Abstract
Characterization of the binding of a tumor necrosis factor (TNF) ligand to its receptor(s) is pivotal to understand how these proteins initiate signal transduction pathways. Unfortunately, kinetic elucidation of these interactions is strongly hampered by the multivalent nature of the binding partners. The interaction between TNF-related apoptosis-inducing ligand and its death receptors was analyzed using in-depth applications of surface plasmon resonance technology. Variations in receptor density and sensor chip type allowed us to manipulate the stoichiometry of the formed complex, and the rate constants describing the binding of trimeric TNF-related apoptosis-inducing ligand to only one receptor molecule were determined. Remarkably, the affinity of this trimer-monomer complex is in the picomolar range, and its dissociation very slow. Further analysis showed that the second and third receptor molecules bind with lower affinity to the preformed trimer-monomer complex. This together with results obtained with receptor activator of NF-κB ligand and B cell-activating factor strongly suggests that the binding of TNF family ligands to their receptors is initiated via the formation of a trimer-monomer complex that is sufficiently stable to allow binding of two additional receptor molecules. These results suggest that avidity does not play a significant role and thus provide new insight in how TNF ligands form the biologically important complexes with their receptors.
Cytokines are signaling molecules involved in a range of biological processes and diseases. Cytokines and receptors belonging to the TNF superfamily have been a subject of interest for developing novel therapies for numerous diseases (1). These cytokines e.g. TNFα, TRAIL,1 RANKL, and BAFF are type II transmembrane proteins, and the extracellular C-terminal moiety can be released by specific proteases to form a soluble and active protein consisting of three subunits of each ∼20 kDa (2). As one of the most promising anticancer therapeutic candidates, recombinant human TRAIL (rhTRAIL; comprising amino acids 114–281) is able to kill a variety of cancer cells but not healthy cells. Currently, rhTRAIL is being tested in clinical phase II studies as an anticancer biopharmaceutical (3).
Despite good progress on structural insight, analysis of the interactions between TNF ligand family members and their receptors lacks unambiguous results. This seems mainly caused by the multivalent character of these molecules, i.e. trimeric cytokines but also often dimeric receptor-Fc fusions, which lead to complex kinetic behavior. In a series of experiments, we obtained several indications that surface plasmon resonance (SPR) assays with rhTRAIL WT and receptor-specific variants (4–6) offer opportunities to establish a better characterization of their interactions with receptor molecules. For this, we meticulously applied SPR technology to elucidate the dynamics of complex formation of this important group of cytokines. We used rhTRAIL WT; a DR5-specific mutant, rhTRAILD269H/E195R; and death receptors DR4 and DR5 to develop the method, but we show with examples of other members of the TNF family that the method is generally applicable. Apart from presenting a method allowing unambiguous affinity determination, our results demonstrate that the binding mechanism of these cytokines is initiated via a high affinity interaction with the first receptor molecule, bringing the cytokine to the membrane.
EXPERIMENTAL PROCEDURES
Materials
SPR buffers, regeneration solutions, and sensor chips were purchased from GE Healthcare. Chemicals unless otherwise stated were from Sigma. The following proteins were purchased: protein A from Staphylococcus aureus (Sigma); receptor-Fc fusion molecules of BCMA, DR4, DR5, RANK, TACI, TNFR1, and TNFR2 (R&D Systems, Oxon, UK); BAFF and TNFα (PeproTech EC, London, UK); and RANKL (R&D Systems). Monomeric receptor DR5, hereafter named DR5, was purchased from PeproTech EC. All rhTRAIL molecules were produced and purified in house as described previously (4). The gene encoding FLAG-rhTRAIL was constructed by introducing the sequence encoding MDYKDDDDKHM (FLAG sequence underlined) directly N-terminally of the rhTRAIL 114–281 sequence in pET15b. FLAG-rhTRAIL was produced in Escherichia coli BL21 (DE3) and purified using the following three chromatographic steps: nickel affinity using HisTrap, ion exchange using Q-Sepharose FF, and gel filtration using Superdex 75 (GE Healthcare). Concentrations of all in house-produced proteins were determined by the Coomassie Plus Protein Assay using bovine serum albumin as standard (PerBio Science, Etten-Leur, The Netherlands).
Binding Cytokine to Captured Receptor
To capture receptor-Fc molecules to the surface of a sensor chip, protein A was directly immobilized to the chip surface of all flow cells in a Biacore 3000 instrument using a solution of 70 μg/ml protein A in 10 mm NaAc, pH 4.5, and the primary amine coupling was performed according to the protocol of the supplier (GE Healthcare). Typical values for protein A immobilization were 5000–20,000 RU for CM5 sensor chips, 800–1200 RU for CM4 sensor chips, and 400–800 RU for C1 sensor chips (GE Healthcare). After some regeneration rounds using 10 mm glycine, pH 1.5–2.0 (which were incorporated to arrive at a reproducible binding capacity of protein A), the chip was ready for use.
SPR methods were written with Biacore 3000 control software v.3.2. Experiments were carried out at 37 °C using a flow rate of 50 μl/min and HBS-P as running and dilution buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.005% (v/v) surfactant P20; GE Healthcare). The method comprised multiple cycles with injections of varying concentrations of the cytokine. Because each sensor chip contains four flow cells, flow cell 1 was used as control, and the other three flow cells were loaded with different densities of a receptor-Fc or with different receptor-Fc molecules. A volume of 100–150 μl of cytokine was injected over all flow cells. After recording the association and dissociation phases, the surface of the chip was regenerated by two injections of 10 mm glycine, pH 1.5–2.0 (30 s), and the next cycle could be started. To obtain a full set of data, at least eight concentrations of cytokine were used. Injections of run buffer were incorporated at regular intervals as internal control.
