Abstract
Precise annotation of genes or open reading frames is still a difficult task that results in divergence even for data generated from the same genomic sequence. This has an impact in further proteomic studies, and also compromises the characterization of clinical isolates with many specific genetic variations that may not be represented in the selected database. We recently developed software called multistrain mass spectrometry prokaryotic database builder (MSMSpdbb) that can merge protein databases from several sources and be applied on any prokaryotic organism, in a proteomic-friendly approach. We generated a database for the Mycobacterium tuberculosis complex (using three strains of Mycobacterium bovis and five of M. tuberculosis), and analyzed data collected from two laboratory strains and two clinical isolates of M. tuberculosis. We identified 2561 proteins, of which 24 were present in M. tuberculosis H37Rv samples, but not annotated in the M. tuberculosis H37Rv genome. We were also able to identify 280 nonsynonymous single amino acid polymorphisms and confirm 367 translational start sites. As a proof of concept we applied the database to whole-genome DNA sequencing data of one of the clinical isolates, which allowed the validation of 116 predicted single amino acid polymorphisms and the annotation of 131 N-terminal start sites. Moreover we identified regions not present in the original M. tuberculosis H37Rv sequence, indicating strain divergence or errors in the reference sequence. In conclusion, we demonstrated the potential of using a merged database to better characterize laboratory or clinical bacterial strains.
The annotation of a genomic DNA sequence with a list of the predicted translated protein repertoire represents the fundamental basis for identification of peptide mass spectra in proteomics (1–3). Therefore, the protein identification capacity of proteomic experiments is dependent on a correct interpretation and definition of the genome being studied. High-throughput genome sequencing technology has led to an exponential increase in the capacity to generate complete genomic data for diverse organisms (4). According to the Genome Online Database (http://www.genomesonline.org/gold_statistics.htm) (5, 6), there were 1020 completed bacterial genomes available during preparation of this article. The accelerated rate of genomic sequencing projects and the translation of this information into protein data sets represent a welcome boost for the establishment and development of proteomic studies for several organisms. However, such a vast amount of sequence data from diverse genomes has made open reading frame predictions dependent on automatic computational analysis. It has been pointed out that several errors have been introduced during the first stages of nucleotide sequencing and open reading frame predictions, and this has a major impact on subsequent studies (for a review, see (7)).
This point can be demonstrated by comparing and testing different gene annotations obtained from the same genomic sequence. For example, the M. tuberculosis H37Rv gene annotations from the Sanger Institute and the JCV Institute differ in 15% of the annotated genes, whereas for genes identified by both annotations, different start codons were designated for 50% of them (8). Another species, like Mycobacterium leprae, represents an even more extreme example, with a 300% difference in number of annotated genes (9). Not surprisingly, comparison of protein identifications using primary or secondary annotations clearly illustrates the limitations encountered if only one of the annotations is taken into consideration for analysis of experimental proteomic data. This shows that even for well characterized genomes there is no consensus on how to determine an exact gene number, and consequently also the identity and structure of the genes.
Many efforts have been put into creation of more suitable, proteomics-friendly databases. These databases contain compilations of protein entries from different sources such as SwissProt (10), a manually curated protein database. For example, the International Protein Index (IPI)1 (11) has been well accepted and used by the proteomic community. The IPI approach retrieves individual entries from different sources, and through clustering of sequences based on sequence similarities and cross-references, redundant proteins are reported only once and unique entry information is also added. Current IPI versions provide highly reliable genome coverage with a low redundancy level. However, IPI is only available for seven species (human, mouse, rat, zebra fish, Arabidopsis, chicken, and cow), and does not include variations based on single amino acid polymorphisms (SAPs). Concatenated databases such as NCBInr have good organism coverage and also contain gene entries from uncompleted genomes. However, database size and entry redundancy might limit data analysis.
SAPs and other types of sequence variations can be included in proteomics-friendly databases by adding the portion of the sequence containing the mutation(s) after the main entry sequence, but separated by the letter “J” to serve as a neutral symbol (12). This approach does not increase the redundancy of entries and therefore does not compromise the statistics of the analysis. By applying this “MS-friendly” method to the IPI human database, these authors were able to identify many SAPs and processed N-terminal peptides in a breast cancer cell line (MCF7). This approach has an obvious potential for the analysis of prokaryotic clinical strains because the characterization of proteins with polymorphisms is suboptimal if the variants are not represented in the publicly available database of a particular pathogen. In many cases antibiotic resistance is caused by single nucleotide polymorphisms (SNP)s, and recent data also suggests that microbial virulence can be related to SNPs (13). It is therefore crucial to provide protein databases that can compensate for annotation errors and cover genetic variations among closely related organisms that provide readily identifiable unique observations. Although the same can be achieved by merely concatenating databases of interest, database size and data analysis will become an issue as more genomic information is made available.
We have therefore developed a software for prokaryotic genomes termed Multistrain Mass Spectrometry prokaryote database builder (MSMSpdbb) to perform clustering of homologous protein entries from different sources (14). This is similar to what is achieved by the IPI format, but we also add information about different translational start site (TSS) choices, SAPs and other mutations in line with what was reported for eukaryotic organisms (12). We used the publicly available nonredundant M. tuberculosis complex protein database, and analyzed mass spectrometry data collected from different fractions of the M. tuberculosis H37Rv and M. tuberculosis H37Ra laboratory strains, plus samples from two clinical M. tuberculosis Beijing isolates. Our data demonstrate the potential for MSMSpdbb-generated databases to identify relevant SAPs, as well as identification of proteins annotated in only a subset of the genomes. Furthermore, we have found a highly abundant protein from the ESAT-6 family in the M. tuberculosis H37Rv ATCC 27294 strain that is encoded in an area of the genome sequence not described within the original M. tuberculosis H37Rv genomic sequence (a non-ATCC strain) (15).
