myelin, and substance P are Ac-IEAEKGR, Ac-ASQKRPSQRHG, and RPKPQQF, respectively.
In addition to interlinked peptides, dead end-modified peptides were analyzed. Fig. 2J displays the MS spectrum of the DN-modified substance P (γDN; m/z 538.762+). As predicted in Fig. 1D, MS/MS analysis of γDN led to two expected fragments, the alkene- (γA; m/z 478.032+) and sulfenic acid (γS, m/z 502.952+)-containing peptide fragments, representing the characteristic feature of dead end-modified peptides. The fragment ions carry the same charge state as the parent ion, and MS3 analysis of the γA fragment confirmed its sequence unambiguously (Fig. 2L). Taken together, the results clearly demonstrate that the new MS-cleavable bonds in DSSO are labile and can be preferentially fragmented prior to peptide bond breakage, and the desired fragmentation is independent of peptide charge states and sequences.
Characterization of DSSO-cross-linked Peptides of Model Proteins by MSn Analysis
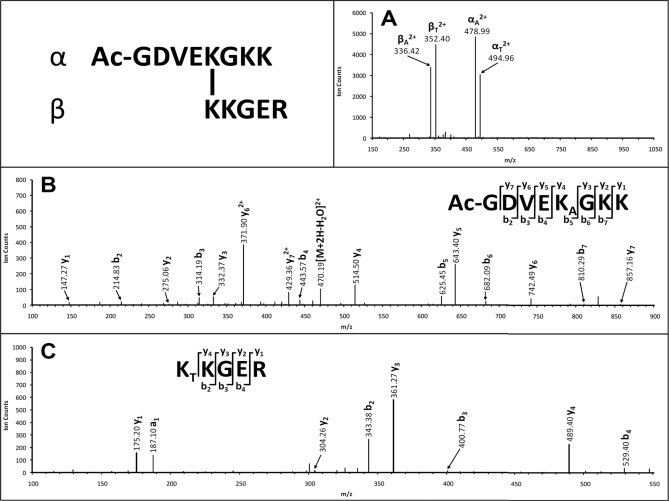
We next evaluated the applicability of DSSO for protein cross-linking under physiological conditions. Model proteins cytochrome c (1, 19–21, 31, 41–43) and ubiquitin (18, 38) have been extensively utilized to test various new cross-linking strategies because they have a relatively large number of lysine residues accessible for cross-linking. Based on our previous work (21), cytochrome c was cross-linked with a 10-fold excess of DSSO. The cytochrome c cross-linking efficiency using DSSO was comparable with the efficiency using disuccinimidyl glutarate or our previously developed azide-disuccinimidyl glutarate cross-linkers (21), indicating that DSSO is as effective for protein cross-linking reactions. The DSSO-cross-linked cytochrome c was then digested with trypsin and analyzed by LC MSn. Three types of cross-linked peptides of cytochrome c (i.e. interlink, intralink, and dead end) were observed. Fig. 3A displays the MS/MS spectrum of a tryptic peptide of cytochrome c with m/z 419.97164+ in which only four abundant fragment ions (m/z 336.422+, 352.402+, 478.992+, and 494.962+) were detected, suggesting this peptide as a potential heterodimeric interlinked peptide (α-β). Two possible fragment pairs, αA/βS/T and αS/T/βA, are thus expected in which S/T means either S- or T-containing fragment ions will be observed. Using the mass relationship between the pairs and the parent ion of interlinked peptides (Fig. 1F, Equations 1, 2, and 5), we identified two fragment pairs as αA/βT (478.992+/352.402+) and αT/βA (494.962+/336.422+), confirming that this peptide is a heterodimeric interlinked peptide (α-β). Mass mapping of the parent ion (m/z 419.97164+) by MS-Bridge revealed that it matches to an interlinked peptide [Ac-GDVEKGKK interlinked to KKGER] with an error of 0.48 ppm. The fragment ions αA (m/z 478.992+) and βT (m/z 352.402+) were further subjected to MS3 sequencing, and their MS3 spectra are illustrated in Fig. 3, B and C. Based on the series of y (i.e. y1–7) and b (i.e. b2–7) ions, the sequence of the MS/MS fragment ion αA (m/z 478.992+) was unambiguously identified as Ac-GDVEKAGKK in which Lys at the fifth position from the N terminus was determined to be modified with the alkene moiety. MS3 analysis of the corresponding fragment pair ion βT (m/z 352.402+) determined its sequence as KTKGER. Although there are two lysine residues in the sequence, the occurrence of y4 and a1 ions indicates that the first N-terminal Lys is modified with an unsaturated thiol moiety. Taken together, the identity and cross-linking site of the interlink peptide [Ac-GDVEKGKK interlinked to KKGER] was determined unambiguously.
Fig. 3.
MSn analysis of DSSO heterodimeric interlinked peptide of cytochrome c (α-β: Ac-GDVEKGKK interlinked to KKGER). A, MS/MS spectrum of [α-β]4+ (m/z 419.97164+) in which two fragment pairs were observed: αA (m/z 478.992+)/βT (m/z 352.402+) and αT (m/z 494.962+)/βA (m/z 336.422+). B, MS3 spectrum of αA (m/z 478.992+) in which detection of y1–y7 and b2–b7 determined the sequence unambiguously as Ac-GDVEKAGKK. C, MS3 spectrum of βT (m/z 352.402+) in which detection of y1–y4, a1, and b2–b7 ions determined the sequence unambiguously as KTKGER. KA is modified with the alkene moiety, and KT is modified with the unsaturated thiol moiety.