The concentration of receptor was adjusted to the desired density. Importantly, to have a homogeneous surface of captured receptor, the solution of receptor-Fc was injected at a relatively high flow rate (>30 μl/min). Correction of all binding curves was performed by so-called double referencing, i.e. subtraction of the data of the “empty” flow cell 1 followed by subtraction of the data from a run buffer injection cycle as described before (7). Waiting steps of several minutes were incorporated to avoid signal instability caused by changing the flow path. Sensor chips with immobilized protein A could be used for multiple experiments and were discarded when regeneration as described above was no longer effective.
Besides protein A, additional capturing molecules were tested, e.g. monoclonal mouse anti-human IgG (Fc) antibody as part of the human antibody capture kit (GE Healthcare); the monovalent IgG binding domain of Staphylococcal protein A, Z-fragment (8), supplied by Dr. P. A. Nygren; and monovalent camelid-derived human Fc ligand antibody (BAC, Naarden, The Netherlands). The results with these proteins were similar with respect to stoichiometry of the formed complexes between rhTRAIL and receptor-Fc. The camelid-derived human Fc ligand antibody showed bleeding of the captured receptor-Fc molecules and was discarded from further experiments. The Z-fragment and the mouse monoclonal anti-human IgG antibody were used for experiments in parallel to protein A, but because the results were similar to those obtained with protein A, the results have not been mentioned separately.
Binding of Monomeric DR5 Receptor to Captured FLAG-rhTRAIL
For the binding of DR5 to FLAG-rhTRAIL, rabbit anti-FLAG antibody (Sigma) was captured to a density of 350 RU by protein A that was directly immobilized by standard amine coupling to the surface of a CM5 chip as described above.
Molecular Weights and Calculations of Binding Ratios
If molecule A binds to immobilized molecule B to form complex AB, the SPR signal that can maximally be reached is dependent on the amount of captured B (Capt) and the respective molecular weights (MW): Rmax = (MWA/MWB) × Capt. Similarly, the maximum response for formation of complex ABn is: Rmax = (MWA/(n × MWB)) × Capt.
For rhTRAIL WT, we used the molecular weight calculated for our trimeric protein (amino acids 114–281): MWA = 3 × 19,500 = 58,500. The molecular weight of FLAG-TRAIL was calculated to be 3 × 20,800 = 62,400. The bacterially produced BAFF has a molecular weight of 3 × 17,000 = 51,000. Monomeric DR5 (PeproTech EC) has a molecular weight of 14,900. For all proteins produced in mammalian cells, we used the molecular weight of the (glycosylated) protein that was given by the supplier and based on SDS-polyacrylamide gel electrophoresis under reducing conditions. The values are 3 × 35,000 = 105,000 for trimeric RANKL, 46,000 for the monomeric DR5-Fc, 46,000 for DR4-Fc, 60,000 for RANK-Fc, 45,000 for BCMA-Fc, and 47,000 for TACI-Fc.
RESULTS
Complex Binding Behavior of rhTRAIL and Its Death Receptors at High Receptor Density
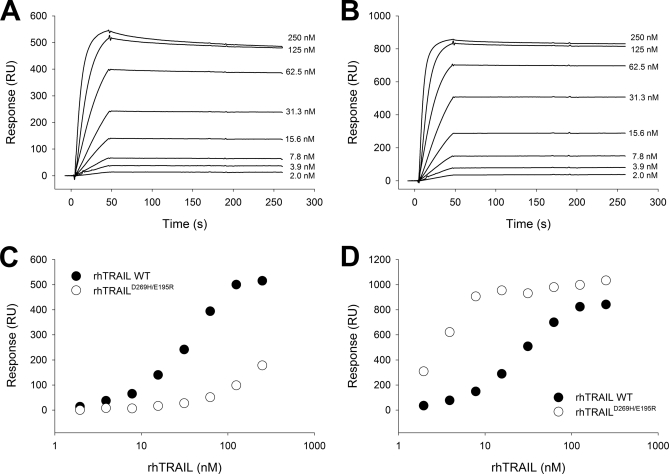
In many studies reported up to now, research groups have used an equilibrium approach to determine the affinity of the cytokine to its receptor. For this, receptor binding responses at the end of an injection of rhTRAIL are measured at a range of concentrations, and the binding isotherm is plotted to reveal the dissociation constant. In Fig. 1, the sensorgrams of rhTRAIL WT binding to captured DR4- and DR5-Fc (Fig. 1, A and B) and the corresponding binding isotherms (Fig. 1, C and D) are shown. We have used the term pre-steady state to describe an affinity value determined by this method (6) because it is clear that at lower rhTRAIL concentrations no equilibrium is reached. Additional experiments showed that increasing injection volumes resulted in increasing apparent affinity values (data not shown). A simulation shows that the applied injection time dictates the outcome of this value (supplemental Fig. 1).