EXPERIMENTAL PROCEDURES
Generation of a Database for the M. tuberculosis Complex
The protein database in FASTA format was generated by in-house developed software named MSMSpdbb (14). Genomic sequences of M. tuberculosis strains: CDC1551 (16), F11, H37Ra (17), H37Rv (15), and KZN1435, as well as the M. bovis strains: AF2122/97 (18), BCG Pasteur 1173P2, and BCG Tokyo 172 (19), together with annotated protein information were used as a basis for the database. Only primary annotations were used. i.e. gene annotations consequently performed by independent groups were not considered. Only protein products larger than 50 amino acids were considered during stop-to-stop translation. Peptides describing different translational start site choices or sequence differences across strains were only added if the peptide sequence was longer than seven amino acids and shorter than 35. Peptides containing amino acid ambiguities, i.e. containing an X symbol because of not confirmed nucleotide determination in the genome sequence, were not added. Whenever proteins from different strains were clustered, the accession number and description to use for the entry was retrieved in a prioritized manner where M. tuberculosis H37Rv had highest priority. Translated entries, which did not cluster with any currently annotated genes, were discarded for the annotated-only database option of MSMSpdbb. It is worth mentioning that MSMSpdbb protein entries will be larger. Consequently, calculations for sequence coverage, molecular weight, and sequence size as given in the final results are reported relative to the complete protein entry.
Data Collection
All M. tuberculosis H37Rv data collected for this work was derived from the M. tuberculosis H37Rv ATCC27294 strain. High-resolution mass spectrometry data collected in the last 2 years by our group was submitted for analysis with our in-house M. tuberculosis complex database. The samples included: M. tuberculosis H37Rv culture filtrate fraction (8), M. tuberculosis H37Rv and M. tuberculosis H37Ra membrane fractions (20), M. tuberculosis H37Rv whole cell lysates (collected for this work), and whole cell lysates of two different clinical Beijing isolates (21). In total, 142 MS acquisition files were analyzed. See Supplementary file S1 for data acquisition statistics (“Summary” sheet).
Gel Electrophoresis and In-Gel Digestion of Proteins
Fifty micrograms of protein sample was mixed with 15 μl sodium-dodecyl-sulfate loading buffer containing 10 mm dithiotreitol, and boiled for 5 min before separation using 4%–12% SDS-PAGE (NuPAGE kit, Invitrogen, Carlsbad, CA). The protein bands were visualized with Coomassie Brilliant Blue R-250 staining kit (Invitrogen). Protein lanes were excised and washed twice with 50% acetonitrile (ACN) at room temperature, until excess Coomassie Brilliant Blue and sodium-dodecyl-sulfate were removed. The proteins were then reduced using 10 mm dithiotreitol in water at 58 °C for 1 h, and alkylated with 55 mm iodacetamide in 100 mm NH4HCO3 pH 8.0 for 45 min at RT. The gel pieces were washed twice 50 mm NH4HCO3, followed by gel dehydration by 100% ACN. Gel pieces were rehydrated in 30 μl 50 mm NH4HCO3 containing 0.125 μg of sequence-grade trypsin (Promega, Madison, WI) overnight at 37 °C. Trypsin reaction was quenched using 1% trifluoroacetic acid. The digested peptides were eluted by incubating the gel pieces with 50 μl 50% ACN for 20 min at room temperature twice, plus a final wash with 100% ACN for 10 min. Peptide mixtures were then desalted using STAGE-tips packed with C18 resin (3 m, USA) (22).
Mass Spectrometry
All experiments were performed on a Dionex Ultimate 3000 nano-LC system (Sunnyvale, CA) connected to a linear quadrupole ion trap - Orbitrap (LTQ-Orbitrap) mass spectrometer (ThermoElectron, Bremen, Germany) equipped with a nanoelectrospray ion source. For liquid chromatography separation we used an Acclaim PepMap 100 column (C18, 3 μm, 100 Å) (Dionex, Sunnyvale, CA) capillary of 12 cm bed length. The flow rate was 0.3 μl/min for the nano column, and the solvent gradient was 7% B to 40% B in 87 min, then 40%–80% B in 8 min. Solvent A was aqueous 2% ACN in 0.1% formic acid, whereas solvent B was aqueous 90% ACN in 0.1% formic acid.
The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full scan MS spectra (from m/z 300 to 2000) were acquired in the Orbitrap with resolution r = 60,000 at m/z 400 (after accumulation to a target of 1,000,000 charges in the LTQ). The method used allowed sequential isolation of the most intense ions, up to six, depending on signal intensity, for fragmentation on the linear ion trap using collisionally induced dissociation at a target value of 10,000 charges.
For accurate mass measurements the lock mass option was enabled in MS mode and the polydimethylcyclosiloxane ions generated in the electrospray process from ambient air were used for internal recalibration during the analysis (23). Target ions already selected for MS/MS were dynamically excluded for 60 s. General mass spectrometry conditions were: electrospray voltage, 1.5 kV; no sheath and auxiliary gas flow. Ion selection threshold was 500 counts for MS/MS, and an activation Q-value of 0.25 and activation time of 30 ms were also applied for MS/MS.