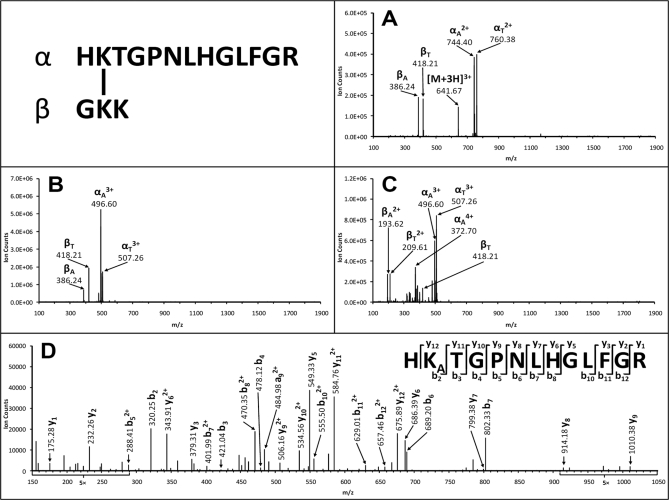
Fig. 4, A–C, display MS/MS spectra of triply (m/z 641.67303+), quadruply (m/z 481.50694+), and quintuply (m/z 385.40705+) charged ions of a cytochrome c cross-linked peptide. The MS/MS spectrum of the triply charged ion (m/z 641.67303+) resulted in four dominant fragment ions (m/z 386.24, 418.21, 744.402+, and 760.382+), which were determined as the two fragment pairs αA/βT (744.402+/418.21) and αT/βA (760.382+/386.24), indicating that this peptide is a heterodimeric interlinked peptide. The same characteristic fragment pairs, i.e. αA/βT and αT/βA, were also identified but with different charges in the MS/MS spectra of the quadruply (m/z 481.50694+) and quintuply (m/z 385.40705+) charged parent ions, respectively (Fig. 4, B and C). It is noted that some charge distribution of fragment ions was observed in the pairs (Fig. 4C) due to the high charge state of the parent ion. Nevertheless, the dominant ions are the characteristic fragment ions of the interlinked peptide. MS3 analysis of the αA (m/z 496.603+) fragment revealed its sequence identity unambiguously as HKATGPNLHGLFGR in which the Lys at the second position from the N terminus was modified with the alkene moiety (Fig. 4D). In combination with the MS-Bridge result, the interlinked peptide was identified as [HKTGPNLHGLFGR interlinked to GKK]. These results demonstrate that preferred fragmentation of the C–S bonds in DSSO-interlinked peptides of cytochrome c occurs as expected and is independent of peptide charge states and sequences.
Fig. 4.
MSn analysis of DSSO heterodimeric interlinked peptide of cytochrome c (α-β: HKTGPNLHGLFGR interlinked to GKK). This peptide was detected in MS as triply charged [α-β]3+ (m/z 641.67303+), quadruply charged [α-β]4+ (m/z 481.50694+), and quintuply charged [α-β]5+ (m/z 385.40705+) ions. A, MS/MS spectrum of [α-β]3+ (m/z 641.67303+) in which two fragment pairs were observed: αA (m/z 744.402+)/βT (m/z 418.21) and αT (m/z 760.382+)/βA (m/z 386.24). B, MS/MS spectrum of [α-β]4+ (m/z 481.50694+) in which two fragment pairs were observed: αA (m/z 496.603+)/βT (m/z 418.21) and αT (m/z 507.263+)/βA (m/z 386.24). C, MS/MS spectrum of [α-β]5+ (m/z 385.40705+) in which two fragment pairs were observed: αA/βT (m/z 496.603+/209.612+ and 372.704+/418.21) and αT (m/z 507.263+)/βA (m/z 193.622+). D, MS3 spectrum of αA fragment (m/z 496.603+) in which detection of a series of y and b ions determined its sequence unambiguously as HKATGPNLHGLFGR. KA is modified with the alkene moiety.
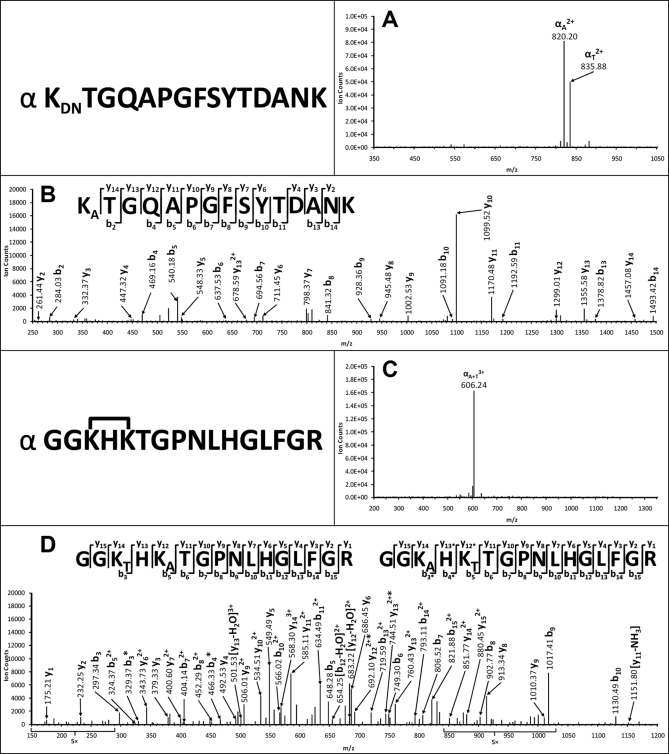
To understand how dead end-modified peptides of cytochrome c behave in MSn analysis, Fig. 5A illustrates the MS/MS spectrum of a selected dead end-modified peptide (m/z 880.89752+). As shown, two major fragment ions (m/z 820.202+ and 835.882+) were detected, and they are 122 and 90 Da less than the parent ion, respectively. Such mass differences between the parent ion and its fragment ions fit well with those predicted for DSSO dead end-modified peptides (Fig. 1F, Equation 3), identifying the ion m/z 820.202+ as αA and 835.882+ as αT fragment. MS3 analysis of the αA fragment (m/z 820.202+) (Fig. 5B) as well as the MS-Bridge result of the parent ion (m/z 880.89752+) identified its sequence as KDNTGQAPGFSYTDANK.