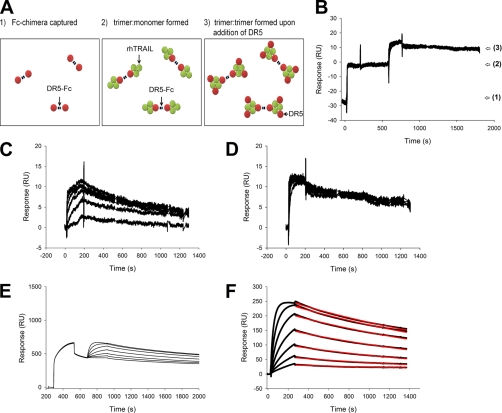
Fig. 1.
Formation of trimer-trimer complex by binding rhTRAIL to immobilized DR5. A CM5 sensor chip was prepared by chemical coupling of protein A. A series of cycles was programmed, comprising capturing DR4-Fc (A) or DR5-Fc (B) to a density of 1205 and 1850 RU, respectively, followed by an injection of rhTRAIL protein and finally regeneration (A–D). Eight concentrations of rhTRAIL WT were used, ranging from 2 to 250 nm (lower to upper curves). Pre-steady state analysis was performed as described (4) by reading response values 30 s after the end of the injection and plotting these as a function of the rhTRAIL concentrations injected to obtain the binding curves of rhTRAIL to DR4-Fc (C) or DR5-Fc (D). As a comparison, pre-steady state values are also shown for rhTRAILD269H/E195R.
While performing the SPR pre-steady state approach, we noticed that despite the nearly identical molecular weights of rhTRAIL WT and DR5-specific variant rhTRAILD269H/E195R the binding to DR5-Fc showed a different maximal value (Fig. 1D). This strongly suggested that the stoichiometry of the receptor complexes formed by these TRAIL proteins differs. More precisely, rhTRAILD269H/E195R seems to be able to form lower complexes, i.e. complexes of trimeric rhTRAIL with less than three receptor molecules bound. In an SPR assay, formation of such a complex results in a higher Rmax value because more rhTRAIL molecules can bind to the same number of captured receptors. Additional analyses at high receptor density confirmed this interpretation (supplemental Fig. 2). We therefore decided to look into the influence of receptor density on complex formation.
Density and Type of Sensor Chip Determine Stoichiometry of Complex between TRAIL and Its Death Receptors
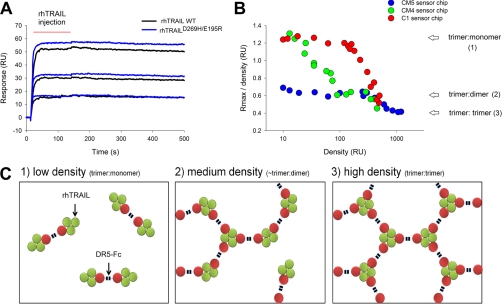
As shown above, we observed a difference in Rmax values at a high density of receptor-Fc, indicating that rhTRAILD269H/E195R has a stronger ability to form lower complexes than WT. By lowering the receptor density to a level where only the lowest complex can be formed, binding of each TRAIL protein should result in an identical Rmax value. This was indeed achieved with two types of sensor chips, CM4 (long dextran chains with a lower degree of carboxymethylation as compared with CM5) and C1 (no dextran chains). The experiment using a CM4 sensor chip is depicted in Fig. 2A. At the lowest density of receptor DR5-Fc, 13 resonance units (RU), identical responses were obtained after injection of 500 nm rhTRAIL WT and rhTRAILD269H/E195R, whereas already at a slightly higher density (24 RU), a difference could be observed between the two TRAIL proteins. This difference increased with the density of immobilized receptor. The capture method in which the receptor-Fc molecules are bound by immobilized protein A allowed us to calculate that at the lowest density of 13 RU solely the trimer-monomer complex was formed (expected response was 13 × 58.5/46 = 16.5 RU), whereas at 24 RU of receptor, higher complexes also were formed. Importantly, this calculation demonstrates that each of the monomers of the receptor-Fc molecule, despite the bidentate nature of the Ig fusion protein, cannot bind to the same rhTRAIL molecule (Fig. 2C).
Fig. 2.
Stoichiometry of formed complexes of rhTRAIL and DR5-Fc as a function of receptor density on different sensor chips. A, binding of 500 nm rhTRAIL WT (black) and rhTRAILD269H/E195R (blue) to low densities (13, 24, and 35 RU; bottom to top) of captured receptor DR5-Fc on a CM4 sensor chip. B, calculated ratio Rmax/density as a function of the density of captured receptor on different sensor chips, CM5 (blue circle), CM4 (green circle), and C1 (red circle), as determined with rhTRAIL WT and DR5-Fc. The arrows indicate the ratios for formation of the different possible rhTRAIL-receptor complexes, trimer-monomer (3:1), trimer-dimer (3:2), and trimer-trimer (3:3), that were calculated using the molecular weights of the components. A trimer-trimer complex was obtained on the CM5 chip (>1000 RU receptor), whereas the trimer-monomer complex was formed at low density on CM4 (<15 RU) or C1 (<100 RU) sensor chips. A schematic view of the trimer-monomer, trimer-dimer, and the trimer-trimer complexes on the surface of a sensor chip is shown (C).