Protein Identification
All acquired data was processed and analyzed using MaxQuant (version 1.0.13.8), a software specifically developed for data acquired using high-resolution instrumentation (24). MS/MS peak lists from individual 142 RAW files were generated using Quant.exe tool from the MaxQuant package, using default extract_msn (Thermo) parameters. Protein identification was performed by searching the data against M. tuberculosis database generated by the MSMSpdbb script. The database was also modified to contain reversed sequences of all entries as a control of false-positive identifications during analysis. Common contaminants, such as keratins, BSA, trypsin, were also added to the database. We used MASCOT Daemon for submission of multiple searches on a local Mascot server v2.2 (Matrix Science). Our Mascot program was slightly modified to recognize the letter codes “O” and “J” as an amino acid of mass 0.00. In addition a special trypsin cleavage rule was created that recognized the C- and N-terminal flanks of “O” and “J” as cleavage sites. The search parameters were: Enzyme specificity: trypsin with no proline restriction, modified to recognize O and J codes; Maximum missed cleavages: three; carbamidomethyl (C) as fixed modification; N-acetyl (Protein), oxidation (M), pyro-glu (Gln to pyro-Glu) and pyro-glu (Glu to pyro-Glu) as variable modifications; peptide mass tolerance of ±15 parts per million; MS/MS mass tolerance of 0.5 Da. The M. tuberculosis complex database from MSMSpdbb (14) contained 9290 entries (4630 forward sequences, 4630 reverse sequences, 30 most common contaminants).
MaxQuant report identifications as “Protein groups,” where proteins entries sharing all or part of the identified peptides are reported as a single protein hit including all members in the family. If just part of the peptides identified is shared by two or more entries, the group is reported as one, and the “leading” name given is the one of the protein with more unique observations. The presence of a unique peptide sequence to a “protein group” is mandatory for the group to be considered. Protein group identification and validation was performed by the Identify.exe option from MaxQuant under the following parameters: peptide and protein false discovery rate: 0.01 (1%), minimal peptide length was seven, and to guarantee a high confidence identification rate, the maximal allowed posterior error probability (PEP) was set to 0.15; minimal number of unique peptides per protein: one. Overall, the average mass accuracy of the identified peptides was 400 parts per billion.
Validation of De Novo DNA Sequencing Data
The DNA of the clinical M. tuberculosis Beijing high virulent strain was isolated according to the method of van Soolingen (25). DNA was sheared and sequenced on a GS FLX system (Roche, 454 Life Sciences, Branford, CT). 100,070,772 high-quality bases in sequencing reads of on average 250 bases were collected, with 22.7 fold coverage. The peptide sequences, that were identified in the high virulent isolate as described above, were subjected to an automated tBLASTN search (BLAST 2.2.17, (26)) against the collection of short sequencing reads. The peptides were considered to be confirmed if at least one BLAST hit without a mismatch and coverage of 100% was detected in the nucleotide read collection.
RESULTS
Structure and Redundancy of the Annotated M. tuberculosis Complex Database
The clustered database of the annotated proteins of the M. tuberculosis complex contained 4630 entries. From these, the MSMSpdbb software inserted 6636 new “J” peptides, representing SAPs and other sequence variations. The ratio between number of entries and number of sequence variations was 1:1.4, which indicates a high level of sequence similarity among the members of the M. tuberculosis complex. The number of “O” peptides representing predicted TSS choices was 10,158 (1:2.2), indicating that the annotations gave discrepant TSS choices for most of the entries. The database contained 60 sequence ambiguities (represented as “X”). These were mainly derived from protein sequences of the clinical M. tuberculosis isolate CDC1551. Peptides containing such ambiguities were not used for redundancy calculation or appended as “J” or “O” peptides to the main sequence.
One of the issues that might be raised against database merging and protein clustering is the fact that the merging itself will generate additional redundancy at the peptide level, even if the clustered proteins are not redundant per se. With this limitation in mind, we performed a redundancy check of the tryptic peptides present in the M. tuberculosis H37Rv database, as well as the merged and clustered M. tuberculosis complex database generated by MSMSpdbb (14). Our data shows that the M. tuberculosis H37Rv database has 62,173 tryptic peptides with seven amino acids or more. From those, only 478 tryptic peptides were observed in two entries or more. This means that, if a protein is identified based on a single peptide hit, the chance of ambiguous gene identification because of sequence redundancy is only 0.81%. In addition, the protein groups that share identical tryptic peptides are very limited and can be easily distinguished. For example, 54 out of 60 peptides that are repeatedly observed in four entries or more belong to more than 40 transposases present in the M. tuberculosis H37Rv genome. Of the remaining six peptides, four were observed in ESAT-6-like proteins.
When the clustered M. tuberculosis complex database was analyzed, similar results were observed. From 81,574 tryptic peptides with seven amino acids or more, 734 were observed in more than one entry in two or more genomes. This indicates a redundancy rate of 0.91%. More than one fifth of the redundant peptides were derived from the highly homologous transposases encoded in several locations throughout the genomes. Because MSMSpdbb cluster and report only one copy of the homolog genes, it is evident that peptide redundancy will rather decrease than increase as compared with one of the isolated databases. In addition, protein identifications will be based in unique peptides for 99.09% of the entries in the database.