Fig. 5.
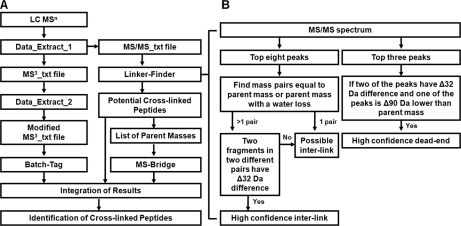
MSn analysis of DSSO dead end-modified peptide (A and B) and intralinked peptide of cytochrome c (C and D). A, MS/MS spectrum of a dead end-modified peptide (αDN; m/z 880.89752+, KDNTGQAPGFSYTDANK) in which two fragment ions were determined as αA (m/z 820.202+) and αT (m/z 835.882+). B, MS3 spectrum of αA (m/z 820.202+) in which detection of a series of y and b ions determined its sequence unambiguously as KATGQAPGFSYTDANK. C, MS/MS spectrum of an intralinked peptide (αintra; m/z 611.98023+, GGK*HK*TGPNLHGLFGR) in which one fragment ion was observed and determined as αA+T (m/z 606.243+). D, MS3 spectrum of αA+T (m/z 606.243+) in which detection of a series of y and b ions determined the presence of a mixture of GGKAHKTTGPNLHGLFGR and GGKTHKATGPNLHGLFGR. KA is modified with the alkene moiety, and KT is modified with the unsaturated thiol moiety.
As discussed above (Fig. 1D), we predicted that MS/MS analysis of the intralinked peptide (αintra) would lead to either a fragment ion (αA+S) containing one LysA and one LysS with the same mass as the parent ion or a fragment ion (αA+T) containing one LysA and one LysT with a mass 18 Da less than that of the original parent ion. Fig. 5C displays the MS/MS spectrum of a cytochrome c tryptic peptide with m/z 611.98023+ in which only one major fragment ion (m/z 606.242+) was detected with a mass 18 Da less than that of the parent ion. This suggests that the peptide is potentially an intralinked peptide of cytochrome c, and its MS/MS fragment ion (m/z 606.242+) can be labeled as αA+T. Mass mapping of the parent ion m/z 611.98023+ using MS-Bridge matched to an intralinked peptide, GGK*HK*TGPNLHGLFGR, where the two N-terminal Lys* residues are linked. Because the CID-induced C–S bond breakage can occur at either side of the sulfoxide, a mixture of two fragments with identical masses but with A or T moieties at either Lys can be generated. Fig. 5D illustrates the MS3 spectrum of the MS/MS fragment ion (m/z 606.243+) with a series of y and b ions, confirming its identity as GGKTHKATGPNLHGLFGR and/or GGKAHKTTGPNLHGLFGR. The detection of y13 (760.432+) and b3 (297.34) ions indicates the presence of the peptide fragments from the sequence of GGKTHKATGPNLHGLFGR, and the detection of b3* (329.37), b4* (466.33), y12* (692.102+), and y13* (744.512+) identified the peptide fragments from the GGKAHKTTGPNLHGLFGR sequence.
Development of Integrated Work Flow for Fast and Accurate Identification of DSSO-cross-linked Peptides by LC MSn
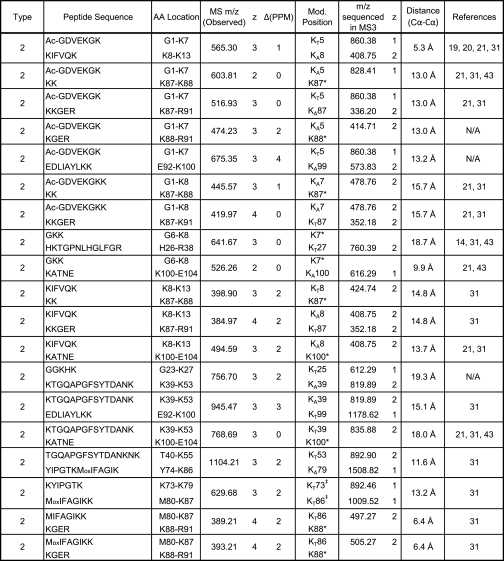
To facilitate data analysis for the identification of DSSO-cross-linked peptides from complex mixtures, we developed an integrated work flow for processing LC MSn data acquired by the LTQ-Orbitrap XL mass spectrometer (Fig. 6A). During LC MSn analysis, three types of data were collected, i.e. MS, MS/MS, and MS3 spectra, in which MS and MS/MS scans were acquired in FT mode to allow accurate mass measurement and charge determination of both parent ions in MS and their fragment ions in MS/MS spectra. MS3 was obtained in the LTQ to achieve the highest sensitivity. As shown, the first data extraction step is to generate the text files containing peak lists of MS/MS and MS3 data, respectively. Based on the unique MS/MS fragmentation profiles of DSSO-cross-linked peptides and the defined mass relationships between parent ions and their fragment ions (Fig. 1), the Link-Finder program was developed to automatically search MS/MS data to identify putative DSSO-cross-linked peptides (Fig. 6B). As discussed above, the interlinked products produce distinct MS/MS spectra with two pairs of dominant peptide fragments (αA/βS/T and αT/S/βA). For each MS/MS scan, among the top eight most abundant peaks, if there is a fragment pair with a mass sum equal to their parent mass with or without a water loss (−18 Da), the parent ion will be categorized as a possible interlinked peptide. If two of those pairs can be found, and the mass difference between any two fragments from the two distinct pairs is 32 Da, i.e. the mass difference between the thiol and alkene moieties, then it is almost certain that the parent ion is a true interlinked product. The dead end product typically has two major fragment ions representing the parent peptide attached with either a thiol or an alkene moiety. Among the top three peaks, if there are two peaks with mass difference of 32 Da, and one of them is 90 Da less than the parent mass, then it is categorized as a high confidence dead end peptide. Using the Link-Finder program, a list of parent ions were identified as putative interlinked or dead end-modified peptides. The generated list of parent ion masses was then subjected to MS-Bridge to identify putative cross-linked peptides of all types by mass matching with high mass accuracy (<10 ppm).