To further explore the relationship of the stoichiometry of the formed TRAIL-receptor complexes with the density of captured receptor and the sensor chip type, SPR measurements were performed with the sensor chips CM5, CM4, and C1 at different densities of captured receptor DR5-Fc. Each Rmax value was determined with a concentration series of 1–250 nm rhTRAIL WT. The ratio of Rmax/receptor density was calculated and plotted as a function of the density of captured receptor (Fig. 2B). Clearly, the combination of the type of sensor chip and the density of the captured receptor determines the stoichiometry of the formed complex.
Kinetic Analysis of Trimer-Monomer Complex of rhTRAIL and Its Death Receptors
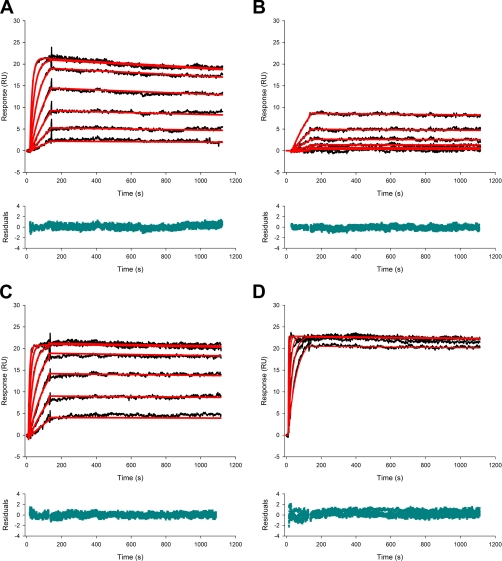
Our results allowed us to choose conditions in favor of trimer-monomer complex formation and to analyze the data with a 1:1 binding model. Fig. 3 shows the response after injecting different concentrations of rhTRAIL WT and rhTRAILD269H/E195R to 16 RU of captured DR4-Fc and DR5-Fc on a CM4 sensor chip. These data could be fitted with a 1:1 Langmuir model with high accuracy using BIAevaluation 4.1.
Fig. 3.
Sensorgrams for trimer-monomer complex formation of rhTRAIL WT or rhTRAILD269H/E195R and death receptor DR4-Fc or DR5-Fc. Binding of rhTRAIL WT (A and C) or rhTRAILD269H/E195R (B and D) to 16 RU of DR4-Fc (A and B) or 16 RU of DR5-Fc (C and D) captured by protein A on the surface of a CM4 sensor chip was followed at seven concentrations of TRAIL (3.91–250 nm; bottom to top) using a flow rate of 50 μl/min at 37 °C in HBS-P. The data were fitted with the 1:1 Langmuir model using BIAevaluation 4.1. The experimental curves are indicated in black, the fitted curves are in red; below every sensorgram, the residuals of the fitted curves are shown.
Association and dissociation rate constant values were obtained by global fitting after which the corresponding dissociation constant KD was calculated (Table I). rhTRAIL WT had a nearly 6-fold higher affinity for DR5-Fc compared with DR4-Fc, caused by a higher association rate constant and a smaller dissociation rate constant.
Table I. Overview of rate constants for trimer-monomer complex formation between rhTRAIL WT and its death receptors DR4 and DR5 in comparison with rhTRAILD269H/E195R.
Mean values of several determinations (3 ≤ n ≤ 10) are indicated with the standard deviation.
| ka | kd | KD | |
|---|---|---|---|
| m−1s−1 | s−1 | nm | |
| DR4-Fc | |||
| rhTRAIL WT | (6.3 ± 1.2)·105 | (1.1 ± 0.3)·10−4 | 0.17 ± 0.03 |
| rhTRAILD269H/E195R | (0.18 ± 0.1)·105 | (0.53 ± 0.4)·10−4 | 2.9 ± 1.7 |
| DR5-Fc | |||
| rhTRAIL WT | (11.9 ± 1.8)·105 | (0.36 ± 0.20)·10−4 | 0.03 ± 0.02 |
| rhTRAILD269H/E195R | (51.2 ± 4.2)·105 | (0.63 ± 0.18)·10−4 | 0.012 ± 0.005 |
Remarkably, the differences between rhTRAIL WT and rhTRAILD269H/E195R for each receptor were found in their association rate constants and hardly in their dissociation rate constants. rhTRAILD269H/E195R had a nearly 5-fold higher association rate constant for DR5 than rhTRAIL WT but a 35-fold lower association rate constant for DR4 when compared with rhTRAIL WT. Combination of these changes makes this mutant highly selective for DR5 as demonstrated before with pre-steady state SPR and cell assays (6).
Kinetic System Is Generally Applicable to TNF Cytokines
In Fig. 4A, the binding of RANKL to three densities of captured RANK-Fc is shown. As for TRAIL, the stoichiometry of the formed complex of RANKL and RANK depended on the density of the captured receptor with Rmax/density ratios of 0.51, 0.55, and 1.15 at captured receptor densities of 255, 150, and 40 RU, respectively. These ratios correspond well with ratios found for rhTRAIL WT complexes with DR5-Fc (Fig. 2B). The stoichiometry of the corresponding complexes was more difficult to calculate because RANKL and RANK-Fc are both produced in mammalian cells and show large deviations in calculated and experimentally determined molecular weights. At 40 RU of receptor, binding of RANKL may still result in a mixed population of trimer-monomer and trimer-dimer complexes. However, when immobilizing only 13.4 RU of RANK-Fc to a CM4 sensor chip, a maximum was reached upon binding of increasing concentrations of RANKL (Fig. 4B), indicating strongly that the trimer-monomer complex is formed. The data could be accurately fitted to a 1:1 Langmuir model, resulting in a dissociation constant (KD) of 0.67 nm with ka = 4.9·105 m−1 s−1 and kd = 3.3·10−4 s−1. A similar analysis was performed for the trimer-monomer binding of BAFF to BCMA-Fc and TACI-Fc, resulting in KD values of 0.73 nm for BCMA-Fc and 1.4 nm for TACI-Fc.