Proteomic Analysis
Using M. tuberculosis laboratory and clinical strains, we performed high-resolution proteomic screens with a LTQ-Orbitrap mass spectrometer. In total, 2561 different proteins were identified in this study. Supplemental Data S1 contains the protein lists and Supplemental Data S2 the peptide lists output of all fractions/samples analyzed in this work. Each file contains information about protein mass, number of peptides per identified protein, peptide length, observed charge, observed mass over charge ratio, measured peptide mass (Da), Mascot Score, PEP values, the presence of modifications (as N-terminal acetylation or Met oxidation), and the error of the observed/theoretical mass in parts per million. All this can be accessed in order to verify data quality and representation. Supplemental Data S3 reports three lists: The list named “Full proteins” represent all of the identified protein entries from all of the analyzed fractions. The second list “SAPs” reports all SAPs identified in our work, and the third list “TSS” shows all TSS choices. The importance of this is discussed below.
Validation of TSS Choices and Nonpredicted Coding Sequences
The MSMSpdbb software was developed to build stop-to-stop codon translations from genomic data. Therefore, predicted TSS choices for previously annotated genes had to be artificially inserted in the protein sequence of the FASTA file. Fig. 1A and Fig. 2A illustrate examples of sequence entries used in the M. tuberculosis complex database. Note that the predicted TSSs are inserted in the original sequence after an “O” letter (in bold). Expected N-terminal peptides are considered with a Met start, or with a start with the amino acid at position +2, because Met processing is also possible.
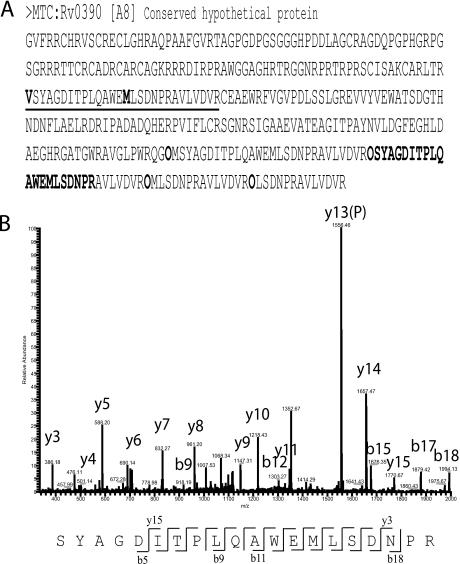
Fig. 1.
TSS choice validation for protein Rv0390. A, The entry in FASTA format is shown. Underlined region delimit the expected tryptic peptides of two predicted TSS choices, a valine and a methionine (bold, underlined). These tryptic peptides were artificially added in the end of the entry after the letter code “O” (bold). B, Fragmentation profile of peptide SYAGDITPLQAWEMLSDNPR.
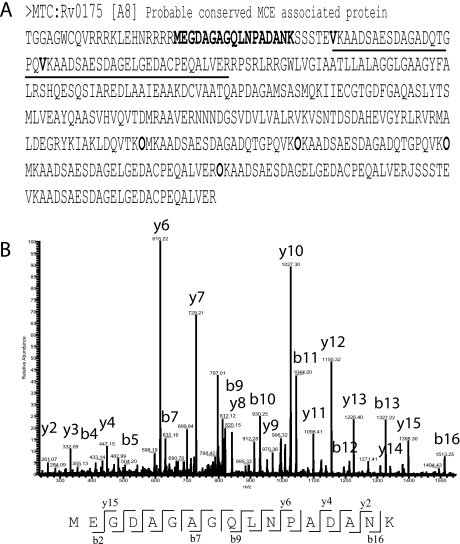
Fig. 2.
Identification of regions predicted as noncoding. A, The entry in FASTA format is shown, with the predicted N-terminal tryptic peptide underlined. The sequence in bold is present in a region initially predicted as noncoding in all eight genomes used in this work. B, The fragmentation pattern of sequence MEGDAGAGQLNPADANK is shown, indicating that this region is indeed coding. This amino acid sequence is not present in any other gene of the database.
In total, we validated 367 peptides preceded by “O,” which confirms one of the TSS choices used for the corresponding genes. Only one TSS choice was confirmed per gene. They represent ∼8% of the genes present in the M. tuberculosis complex database. Fig. 1 exemplifies one of these cases. In Fig. 1A, the sequence of the stop-to-stop codon region containing the gene Rv0390 [A8] (present in all of the eight genomes) is shown. This gene had two predicted TSS choices in all the genomes used, one at V121 and another at M135. In Fig. 1B, the MS/MS fragmentation pattern of peptide SYAGDITPLQAWEMLSDNPR is shown. In the original sequence, this peptide is preceded by a valine and does not constitute a tryptic peptide, which demonstrates why the peptide had to be appended with the “O” code (see Methods).
Additionally, we were able to identify a few examples of peptide products from genomic regions initially predicted to be noncoding. Fig. 2A shows the entry sequence format of protein Rv0175 [A8]. This protein had two possible predicted TSSs, one at V44 and one at V63. However, we identified the peptide MEGDAGAGQLNPADANK which is present from position M22 to K38. The presence of an arginine in position 21 in the sequence does not allow us to discriminate between M22, V9, or L14 as the possible TSS choice, because the identified peptide is also a normally occurring tryptic peptide.