Fig. 6.
A, integrated data analysis work flow for identifying DSSO-cross-linked peptides by LC MSn. B, the work flow for the Link-Finder program.
For MS3 data, only the original parent ion observed in the MS scan is listed as the precursor ion during database searching. To extract the MS3 parent ion (fragment ions in MS/MS) for a Batch-Tag search, the second data extraction step is carried out using in-house scripts to generate a modified MS3_txt file. The Batch-Tag search result provides high confidence identification of single peptide fragments generated in MS/MS that are initially cross-linked. Finally, the results from three different types of searches, i.e. Batch-Tag (MS3 data), Link-Finder (MS/MS data), and MS-Bridge (MS data), are integrated using in-house scripts within the Link-Finder program to obtain accurate and reliable identification of cross-linked peptides. Among them, MS3 sequencing with Batch-Tag searching is essential for unambiguous identification of cross-linking sites.
Identification of DSSO-cross-linked Peptides of Model Proteins by Automated Database Searching
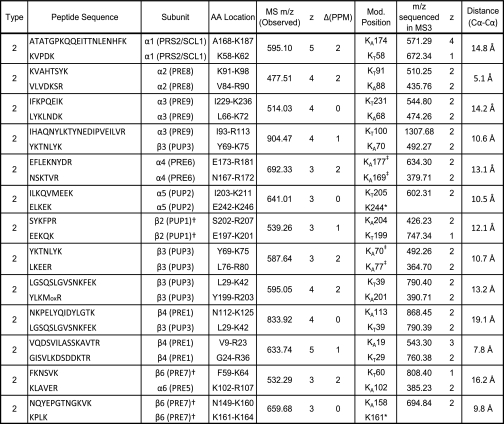
The newly developed integrated work flow was first used to identify DSSO-cross-linked peptides of cytochrome c. In total, 19 interlinked peptides were unambiguously identified and are summarized in Table I (for details, see supplemental Table 1 and supplemental Fig. 1). Cross-linked peptides have characteristic fragment pairs in MS/MS spectra and were identified by the Link-Finder program. In addition, one or two MS/MS fragment pair ions were sequenced by MS3 to provide unambiguous identification. Moreover, all of the parent masses fit well with cross-linked peptides identified by the MS-Bridge program with high mass accuracy. In comparison with reported cross-linking studies of cytochrome c (14, 19–21, 31, 41–43), three novel interlinks were identified in this work. Besides the interlinked peptides, seven intralinked and eight dead end peptides were also identified (see supplemental Table 1). For the dead end-modified peptides, each has a dead end fragment pair, and at least one of the fragment ions was sequenced, which correlates very well with the MS-Bridge and Batch-Tag results. The intralinked peptides were mainly identified by Batch-Tag and MS-Bridge results.
Table I. Summary of DSSO-interlinked peptides of cytochrome c identified by LC MSn.
All of the interlinked peptides displayed characteristic fragment pairs and were identified by Batch-Tag, MS-Bridge, and Link-Finder. AA, amino acid; Mod., modification.
* Peptide fragments containing these sites were not sequenced by MS3.
‡ They were identified from different fragment pair ions by MS3.
In addition to products with one cross-link (i.e. types 0, 1, and 2), peptides containing two cross-links were also identified using this integrated work flow. In this work, 11 non-redundant DSSO-cross-linked peptides with two links (e.g. one interlink with one dead end, one interlink with one intralink, or one intralink with one dead end) were identified and are summarized in supplemental Table 1. This type of information is not commonly reported because peptide sequencing of multilinked peptides is highly complicated. This demonstrates the ability of our new cross-linking strategy for identifying such complex products.
Based on the crystal structure of bovine heart cytochrome c (Protein Data Bank code 2B4Z) (44), we calculated the distances between α carbons of the identified cross-linked lysine residues (Table I and supplemental Table 1). Among the 26 non-redundant interlinked lysines in cytochrome c identified in this work (excluding linkages between two adjacent lysines), all of the linkages had distances between their α carbons within the range of 5.3–19.3 Å. This is consistent not only with the length of a fully expanded DSSO (10.1-Å spacer length) and two lysine side chains but also with the previous results using similar lengths of NHS ester cross-linkers (21, 31, 43, 45). The results suggest that our cross-linking conditions did not induce significant disturbance to cytochrome c structural conformations.
In addition to cytochrome c, the same strategy was successfully applied to identify DSSO-cross-linked peptides of ubiquitin. Using the same analysis strategy, three interlinked, one intralinked, and five dead end peptides were identified as summarized in supplemental Table 2 and supplemental Fig. 1. Based on the crystal structure of bovine ubiquitin (Protein Data Bank code 1AAR), all of the identified inter/intralinked lysines in ubiquitin had distances between their α carbons within the range of 6–18 Å. The identified cross-linked lysines are consistent with the known structure of ubiquitin and previous reports (18, 38). It is interesting to note that one of the identified interlinked peptides is [LIFAGK48QLEDGR interlinked to LIFAGK48QLEDGR], which is a cross-link formed between the ubiquitin dimer. Residue Lys48 is located at a hydrophobic patch important for protein interactions, and Lys48 is also an in vivo chain linkage site for polyubiquitination required for ubiquitin/ATP-dependent proteasomal degradation (46). The same Lys48-Lys48 cross-link was identified previously using an alkyne-tagged NHS ester but only after selective enrichment coupled with CID and electron transfer dissociation analyses (18). In comparison, we were able to identify the Lys48-interlinked peptide without any enrichment, thus further demonstrating the effectiveness of our approach to identify DSSO-cross-linked peptides from complex mixtures.