Fig. 4.
Sensorgrams of binding of RANKL to RANK-Fc on CM4 sensor chip. A, binding of 500 nm RANKL to RANK-Fc, captured by protein A at different densities (40, 150, and 255 RU) on the surface of a CM4 sensor chip. B, binding of 3.9–500 nm (bottom to top) RANKL to 13.4 RU of captured RANK-Fc to form complex AB was followed using a flow rate of 50 μl/min at 37 °C in HBS-P. The data were analyzed by global fitting using the 1:1 Langmuir model. Experimental data are indicated by the black lines, and the fitted curves are in red.
Binding of DR5 to Preformed Trimer-Monomer Complex of rhTRAIL WT and DR5-Fc
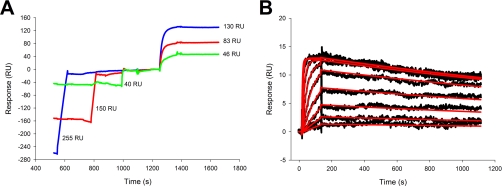
We used the ability to form a stable trimer-monomer to analyze the binding of the second and third receptor molecule. In Fig. 5, capturing of DR5-Fc was followed by binding of rhTRAIL to form a trimer-monomer complex; thereafter, 94–3000 nm DR5 was injected.
Fig. 5.
Binding of DR5 to preformed trimer-monomer complex of rhTRAIL and DR5-Fc or to captured FLAG-rhTRAIL. A trimer-monomer complex was formed by binding 500 nm rhTRAIL to a low density of DR5-Fc captured by protein A on a CM4 sensor chip and followed by an injection of DR5 (concentrations, 94–3000 nm; bottom to top). Binding of ∼26 RU rhTRAIL to form a trimer-monomer with DR5-Fc was expected to allow binding of two molecules of DR5, which would lead to a response of 26 × 2 × 14.9/58.5 = 13.2 RU as can also be measured. The different complexes are schematically represented in A and also indicated with arrows in B. The data for binding of DR5 to the complex of rhTRAIL WT or rhTRAILD269H/E195R with DR5-Fc were processed by double correction as described (C and D, respectively). In another setup, FLAG-tagged rhTRAIL WT was captured by anti-FLAG antibody on a CM5 sensor chip. A series of cycles was performed, each with a sequence as follows: capture of anti-FLAG antibody by immobilized protein A to bind FLAG-tagged rhTRAIL whereafter a concentration of DR5 was injected finally followed by regeneration of the chip surface (E). Responses due to binding of a series of concentrations of DR5 (8.7–1000 nm; bottom to top) to FLAG-rhTRAIL were corrected with data of an empty lane and of buffer injection (F). The dissociation phase was fitted separately to a 1:1 Langmuir model (experimental curves are in black; fitted curves with a dissociation rate constant of 4.4·10−4 s−1 are in red).
This experiment delivers further proof of formation of the trimer-monomer because it was calculated that two molecules of DR5 are bound per preformed trimer-monomer complex. For WT, the maximum response was barely reached at 3000 nm DR5 (Fig. 5C), indicating that the second and third receptor binding sites have a lower affinity compared with the binding of the first receptor molecule by rhTRAIL. Although no satisfactory fit could be obtained with available software (using model heterogeneous ligand-parallel reactions), both the association and dissociation rate constants seem to be affected. As expected, binding of DR5 to the preformed complex of rhTRAILD269H/E195R was characterized by faster association compared with rhTRAIL WT (Fig. 5D).
Binding of DR5 to Captured FLAG-rhTRAIL
In an alternative approach to analyze the dynamics of TRAIL-receptor complex formation, we bound FLAG-tagged rhTRAIL via anti-FLAG antibody captured by protein A and injected DR5 (Fig. 5E). To correct for bleeding of FLAG-TRAIL from the antibody (Fig. 5E), the data from a cycle in which buffer was injected instead of DR5 were subtracted. The corrected sensorgram is shown in Fig. 5F. The ratio of captured FLAG-tagged rhTRAIL (420 RU) and the binding of DR5 (250 RU at 1000 nm) are in agreement with 2.5 receptor molecules bound to each captured FLAG-rhTRAIL. The discrepancy with the expected three molecules of DR5 per FLAG-rhTRAIL is most likely caused by the bleeding of FLAG-rhTRAIL from the surface and by the fact that the Rmax value is not yet attained with 1000 nm DR5. Under the assumption that the three receptor binding sites on rhTRAIL are identical, dissociation of the receptor molecules from the trimer-trimer complex should show a single exponential behavior. We fitted the dissociation phase separately with BIAevaluation assuming a monoexponential decay (Fig. 5F). Interestingly, the sensorgram deviates from the fitted dissociation curves, suggesting that the three receptor binding sites of rhTRAIL are not identical, having dissociation rate constants that may differ up to 1 order of magnitude.