Identification of M. tuberculosis H37Rv Primary Unannotated Genes
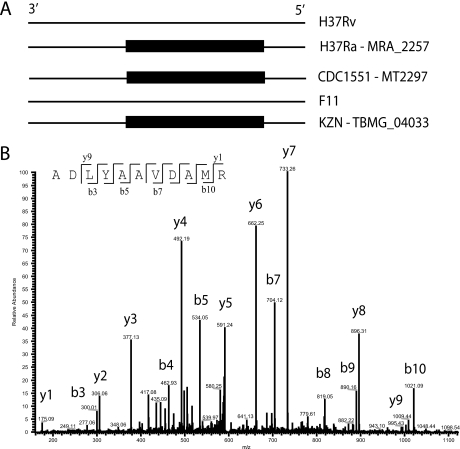
The sequence clustering provided by MSMSpdbb also allows the reporting of annotated genes that are specific to one or some of the genomes, thereby reducing annotation error discrepancies. For example, the annotated protein MT2297 [A3] is annotated in the M. tuberculosis strains: CDC1551, H37Ra (entry MRA_2257), and KZN1435 (entry TBMG_04033) (Fig. 3A, black boxes schematically representing the gene presence). It is important to note that, whereas the M. tuberculosis H37Rv and M. tuberculosis F11 annotation efforts did not consider that region in the genome as coding, both strains possess the region nonetheless. We identified three peptides of this protein, with Mascot scores from 54 to 81, in membrane fractions and whole cell lysate fractions of M. tuberculosis H37Rv strains (Fig. 3B shows the fragmentation pattern of one of those). This indicates that the gene was not correctly annotated for that genome. In total, we identified 24 genes in M. tuberculosis H37Rv or in the clinical isolates, which were not correctly predicted by the M. tuberculosis H37Rv annotation (Genolist) and would be discarded if our database was not used.
Fig. 3.
Identification of M. tuberculosis H37Rv unannotated genes. A, Schematic representation of the genomic region containing the gene MT2297 from the M. tuberculosis CDC1551 strain. Black boxes indicate gene annotation. In M. tuberculosis H37Rv and M. tuberculosis F11 genomes, the gene is not annotated but the genomic region is nonetheless present. B, Fragmentation pattern of peptide ADLYAAVDAMR from MT2297, present in M. tuberculosis H37Rv fractions.
Identification of SAPs in Clinical Isolates
We used both laboratory and clinical strains to determine the presence of SAPs. In total, we identified 280 SAPs (Supplemental Data S3), and mapped their occurrence in the 8 M. tuberculosis genomes (which were classified in a presence/absence matrix).
We analyzed two M. tuberculosis Beijing strains, one high virulent and one low virulent, in order to characterize divergence of clinical strains. From the 280 observed SAPs among the four strains used in this work, 153 were found in the clinical isolates. Interestingly, analysis of the SAPs in the Beijing strains indicates that they diverge as compared with the other clinical M. tuberculosis strains: F11, CDC1551, and KZN (Supplemental Data S4). The Beijing strains had 34 SAPs that were absent from at least one of the M. tuberculosis clinical genomes (CDC1551, F11, and KZN1435). From those, 17 SAPs were not present in M. tuberculosis CDC1551. In addition, the Beijing strains also had eight SAPs that were only observed in the M. bovis strains. However, a proper comparison should be performed by distance matrix analysis once we have the completed genomic sequences of the high virulent and low virulent Beijing strains used in this work.
Validation of SAPs and Prediction of Translational Start Sites: Application to DNA De Novo Sequencing
To demonstrate the usefulness of our approach to de novo sequencing, we compared the 249 peptides identified in the M. tuberculosis Beijing high virulent isolate to a collection of high-throughput DNA sequencing reads of the same isolate. We validated 247 peptides that were found by comparison to a collection of DNA reads covering all six reading frames. The comparison allowed the determination of 131 N-terminal start sites. Additionally, 116 SAPs were confirmed. Only two out of 249 peptides (false discovery rate of 0.80%) were absent in the translations of the six reading frames, which is in accordance with the 1% false discovery rate reported in the proteomic analysis. Both these peptides might represent possible unexpected SAPs, because of variation in bacterial subpopulations. The SAP from the protein Rv2935 is a possible example of this, because both expected and unexpected polymorphic variants were discovered in the low virulent and high virulent isolates with good Mascot scores. For the variant observed in protein Rv3646c, however, a closer analysis of the identified peptide revealed that the Mascot score might indicate it is a false-positive identification in our database. This peptide is shown in red in Supplemental Data S3.
Peptide Evidence Demonstrating Strain Divergence
When searching against the M. tuberculosis complex database, we made two observations that provide further evidence to support that the original M. tuberculosis H37Rv used for the genomic sequence has a divergent genetic background when compared with the ATCC strain, or even to M. tuberculosis H37Ra or M. tuberculosis CDC1551. First, we identified a SAP in the protein Rv2037c, which is present in all eight genomes, except M. tuberculosis H37Rv. In this example, a cysteine present in M. tuberculosis H37Rv is substituted to tyrosine in the peptide FLANWNYADYLADCGGPFTPSL.