Structural Elucidation of Yeast 20 S Proteasome Complex Using DSSO Cross-linking
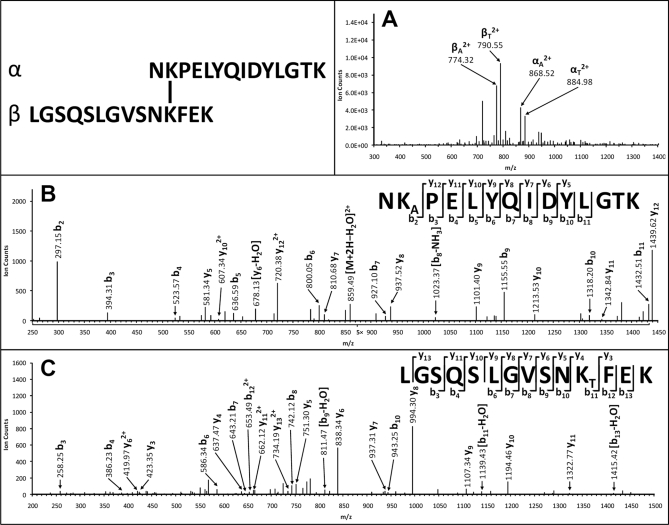
The ubiquitin-proteasome degradation pathway plays an important role in regulating many biological processes (46). The 26 S proteasome complex is the macromolecular machine responsible for ubiquitin/ATP-dependent protein degradation, and it is composed of two subcomplexes: the 20 S core particle and the 19 S regulatory complex. To date, only the crystal structure of the 20 S proteasome complex has been resolved. However, structures of the 19 and 26 S complexes remain elusive, thus hindering the understanding of the structure and functional relationship of the 26 S proteasome complex. To develop an effective cross-linking strategy to elucidate structures of the 19 and 26 S proteasome complexes, we therefore investigated the structure of the yeast 20 S proteasome complex using the DSSO cross-linking approach. The cross-linking of the 20 S proteasome complex was carried out in PBS buffer under conditions allowing efficient cross-linking of all subunits as based on one-dimensional SDS-PAGE (supplemental Fig. 2). The tryptic digest of the cross-linked proteasome complex was subjected to LC MSn analysis, and the data were analyzed using the integrated work flow described above (Fig. 6). In total, 13 unique interlinked peptides were identified, including 10 intrasubunit and three intersubunit heterodimeric interlinks, as summarized in Table II (for details, see supplemental Table 3), that were determined unambiguously by integration of Link-Finder, Batch-Tag (MS3 sequencing; see supplemental Fig. 3), and MS-Bridge (mass mapping of the cross-linked peptides) results. As an example, Fig. 7A displays the MS/MS spectrum of a DSSO heterodimeric interlinked peptide α-β (m/z 833.92314+) of the yeast 20 S proteasome complex in which two fragment pairs were detected and determined as αA/βT (868.522+/790.552+) and αT/βA (884.982+/774.322+). MS3 analysis of the αA fragment (m/z 868.522+) identified the α chain unambiguously as NKAPELYQIDYLGTK, which matched to 20 S subunit β4. In this sequence, LysA is modified with the alkene moiety. In addition, MS3 analysis of the βT fragment (m/z 790.552+) identified the β chain unambiguously as LGSQSLGVSNKTFEK, which matched to 20 S subunit β3. Here LysT is modified with an unsaturated thiol moiety. Mass mapping by MS-Bridge further confirmed this intersubunit (β4-β3) interlinked peptide as [NKPELYQIDYLGTK interlinked to LGSQSLGVSNKFEK].
Table II. Summary of DSSO-interlinked peptides of the yeast 20 S proteasome complex identified by LC MSn.
All of the interlinked peptides displayed characteristic fragment pairs and were identified by Batch-Tag, MS-Bridge, and Link-Finder. AA, amino acid; Mod., modification.
† Mature sequence from crystal data was used for data analysis.
‡ They were identified from different fragment pair ions by MS3.
* Peptide fragments containing these sites were not sequenced by MS3.
Fig. 7.
MSn analysis of DSSO heterodimeric interlinked peptide of the yeast 20 S proteasome complex (α-β: NKPELYQIDYLGTK interlinked to LGSQSLGVSNKFEK) with intersubunit link between 20 S subunit β4 and β3. A, MS/MS spectrum of [α-β]4+ (m/z 833.92314+) in which two fragment pairs were detected and determined as αA (m/z 868.522+)/βT (m/z 790.552+) and αT (m/z 884.982+)/βA (m/z 774.322+). B, MS3 spectrum of αA (m/z 868.522+) in which detection of a series of y and b ions determined its sequence unambiguously as NKAPELYQIDYLGTK. C, MS3 spectrum of βT (m/z 790.552+) in which detection of a series of y and b ions determined its sequence unambiguously as LGSQSLGVSNKTFEK. KA is modified with the alkene moiety, and KT is modified with the unsaturated thiol moiety.
In addition, 21 dead end-modified peptides were identified by multiple lines of evidence as illustrated in supplemental Table 3. The fragmentation behavior for the dead end-modified peptides of the 20 S subunits is the same as that of cytochrome c, showing two distinct dead end pairs in MS/MS spectra. This is illustrated with an example shown in supplemental Fig. 4.