DISCUSSION
One of the standard technologies to analyze the affinity of a cytokine to its receptor(s) is SPR. SPR is a powerful technique for which no labeling of molecules is necessary, but experimental design and data analysis remain an important issue (9). To our knowledge, in all SPR studies on TNF family ligands and their receptors, CM5 sensor chips have been used with medium to high densities of receptor often directly immobilized by primary amine coupling (4–6, 10–17). In these studies, formation of the trimer-trimer complex, i.e. three receptor molecules to one trimeric TRAIL molecule, is assumed but not demonstrated. In a few SPR studies published (11, 12), rate constants for rhTRAIL were determined by applying the 1:1 Langmuir model, which does not take into account the complexity of the binding of a multivalent molecule binding to immobilized receptor. Indeed, the trivalent character of TNF cytokines and the bidentate nature of receptor-Fc fusion proteins are a serious problem for kinetic elucidation of the interactions.
As an alternative method for SPR affinity determination, a steady state approach can be followed where equilibrium values are measured at varying concentrations, and the resulting binding isotherm reveals the affinity. When applying this approach for TRAIL, we realized that at low concentrations of rhTRAIL no equilibrium was reached (Fig. 1, A and B). In earlier studies, we therefore used the term “pre-steady state” affinities (4–6). We observed that an increase of the injection time led to higher pre-steady state responses at low rhTRAIL concentrations and resulted in lower apparent KD values (data not shown). This is in agreement with a simulation of binding curves calculated for different injection volumes (supplemental Fig. 1), demonstrating that the derived apparent affinity values can deviate up to 2 orders of magnitude. Clearly, the extremely long injection times needed to reach equilibrium at low concentrations are impossible to achieve with SPR and most other techniques, leaving no room for improvement of this approach.
Interestingly, despite their nearly identical molecular weight, rhTRAIL mutant and WT proteins did not reach the same maximum SPR response at medium to high receptor density (Fig. 1, C and D) as also observed before (4, 6). Capturing the receptor molecules via protein A allowed us to calculate the stoichiometry of the formed complexes. Only at very low densities of DR5-Fc were the Rmax values for the DR5-selective and WT rhTRAIL proteins identical (Fig. 2A). We could calculate that under these conditions each molecule of trivalent rhTRAIL bound to one molecule of DR5-Fc receptor, thus forming a trimer-monomer complex. These identical Rmax values could only be measured when using CM4 or C1 sensor chips, both of which have less active groups than the standard CM5 sensor chips. Clearly, the density of receptor and the type of sensor chip determine the stoichiometry of the formed complexes (Fig. 2B). The formation of trimer-monomer complexes demonstrates that each monomer of DR5 in the Fc fusion protein binds to a different rhTRAIL molecule (Fig. 2C).
With these results, rhTRAIL could be defined as the only multivalent protein, and under the proper conditions, simple kinetics of one rhTRAIL molecule binding to one receptor molecule could be applied. Indeed, data obtained while injecting rhTRAIL proteins to very low densities of captured DR4-Fc or DR5-Fc could be fitted with the 1:1 Langmuir binding model, resulting in the association and dissociation rate constants for the binding of one receptor molecule to the trimeric rhTRAIL molecule (Fig. 3 and Table I). The dissociation rate constants are very low, indicating that the first binding event is kinetically very important to enable formation of the proapoptotic cytokine-receptor complex.
The trimer-monomer complex of variant rhTRAILD269H/E195R showed a 2.5-fold larger affinity for DR5-Fc compared with WT (Table I), which is in agreement with earlier derived differences (6). This higher affinity for DR5 is caused by a significant increase in association rate constant. Unexpectedly, its more than 30-fold lower affinity for DR4-Fc is not caused by a higher dissociation rate of the complex but by a severe lowering of the association rate constant.
The affinity of the trimer-monomer complex of rhTRAIL WT and DR5-Fc molecule (KD = 0.03 nm; Table I) is higher than affinities reported in the complex in the previous studies. However, it should be realized that other determinations report overall affinities and not only the first binding event. The relatively high pre-steady state affinity value of 2.5 nm that we determined earlier for rhTRAIL WT and DR5-Fc (6) can be explained by the fact that apparent KD values can be substantially higher when equilibrium at low concentrations is not reached (supplemental Fig. 1). In other experiments, KD values of 0.51 and 0.76 nm were determined via rate constants using a 1:1 model despite the fact that high receptor densities were used and thus mostly likely a trimer-trimer complex was formed (10, 11). Interestingly, isothermal titration calorimetry resulted in a KD value below the instrumental limit of 1 nm (18). Taken together, the overall affinity of DR5 for rhTRAIL does not seem to be higher than the affinity describing binding of the first receptor molecule, strongly suggesting that the affinity of rhTRAIL for its receptor is dominated by the first binding event and that the two other receptor binding sites have lower affinities. Differences in affinity for the three binding sites are also suggested by binding of DR5 to preformed trimer-monomer (Fig. 5, A–D). The need for very high receptor concentrations to reach an Rmax value indicates that the second and third receptor molecules bind with lower affinity. In addition, binding and dissociation of DR5 to captured FLAG-rhTRAIL showed heterogeneous behavior, pointing at deviations between the three receptor binding sites on rhTRAIL (Fig. 5F). In agreement, crystal structures of the trimer-trimer complex of rhTRAIL and DR5 (19, 20) also show structural differences between the three receptor binding sites, and significant differences in binding energy were calculated (supplemental Fig. 3). The interpretation that the second and third receptor molecules bind with lower affinities may not be as surprising when realizing that upon binding of the first receptor molecule the trimer-monomer complex is trapped in the same two-dimensional plane as the receptor molecules with relatively high local concentrations of receptor.