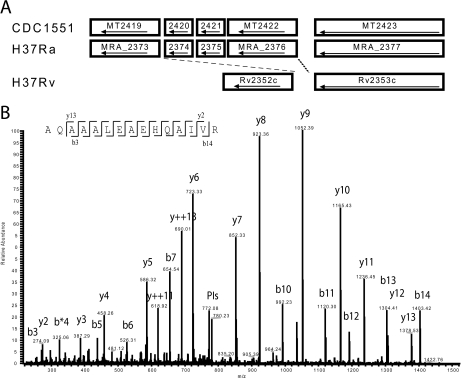
In addition, multiple peptides were observed, matching the protein entry MT2420 [A2], an ESAT-6-like protein that is only described in the M. tuberculosis CDC1551 and M. tuberculosis H37Ra genomes (Fig. 4A). Fig. 4A also illustrate M. tuberculosis H37Rv representation of the same region, based on the original genome sequence. In the original M. tuberculosis H37Rv, the DNA region containing MT2420 and two other genes (MT2421 and MT2422), is not supposed to exist, illustrating a possible deletion of that region in M. tuberculosis H37Rv. However, Fig. 4B shows the MS/MS profile of one of these peptides of m/z of 789.42, identified as AQAAALEAEHQAIVR with a Mascot score of 118 and 0.9 ppm mass accuracy. The four peptides identified for MT2420 in our work are unique for this entry, and are not present in any other entry of our M. tuberculosis complex database. This identification suggests the existence of the DNA region containing the MT2420 “deletion” in the ATCC strain of M. tuberculosis H37Rv.
Fig. 4.
Missing region of the M. tuberculosis H37Rv genome. A, Alignment of selected gene sequences from M. tuberculosis CDC1551, H37Ra, and H37Rv genomes, illustrating a deletion region in M. tuberculosis H37Rv that includes genes MT2420/MRA_2374 to MT2422/MRA_2376. Interestingly, M. tuberculosis H37Rv and M. tuberculosis H37Ra share the same ancestor, but the M. tuberculosis H37Ra genome sequence share more similarities with the M. tuberculosis CDC1551 genome than with the original M. tuberculosis H37Rv genome sequence. B, Fragmentation pattern of peptide AQAAALEAEHQAIVR from MT2420, found in M. tuberculosis H37Rv (ATCC27294) whole cell lysates, indicating that the deletion reported in the original M. tuberculosis H37Rv sequencing effort is incorrect. MS/MS information tables (MaxQuant output) are openly available at www.proteomecommons.org under the Hash code: dXuxNwU84QKYzzkLfmpU8Mcv6p277wRTOWXjRuWEH/WkkdAyYT/DeWm3ILF43l3lLZF7MMchNwPBwWa6G16fo6KhRrIAAAAAAAAC/w = = All RAW files used in this work can be downloaded using the Hash code: EIH2o0QZ9mMIXgurpLpJ34rgf1PQHXKOIa0EUOX0NIZ+bJdOOsdkXvcCQ9N5ZUqtlAEDZ/TQaoPn/uTOvpR5SPQuAyAAAAAAAAB0Cw = =.
DISCUSSION
The current overwhelming number of complete and shotgun genomic sequences available in Genbank has boosted genetic and genomic studies (for a review, see (27)). It has also facilitated the visualization of genetic divergence among closely related organisms, exemplified by the characterization of horizontal gene transfers and SNPs, for example. However, limitations with gene annotations from individual sources can generate discrepancies, and such discrepancies will interfere with the generation of an accurate protein database to be used by proteomic approaches. As previously demonstrated (8), the generation of databases with one in-silico approach or without a good integration among other annotations can result in less efficient characterization of an organism's proteome.
To avoid such discrepancies, we recently developed a software to select genomic information and annotations from different sources, merge them and generate a protein FASTA database report with nonredundant information. We created a database including the publicly available M. bovis and M. tuberculosis genomic data. Initial analysis of M. tuberculosis H37Rv whole cell lysates showed that proteomic data analyzed using the database generated by our script could identify unannotated genes of M. tuberculosis H37Rv as well as peptides with SAPs (14). Herein, we expanded the evaluation to a deep analysis of laboratory and clinical strains. In total we identified 2561 proteins, comprising ∼63% of the genes predicted by Genolist, which contains the updated version of the annotation initially presented by the M. tuberculosis genome sequencing project (15). Within these identifications, we were able to determine genetic features such as the most accurate TSS prediction for a gene, gene annotation discrepancies, etc. This could be performed in a single analysis using one database. The identification of SAPs in a clinical strain of M. tuberculosis allowed the confirmation of SAPs predicted by high-throughput sequencing. The determination of start sites demonstrates the usefulness of the database for annotation and validation of de novo sequencing projects.
Validation and interpretation of genomic information through proteomic analysis can provide many advances to diverse research fields. For example, the improvement of TSS annotation and assignment can result in better identification of promoter regions and other structural entities or regulatory motifs (28, 29). This can consequently result in a better understanding of protein function and transcriptional regulation. We were able to confirm not only 367 TSS choice divergences (representing 9% of the primary M. tuberculosis H37Rv annotation), but we also identified peptides from regions considered to be upstream of the predicted TSS choice. These observations can be used to update database files to show only the confirmed N-terminals. This information can also be used to improve prediction algorithms. As supplemental material, we are making public an improved version of our M. tuberculosis complex database, where the protein sequence entries containing the 367 confirmed TSS choices were modified accordingly so that the sequences of these proteins start at the correct amino acid and predicted but inaccurate TSS choices previously added as “O” peptides are deleted from the entry (Supplemental Data S5).