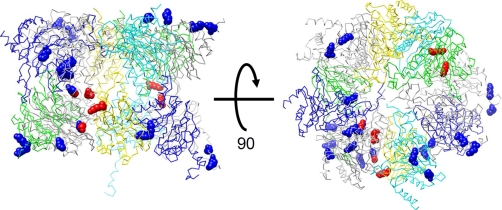
The experimentally determined structure of the yeast 20 S proteasome holocomplex was utilized (Protein Data Bank code 1RYP) to assess the cross-linked lysine pairs identified in this study. For each identified cross-link, the distance between the α carbons was calculated, and the results are summarized in Table II. Considering the spacer length of DSSO and lysine side chains, the theoretical upper limit for the distance between the α carbon atoms of paired lysines is ∼26 Å. Our reported distances are within this upper limit, providing some evidence that the proteasome cross-links are formed in the native state. The quaternary proteasome structure is formed by four stacked seven-member rings in the order αββα. The side view and basal view of the arrangement among one set of the symmetric αβ rings and their subunits are shown in Fig. 8. The α carbon trace is shown for all subunits, and the cross-linked lysines are shown in space-filling representation. Lysines forming intrasubunit cross-links appear in blue, and those forming intersubunit cross-links appear in red. The images in Fig. 8 were generated using UCSF Chimera visualization software (47).
Fig. 8.
Mapping identified DSSO-interlinked lysines onto crystal structure of yeast 20 S proteasome. The lysines forming intrasubunit cross-links appear space-filled in blue, and those forming intersubunit cross-links appear space-filled in red.
DISCUSSION
We have presented a novel cross-linking strategy for structural analysis of model proteins and the yeast 20 S proteasome complex by combining a newly designed MS-cleavable cross-linker, DSSO, with an integrated data analysis work flow. This approach is effective and facilitates fast and accurate identification of DSSO-cross-linked peptides by LC MSn. The new MS-cleavable cross-linker DSSO is attractive for cross-linking studies of protein complexes for a number of reasons: 1) It can be easily synthesized and can cross-link protein complexes effectively at submicromolar concentrations (∼1 μm); 2) It has two symmetric CID-labile C–S bonds that preferentially fragment prior to peptide backbone breakage; 3) The CID-induced cleavage of interlinked peptides is specific and independent of peptide charges and sequences; 4) DSSO-cross-linked peptides can generate characteristic fragmentation patterns in MS/MS spectra that are unique to different types of cross-linked peptides for easy identification; 5) There are unique mass and charge relationships between MS/MS peptide fragment ions and their parent ions, permitting automated data processing. In comparison with existing MS-cleavable cross-linkers (16, 34–36, 38), the DSSO cross-linker can provide a specific and selective fragmentation of cross-linked peptides for identification. The fragmentation patterns of DSSO-cross-linked peptides are similar to those of fixed charge sulfonium ion-containing cross-linked model peptides developed by Lu et al. (37). Although DSSO does not carry a fixed charge, our results demonstrated that the preferential cleavage of the C–S bond adjacent to the sulfoxide in DSSO is as effective as cleavage of the C–S bond in the sulfonium ion-containing cross-linker (i.e. S-methyl 5,5′-thiodipentanoylhydroxysuccinimide) (37). However, fragmentation of the sulfonium ion-containing cross-linked peptide requires the formation of a five-member ring with the sulfonium ion and the amide of the linker such that it is not feasible to change spacer lengths in these cross-linkers. In contrast, the simple fragmentation mechanism gives DSSO the flexibility of changing its spacer lengths to accommodate cross-linking lysines at different distances while maintaining the symmetry of the linker with easily interpretable fragmentation patterns. In addition, DSSO has better potential for studying protein interactions by in vivo cross-linking. It is well known that cross-linking study of protein complexes is extremely challenging because of the inherent limitations of current cross-linkers. With the improvement on database searching of non-cleavable interlinked peptides, it is possible to identify cross-linked peptides of protein complexes using non-cleavable cross-linkers (12, 13). However, this requires a special program for data interpretation, and the false positive rate of identifying interlinked sequences is higher than that of identifying single sequences. Here we have demonstrated the feasibility of using the novel DSSO cross-linking strategy to study the structure of the yeast 20 S proteasome complex. This work represents a major advancement in structural elucidation of multisubunit protein complexes with improved data analysis and accuracy as such application of MS-cleavable cross-linkers has not been reported before.
In addition to the design of this novel MS-cleavable linker, we developed an integrated data analysis work flow to achieve fast, easy, and accurate identification of cross-linked peptides and the cross-linking sites. Identification of DSSO-cross-linked peptides from complex mixtures was accomplished with high confidence by integrating data analyses of three different data sets, MS, MS/MS, and MS3 data. Because of the difficulty in interpreting MS/MS spectra of unseparated interlinked peptides, many of previously reported interlinked products were determined only based on parent masses. In contrast, all of the interlinked peptides of cytochrome c, ubiquitin, and the yeast 20 S proteasome complex were identified in this work with three lines of evidence, including characteristic fragmentation pairs (Link-Finder), peptide sequence determination by MS3 sequencing (Batch-Tag), and mass mapping (MS-Bridge). This procedure permits the identification of cross-linked peptides with high accuracy, reliability, and speed. It is important to note that existing database search programs can be easily adapted for analyzing DSSO-cross-linked peptides; thus, a broad application of the DSSO-based cross-linking strategy is foreseeable. Furthermore, cross-linked peptides of cytochrome c with two links can be identified, suggesting the capability of the new cross-linking strategy for identifying more complex cross-linked products.