In the absence of a functional method to analyze the kinetics of the trimer-trimer complex formation, we strongly advise using the trimer-monomer complex for the characterization of the dynamics of a cytokine-receptor pair. We could demonstrate that the method of analysis is applicable to other cytokines. For RANKL-RANK-Fc, formation of the trimer-monomer is characterized by an affinity of 0.7 nm (Fig. 4), whereas BAFF with TACI-Fc or BCMA-Fc resulted in affinities of 1.4 and 0.7 nm, respectively. For others, such as TNFα and TNFR1-Fc or TNFR2-Fc, the responses were very low (data not shown), making it impossible to calculate the stoichiometry of the formed complex. Because the dissociation of the complexes did not show heterogeneity, the most likely explanation is that part of the receptor is not capable to bind the cytokine. Whether this is due to glycosylation or partially unfolded protein remains unclear. Interestingly, all analyzed trimer-monomer complexes of TNF family cytokine-receptor pairs are characterized by a slow dissociation. Our results indicate that the concept of avidity does not play an important role for TNF ligands. It seems that a strong binding of the cytokine to the first receptor molecule is important to bring it to the surface of the cell. The lack of significant dissociation will allow the second and third receptor molecule to bind and form the biologically active trimer-trimer complex. From this perspective, our results indicate that preformed oligomeric structures of receptors (21, 22) are not a prerequisite for formation of trimer-trimer complexes, although they will facilitate the signaling. Similarly, recruitment of death receptors into lipid rafts would increase the local density of receptor molecules and help to rapidly form the biologically active complex, thus making the cell more susceptible to TRAIL-mediated apoptosis (23). For drug development, it is obvious that (receptor-specific) mutants of cytokines can be much better characterized by our method. Last but not least, our results suggest that a mono- or dimeric variant of a cytokine with higher receptor affinity than WT can have a strong antagonistic activity, which would not be easily achieved when avidity plays a more dominant role. In summary, our method allows for the first time unambiguous kinetic analysis of the interactions between TNF ligands and their receptors, which helps to better understand how these molecules initiate their signaling cascade.
Acknowledgments
We thank Dr. P. A. Nygren for the gift of purified Z-fragment. In addition, we are grateful to Dr. N. de Mol and Dr. R. Karlsson for contributions in the discussion on kinetic analysis using SPR. The SPR equipment is part of the Groningen Proteomics Facility.
Footnotes
* This work was supported by the European Union Sixth Framework Program Grant LSH-2005-2.2.0-2 and by the Dutch Technology Foundation STW, the applied science division of the Netherlands Organization for Scientific Research NWO, and the technology program of the Ministry of Economic Affairs.
 This article contains supplemental Figs. 1–3.
This article contains supplemental Figs. 1–3.
1 The abbreviations used are:
- TRAIL
- TNF-related apoptosis-inducing ligand
- RANKL
- receptor activator of NF-κB ligand
- BAFF
- B cell-activating factor
- SPR
- surface plasmon resonance
- rhTRAIL
- recombinant human TRAIL
- RANK
- receptor activator of NF-κB
- BCMA
- B cell maturation
- RU
- resonance units
- TACI
- transmembrane activator and CAML-interactor.
REFERENCES
- 1. Younes A., Kadin M. E. (2003) Emerging applications of the tumor necrosis factor family of ligands and receptors in cancer therapy. J. Clin. Oncol. 21, 3526–3534 [DOI] [PubMed] [Google Scholar]
- 2. Bodmer J. L., Schneider P., Tschopp J. (2002) The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27, 19–26 [DOI] [PubMed] [Google Scholar]
- 3. Holoch P. A., Griffith T. S. (2009) TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur. J. Pharmacol. 625, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reis C. R., van der Sloot A. M., Szegezdi E., Natoni A., Tur V., Cool R. H., Samali A., Serrano L., Quax W. J. (2009) Enhancement of Antitumor Properties of rhTRAIL by Affinity Increase toward Its Death Receptors (dagger). Biochemistry 48, 2180–2191 [DOI] [PubMed] [Google Scholar]
- 5. Tur V., van der Sloot A. M., Reis C. R., Szegezdi E., Cool R. H., Samali A., Serrano L., Quax W. J. (2008) DR4-selective tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) variants obtained by structure-based design. J. Biol. Chem. 283, 20560–20568 [DOI] [PubMed] [Google Scholar]
- 6. van der Sloot A. M., Tur V., Szegezdi E., Mullally M. M., Cool R. H., Samali A., Serrano L., Quax W. J. (2006) Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 8634–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Myszka D. G. (2000) Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 323, 325–340 [DOI] [PubMed] [Google Scholar]