One of the major limitations when characterizing the proteome of clinical bacterial isolates is that it is hard to determine which of the available sequenced genome databases is the most appropriate to use. This is the case because most, if not all, protein search engines use the mass and the sequence information of the experimental data (30, 31). Even slight mass variations resulting from a mutation of a lysine by a glutamine (i.e. equal to 0.036 Da) can be reliably measured by mass spectrometry instruments able to achieve precision at the sub-ppm level. In such cases, using a database that contains described SAPs or other variations are highly desirable, but proper attention to identifications containing SAP mass shifts, which are isobaric to post-translational shifts, (such as methionine oxidation) should always be given, to avoid false-positives. Furthermore, it can consequently be assumed that the usage of genome sequencing of clinical strains is more appropriate in order to characterize not-sequenced clinical strains. However, our data suggests that a randomly picked strain of epidemiological interest had a higher SAP divergence with known clinical strains, and higher similarity with the M. tuberculosis H37Rv and M. tuberculosis H37Ra laboratory strains. Curiously, M. tuberculosis is largely assumed to be a pathogen with a low rate of polymorphisms (32, 33), but our data indicates that the genetic background of clinical isolates of M. tuberculosis is variable. This might be more evident when additional complete genomic information is made available for M. tuberculosis strains (34, 35).
Finally, the most striking of our observations was the identification of the gene MT2420, which is not present in M. tuberculosis H37Rv but is annotated in the M. tuberculosis CDC1551 and M. tuberculosis H37Ra genomes. Recently, the genomic sequence of M. tuberculosis H37Ra was released, as well as its comparison with the M. tuberculosis H37Rv and M. tuberculosis CDC1551 genomic sequences (17). Surprisingly the H37Ra genome, which is derived from the same ancestral strain as M. tuberculosis H37Rv, showed a much higher sequence similarity with the M. tuberculosis CDC1551 clinical strain. This includes many SNPs and even insertions and deletions that were commonly observed in M. tuberculosis H37Ra and M. tuberculosis CDC1551, but absent in M. tuberculosis H37Rv. When selected domains of the M. tuberculosis H37Rv (ATCC27294) genome were resequenced, it became clear that the ATCC27294 sequence had the same SNPs as observed in M. tuberculosis H37Ra. These observations have raised concern about the genetic background of the strain used in the first M. tuberculosis genome sequencing project (15). Our identification of a protein product from a region that is supposed not to exist in M. tuberculosis H37Rv supports recent DNA sequence-based findings indicating that this region of the M. tuberculosis H37Rv whole genome sequence is not representative of the M. tuberculosis H37Rv ATCC reference strain (36). Our discovery of a SAP in Rv2037c, which was supposed to be absent in M. tuberculosis H37Rv, reinforces our conclusion that this inaccuracy is true for multiple regions of the M. tuberculosis H37Rv original sequence. This demonstrates that our database is able to decrease divergence originating from genomic errors or strain divergence, and not only errors originating from divergent open reading frame annotation approaches.
Acknowledgments
Co-operation (Project 64495). We thank the Proteomic Unit of Bergen (PROBE) for analytical support.
Footnotes
* This research was supported by funding from the Norwegian Research Council (Project 175141 and 183418), and South Africa-Norway Programme on Research.
 This article contains supplemental data S1-S5.
This article contains supplemental data S1-S5.
The authors declare no competing financial interests.
1 The abbreviations used are:
- IPI
- International Protein Index
- SAP
- single amino acid polymorphism
- MSMSpdbb
- multistrain mass spectrometry prokaryote database builder
- TSS
- translational start site
- SNP
- single nucleotide polymorphism
- ACN
- acetonitrile.
REFERENCES
- 1. Garrels J. I. (2002) Yeast genomic databases and the challenge of the post-genomic era. Funct. Integr. Genomics 2, 212–237 [DOI] [PubMed] [Google Scholar]
- 2. Rappsilber J., Mann M. (2002) What does it mean to identify a protein in proteomics? Trends Biochem. Sci. 27, 74–78 [DOI] [PubMed] [Google Scholar]
- 3. Ge H., Walhout A. J., Vidal M. (2003) Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends. Genet. 19, 551–560 [DOI] [PubMed] [Google Scholar]
- 4. Overbeek R. (2000) Genomics: what is realistically achievable? Genome Biol.. 1, Comment2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kyrpides N. C. (1999) Genomes OnLine Database (GOLD 1.0): a monitor of complete and ongoing genome projects world-wide. Bioinformatics 15, 773–774 [DOI] [PubMed] [Google Scholar]
- 6. Bernal A., Ear U., Kyrpides N. (2001) Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic. Acids. Res. 29, 126–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brent M. R. (2005) Genome annotation past, present, and future: how to define an ORF at each locus. Genome Res. 15, 1777–1786 [DOI] [PubMed] [Google Scholar]
- 8. de Souza G. A., Målen H., Søfteland T., Saelensminde G., Prasad S., Jonassen I., Wiker H. G. (2008) High accuracy mass spectrometry analysis as a tool to verify and improve gene annotation using Mycobacterium tuberculosis as an example. BMC Genomics 9, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Souza G. A., Søfteland T., Koehler C. J., Thiede B., Wiker H. G. (2009) Validating divergent ORF annotation of the Mycobacterium leprae genome through a full translation data set and peptide identification by tandem mass spectrometry. Proteomics 9, 3233–3243 [DOI] [PubMed] [Google Scholar]