Cross-linking/mass spectrometry has been previously attempted to study the yeast 20 S proteasome complex using Ru(II)(bpy)32+ (tris(2,2′-bipyridyl)ruthenium (II) dication)/ammonium persulfate/light-mediated cross-linking (48) in which multiple subunit interconnectivity has been determined based on MS identification of co-migrated subunits by SDS-PAGE after cross-linking. No cross-linked peptides were identified due to complicated chemistry of the radical-based cross-linking reaction. Therefore, our work describes the first successful use of a cross-linking/mass spectrometry strategy to determine intersubunit and intrasubunit interaction interfaces of the yeast 20 S proteasome complex. Although only 13 interlinked peptides of the yeast 20 S proteasome were identified and reported here, this work presents the first step toward full characterization of proteasome structures using cross-linking/mass spectrometry in the future. The feasibility of using the DSSO-based cross-linking strategy to identify cross-linked peptides of a large protein complex at 1 μm or less concentration is very significant and of great promise to structural studies of protein complexes because purifying protein complexes at high concentrations is technically challenging.
During LC MSn analysis using the LTQ-Orbitrap XL mass spectrometer, collision energy cannot be adjusted on the fly to account for differences in peptide charge states; therefore, compromised collision energy is set during the entire LC MSn run. Thus, there exists a possibility that the collision energy may be too high for the highly charged ions but too low for peptides with lower charges. Future improvement on charge selection and energy adjustment during LC MSn data acquisition may be needed to further enhance the quality of the results. Additionally, optimized peptide separation prior to LC MSn analysis will be necessary to improve the dynamic range of peptide analysis and allow the detection of low abundance cross-linked peptides. Moreover, refinement of the Link-Finder program is needed to improve the identification of intralinked peptides. Lastly, the addition of an affinity tag to the sulfoxide-containing cross-linker will improve detection of cross-linked peptides, which will be the subject of our future study.
In summary, we developed a new MS-cleavable cross-linker DSSO that is applicable for model peptides and proteins and a multisubunit protein complex. The unique MS features of DSSO-cross-linked peptides together with our integrated data analysis work flow for analyzing LC MSn data greatly reduce the time spent identifying cross-linked peptides. Given its simplicity, speed, and accuracy, we believe that this cross-linking strategy will have a broad application in elucidating structures of proteins and protein complexes in the future.
Acknowledgments
We thank members of the Huang and Rychnovsky laboratories for help during this study. We thank Prof. A. L. Burlingame, Peter Baker, and Robert Chalkley at the University of California San Francisco for use of Protein Prospector.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant GM-74830 (to L. H.). This work was also supported by a Department of Education (Graduate Assistance in Areas of National Need) predoctoral fellowship (to D. V.).
 This article contains supplemental Tables 1–3 and Figs. 1–4.
This article contains supplemental Tables 1–3 and Figs. 1–4.
1 The abbreviations used are:
- MSn
- multistage tandem MS
- DSSO
- disuccinimidyl sulfoxide
- HRMS
- high resolution MS
- ES
- electrospray
- NHS
- N-hydroxysuccinimide
- A
- alkene
- S
- sulfenic acid
- T
- unsaturated thiol
- DN
- dead end.
REFERENCES
- 1. Sinz A. (2003) Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 38, 1225–1237 [DOI] [PubMed] [Google Scholar]
- 2. Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 3. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sinz A. (2010) Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal. Bioanal. Chem. 397, 3433–3440 [DOI] [PubMed] [Google Scholar]
- 5. Vasilescu J., Guo X., Kast J. (2004) Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 6. Guerrero C., Tagwerker C., Kaiser P., Huang L. (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 7. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 8. Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. (2008) Characterization of the proteasome interaction network using a Qtax-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaake R. M., Milenkoviæ T., Przulj N., Kaiser P., Huang L. (2010) Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the Qtax strategy. J. Proteome Res. 9, 2016–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Back J. W., de Jong L., Muijsers A. O., de Koster C. G. (2003) Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 331, 303–313 [DOI] [PubMed] [Google Scholar]
- 11. Rappsilber J., Siniossoglou S., Hurt E. C., Mann M. (2000) A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem. 72, 267–275 [DOI] [PubMed] [Google Scholar]
- 12. Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6, 2200–2211 [DOI] [PubMed] [Google Scholar]
- 13. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. (2003) Ms2assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 [DOI] [PubMed] [Google Scholar]
- 15. Trester-Zedlitz M., Kamada K., Burley S. K., Fenyö D., Chait B. T., Muir T. W. (2003) A modular cross-linking approach for exploring protein interactions. J. Am. Chem. Soc. 125, 2416–2425 [DOI] [PubMed] [Google Scholar]
- 16. Tang X., Munske G. R., Siems W. F., Bruce J. E. (2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 [DOI] [PubMed] [Google Scholar]
- 17. Chu F., Mahrus S., Craik C. S., Burlingame A. L. (2006) Isotope-coded and affinity-tagged cross-linking (ICATXL): an efficient strategy to probe protein interaction surfaces. J. Am. Chem. Soc. 128, 10362–10363 [DOI] [PubMed] [Google Scholar]
- 18. Chowdhury S. M., Du X., Toliæ N., Wu S., Moore R. J., Mayer M. U., Smith R. D., Adkins J. N. (2009) Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 81, 5524–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kasper P. T., Back J. W., Vitale M., Hartog A. F., Roseboom W., de Koning L. J., van Maarseveen J. H., Muijsers A. O., de Koster C. G., de Jong L. (2007) An aptly positioned azido group in the spacer of a protein cross-linker for facile mapping of lysines in close proximity. Chembiochem 8, 1281–1292 [DOI] [PubMed] [Google Scholar]