- 8. Andersson K., Gülich S., Hämäläinen M., Nygren P. A., Hober S., Malmqvist M. (1999) Kinetic characterization of the interaction of the Z-fragment of protein A with mouse-IgG3 in a volume in chemical space. Proteins 37, 494–498 [DOI] [PubMed] [Google Scholar]
- 9. Rich R. L., Myszka D. G. (2010) Grading the commercial optical biosensor literature-class of 2008: ‘the mighty binders’. J. Mol. Recognit. 23, 1–64 [DOI] [PubMed] [Google Scholar]
- 10. Emery J. G., McDonnell P., Burke M. B., Deen K. C., Lyn S., Silverman C., Dul E., Appelbaum E. R., Eichman C., DiPrinzio R., Dodds R. A., James I. E., Rosenberg M., Lee J. C., Young P. R. (1998) Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J. Biol. Chem. 273, 14363–14367 [DOI] [PubMed] [Google Scholar]
- 11. Gasparian M. E., Chernyak B. V., Dolgikh D. A., Yagolovich A. V., Popova E. N., Sycheva A. M., Moshkovskii S. A., Kirpichnikov M. P. (2009) Generation of new TRAIL mutants DR5-A and DR5-B with improved selectivity to death receptor 5. Apoptosis 14, 778–787 [DOI] [PubMed] [Google Scholar]
- 12. Hymowitz S. G., O'Connell M. P., Ultsch M. H., Hurst A., Totpal K., Ashkenazi A., de Vos A. M., Kelley R. F. (2000) A unique zinc-binding site revealed by a high-resolution X-ray structure of homotrimeric Apo2L/TRAIL. Biochemistry 39, 633–640 [DOI] [PubMed] [Google Scholar]
- 13. Kelley R. F., Totpal K., Lindstrom S. H., Mathieu M., Billeci K., Deforge L., Pai R., Hymowitz S. G., Ashkenazi A. (2005) Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J. Biol. Chem. 280, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 14. Lee H. W., Lee S. H., Lee H. W., Ryu Y. W., Kwon M. H., Kim Y. S. (2005) Homomeric and heteromeric interactions of the extracellular domains of death receptors and death decoy receptors. Biochem. Biophys. Res. Commun. 330, 1205–1212 [DOI] [PubMed] [Google Scholar]
- 15. Mukai Y., Nakamura T., Yoshioka Y., Shibata H., Abe Y., Nomura T., Taniai M., Ohta T., Nakagawa S., Tsunoda S., Kamada H., Yamagata Y., Tsutsumi Y. (2009) Fast binding kinetics and conserved 3D structure underlie the antagonistic activity of mutant TNF: useful information for designing artificial proteo-antagonists. J. Biochem. 146, 167–172 [DOI] [PubMed] [Google Scholar]
- 16. Mukai Y., Shibata H., Nakamura T., Yoshioka Y., Abe Y., Nomura T., Taniai M., Ohta T., Ikemizu S., Nakagawa S., Tsunoda S., Kamada H., Yamagata Y., Tsutsumi Y. (2009) Structure-function relationship of tumor necrosis factor (TNF) and its receptor interaction based on 3D structural analysis of a fully active TNFR1-selective TNF mutant. J. Mol. Biol. 385, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 17. Shibata H., Yoshioka Y., Ohkawa A., Minowa K., Mukai Y., Abe Y., Taniai M., Nomura T., Kayamuro H., Nabeshi H., Sugita T., Imai S., Nagano K., Yoshikawa T., Fujita T., Nakagawa S., Yamamoto A., Ohta T., Hayakawa T., Mayumi T., Vandenabeele P., Aggarwal B. B., Nakamura T., Yamagata Y., Tsunoda S., Kamada H., Tsutsumi Y. (2008) Creation and X-ray structure analysis of the tumor necrosis factor receptor-1-selective mutant of a tumor necrosis factor-alpha antagonist. J. Biol. Chem. 283, 998–1007 [DOI] [PubMed] [Google Scholar]
- 18. Truneh A., Sharma S., Silverman C., Khandekar S., Reddy M. P., Deen K. C., McLaughlin M. M., Srinivasula S. M., Livi G. P., Marshall L. A., Alnemri E. S., Williams W. V., Doyle M. L. (2000) Temperature-sensitive differential affinity of TRAIL for its receptors. DR5 is the highest affinity receptor. J. Biol. Chem. 275, 23319–23325 [DOI] [PubMed] [Google Scholar]
- 19. Cha S. S., Sung B. J., Kim Y. A., Song Y. L., Kim H. J., Kim S., Lee M. S., Oh B. H. (2000) Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity. J. Biol. Chem. 275, 31171–31177 [DOI] [PubMed] [Google Scholar]
- 20. Hymowitz S. G., Christinger H. W., Fuh G., Ultsch M., O'Connell M., Kelley R. F., Ashkenazi A., de Vos A. M. (1999) Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol. Cell 4, 563–571 [DOI] [PubMed] [Google Scholar]
- 21. Chan F. K. (2000) The pre-ligand binding assembly domain: a potential target of inhibition of tumour necrosis factor receptor function. Ann. Rheum. Dis. 59, Suppl. 1, i50–i53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siegel R. M., Frederiksen J. K., Zacharias D. A., Chan F. K., Johnson M., Lynch D., Tsien R. Y., Lenardo M. J. (2000) Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288, 2354–2357 [DOI] [PubMed] [Google Scholar]
- 23. Pennarun B., Meijer A., de Vries E. G., Kleibeuker J. H., Kruyt F., de Jong S. (2010) Playing the DISC: turning on TRAIL death receptor-mediated apoptosis in cancer. Biochim. Biophys. Acta 1805, 123–140 [DOI] [PubMed] [Google Scholar]