- 10. Boeckmann B., Bairoch A., Apweiler R., Blatter M. C., Estreicher A., Gasteiger E., Martin M. J., Michoud K., O'Donovan C., Phan I., Pilbout S., Schneider M. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids. Res. 31, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 12. Schandorff S., Olsen J. V., Bunkenborg J., Blagoev B., Zhang Y., Andersen J. S., Mann M. (2007) A mass spectrometry-friendly database for cSNP identification. Nature Methods 4, 465–466 [DOI] [PubMed] [Google Scholar]
- 13. Garcia Pelayo M. C., Uplekar S., Keniry A., Mendoza Lopez P., Garnier T., Nunez Garcia J., Boschiroli L., Zhou X., Parkhill J., Smith N., Hewinson R. G., Cole S. T., Gordon S. V. (2009) A comprehensive survey of single nucleotide polymorphisms (SNPs) across Mycobacterium bovis strains and M. bovis BCG vaccine strains refines the genealogy and defines a minimal set of SNPs that separate virulent M. bovis strains and M. bovis BCG strains. Infect. Immun. 77, 2230–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Souza G. A., Arntzen M. Ø., Wiker H. G. (2010) MSMSpdbb: providing protein databases of closely related organisms to improve proteomic characterization of prokaryotic microbes. Bioinformatics 26, 698–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 16. Fleischmann R. D., Alland D., Eisen J. A., Carpenter L., White O., Peterson J., DeBoy R., Dodson R., Gwinn M., Haft D., Hickey E., Kolonay J. F., Nelson W. C., Umayam L. A., Ermolaeva M., Salzberg S. L., Delcher A., Utterback T., Weidman J., Khouri H., Gill J., Mikula A., Bishai W., Jacobs, W. R., Venter J. C., Fraser C. M. (2002) Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184, 5479–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng H., Lu L., Wang B., Pu S., Zhang X., Zhu G., Shi W., Zhang L., Wang H., Wang S., Zhao G., Zhang Y. (2008) Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS. ONE 3, e2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garnier T., Eiglmeier K., Camus J. C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Simon S., Harris B., Atkin R., Doggett J., Mayes R., Keating L., Wheeler P. R., Parkhill J., Barrell B. G., Cole S. T., Gordon S. V., Hewinson R. G. (2003) The complete genome sequence of Mycobacterium bovis. Proc. Natl Acad. Sci. USA 100, 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seki M., Honda I., Fujita I., Yano I., Yamamoto S., Koyama A. (2009) Whole genome sequence analysis of Mycobacterium bovis bacillus Calmette-Guerin (BCG) Tokyo 172: a comparative study of BCG vaccine substrains. Vaccine 27, 1710–1716 [DOI] [PubMed] [Google Scholar]
- 20. Målen H., Pathak S., Søfteland T., de Souza G. A., Wiker H. G. (in press) Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiology. 10, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Souza G. A., Fortuin S., Aguilar D., Pando R. H., McEvoy C. R., van Helden P. D., Koehler C. J., Thiede B., Warren R. M., Wiker H. G. (in press) Using a label-free proteomic method to identify differentially abundant proteins in closely related hypo- and hyper-virulent clinical Mycobacterium tuberculosis Beijing isolates. Mol. Cell Proteomics 10.1074/mcp.M900422-MCP900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 23. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. van Soolingen D., Arbeit R. D. (2001) Dealing with variation in molecular typing of Mycobacterium tuberculosis: low-intensity bands and other challenges. J. Med. Microbiol. 50, 749–751 [DOI] [PubMed] [Google Scholar]
- 26. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol.. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 27. Mardis E. R. (2008) The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141 [DOI] [PubMed] [Google Scholar]
- 28. Salgado H., Moreno-Hagelsieb G., Smith T. F., Collado-Vides J. (2000) Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. U S A 97, 6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edwards M. T., Rison S. C., Stoker N. G., Wernisch L. (2005) A universally applicable method of operon map prediction on minimally annotated genomes using conserved genomic context. Nucleic Acids Res. 33, 3253–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yates J. R., 3rd, Eng J. K., McCormack A. L. (1995) Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 67, 3202–3210 [DOI] [PubMed] [Google Scholar]
- 31. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 32. Sreevatsan S., Pan X., Stockbauer K. E., Connell N. D., Kreiswirth B. N., Whittam T. S., Musser J. M. (1997) Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl Acad. Sci. U S A 94, 9869–9874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Musser J. M., Amin A., Ramaswamy S. (2000) Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hershberg R., Lipatov M., Small P. M., Sheffer H., Niemann S., Homolka S., Roach J. C., Kremer K., Petrov D. A., Feldman M. W., Gagneux S. (2008) High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS. Biol. 6, e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niemann S., Köser C. U., Gagneux S., Plinke C., Homolka S., Bignell H., Carter R. J., Cheetham R. K., Cox A., Gormley N. A., Kokko-Gonzales P., Murray L. J., Rigatti R., Smith V. P., Arends F. P., Cox H. S., Smith G., Archer J. A. (2009) Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS. ONE. 4, e7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McEvoy C. R., van Helden P. D., Warren R. M., Gey van Pittius N. C. (2009) Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC. Evol. Biol. 9, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]