- 20. Nessen M. A., Kramer G., Back J., Baskin J. M., Smeenk L. E., de Koning L. J., van Maarseveen J. H., de Jong L., Bertozzi C. R., Hiemstra H., de Koster C. G. (2009) Selective enrichment of azide-containing peptides from complex mixtures. J. Proteome Res. 8, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vellucci D., Kao A., Kaake R. M., Rychnovsky S. D., Huang L. (2010) Selective enrichment and identification of azide-tagged cross-linked peptides using chemical ligation and mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 1432–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collins C. J., Schilling B., Young M., Dollinger G., Guy R. K. (2003) Isotopically labeled crosslinking reagents: resolution of mass degeneracy in the identification of crosslinked peptides. Bioorg. Med. Chem. Lett. 13, 4023–4026 [DOI] [PubMed] [Google Scholar]
- 23. Petrotchenko E. V., Olkhovik V. K., Borchers C. H. (2005) Isotopically coded cleavable cross-linker for studying protein-protein interaction and protein complexes. Mol. Cell. Proteomics 4, 1167–1179 [DOI] [PubMed] [Google Scholar]
- 24. Petrotchenko E. V., Borchers C. H. (2010) ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinformatics 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sinz A., Wang K. (2004) Mapping spatial proximities of sulfhydryl groups in proteins using a fluorogenic cross-linker and mass spectrometry. Anal. Biochem. 331, 27–32 [DOI] [PubMed] [Google Scholar]
- 26. Back J. W., Hartog A. F., Dekker H. L., Muijsers A. O., de Koning L. J., de Jong L. (2001) A new crosslinker for mass spectrometric analysis of the quaternary structure of protein complexes. J. Am. Soc. Mass Spectrom. 12, 222–227 [DOI] [PubMed] [Google Scholar]
- 27. Chu F., Baker P. R., Burlingame A. L., Chalkley R. J. (2010) Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol. Cell. Proteomics 9, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao Q., Xue S., Shaffer S. A., Doneanu C. E., Goodlett D. R., Nelson S. D. (2008) Minimize the detection of false positives by the software program DetectShift for 18O-labeled cross-linked peptide analysis. Eur. J. Mass Spectrom. 14, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. (2008) Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal. Chem. 80, 8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee Y. J., Lackner L. L., Nunnari J. M., Phinney B. S. (2007) Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J. Proteome Res. 6, 3908–3917 [DOI] [PubMed] [Google Scholar]
- 32. Nadeau O. W., Wyckoff G. J., Paschall J. E., Artigues A., Sage J., Villar M. T., Carlson G. M. (2008) CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins. Mol. Cell. Proteomics 7, 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang L., Tang X., Weisbrod C. R., Munske G. R., Eng J. K., von Haller P. D., Kaiser N. K., Bruce J. E. (2010) A photocleavable and mass spectrometry identifiable cross-linker for protein interaction studies. Anal. Chem. 82, 3556–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang H., Tang X., Munske G. R., Tolic N., Anderson G. A., Bruce J. E. (2009) Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell. Proteomics 8, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soderblom E. J., Goshe M. B. (2006) Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78, 8059–8068 [DOI] [PubMed] [Google Scholar]
- 36. Soderblom E. J., Bobay B. G., Cavanagh J., Goshe M. B. (2007) Tandem mass spectrometry acquisition approaches to enhance identification of protein-protein interactions using low-energy collision-induced dissociative chemical crosslinking reagents. Rapid Commun. Mass Spectrom. 21, 3395–3408 [DOI] [PubMed] [Google Scholar]
- 37. Lu Y., Tanasova M., Borhan B., Reid G. E. (2008) Ionic reagent for controlling the gas-phase fragmentation reactions of cross-linked peptides. Anal. Chem. 80, 9279–9287 [DOI] [PubMed] [Google Scholar]
- 38. Gardner M. W., Vasicek L. A., Shabbir S., Anslyn E. V., Brodbelt J. S. (2008) Chromogenic cross-linker for the characterization of protein structure by infrared multiphoton dissociation mass spectrometry. Anal. Chem. 80, 4807–4819 [DOI] [PubMed] [Google Scholar]
- 39. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 40. Reid G. E., Roberts K. D., Kapp E. A., Simpson R. I. (2004) Statistical and mechanistic approaches to understanding the gas-phase fragmentation behavior of methionine sulfoxide containing peptides. J. Proteome Res. 3, 751–759 [DOI] [PubMed] [Google Scholar]
- 41. Pearson K. M., Pannell L. K., Fales H. M. (2002) Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun. Mass Spectrom. 16, 149–159 [DOI] [PubMed] [Google Scholar]
- 42. Dihazi G. H., Sinz A. (2003) Mapping low-resolution three-dimensional protein structures using chemical cross-linking and Fourier transform ion-cyclotron resonance mass spectrometry. Rapid. Commun. Mass Spectrom. 17, 2005–2014 [DOI] [PubMed] [Google Scholar]
- 43. Guo X., Bandyopadhyay P., Schilling B., Young M. M., Fujii N., Aynechi T., Guy R. K., Kuntz I. D., Gibson B. W. (2008) Partial acetylation of lysine residues improves intraprotein cross-linking. Anal. Chem. 80, 951–960 [DOI] [PubMed] [Google Scholar]
- 44. Mirkin N., Jaconcic J., Stojanoff V., Moreno A. (2008) High resolution x-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins 70, 83–92 [DOI] [PubMed] [Google Scholar]
- 45. Kruppa G. H., Schoeniger J., Young M. M. (2003) A top down approach to protein structural studies using chemical cross-linking and Fourier transform mass spectrometry. Rapid Commun. Mass Spectrom. 17, 155–162 [DOI] [PubMed] [Google Scholar]
- 46. Pickart C. M., Cohen R. E. (2004) Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 47. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Denison C., Kodadek T. (2004) Toward a general chemical method for rapidly mapping multi-protein complexes. J. Proteome Res. 3, 417–425 [DOI] [PubMed] [Google Scholar]